Abstract
The winged-helix transcription factor Foxh1 is an essential regulator of Nodal signaling during the key developmental processes of gastrulation, anterior-posterior (A-P) patterning, and the derivation of left-right (L-R) asymmetry. Current models have Foxh1 bound to phospho-Smad2/3 (pSmad2/3) as a central transcriptional activator for genes targeted by Nodal signaling including Nodal itself, the feedback inhibitor Lefty2, and the positive transcriptional effector Pitx2. However, the conserved Engrailed homology-1 (EH1) motif present in Foxh1 suggests that modulated interaction with Groucho (Grg) co-repressors would allow Foxh1 to function as a transcriptional switch, toggling between transcriptional on and off states via pSmad2-Grg protein-switching, to ensure the properly timed initiation and suppression, and/or amplitude, of expression of Nodal and its target genes. We minimally mutated the Foxh1 EH1 motif, creating a novel Foxh1mEH1 allele to test directly the contribution of Foxh1-Grg–mediated repression on the transient, dynamic pattern of Nodal signaling in mice. All aspects of Nodal and its target gene expression in Foxh1mEH1/mEH1 embryos were equivalent to wild type. A-P patterning and organ situs in homozygous embryos and adult mice were also unaffected. The finding that Foxh1-Grg–mediated repression is not essential for Nodal expression during mouse embryogenesis suggests that other regulators compensate for the loss of repressive regulatory input that is mediated by Grg interactions. We suggest that the pervasive inductive properties of Nodal signaling exist within the context of a strongly buffered regulatory system that contributes to resilience and accuracy of its dynamic expression pattern.
Keywords: Nodal, Foxh1, Groucho, Transcriptional regulation, Embryonic patterning
1. Introduction
Nodal is a key regulator of body axis formation and patterning in the developing embryo of a broad spectrum of animal species. Among vertebrates, Nodal signaling specifically plays conserved and essential roles in germ layer specification, gastrulation, anterior-posterior (A-P) axis formation, and left-right (L-R) patterning (Lu et al., 2001; Schier and Shen, 1999). As a morphogen belonging to the transforming growth factor β (TGFβ) superfamily, Nodal signals through the type I (Alk4/5/7) and type II (ActRIIA/B) serine/threonine receptors, with the aid of the EGF-CFC family of co-receptors (Crypto/Cryptic in mouse) (Schier, 2009; Wu and Hill, 2009). The downstream phosphorylation of the signal transducers Smad2 and Smad3 leads to interactions with Smad4 and nuclear translocation, and binding of the forkhead box transcription factor Foxh1, to initiate transcription of downstream target genes. Cell fate determination is greatly influenced by the level of Nodal signaling, and therefore dynamic but precise regulation of Nodal signaling duration and amplitude is essential for proper embryogenesis (Schier, 2009).
Foxh1 is essential to the initiation and maintenance of the Nodal autoregulatory circuit by which Nodal signaling initiates the expression of Nodal itself, its feedback inhibitor Lefty2, and the homeodomain transcription factor Pitx2, an important effector of Nodal signaling (Norris et al., 2002; Saijoh et al., 2000; Shiratori et al., 2001). Foxh1 binds conserved sequences in a cis-regulatory region known as an asymmetric enhancer element (ASE; named for its ability to drive expression on the left but not right side of the embryo) that is present in all three Nodal autoregulatory circuit genes (Nodal, Lefty2, Pitx2) (Meno et al., 2001; Saijoh et al., 1999; Saijoh et al., 2000; Shiratori et al., 2001). The ASE, along with another enhancer, drives Nodal expression during early patterning events and then initiates the expression of Nodal, Lefty2, and Pitx2 solely within the left lateral plate mesoderm (L LPM) during L-R patterning (Adachi et al., 1999; Norris and Robertson, 1999; Saijoh et al., 1999; Shiratori et al., 2001). Deletion of the Foxh1 binding-sites, or deletion of the ASE as a whole in mouse, leads to decreased Nodal expression in the epiblast and complete loss of Nodal, Lefty2, and Pitx2 expression in the L LPM (Adachi et al., 1999; Norris and Robertson, 1999; Norris et al., 2002; Saijoh et al., 2000; Shiratori et al., 2001). Foxh1 binding sites play a conserved role in the ASE as deletion of these sites also attenuated Nodal signaling in other species (Osada et al., 2000). Furthermore, the majority of embryos with a global deletion of Foxh1 fail to orient the A-P axis appropriately, elongate the primitive streak, or form a node; together these defects cause embryonic lethality (Hoodless et al., 2001; Yamamoto et al., 2001). Embryos in which Foxh1 was conditionally inactivated within the L LPM failed to express Nodal, Lefty2, and Pitx2 in that tissue and exhibited right isomerism (Yamamoto et al., 2003).
The pSmad2/3-Smad4 complex binds to the Smad interaction domain (SID) on the C-terminus of Foxh1 (Chen et al., 1997; Weisberg et al., 1998). In addition to the SID, Foxh1 contains another more N-terminally located co-factor interaction motif—the 8 amino-acid Engrailed homology-1 (EH1) motif—that is recognized and bound by the Groucho/Groucho-related-gene/Transducin-like enhancer of split (Gro/Grg/TLE) family of co-repressors (Yaklichkin et al., 2007a). The Gro/Grg/TLE protein family comprises four full-length members: TLE1-4 in human and originally termed Grg1-4 in mouse; the latter nomenclature we will use hereafter. Each member contains a conserved C-terminal WD-repeat domain that mediates interactions with transcription factors by recognizing two classes of motifs: the full EH1 or a smaller tetrapeptide (WRPW). It is currently unclear as to the direct mode by which Grg proteins repress transcription, but likely multiple mechanisms are used, and are dependent on biological context. One reported mechanism is the recruitment of histone deacetylases (HDACs) to Groucho-bound loci, promoting a closed chromatin conformation (Jennings and Ish-Horowicz, 2008).
The presence of both the EH1 motif and SID suggests that Foxh1 acts as a transcriptional switch, toggling between activator or repressor states by competitive partner-switching between pSmad2/Smad4 and Grg. This is an attractive mechanism for rapid and precise transcriptional control of Nodal signaling, for example, during the dynamically changing expression pattern of Nodal in both the epiblast and L LPM. At embryonic day (E) 5.5, Foxh1-dependent Nodal expression is initially throughout the epiblast and visceral endoderm, and then resolves to the prospective posterior side where it is expressed within the elongating primitive streak, becoming terminated by E7.5 (expression in the node at this time is Foxh1-independent) (Norris and Robertson, 1999; Adachi et al. 1999). Expression reinitiates exclusively in the L LPM at the 2–3 somite-stage. Within hours, Nodal expression, as well as the expression of its target and feedback inhibitor Lefty2, traverses the L LPM in a wave-like pattern (for data from frog embryos: Ohi and Wright, 2007), initiating in the posterior L LPM adjacent to the node and then rapidly expanding toward the anterior boundary of the LPM before becoming extinguished by 6–8 somite stage (Collignon et al., 1996; Lowe et al., 1996; Meno et al. 1996; Norris et al., 2002). During stages of L-R patterning in mouse, Foxh1, as well as Grg3 and Grg4, are expressed bilaterally in the LPM (Koop et al., 1996; Leon and Lobe, 1997; Weisberg et al., 1998). Colocalization of the proposed switch components in the R LPM, which does not express Nodal but is competent to respond to Nodal signaling (Ohi and Wright, 2007; Yamamoto et al., 2003), as well as in the L LPM that expresses Nodal under rapidly changing spatiotemporal control, further supports the idea that Foxh1 is a transcriptional switch that participates in repressing Nodal transcription. Foxh1 bifunctionality that is conveyed by interchangeable binding of pSmad2 and Grg could facilitate rapid and precise tailoring of target gene transcription to varying thresholds of external instructive signals, as compared to mechanisms that rely on distinct complexes for transcriptional activation and repression (Cinnamon and Paroush, 2008).
Recent work in Xenopus fits the proposal of FoxH1 as such a transcriptional switch with respect to regulating Nodal expression, and with pSmad2 and Grg4 acting as co-activator and co-repressor, respectively. Overexpression of Grg4 in Xenopus embryos strongly reduced Nodal-dependent transcriptional activation, and chromatin-immunoprecipitation (ChIP) showed Grg4 occupancy at the Nodal ASE that depended upon the EH1 motif of FoxH1. ASE occupancy by Grg4 was greatly decreased with Nodal or Smad2 overexpression (D. S. Kessler, personal communication). These results suggest that pSmad2 displaces Grg4 in response to Nodal signaling, and that complementary Grg4 displacement of pSmad2 causes transcriptional repression.
Transcriptional switching has been documented to control target gene transcription in other developmental signaling pathways such as Notch and Wnt (Bray and Furriols, 2001; Daniels and Weis, 2005). For Wnt signaling, β-catenin and Grg co-repressors have overlapping binding domains on Tcf/Lef and compete with each other for occupancy at the locus (Daniels and Weis, 2005). The EH1 motif and SID do not overlap in Foxh1 and it is not known if pSmad2 and Grg engage in direct steric competition in the complete transcriptional complex. Alternatively, unknown components of the transcriptional complex might promote removal of pSmad2/Smad4 and/or Grg, leading to a mutual interference model that is highly dependent upon the levels of activator or repressor.
Knowledge about mechanisms regulating transcriptional repression of Nodal expression is greatly lacking. To determine if Foxh1 is conserved as a transcriptional switch and to identify the developmental consequences of removing the Foxh1-Grg repression arm of this switch, we developed a novel mouse model carrying a new mutant allele, Foxh1mEH1, in which the interaction between Foxh1 and Grg is perturbed. Manipulations made to the Foxh1mEH1 locus were carefully controlled for, by deriving a control line carrying identical insertions while maintaining a wild-type EH1 motif. This study is the first in mouse to address the role of Foxh1 as a transcriptional-switch repressor of Nodal expression. Our findings support the idea that the mammalian embryo has evolved a robust and potentially diverse set of mechanisms to control-Nodal signaling levels and ensure successful germ-layer development and embryonic patterning.
2. Results
2.1. Derivation of Foxh1mEH1/mEH1 mice
In an effort to disrupt binding between Foxh1 and Grg co-repressors, we mutated the Foxh1 EH1 motif, FSIKSLLG. Before mutating the motif in mouse, three different EH1 mutations (eight amino-acid deletion, Ala substitution AAAAAALG, or point mutation ESIKSLLG) were tested for their effect on the production and stability of Foxh1. Western blot analyses on lysates collected from Xenopus embryos that were injected with RNA encoding these Foxh1 variants confirmed that the EH1 mutations did not destabilize or over-stabilize Foxh1 (data not shown). The single amino-acid substitution of F-to-E was chosen because it introduced the fewest Foxh1 sequence alterations and would, therefore, be least likely to affect the overall folding or activator functions of Foxh1 (Fig. 1A), an important consideration when moving forward into the derivation of mutant mouse strains. This minimal mutation has been shown to fully disrupt the interaction between EH1-motif-containing proteins and Grg co-repressors in numerous species (Jennings et al., 2006; Jiménez et al., 1999; Yaklichkin et al., 2007b). Also, the phenylalanine (F) residue, which binds in a hydrophobic recess of the WD-repeat domain of Grg proteins Jennings et al., 2006), is 100% conserved in all EH1 motifs of vertebrate Fox proteins (Yaklichkin et al., 2007a).
Fig. 1.
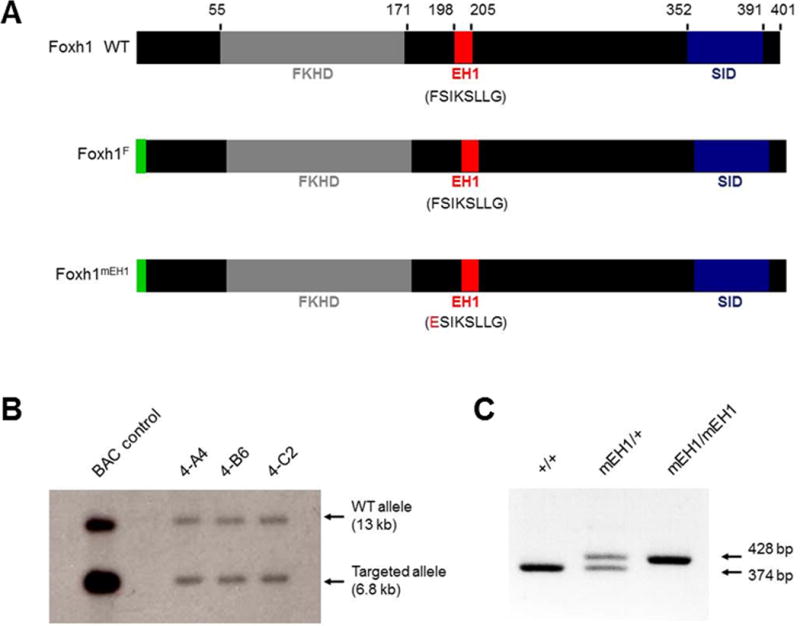
Gene targeting and manipulation of Foxh1 EH1 domain. (A) The locations of FLAG tag (green), forkhead DNA-binding domain (gray), EH1 motif (red), and Smad-interacting domain (blue) in wild-type and variant Foxh1 proteins. Numbers denote amino acid position. Altered EH1 amino acids in Foxh1mEH1 are in red. (B) Southern blot analysis showing correct targeting of Foxh1 locus in three independent mESC clonal cell lines heterozygous for Foxh1LCA allele. (C) PCR analysis of offspring derived from Foxh1mEH1 chimeras detects two bands, 428 bp and 374 bp, for the Foxh1mEH1 and wild-type Foxh1 alleles, respectively.
A single FLAG tag was added to the N-terminus of the mutant EH1 Foxh1 allele (Foxh1mEH1; notation of FLAG tag was avoided in this abbreviated nomenclature for simplicity) to aid in detecting Foxh1mEH1 protein over wild-type Foxh1, and to facilitate future chromatin-binding and target gene studies. The N-terminus was tagged in order to avoid affecting the conformation of, and binding of other factors to, the more C-terminally located SID. The N-terminal tag did not affect the production or stability of Foxh1 via western-blot analysis of lysates collected from Xenopus embryos injected with RNA encoding FLAG-tagged Foxh1 (data not shown). A control mouse line, Foxh1F, which is identical to Foxh1mEH1 except for its wild-type EH1 motif, was derived in parallel, for the most rigorous testing that any abnormal phenotype observed in Foxh1mEH1 mice did in fact arise because of the EH1 mutation and not the FLAG tag or residual exogenous sequences (Lox, FRT, or restriction enzyme sites) inserted into the Foxh1 locus that remain after gene engineering (Fig. 1A; supplementary material Fig. S1).
Prior to deriving the Foxh1mEH1 and Foxh1F mouse strains by inserting them into the Foxh1LCA in mouse ES cells, the mesoderm-inducing and Grg-binding potential of each protein was characterized to confirm that the exogenous sequences did not disrupt Foxh1 activator functions, and that the EH1 mutation abrogated the binding to Grg co-repressors. RNA encoding Foxh1F, Foxh1mEH1, or true wild-type Foxh1 was injected into the animal region of 1-cell Xenopus embryos. Injected embryos were cultured until the start of gastrulation at which time animal caps were cut, cultured overnight, and assessed next day for their elongation, as a measure of the induction, via the Nodal signaling pathway, of mesendoderm and other tissues that undergo convergent extension. Compared to caps from uninjected embryos, which rounded up and became atypical epidermis, caps from all three injection conditions had elongated similarly (data not shown), indicating activation of the mesendoderm-inducing pathways currently held to be controlled by threshold-dependent Nodal signaling (for example: Jones et al., 1995). In addition, quantitative real-time PCR (qRT-PCR) analysis of these “induced animal caps” showed increased relative expression of the mesendodermal markers and Nodal target genes brachyury and goosecoid, as well as the mesodermal derivative muscle actin marker, compared to control caps from uninjected embryos. Conversely, animal caps from embryos producing Foxh1mEH1, Foxh1F, or wild-type Foxh1 had decreased relative expression of the ectodermal marker epidermal keratin compared to uninjected controls (data not shown). Both the animal cap and qRT-PCR assays indicated that addition of the FLAG tag and EH1 mutation did not compromise the ability of Foxh1to cause mesendoderm induction.
To verify that the F-to-E mutation disrupted Foxh1-Grg binding, co-immunoprecipitation (co-IP) assays were performed with homogenized Xenopus gastrula embryos that had been injected at the 1-cell stage with RNA encoding Foxh1F or Foxh1mEH1 alone, or in combination with Myc-tagged Xenopus Grg4 (MycGrg4). Foxh1F and MycGrg4 were reproducibly co-immunoprecipitated with an antibody against Foxh1, but the ability of MycGrg4 to co-IP with Foxh1mEH1 was absent or prominently reduced, even under these overexpression conditions, indicating that the EH1 mutation greatly disrupted Foxh1-Grg4 physical interactions (n = 3 independent injection experiments and western blots) (supplementary material Fig. S2). As argued in the introduction above, the high conservation of the WD-repeat domain among Grg family members means that this mutation likely also disrupts any interactions with other Grg proteins. Based on published in situ hybridization expression patterns, Grg3 and Grg4 are the most likely candidates to function in the transcriptional switch because they, like Foxh1, are expressed symmetrically within the LPM during stages of asymmetric Nodal signaling (Koop et al., 1996; Leon and Lobe, 1997; Weisberg et al., 1998).
A loxed cassette acceptor (LCA) allele of the Foxh1 locus (supplementary material Fig. S1) was established in ES cells, to allow insertion of Foxh1mEH1 and Foxh1F alleles into the endogenous locus through the highly efficient method of recombinase-mediated cassette exchange (RMCE). Analysis of Foxh1 genomic organization across multiple species helped to avoid potentially conserved cis-regulatory regions and locate the least disruptive points for inserting variant Lox sites into the Foxh1 locus. Foxh1 and its neighboring gene Kifc2, a kinesin superfamily member expressed exclusively in neural tissue, are reported to share an overlapping 3′ untranslated region (UTR) (Liu et al., 1999). The lack of apparent phenotype in Kifc2−/− suggests that Kifc2 is dispensable for normal development and behavior (Yang et al., 2001); therefore, a Lox2272 site was placed within its 3′ UTR. Correct targeting of the Foxh1 locus was verified by Southern-blot analysis of BamHI-digested genomic DNA with two probes, a 3′ probe external to the 3′ homology arm and a 5′ probe requiring derivation from within the 5′ homology arm because a repetitive short interspersed nuclear element (SINE) lies just outside the homology arm (Fig. 1B; supplementary material Fig. S1). Chimeras were generated with the Foxh1LCA mESC line, and Southern blotting and PCR analysis confirmed Foxh1LCA germline transmission from them (Fig. 1C). Embryos homozygous for the Foxh1LCA allele resembled Foxh1−/− embryos (data not shown), further verifying that wild-type Foxh1 was correctly targeted and replaced.
The Foxh1mEH1 and Foxh1F RMCE replacement constructs were electroporated into the Foxh1LCA mESC line, and correctly recombined clonal cell lines were injected into C57BL/6 blastocysts to generate Foxh1mEH1 and Foxh1F chimeras. Mice homozygous for the Foxh1mEH1 or Foxh1F allele were born at Mendelian ratio, survived to adulthood, and bred normally. The Foxh1 expression pattern was unchanged in Foxh1F/F and Foxh1mEH1/mEH1 embryos compared to wild-type embryos, indicating that the EH1 mutation and FLAG tag did not disrupt Foxh1 transcription (supplementary material Fig. S3). Foxh1F/F embryos were molecularly and morphologically identical to wild type, therefore Foxh1F/F and wild-type animals were used interchangeably as control embryos.
2.2. Gastrulation and A-P patterning are unaffected in Foxh1mEH1/mEH1 embryos
Since Foxh1-dependent Nodal signaling is essential for proper gastrulation and A-P patterning, E6.75-7.25 Foxh1mEH1/mEH1 embryos were analyzed for defects in cell-lineage allocation and axis orientation. Previously characterized Nodal gain-of-function mutants, such as Lefty2−/− embryos, present with an expanded primitive streak and excess mesoderm (Meno et al., 1999). If Foxh1-Grg–mediated repression is an important negative regulator of Nodal signaling at this stage, loss of the Grg interaction would be expected to cause increased and/or prolonged Nodal signaling within the epiblast, resulting in defects similar to those seen in Lefty2−/− embryos. However, Foxh1mEH1/mEH1 embryos appeared morphologically normal (Fig. 2; see supplementary material Table S1 for n values). Any size difference between wild-type and Foxh1mEH1/mEH1 embryos is attributed to the size and stage variation routinely seen within and between litters at these early developmental stages (Downs and Davies, 1993). The morphological observations were confirmed molecularly by in situ hybridization analyses of Nodal, its downstream targets, and early patterning genes. No spatial or temporal defects in the expression of Nodal (Fig. 2A,B) or its downstream target Lefty2 (Fig. 2C,D) were observed. Otx2 expression, which begins in the visceral endoderm and shifts anteriorly to mark the anterior ectoderm as gastrulation progresses (Zakin et al., 2000), was not altered in mutant embryos, suggesting that regionalized lineage allocation was normal in these embryos (Fig. 2G,H). Additionally, expression of the mesoderm and primitive-streak markers, and targets of Foxh1-dependent transcription, Brachyury/T and Goosecoid were unaffected in Foxh1mEH1/mEH1 embryos, confirming that the primitive streak was correctly patterned (Fig. 2E,F,I,J). Finally, the definitive endoderm marker Foxa2, whose expression is dependent on Foxh1 (Hoodless et al., 2001) and is disrupted in both loss- and gain-of-function Nodal mutants (Perea-Gomez et al., 2002; Vincent et al., 2003), was clearly expressed in the axial midline of early somite mutant embryos (Fig. 2K,L). Combined, these results show that early cell specification and patterning were not altered in Foxh1mEH1/mEH1 embryos, and that Foxh1-Grg–mediated repression is not a primary regulator of Nodal signaling at this stage.
Fig. 2.
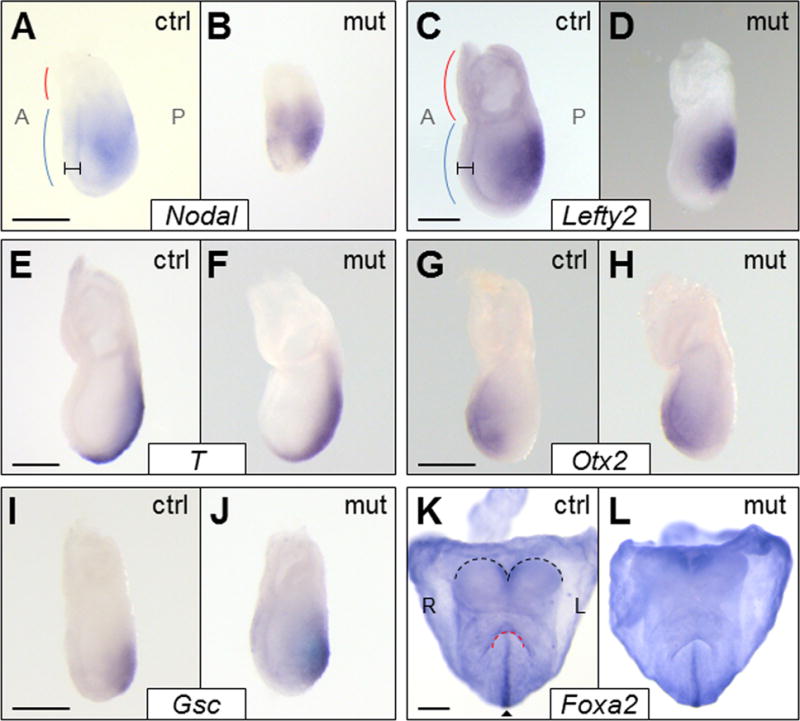
A-P patterning and gastrulation were unaffected in Foxh1mEH1/mEH1 embryos. Expression of multiple A-P patterning genes was examined by whole-mount in situ hybridization in control (ctrl represents wild-type or Foxh1F/F) or Foxh1mEH1/mEH1 (mut) E6.75-E7.25 embryos. The expression patterns of Nodal (A,B), Lefty2 (C,D), Brachyury/T (E,F), Otx2 (G,H), and Gsc (I,J) were similar between control and Foxh1mEH1/mEH1 embryos. Red and blue lines mark representative location of extraembryonic tissue and embryo proper, respectively, in (A) E6.75 and (C) E7.25 embryos. Hash marks denote definitive endoderm. (K,L) Foxa2 expression was unaffected in Foxh1mEH1/mEH1, confirming midline (arrowhead) integrity in mutant embryos. Black and red lines outline headfolds and foregut pocket, respectively. Embryo orientations: (A–J) Lateral views. (K–L) Anterior views. Scale bars: 200 μm. A, anterior; L, left; P, posterior; R, right.
2.3. L-R patterning is unaffected in Foxh1mEH1/mEH1 embryos
We next investigated if Foxh1-Grg–mediated repression regulates Nodal signaling during stages of L-R patterning. As discussed in the Introduction, Nodal expression, as well as that of Lefty2 and Pitx2, is strikingly left-sided, quickly moving through the L LPM in a dynamic posterior-to-anterior wave of expression (Collignon et al., 1996; Lowe et al., 1996; Meno et al., 1996; Ohi and Wright, 2007). Bilateral expression of Foxh1, Smad2/4, and Groucho co-repressors in the LPM (Koop et al., 1996; Leon and Lobe, 1997; Waldrip et al., 1998; Weisberg et al., 1998) suggests that Foxh1 transcriptional switching may regulate the spatiotemporal expression of Nodal circuit genes by initiating their expression in the L LPM and suppressing it in the R LPM. If the Foxh1-Grg repression arm negatively regulated Nodal signaling, it was predicted that disrupting this interaction would result in premature, prolonged, and/or R-sided expression of Nodal and its downstream targets. In all Foxh1mEH1/mEH1 embryos assayed, Nodal was solely L-sided, and not detected in the L LPM prior to 2–3 somites or beyond 8 somites (Fig. 3A,B; supplementary material Fig. S4A–D). As expected, Nodal expression in the node, which is controlled by a Foxh1-independent enhancer, was not altered (data not shown). In accordance with Nodal expression, no spatial or temporal defects were detected in Lefty2 and Pitx2 expression (Fig. 3C–F; supplementary material Fig. S4E–L). From these results we conclude that loss of Foxh1-Grg–mediated repression during L-R patterning can be overcome without compromising development.
Fig. 3.
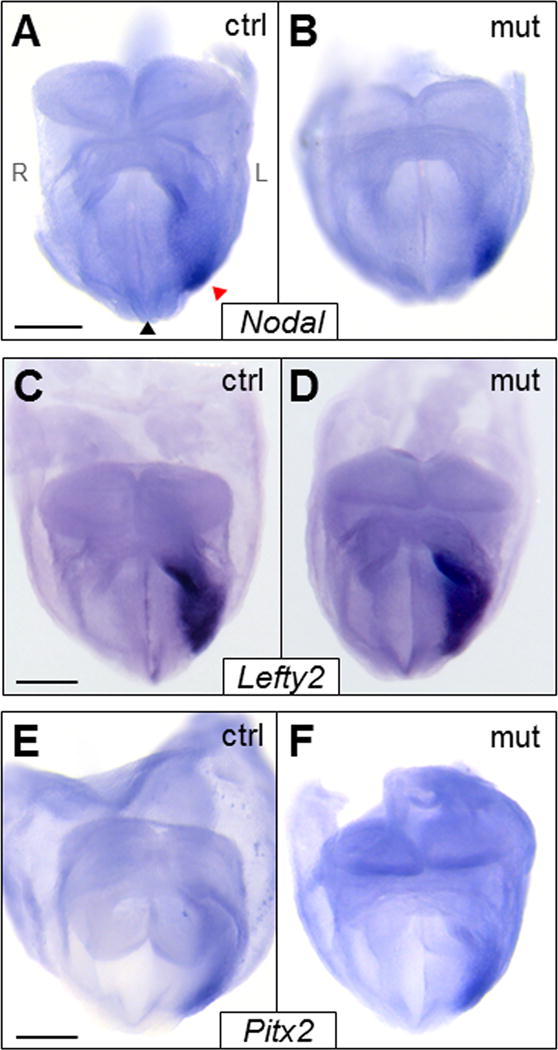
Expression of L-R patterning genes was unaffected in Foxh1mEH1/mEH1 embryos. (A–F) Whole-mount in situ hybridization analysis of L-R patterning genes (A,B) Nodal, (C,D) Lefty2, and (E,F) Pitx2 in the left LPM of control or Foxh1mEH1/mEH1 embryos at E8.25. No aberrant expression of L-R patterning genes was detected in mutant embryos. Black and red arrowheads indicate midline and LPM, respectively. Embryos orientations: (A–F) anterior views. Scale bars: 200 μm. L, left; R, right.
2.4. Foxh1mEH1/mEH1 embryos display normal L-R situs
Nodal signaling (Nodal, Lefty2, Pitx2) in the L LPM directs proper placement and anatomy of the visceral organs (Nakamura and Hamada, 2012). Alterations in Nodal expression, and thus signaling, such as right-sided or bilateral expression, can lead to multiple heart abnormalities, pulmonary isomerism, intestinal malrotation, and misplacement of visceral organs with respect to the midline. Heart abnormalities commonly observed in Nodal signaling mutants include dextrocardia, reversed heart looping, transposition of the great arteries, and ventricular septation (Bisgrove and Yost, 2001). Analysis of heart morphology and placement in mutant E9.5–E10.5 embryos revealed, in all embryos analyzed, stereotypically normal rightward looping and correct cardiac positioning left of midline (Fig. 4A,B). Functional assessment of heart morphology for septal and vessel-transposition defects was done by injecting blue and yellow liquid latex, respectively, into the left and right ventricles of E15.5 Foxh1mEH1/mEH1 embryos to highlight the vasculature and aid in detecting cardiovascular defects. As seen in wild-type embryos, the pulmonary trunk passed ventrally to the aorta in all Foxh1mEH1/mEH1 embryos analyzed, indicating no transposition of the great arteries. Also, no ventricular septations were identified, as mixing of the yellow and blue latexes was not observed in the ventricles of these embryos (Fig. 4C,D). Overall morphology of visceral organs (lungs, liver, gut, stomach) appeared normal in Foxh1mEH1/mEH1 embryos (Fig. 4E–H). These results corroborate our early findings that showed no molecular defects in L-R patterning. Together, these data show that major disruption of the Foxh1-Grg interaction alone does not alter Nodal signaling in the mouse, and that, possibly, the role Foxh1-Grg–mediated repression plays in regulating Nodal transcription is masked by a highly buffered regulatory system, which compensates for its loss in an effort to maintain the spatiotemporal dynamics of Nodal signaling.
Fig. 4.
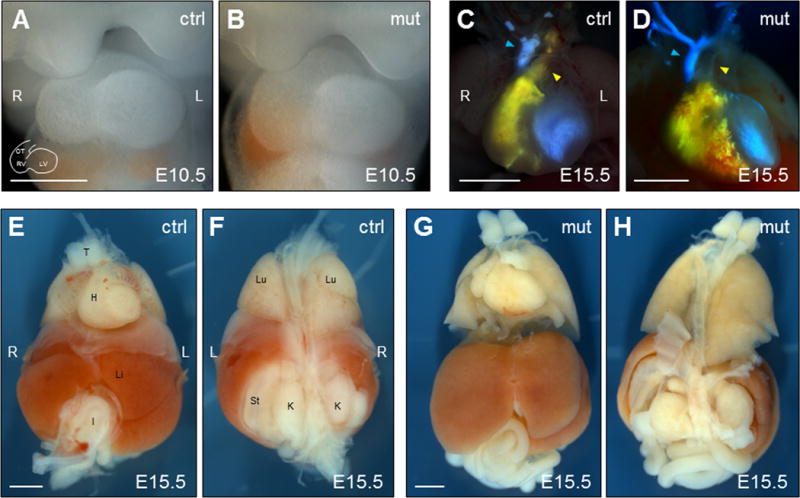
Internal organ situs and morphology was normal in Foxh1mEH1/mEH1 embryos. (A,B) Normal dextral looping of the heart was observed in E10.5 Foxh1mEH1/mEH1 embryos. (C,D) Different colors of liquid latex were injected into the right (yellow) and left (blue) ventricles of E15.5 control and Foxh1mEH1/mEH1 embryos to highlight potential defects in ventricular septation, formation and positioning of the great arteries and aortic arch. No latex mixing, which would have indicated septation defects, was observed in Foxh1mEH1/mEH1 embryos. Also, the pulmonary trunk (yellow arrowhead) always appeared ventral to the aorta (blue arrowhead), and aortic branching was equivalent to control embryos. (E–H) Overall organ placement and morphology was unaffected in Foxh1mEH1/mEH1 embryos at E15.5. Embryo orientations: (A–E, G) Ventral views. (F, H) Dorsal views. Scale bars: (A,B) 500 μm; (C–H) 1 mm. H, heart; I, intestine; K, kidney; L, left; Li, liver; Lu, lung; LV; left ventricle; OT, outflow tract; RV, right ventricle; St, stomach; T, thymus.
3. Discussion
This is the first study to use a precise amino-acid alteration to address the role of Foxh1-Grg–mediated repression in regulating Nodal signaling during early mouse development. We altered the endogenous Foxh1 locus so that our experiments were performed under endogenous levels of gene and protein expression, rather than in an overexpression context, providing the most rigorous way to assay the direct influence of Grg-based repression acting through Foxh1. The minor modifications made to create the Foxh1mEH1 allele were carefully controlled for by the parallel construction of the “wild-type” Foxh1F allele (note that both proteins also contained an N-terminal FLAG tag). We conclude that the blocked interaction between Foxh1 and Grg can be accommodated by adaptive gene regulatory mechanisms, such that there are no major effects on Nodal signaling at the level of target gene readout, or on overall body axis formation, anterior-posterior or left-right patterning. Our results suggest that there is substantial robustness in the mechanisms that have evolved to stabilize Nodal regulatory systems and lead to successful germ-layer development and embryonic patterning.
3.1. Buffered regulatory system potentially compensates for loss of Foxh1-Grg–mediated repression
Cell fate determination is qualitatively influenced by the level and duration of Nodal signaling. Experiments in Xenopus embryos showed that higher Nodal/Activin signaling induced the mesendoderm marker gsc, while lower levels induced the pan-mesodermal marker xbra (Agius et al., 2000; Gurdon et al., 1994; Jones et al., 1995). Dose-dependent responses also control mouse development (Robertson, 2014). In Nodal null embryos, little mesodermal differentiation occurs and there is subsequent failure to gastrulate, causing developmental arrest (Conlon et al., 1994; Zhou et al., 1993). Mesoderm induction is restored in mutants with reduced Nodal expression, but definitive endoderm is still absent (Norris et al., 2002; Vincent et al., 2003). Left-right patterning also depends upon the level of the Nodal signaling component Pitx2, a transcriptional effector that is essential for asymmetric organogenesis (Nakamura and Hamada, 2012). Experiments with Pitx2 revealed a progressive gene-dosage requirement in organ formation and L-R patterning: lung and gut required the highest level of Pitx2 expression, followed by the heart, with the stomach requiring the least (Gage et al., 1999; Liu et al., 2001). Because proper embryonic patterning depends upon the precise titration of Nodal signaling, it is likely that buffered regulatory systems were developed to provide stability but also rheostat-like level control. The employment of such a system may be one explanation for the lack of phenotype in FoxhmEH1/mEH1 embryos.
Tissue-specific expression of Nodal is, in part, positively regulated by five different enhancers that drive expression in the epiblast, node, and LPM (Adachi et al., 1999; Papanayotou et al., 2014; Saijoh et al., 2005; Vincent et al., 2003). The dynamic expression in the epiblast and LPM is also strongly affected by a self-enhancement and lateral-inhibition (SELI) system through which Nodal enhances its own expression with a positive feed-forward loop that activates through the ASE. Nodal then rapidly initiates expression of its feedback antagonist Lefty2 (Nakamura et al., 2006). Lefty2 molecules, which have an inherent ability to travel faster and farther than Nodal, and have greater stability, help terminate Nodal expression in the L LPM, as well as prevent the Nodal autoregulatory loop from fully initiating in the R LPM (Marjoram and Wright, 2011; Müller et al., 2012; Nakamura et al., 2006). Lefty2 inhibits Nodal signaling by either binding directly to the Nodal dimer, or competitively blocking access to the obligate EGF-CFC co-receptors (Chen and Shen, 2004). Such inhibition by Lefty2 prevents phosphorylation and prevents Smad2 shuttling to the nucleus, shutting down Foxh1-dependent transcription of target genes.
We speculate that the Nodal locus could be more “open” in Foxh1mEH1/mEH1 embryos because of impaired interaction with Grg co-repressors. However, attenuation of Smad2 phosphorylation by Lefty2 inhibition at the cell surface should still occur in mutant embryos, which would dampen the potential over-activation of Nodal expression. Thus, Lefty2 would be an especially important component of the buffered regulatory system that compensates for the reduction/loss of Foxh1-Grg–mediated repression in Foxh1mEH1 mutant mice, preventing a mutant phenotype. Such buffering could be less apparent in the overexpression studies in Xenopus that implicate Foxh1 as a repressor of Nodal transcription (D. S. Kessler, personal communication), as compared to this study with Foxh1mEH1 being expressed from the endogenous locus.
That a buffered system exerts meticulous control over Nodal signaling is suggested by the multiple Nodal signaling loss- and gain-of-function mutations that display incomplete penetrance or variable expressivity. For example, the severity of defects in Foxh1−/− embryos varies greatly, from completely failed development of the embryo proper, to a less severe phenotype without midline structures (Hoodless et al., 2001; Yamamoto et al., 2001). Similarly, embryos lacking the co-receptor Cryptic have numerous defects in asymmetric organogenesis, but the specific organs affected differ among mutant embryos (Yan et al., 1999). Incomplete penetrance is seen in embryos that lack Lefty2 in the LPM, with two-thirds of the animals dying a few days after birth from cardiac defects, which were not detected in mutants that survived to adulthood (Meno et al., 2001). Homozygous mutations in the Nodal antagonists Lefty1 and Cerl alone do not cause defects in gastrulation (Meno et al., 1998; Simpson et al., 1999), but the double homozygous Lefty1−/−;Cerl−/− mutants are 100% embryonic lethal (Perea-Gomez et al., 2002). These results demonstrate that loss of a single negative regulator of Nodal signaling can be compensated for, and that removal of additional members of the buffered regulatory system is required to manifest an abnormal phenotype.
Therefore, it is possible that this new Foxh1mEH1/mEH1 condition represents a sensitized state in which relatively subtle future manipulation of other components of the buffered system could reveal patterning defects. Example experiments could test how the loss of Foxh1-Grg–mediated repression is compensated. For example, loss of Lefty2 in the Foxh1mEH1 background may yield patterning deficits that have so far remained hidden under disruption of only the Foxh1-Grg interaction. While our Foxh1mEH1 amino-acid alteration removes a residue that is essential for tight Grg interaction (Jennings et al., 2006), we must remain open to the possibility that the primary explanation for no abnormal phenotype is that some low-level Foxh1mEH1-Grg association is sufficient to effect substantial repression of Nodal transcription. The strength of the data indicating the importance of the phenylalanine residue for Grg interaction strongly suggests that any residual Grg binding might occur not through the highly disrupted EH1 motif, but indirectly through another member of the as-yet-uncharacterized transcriptional complex. An even simpler interpretation is that Grg-mediated repression of Nodal signaling is not conserved in the mouse. The sensitization experiments described above are pertinent and rely on our findings here as the foundation. In addition, experiments investigating the type of epigenetic repression marks present at the Nodal locus in Foxh1mEH1 mutant embryos compared to control embryos could help determine the extent of repression that remains in Foxh1mEH1 embryos.
3.2. Epigenetic regulation of Nodal signaling
The epigenetic landscape provides critical input onto a gene’s transcriptional state, but almost nothing is known about such landscape alterations at the Nodal locus during development, or the degree to which epigenetic modifications contribute to Nodal signaling regulation. It is well documented that pSmad2/Smad4 recruits the co-activators p300 and CREB binding protein (CBP), which use intrinsic histone-acetyltransferase activity to promote an open-chromatin conformation (Attisano and Wrana, 2000). The ability of Foxh1 to interact with factors that recruit HDACs, which are capable of reversing the pSmad2/Smad4-founded markings, further supports Foxh1 transcriptional switching as a plausible epigenetic mode of modulating Nodal signaling.
Recent studies report roles for histone acetylation and methylation in the positive and negative regulation of Nodal signaling in mouse ESCs and in embryos. In mouse ESCs, Nodal-Smad2/3 signaling recruits Jmjd3, a H3k27me3-specific demethylase, to Nodal target-genes to counteract the repressive effect of Polycomb repressive complex 2 (PRC2) (Dahle et al., 2010). On the other hand, studies in Medaka fish report physical interaction of Foxh1with the PRC2 histone-modifier Ezh1, with Ezh1 knockdown producing bilateral Nodal expression and L-R patterning defects (Arai et al., 2010). Because PRC2 can be recruited by Grg co-repressors (Patel et al., 2012), Foxh1, through interaction with Grg, could initiate longer-term transcriptional repression. Also, others have already implied that HDACs could be part of the repression mechanism for Nodal expression in the R LPM (Carneiro et al., 2011).
Understanding whether or not the Nodal locus remains poised or repressed during time points when Nodal is not expressed may provide insight into the flexible regulation of Nodal signaling during embryogenesis, and the specific levels of signaling that transcriptionally activate the various sets of downstream targets. We also speculate that the Grg-mediated long-term repression of Nodal expression might be essential in preventing neoplastic reactivation of the Nodal signaling pathway. The Foxh1F line will be useful for studying the epigenetic regulation occurring at the Nodal locus, and other ASE-containing loci, as well as for identifying novel targets of Foxh1.
4. Materials and Methods
4.1. Derivation of Foxh1mEH1 and Foxh1F mice
The Foxh1LCA, Foxh1F, and Foxh1mEH1 mouse lines were derived with help from Vanderbilt Transgenic Mouse/Embryonic Stem Cell Shared Resource. A 129S6 Foxh1LCA ES cell line was generated through homologous recombination of a targeting vector that was created by BAC recombineering with the BAC bMQ206P16 (BAC PAC Resources at C.H.O.R.I., Oakland, Ca.). Colonies surviving selection were screened by Southern blot analysis (5/137 colonies correctly targeted Foxh1) and karyotyped. Foxh1LCA chimeras were generated to validate germline transmissibility of Foxh1LCA ES cell clones. Exchange vectors for the Foxh1F and Foxh1mEH1 alleles were generated by PCR-based site-directed mutagenesis of the Foxh1 locus. Recombinase-mediated cassette exchange (RMCE) between the Foxh1LCA ES cell clone 4B6 and exchange vectors was used to generate Foxh1F and Foxh1mEH1 ES cell lines. Colonies surviving selection were screened with PCR and karyotyped. One clone for each line was injected into C57BL/6 embryos for generation of chimeras. Following the generation of founder mice for the Foxh1F and Foxh1mEH1 alleles, hygromycin resistance selection cassettes were excised by mating to mice with FLPe (Rodriguez et al., 2000). Flp-mediated deletion was confirmed with the primers 5′ AGC TGC CCA TTG TAG TAG C 3′ and 5′ CAA AGT GAG TTC CAG GAC A 3′. A schematic of the Foxh1LCA, Foxh1F, and Foxh1mEH1 alleles is provided in supplementary material Fig. S1.
4.2. Mouse husbandry
Animal handling was approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee. Foxh1mEH1 and Foxh1F animals were genotyped by PCR (primers 5′ ACT TGG GAA ACC ACT TGG TC 3′ and 5′ TTG ACT CTT GAA CCT CCA GG 3′). Experimental embryos resulted from crosses between mice homozygous for the Foxh1mEH1, Foxh1F, or wild-type Foxh1 allele.
4.3. Xenopus embryo manipulations and microinjections
Plasmids for generating RNA in vitro were generated by inserting cDNAs encoding Foxh1F or Foxh1mEH1 into pCS2+ (from Dr. Dave Turner, University of Michigan). An injection construct encoding Myc-tagged Xenopus Grg4 (MycGrg4) was a gift from Dr. Daniel Kessler, University of Pennsylvania. Capped RNA was produced using the mMessage mMachine kit (Ambion). RNA encoding Foxh1F or Foxh1mEH1 (2.0 ng) was injected alone or in combination with MycGrg4 (2.0 ng) into the animal caps of 1–2 cell Xenopus embryos. Embryos were cultured in 1x Steinberg’s solution until stage 10, when they were frozen and stored at −20°C for use in co-immunoprecipitations (co-IPs). Embryos were staged according to (Nieuwkoop and Faber, 1967).
4.4. Co-immunoprecipitation and immunoblotting
Ten Xenopus embryos were used in each co-IP. Embryos were homogenized (buffer: 20 mM Tris.HCl pH 8.0, 2.0 mM EDTA, 5.0 mM EGTA, 0.5% NP-40, 1x protease inhibitor cocktail [Roche]), incubated on ice 10 minutes, and centrifuged at 2,000 × g for 5 minutes at 4°C. Centrifuged lysates were removed from the pellet and pellicle and incubated with 4 μg rabbit anti-Foxh1 (ab49133, Abcam) overnight at 4°C mixing end-over-end on a hematological mixer. Lysate-antibody mixtures were then incubated with magnetic Dynabeads Protein G (Life Technologies) for 4–5 hours at 4°C on a hematological mixer. After incubation, beads were boiled in 4x sample reducing buffer (0.125 M Tris-HCl pH 6.8, 4% SDS, 20% v/v glycerol, 0.2 M DTT, 0.02% bromophenol blue). Samples were separated on NuPage 10% Bis-Tris gels (Life Technologies), transferred onto PVDF (0.45 μM, Millipore), and blocked with 7–10% non-fat dry milk dissolved in TBS-Tween (0.1%) for 30–45 minutes at room temperature. Membranes were incubated with monoclonal mouse anti-FLAG M2 (1:3,000; 200472, Agilent Technologies) or rabbit anti-Myc (1:3,000; 06-549, Millipore) in blocking solution overnight at 4°C. Specifically bound primary antibodies were detected with HRP-conjugated goat anti-mouse IgG1 (1:7,500; sc2969, Santa Cruz Biotechnology) or Clean-Blot IP detection reagent HRP (1:4,000; 21230, Thermo Scientific) for 45 minutes at room temperature. Amersham ECL Prime western blotting detection reagent (GE Healthcare) was used for chemiluminescence and membranes were exposed to film (GenHunter) for various time periods.
4.5. In situ hybridizations
Embryos were staged based on morphology according to (Downs and Davies, 1993). Whole-mount in situ hybridization was as described (Belo et al., 1997), except antibody was used at 1:5,000, with DIG labeled Nodal, Lefty2, Pitx2, Foxh1, Otx2, Brachyury/T, Gsc, and Foxa2 RNA probes. Developed embryos were imaged with a Leica M165 FC microscope.
4.6. Latex injections
Ventricles of E15.5 embryos were injected with colored, liquid latex (Connecticut Valley Biological Supply) as described (Oh and Li, 1997). Hearts were allowed to beat several times to help circulate latex before fixing whole embryos in 4% paraformaldehyde at 4°C. Embryos were imaged with a Leica M165 FC microscope.
Supplementary Material
Highlights.
Foxh1-mediated repression of Nodal in gastrulation, left-right asymmetry addressed in mouse.
Grg-binding motif (EH1) in Foxh1 altered to block Groucho (Grg) interaction.
EH1 mutation greatly disrupts Foxh1 binding to Grg co-repressors.
Disrupting Foxh1-Grg interaction does not alter Nodal expression or embryogenesis.
Buffered regulatory system likely compensates for reduced Foxh1-Grg binding.
Acknowledgments
We thank Trish Labosky, Yukio Saijoh, and Ken Zaret for in situ hybridization probes, Mark Magnuson for BAC recombineering and RMCE plasmids, Dan Kessler for sharing his data prepublication, and Guoqiang Gu and Wright Lab members for discussion. Use of the Vanderbilt Transgenic Mouse/ESC Shared Resource was funded in part by the Vanderbilt-Ingram Cancer Center Support Grant (CA68485), the Vanderbilt Diabetes Research and Training Center (DK20593), the Vanderbilt Brain Institute, and the Vanderbilt Center for Stem Cell Biology. This research was supported by the National Institutes of Health [RO1GM56238] and the Louise B. McGavock Chair in Cell and Developmental Biology to C.V.E.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi H, Saijoh Y, Mochida K, Ohishi S, Hashiguchi H, Hirao A, Hamada H. Determination of left/right asymmetric expression of Nodal by a left side-specific enhancer with sequence similarity to a Lefty-2 enhancer. Genes Dev. 1999;13:1589–1600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agius E, Oelgeschläger M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–1183. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai D, Katsura H, Shindo N, Matsumoto M, Higashinakagawa T. Polycomb group protein Ezh1 represses Nodal and maintains the left-right axis. Dev Biol. 2010;341:459–463. doi: 10.1016/j.ydbio.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- Belo JA, Bouwmeester T, Leyns L, Kertesz N, Gallo M, Follettie M, De Robertis EM. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech Develop. 1997;68:45–57. doi: 10.1016/s0925-4773(97)00125-1. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Yost HJ. Classification of left-right patterning defects in zebrafish, mice, and humans. Am J Med Genet. 2001;101:315–323. doi: 10.1002/ajmg.1180. [DOI] [PubMed] [Google Scholar]
- Bray S, Furriols M. Notch pathway: Making sense of Suppressor of Hairless. Curr Biol. 2001;11:R217–R221. doi: 10.1016/s0960-9822(01)00109-9. [DOI] [PubMed] [Google Scholar]
- Carneiro K, Donnet C, Rejtar T, Karger BL, Barisone GA, Díaz E, Kortagere S, Lemire JM, Levin M. Histone deacetylase activity is necessary for left-right patterning during vertebrate development. BMC Dev Biol. 2011;11:1–19. doi: 10.1186/1471-213X-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- Chen C, Shen MM. Two modes by which Lefty proteins inhibit Nodal signaling. Curr Biol. 2004;14:618–624. doi: 10.1016/j.cub.2004.02.042. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18:435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Dahle Ø, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract Polycomb-mediated repression at target genes. Sci Signal. 2010;3:1–8. doi: 10.1126/scisignal.2000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. β-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Harger P, Mitchell A, Lemaire P. Activin signaling and response to a morphogen gradient. Nature. 1994;371:487–492. doi: 10.1038/371487a0. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbé E, Attisano L, Rossant J, Wrana JL. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Gene Dev. 2001;15:1257–1271. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 2008;9:205.1–205.7. doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Jiménez G, Verrijzer CP, Ish-Horowicz D. A conserved motif in Goosecoid mediates Groucho-dependent repression in Drosophila embryos. Mol Cell Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BLM, Smith JC, Wright CVE. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development. 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- Koop KE, MacDonald LM, Lobe CG. Transcripts of Grg4, a murine Groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mech Develop. 1996;59:73–87. doi: 10.1016/0925-4773(96)00582-5. [DOI] [PubMed] [Google Scholar]
- Leon C, Lobe CG. Grg3, a murine Groucho-related gene, is expressed in the developing nervous system and in mesenchyme-induced epithelial structures. Dev Dynam. 1997;208:11–24. doi: 10.1002/(SICI)1097-0177(199701)208:1<11::AID-AJA2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Liu B, Dou C, Prabhu L, Lai E. FAST-2 is a mammalian winged-helix protein which mediates transforming growth factor β signals. Mol Cell Biol. 1999;19:424–430. doi: 10.1128/mcb.19.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu W, Lu M, Brown NA, Martin JF. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–2048. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- Lowe LA, Supp DM, Sampath K, Yokoyama T, Wright CVE, Potter SS, Overbeek P, Kuehn MR. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- Lu CC, Brennan J, Robertson EJ. From fertilization to gastrulation: axis formation in the mouse embryo. Curr Opin Genet Dev. 2001;11:384–392. doi: 10.1016/s0959-437x(00)00208-2. [DOI] [PubMed] [Google Scholar]
- Marjoram L, Wright C. Rapid differential transport of Nodal and Lefty on sulfated proteoglycan-rich extracellular matrix regulates left-right asymmetry in Xenopus. Development. 2011;138:475–485. doi: 10.1242/dev.056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGFβ-family member lefty in mouse embryos. Nature. 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. Lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, et al. Mouse Lefty2 and zebrafish Antivin are feedback inhibitors of Nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–298. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- Meno C, Takeuchi J, Sakuma R, Koshiba-Takeuchi K, Ohishi S, Saijoh Y, Miyazaki J, ten Dijke P, Ogura T, Hamada H. Diffusion of Nodal signaling activity in the absence of the feedback inhibitor Lefty2. Dev Cell. 2001;1:127–138. doi: 10.1016/s1534-5807(01)00006-5. [DOI] [PubMed] [Google Scholar]
- Müller P, Rogers KW, Jordan BM, Lee JS, Robson D, Ramanathan S, Schier AF. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science. 2012;336:721–724. doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hamada H. Left-right patterning: conserved and divergent mechanisms. Development. 2012;139:3257–3262. doi: 10.1242/dev.061606. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mine N, Nakaguchi E, Mochizuki A, Yamamoto M, Yashiro K, Meno C, Hamada H. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus Laevis. Amsterdam, The Netherlands: North Holland Publishing Co; 1967. [Google Scholar]
- Norris DP, Robertson EJ. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes Dev. 1999;13:1575–1588. doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Oh SP, Li E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 1997;11:1812–1826. doi: 10.1101/gad.11.14.1812. [DOI] [PubMed] [Google Scholar]
- Ohi Y, Wright CVE. Anteriorward shifting of asymmetric Xnr1 expression and contralateral communication in left-right specification in Xenopus. Dev Biol. 2007;301:447–463. doi: 10.1016/j.ydbio.2006.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S, Saijoh Y, Frisch A, Yeo C, Adachi H, Watanabe M, Whitman M, Hamada H, Wright CVE. Activin/Nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development. 2000;127:2503–2514. doi: 10.1242/dev.127.11.2503. [DOI] [PubMed] [Google Scholar]
- Papanayotou C, Benhaddou A, Camus A, Perea-Gomez A, Jouneau A, Mezger V, Langa F, Ott S, Sabéran-Djoneidi D, Collignon J. A novel Nodal enhancer dependent on pluripotency factors and Smad2/3 signaling conditions a regulatory switch during epiblast maturation. PLoS Biol. 2014;12:e1001890. doi: 10.1371/journal.pbio.1001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Mol Cell. 2012;45:1–11. doi: 10.1016/j.molcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FDJ, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, et al. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Robertson EJ. Dose-dependet Nodal/Smad signals pattern the early mouse embryo. Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.03.028. (in press) [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency delete mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Mochida K, Ohishi S, Hirao A, Hamada H. Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes Dev. 1999;13:259–269. doi: 10.1101/gad.13.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Sakuma R, Yeo C, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata K, et al. Left-Right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Oki S, Tanaka C, Nakamura T, Adachi H, Yan Y, Shen MM, Hamada H. Two Nodal-responsive enhancers control left-right asymmetric expression of Nodal. Dev Dynam. 2005;232:1031–1036. doi: 10.1002/dvdy.20192. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1(5):a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Shen MM. Nodal signaling in vertebrate development. Nature. 1999;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- Shen MM, Schier AF. The EGF-CFC gene family in vertebrate development. Trends Genet. 2000;16:303–309. doi: 10.1016/s0168-9525(00)02006-0. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada W. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by Nodal signaling and maintenance by Nkx2. Mol Cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- Simpson EH, Johnson DK, Hunsicker P, Suffolk R, Jordan SA, Jackson IJ. The mouse Cer1 (Cerberus related or homologue) gene is not required for anterior pattern formation. Dev Biol. 1999;213:202–206. doi: 10.1006/dbio.1999.9372. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Winnier GE, Chen X, Farnsworth CL, Hogan BLH, Whitman M. A mouse homologue of FAST-1 transduces TGFβ superfamily signals and is expressed during early embryogenesis. Dev Dynam. 1998;79:17–27. doi: 10.1016/s0925-4773(98)00160-9. [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS. TGF-β superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Yaklichkin S, Vekker A, Stayrook S, Lewis M, Kessler DS. Prevalence of the EH1 Groucno interaction motif in the metazoan Fox family of transcriptional regulators. BMC Genomics. 2007a;8:1–17. doi: 10.1186/1471-2164-8-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J Biol Chem. 2007b;282:2548–2557. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Meno C, Sakai Y, Shiratori H, Mochida K, Ikawa Y, Saijoh Y, Hamada H. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev. 2001;15:1242–1256. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Mine N, Mochida K, Sakai Y, Saijoh Y, Meno C, Hamada H. Nodal signaling induces the midline barrier by activating Nodal expression in the lateral plate. Development. 2003;130:1795–1804. doi: 10.1242/dev.00408. [DOI] [PubMed] [Google Scholar]
- Yan Y, Gritsman K, Ding J, Burdine RD, Corrales JD, Price SM, Talbot WS, Schier AF, Shen MM. Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev. 1999;13:2527–2537. doi: 10.1101/gad.13.19.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Roberts EA, Goldstein LSB. Functional analysis of mouse C-terminal kinesin motor KifC2. Mol Cell Biol. 2001;21:2463–2466. doi: 10.1128/MCB.21.7.2463-2466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakin L, Reversade B, Virlon B, Rusniok C, Glaser P, Elelouf J, Brûlet P. Gene expression profiles in normal and Otx2−/− early gastrulating mouse embryos. P Natl Acad Sci USA. 2000;97:14388–14393. doi: 10.1073/pnas.011513398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BLM, Kuehn MR. Nodal is a novel TGF-β-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


