Summary
Understanding the basis of bacterial persistence in latent infections is critical for eradication of tuberculosis. Analysis of Mycobacterium tuberculosis mRNA expression in an in vitro model of non-replicating persistence indicated that the bacilli require electron transport chain components and ATP synthesis for survival. Additionally, low μM concentrations of aminoalkoxydiphenylmethane derivatives inhibited both the aerobic growth and survival of non-replicating, persistent M. tuberculosis. Metabolic labeling studies and quantitation of cellular menaquinone levels suggested that menaquinone synthesis, and consequently electron transport, is the target of the aminoalkoxydiphenylmethane derivatives. This hypothesis is strongly supported by the observations that treatment with these compounds inhibits oxygen consumption and that supplementation of growth medium with exogenous menaquinone rescued both growth and oxygen consumption of treated bacilli. In vitro assays indicate that the aminoalkoxydiphenylmethane derivatives specifically inhibit MenA, an enzyme involved in the synthesis of menaquinone. Thus, the results provide insight into the physiology of mycobacterial persistence and a basis for the development of novel drugs that enhance eradication of persistent bacilli and latent tuberculosis.
Introduction
It is estimated that more than one-third of the world’s population is infected with tubercle bacilli and that 5–10% of these individuals develop tuberculosis (TB) at some point in their lifetime. As a result TB infections resulted in a death toll of approximately 1.7 million people this year (Anonymous, 2007). Mycobacterium tuberculosis, the etiological agent of TB, is able to persist inside the host without causing clinical symptoms (latent infection), a condition which may exist throughout the host’s life; however, latent disease may convert to active disease under appropriate conditions.
During latent infection, it is thought that a dynamic balance exists between the host immune response and the pathogen (Stewart et al., 2003;Ulrichs and Kaufmann, 2006;Ulrichs and Kaufmann, 2002;Wayne and Sohaskey, 2001), and that the bacilli are in a quiescent state of non-replicating persistence (NRP) that is refractory to the commonly used anti-TB drugs. When the host/pathogen balance is disturbed, such as when the immune system is compromised, active disease may ensue. It is not clear whether the interaction between host and pathogen is responsible for the maintenance of a latent infection, or if M. tuberculosis actively shuts down metabolic activity and replication. These issues are still subject to debate (Cosma et al., 2003;Ulrichs and Kaufmann, 2006;Orme, 2001) and it has even been suggested that bacterial metabolic activity may be completely shut down in NRP; however, this hypothesis seems unlikely.
In attempts to reproduce NRP in vitro many environmental factors have been manipulated including oxygen tension, nutrient status, pH and nitric oxide levels (Cosma et al., 2003;Wayne and Sohaskey, 2001;Gomez and McKinney, 2004). Of these environmental factors hypoxia has received the most attention and represents a common theme with the immune hypothesis, since it has long been thought that oxygen depletion is a hallmark of some lesions formed in tuberculosis (Wayne and Sohaskey, 2001;Boshoff and Barry, III, 2005;Ulrichs and Kaufmann, 2006). Although considered obligatory aerobes, tubercle bacilli have shown the ability to adapt and persist under reduced oxygen tension. While abrupt depletion of oxygen is deleterious to the overall viability of the bacterial population, gradual depletion, in vitro, leads to progression through at least two NRP stages (Wayne and Sohaskey, 2001), suggesting that establishing NRP requires adaptation. Recently, it has been shown that adaptation of M. tuberculosis to host immunity also involves successive changes in respiratory state. Parallel transcriptional profiling of M. tuberculosis respiratory pathways and ATP synthetic apparatus during mouse respiratory tract infection and the Wayne model of oxygen depletion (Wayne and Hayes, 1996) has identified important similarities consistent with bacterial growth arrest (Shi et al., 2005), suggesting that electron transport and oxidative phosphorylation play critical roles in NRP. A hypothesis supported by the observations that maintenance of ATP homeostasis and proton motive force is important for the survival of M. tuberculosis in NRP (Koul et al., 2008;Rao et al., 2008).
Although there is little functional data, the sequencing of the M. tuberculosis genome (Cole et al., 1998) provided evidence that there could be systems for aerobic, microaerophilic and, perhaps, anoxic electron transport (Wheeler and Blanchard, 2005). The oxidative electron transport system that likely operates in low oxygen conditions has been demonstrated in mycobacteria (Kana et al., 2001). A detailed characterization of an aerobic respiratory chain in M. tuberculosis showed that NADH:menaquinone oxidoreductase is a viable target for anti-tubercular agents (Weinstein et al., 2005), and the cytochrome bc1-aa3 complex terminating in the aa3-type cytochrome C oxidase has been characterized (Matsoso et al., 2005).
Overall, the electron transport chains of mycobacteria appear to be modular in nature, as are those of Escherichia coli (Gennis and Stewart, 1996;Shi et al., 2005). That is, different components of the system can be substituted in the membrane in addition to, or in place of, other components as needed. In general terms, there are three components: 1) substrate-specific dehydrogenases; 2) lipoquinones; 3) terminal oxidoreductases. All of the dehydrogenases are lipoquinone reductases and all of the terminal oxidoreductases are lipoquinol oxidases (Gennis and Stewart, 1996) as is shown schematically in Fig. 1. What becomes immediately obvious from this schematic is that the lipoquinones occupy a central and essential bottleneck in the electron transport chain.
Fig. 1. The electron transport chain in mycobacteria and synthesis of menaquinone.
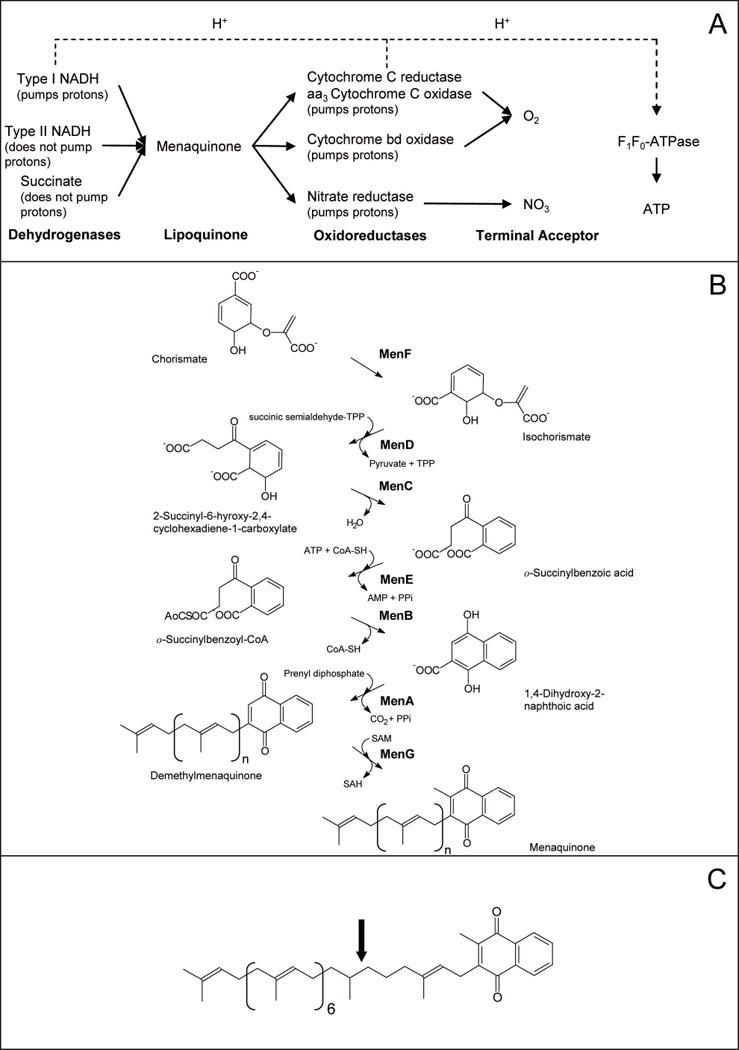
Panel A: Architecture of selected aerobic respiratory pathways in mycobacteria. Panel B: Biosynthetic pathway for menaquinone in E. coli. Enzyme names are indicated in bold. C: Structure of the major menaquinone species found in mycobacteria, MK-9 (II-H2) (Minnikin, 1982); calculated monoisotopic mass = 786.63148. Menaquinones are identified by the length and chemical structure of prenyl chain, the predominant form of menaquinone in mycobacteria has 9 isoprene units with the second one being saturated (indicated by the arrow). Hence this menaquinone is identified as MK-9 (II-H2). The fully unsaturated version is known as MK-9 and has a calculated monoisotopic mass of 784.61583
The lipoquinones involved in the respiratory chains of bacteria consist of menaquinones and ubiquinones (Sherman et al., 1989), while mammals have only ubiquinone. Menaquinones (2-methyl-3-polyprenyl-1,4-naphthoquinones) are the predominant lipoquinones of mycobacteria and other Gram-positive bacteria, whereas Gram-negative bacteria such as E. coli typically utilize both menaquinone and ubiqinone or ubiquinone solely (Collins and Jones, 1981;Embley and Stackebrandt, 1994;Meganathan, 1996;Minnikin, 1982;Pandya and King, 1966).
In E. coli the synthesis of menaquinone is accomplished by seven enzymes, MenA-MenG (Fig. 1). These have been identified due to the availability of the men-mutants, which were generated under aerobic conditions where ubiquinone is utilized as the electron carrier. However, the synthesis of this molecule in pathogenic, Gram-positive organisms has received little attention. Since it has recently been shown that both the electron transport system and ATP synthesis are viable anti-TB drug targets (Andries et al., 2005;Weinstein et al., 2005) it was hypothesized that menaquinone synthesis could be critical for mycobacterial survival and the following studies were undertaken.
Results
Transcripts of genes involved in ATP synthesis and electron transport are abundant under extended periods of oxygen depletion
The total number of transcriptionally active open reading frames was found to be significantly reduced after six months of exposure to gradual oxygen depletion in the Wayne model (NRP) compared to exponentially growing cultures under aerobic conditions. However, when the data were normalized as described in the Experimental Procedures, transcripts that are most abundantly represented in the pool of total mRNA expressed in M. tuberculosis during NRP could be identified (Supplemental Table S1). Using this normalization technique, a gene with an expression index of 1 represents the same proportion of the total mRNA in both conditions, whereas a gene with an expression index of >1 represents a greater proportion of the total mRNA in NRP than in exponential growth. In total, 435 ORFs were found to have an expression index of >1 in non-replicating, persistent bacilli (NRPB) indicating that 10.9% of the M. tuberculosis genes may be preferentially expressed as a proportion of the total mRNA during NRP to support the minimal metabolic activities required for bacterial maintenance. The genes with an expression index of >1 encoded components of lipid metabolism (9%), cell wall metabolism (18%), general metabolism and respiration (28%), unknown or conserved hypothetical ORFs (28%), information pathways (9%), and regulation (5%). Among the genes with the highest expression indices were ORFs encoding products involved in ATP synthesis (with values ranging between 3.1 and 14.8, Supplemental Table S1), coenzyme and NADH metabolism and aerobic and microaerobic respiration. Of these, all of the genes encoding the F1F0-ATP synthase (atpA-atpH), cytochrome C reductase (qcrC, qcrA and qcrB) and the aa3 cytochrome C oxidase (ctaC, ctaD and ctaE), which form a supercomplex in Corynebacterium glutamicum (Niebisch and Bott, 2003) had elevated expression indices (Table 1). Thus, the results suggested that electron transport and ATP synthesis are critical in maintenance of NRP.
Table 1.
Differential expression and essentiality of genes encoding proteins involved in electron transport and oxidative phosphorylation in NRP.
| Function | Expression index > 1 in Wayne model |
|---|---|
|
F1F0 ATP synthase (atpA, atpB, atpC, atpD, atpE, atpF, atpG, and atpH) |
All |
|
Type 1 NADH Dehydrogenase (nuoA, nuoB, nuoC, nuoD, nuoE, nuoF, nuoG, nuoH, nuoI, nuoJ, nuoK, nuoL, nuoM, nuoN) |
nuoC, nuoD, nuoF, nuoG, nuoK, nuoM |
|
Nitrate reductase (narG, narH, narI, narJ) |
None |
|
Type II NADH dehydrogenases (ndh, ndhA) |
None |
|
Succinate dehydrogenase (sdhA sdhB sdhC sdhD) |
None |
|
Cytochrome C reductase (qcrC, qcrA, qcrB) |
All |
|
aa3 cytochrome C oxidase (ctaC, ctaD, ctaE) |
All |
|
Cytochrome bd oxidase (cydA, cydB, cydC, cydD) |
None |
|
Menaquinone synthesis (menA, menB, menC, menD, menE, menF, ubiE (menG)) |
None |
Identification of menaquinone biosynthesis as an antibacterial target in mycobacteria
In other studies, it was determined that a known inhibitor of cholesterol synthesis [specifically oxidosqualene cyclase (OSC)] effectively inhibited the growth of M. tuberculosis (Fig. 2), Mycobacterium bovis BCG and Mycobacterium smegmatis in culture at relatively low μM concentrations (data not shown). Ro 48-8071 is a potent (low nM), orally effective inhibitor of OSC developed by Hoffman La Roche, Inc. (Chugh et al., 2003;Morand et al., 1997), that is effective at lowering plasma cholesterol in hamsters, squirrel monkeys and minipigs (Morand et al., 1997). In order to identify the mycobacterial target inhibited by these oxidosqualene synthesis inhibitors metabolic labeling experiments using M. bovis BCG in the presence of Ro 48-8071 were conducted. Since Ro 48-8071 is known to inhibit cholesterol synthesis, radiolabeled isopentenyl diphosphate and unlabeled geranyl diphosphate and farnesyl diphosphate (precursors of isoprenoid synthesis) were initially used in cell-free and metabolic labeling experiments in the presence and absence of Ro 48-8071. Results indicated that synthesis of a neutral, apolar lipid, as judged by its chromatographic properties, was inhibited. Subsequent metabolic labeling experiments using L-[methyl-14C]methionine, generated similar results (Fig. 2). Thus, the chromatographic properties of the compound and the fact that the material incorporated radioactivity from labeled isoprenoid precursors and methionine suggested that the compound could be menaquinone. For positive identification, lipids were extracted from bacilli, fractionated on silica gel columns and the resulting neutral lipids subjected to preparative TLC. Material corresponding to the radioactively labeled material of interest, which was co-chromatographed on the same TLC plate, was extracted from the silica gel and subjected to HPLC/APCI mass spectrometry. Analysis of the resulting spectra clearly showed the presence of a molecular ion peak in the mass spectra with an m/z value of 787.5, representing the dominant mycobacterial menaquinone [MK-9 (II-H2), see Fig. 1 for mycobacterial menaquinone structure]. In addition, a molecular ion of 785.5, representing the less abundant, fully unsaturated form of mycobacterial menaquinone (MK-9), was also observed. Treatment of M. smegmatis with 40 μM Ro 48-8071 in liquid medium for 8 h resulted in an OD600 that was 50% lower than that seen in matched, untreated controls and, after normalization to OD and recovery of the internal standard, the concentrations of MK-9 (II-H2) and MK-9 in the bacilli were determined to be reduced by 2.5 +/− 0.9 and 3.3 +/− 0.6 fold, respectively.
Fig. 2. Inhibition of bacterial growth and lipid synthesis by Ro 48-8071.
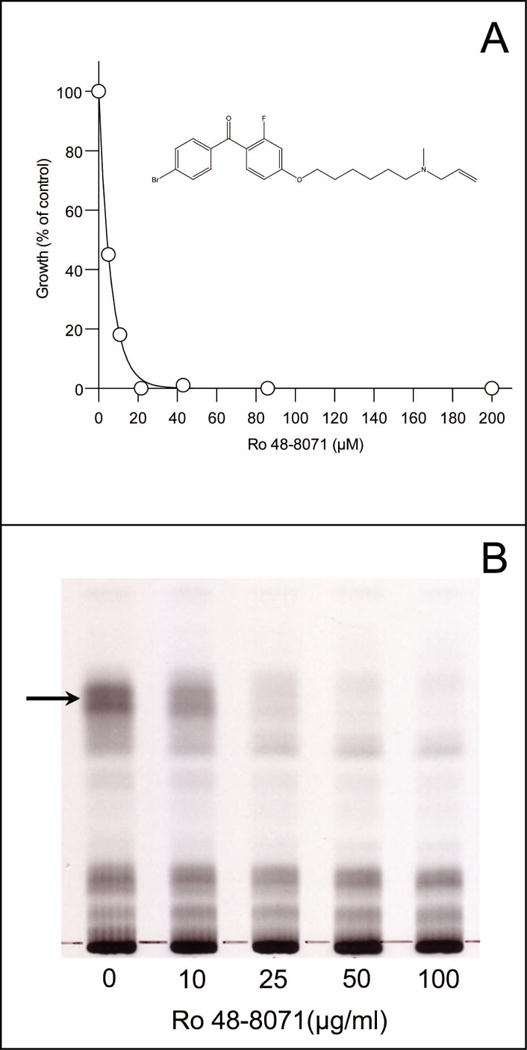
Panel A: Inhibition of M. tuberculosis growth was determined in 96 well plates using 7H9 medium (supplemented with oleic acid, albumin, dextrose and 0.05% Tween 80). Ro 48-8071 was added at the indicated concentrations; the structure of Ro 48-8071 is inset. Panel B: TLC analysis of neutral lipids isolated from M. bovis BCG after labeling with L-[methyl-14C]methionine in the presence of the indicated concentrations of Ro 48-8071. Cells were pre-incubated with Ro 48-8071 for 20 min at 37°C followed by labeling for 2 h. at the same temperature. The arrow indicates the material that was isolated for analysis by mass spectrometry.
To further substantiate the effects of Ro 48-8071 on menaquinone synthesis and electron transport, oxygen consumption in the presence of Ro 48-48071 was determined by following methylene blue decolorization in sealed tubes. Decolorization of methylene blue, a well known redox dye, has been reported to unambiguously demonstrate effects on respiration (Boshoff et al., 2004). Results demonstrated that treatment with Ro 48-8071 inhibited oxygen consumption in M. smegmatis and M. tuberculosis (Fig. 3). Similar results were obtained using trifluoperazine (a phenothiazine based drug, data not shown).
Fig. 3. Oxygen consumption and rescue of bacterial growth and oxygen consumption by menaquinone (Vitamin K2).
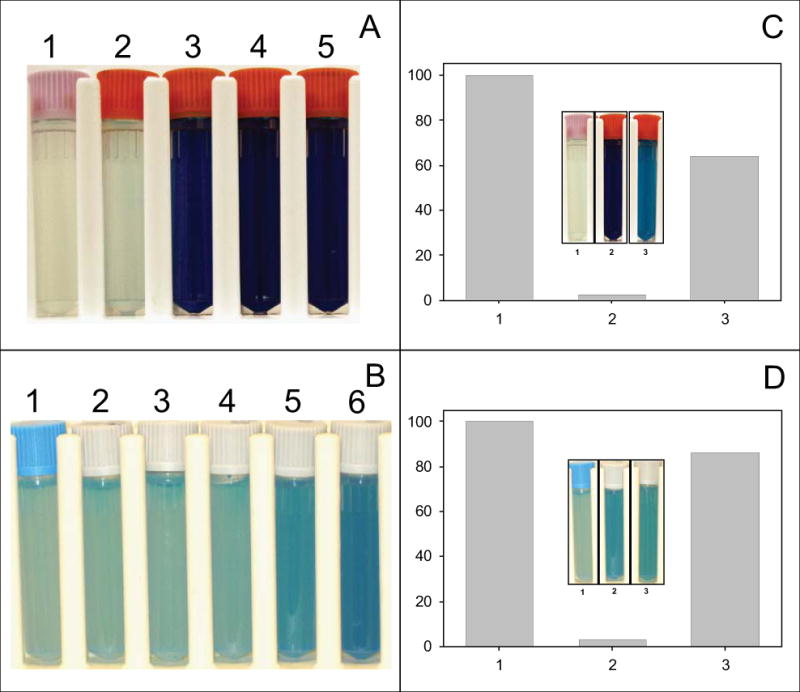
Panel A – M. smegmatis cultures, containing 0.01% methylene blue, treated with Ro 48-8071 at 0, 10, 25, 50 or 100 ug/ml, tubes 1–5 respectively. Cultures were incubated at 37° C for 2 h. Panel B – M. tuberculosis cultures, containing 0.03% methylene blue, treated with Ro 48-8071 at 0, 5, 10, 25, 50 or 100 ug/ml, tubes 1–6 respectively. Cultures were incubated at 37° C for 8 h. Panel C – M. smegmatis was cultured in the presence of 0 μM Ro 48-8071 (lane 1), 20 μM Ro 48-8071, or 20 μM plus 400 μM Vitamin K2 (lane 3) for 24 h. Growth rates were monitored by measuring OD @ 600nm. Inset – M. smegmatis cultures, containing 0.01% methylene blue, were treated with 0 μg/ml Ro48-8071 (tube 1), 50 μg/ml Ro48-8071 (tube 2) or 50 μg/ml Ro48-8071 plus 400 μM Vitamin K2 (tube 3) and incubated at 37° C for 2 h. Panel D – M. tuberculosis was cultured in the presence of 0 μM Ro 48-8071 (lane 1), 20 μM Ro 48-8071, or 20 μM plus 400 μM Vitamin K2 (lane 3) for 12 days; growth rates were monitored by measuring OD @ 600nm. Inset – M. tuberculosis cultures, containing 0.03% methylene blue, were treated with 0 μg/ml Ro48-8071 (tube 1), 50 μg/ml Ro 48-8071 (tube 2) or 50 μg/ml Ro 48-8071 plus 400 μM Vitamin K2 (tube 3) and incubated at 37° C for 8 h. Culture medium was as indicated in Experimental Procedures.
The inhibition of growth and oxygen consumption of M. smegmatis and M. tuberculosis caused by exposure to Ro 48-8071 could be reversed by the addition of menquinone (Vitamin K2) or phylloquinone (Vitamin K1) to the culture medium at 400 μM (Fig. 3) even at concentrations of Ro 48-8071 in large excess of the MIC (supplemental Fig. S1). However, inhibition of respiration by trifluoperazine could not be rescued by menaquinone supplementation (data not shown). The addition of other potential electron donors, such as menadione, N,N,N’,N’-tetramethyl-p-phenylenediamine, 1,4-phenylenediamine dihydrochloride, p-phenylenediamine, plumbagin, or ruthenium red did not rescue growth at any concentration tested. It was also determined that the addition of 1,4-dihydroxy-2-naphthoic acid (DHNA), a precursor of menaquinone (Fig. 1), to the culture medium did not rescue bacterial growth in the presence of Ro 48-8071.
Based on the combined results of the metabolic labeling, depletion and rescue experiments it seemed likely that the target of Ro 48-8071 was an enzyme involved in one of the later steps of menaquinone synthesis, possibly downstream of MenB (Fig. 1). These steps include the ones catalyzed by MenA, which prenylates the naphthoate ring to form demethylmenaquinone, MenG, which methylates the ring structure of demethylmenaquinone, and GrcC1 and/or GrcC2, the prenyl diphosphate synthases that likely synthesize the prenyl diphosphates utilized as substrates by MenA.
Identification of the gene encoding MenA and inhibition of MenA by Ro 48-8071
BLAST searches revealed a single copy of the gene Rv0534c which is annotated as a putative MenA. Rv0534c was amplified from the M. tuberculosis genome, cloned and expressed in E. coli using the pET28a(+) vector. Expression of the protein was confirmed by Western blot using an anti-His antibody. Due to the presence of several membrane spanning domains and typical irreversible loss of activity of aromatic prenyltransferases during solubilization (Brauer et al., 2004) solubilization and purification attempts were unsuccessful. Therefore, to confirm that Rv0534c was responsible for encoding the menA gene, membrane preparations from E. coli strains harboring pET28a(+) containing Rv0534c or empty vector were tested for MenA activity. There was, approximately, an eight-fold increase in the MenA activity in membrane preparations from bacilli expressing Rv0534c over the control.
Ro 48-8071 strongly inhibited MenA activity in membranes isolated from wild-type M. tuberculosis in vitro with an IC50 of 9 μM, a value very close to the MIC of 5 μM (Fig. 4). To demonstrate the specificity of Ro 48-8071 other enzyme activities that also utilize farnesyl diphosphate (FPP) as a sustrate were assayed in the presence and absence of R0 48-8071. Results indicate that the activities of recombinant, purified GrcC1 and GrcC2 were not inhibited by Ro 48-8071, in addition, total prenyl diphosphate synthase activity in the membrane preparations was not inhibited (supplemental Fig. S2). It is possible that MenG is also inhibited by Ro 48-8071; however, this can not be determined as, in the absence of a source of demethylmenaquinone, MenG can only be assayed by coupling to MenA. Overall, the data strongly suggest that MenA is the target of Ro 48-8071 and that this compound inhibits M. tuberculosis growth by preventing prenylation of DHNA to form demethymenaquinone.
Fig. 4. Inhibition of MenA activity by Ro 48-8071.
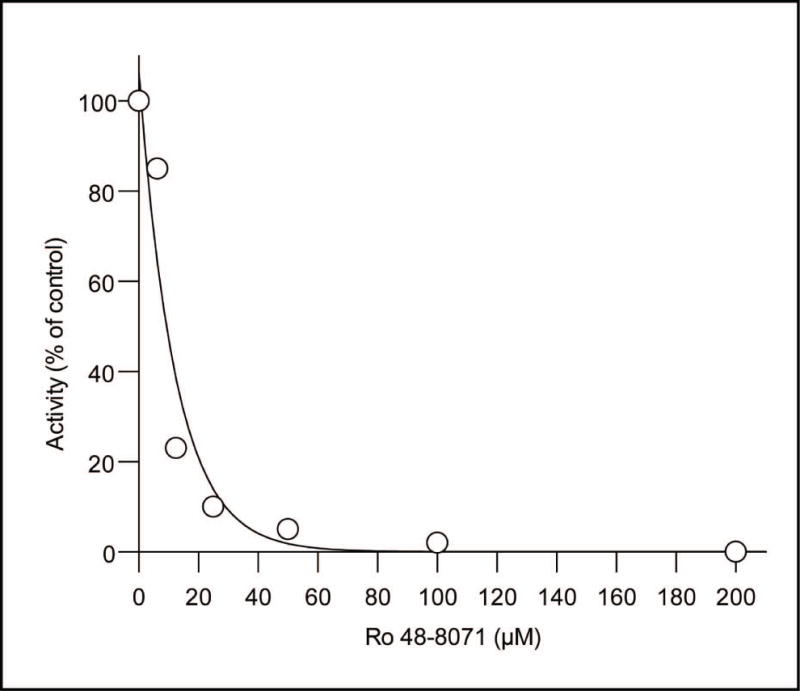
Reaction mixtures contained 500 μM DHNA, 10 μM [3H]farnesyl diphosphate, 5 mM MgCl2 and 0.1% CHAPs in 100 mM Tris-HCl (pH 8.0) and 50–100 μg membrane protein. Ro 48-8071 was added at the indicated concentrations. Reactions were stopped and the product analyzed as described in the Experimental Procedures. Data presented are average values of triplicate determinations.
Ro 48-8071 does not induce the dormancy response in M. tuberculosis
Recently, the two component system DosS/DosT and DosR has been reported to be involved in mycobacterial adaptation to hypoxic conditions (Roberts et al., 2004;Voskuil et al., 2003). It has also been demonstrated that cyanide, a cytochrome C oxidase inhibitor, does not upregulate the Dos regulon in M. tuberculosis (Voskuil et al., 2003;Boshoff et al., 2004), and that lipoquinones act as direct signals for regulation of autophosphorylation of ArcB, a two component system in E. coli (Georgellis et al., 2001). Thus, it seemed possible that inhibition of menaquinone synthesis could have profound effects on entrance to, and maintenance of NRP in M. tuberculosis. In order to address this question, we initiated quantitative real-time PCR (QRT-PCR) experiments to determine the effect of Ro 48-8071 treatment on the transcription of selected genes in M. tuberculosis. The data indicate that exposure to Ro 48-8071 does not induce expression of the sensor kinase dosS, the transcriptional regulator dosR, or hspX, one of the predominant markers of mycobacterial dormancy, suggesting that inhibition of menaquinone synthesis does not induce expression of the dormancy regulon in M. tuberculosis. Interestingly, transcription of menA, menB and menH (annotated as a possible menaquinone methyltransferase) was little changed by treatment with Ro 48-8071 generating mean log2 values of 1.7, 0.55 and 1.5, respectively (Supplemental Fig. S3).
Synthesis and in vitro activity of new MenA inhibitors
A series of approximately 100 new compounds, with the generalized structure shown in Fig. 5, were synthesized (Kurosu et al., 2007) and tested as inhibitors of bacterial growth and MenA activity; results for six representative compounds are shown in Fig. 5; in all cases there was good concordance between MIC and IC50. In order to obtain confirmation, CSU-17, -18 and -20 were synthesized in gram quantities for independent testing, which determined the MIC values for these three compounds against a battery of Mycobacterium spp and strains. In general, the MIC values obtained at CSU and the independent laboratory were in close agreement. The compounds were also tested against other species of pathogenic Gram-positive bacteria and pathogenic Gram-negative bacteria (Kurosu et al., 2007). Only the growth of Gram-positive organisms (including methicillin, vancomycin and linezolid resistant strains) was inhibited, supporting the hypothesis that menaquinone synthesis is the target of the compounds. The effectiveness of two representative compounds (CSU-17 and CSU-20) against M. tuberculosis in NRP phase II) was also tested using the Wayne model (Wayne and Hayes, 1996) with minor modifications (Lenaerts et al., 2005). These experiments indicate that CSU-20 is 320-, 180- and 3-fold more effective in killing non-replicating bacteria than ethambutol, isoniazid or rifampin, respectively (Fig. 6) and suggest that electron transport is required for survival of NRPB.
Fig. 5. MenA inhibitors.
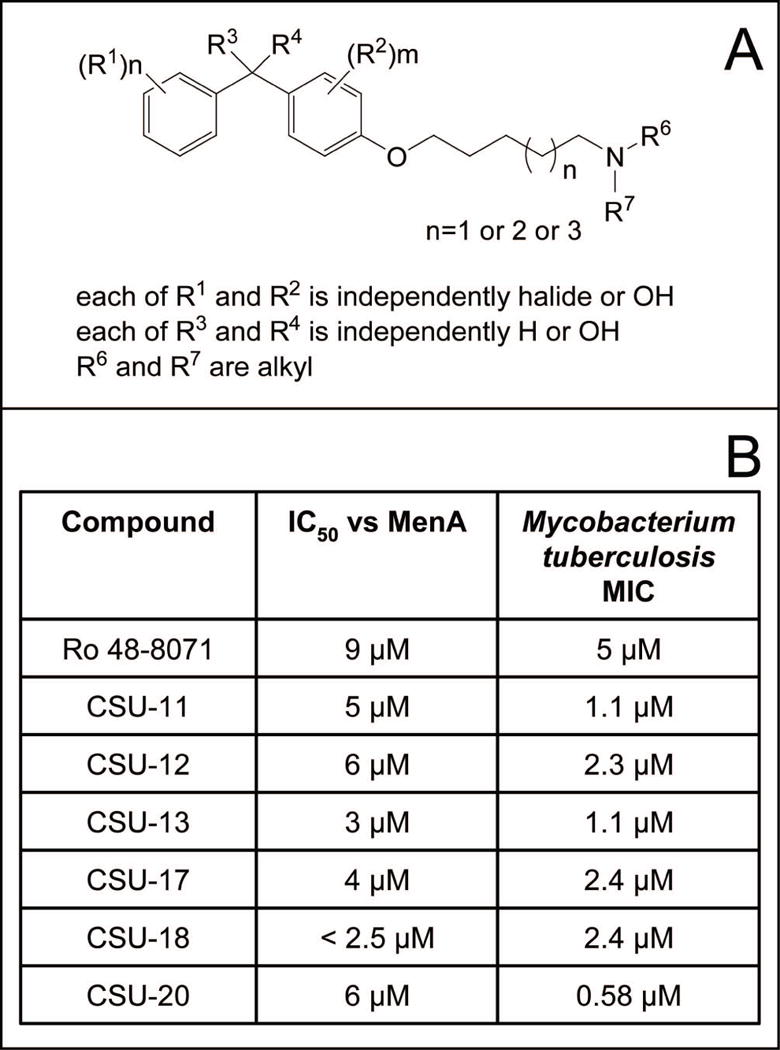
Panel A: General structure of compounds synthesized, synthetic schemes have been previously published (Kurosu et al., 2007). Panel B: Inhibition of MenA activity and M. tuberculosis growth by representative compounds. The structures are shown in supplemental Table S2.
Fig. 6. Recovery of NRP M. tuberculosis after treatment with various compounds.
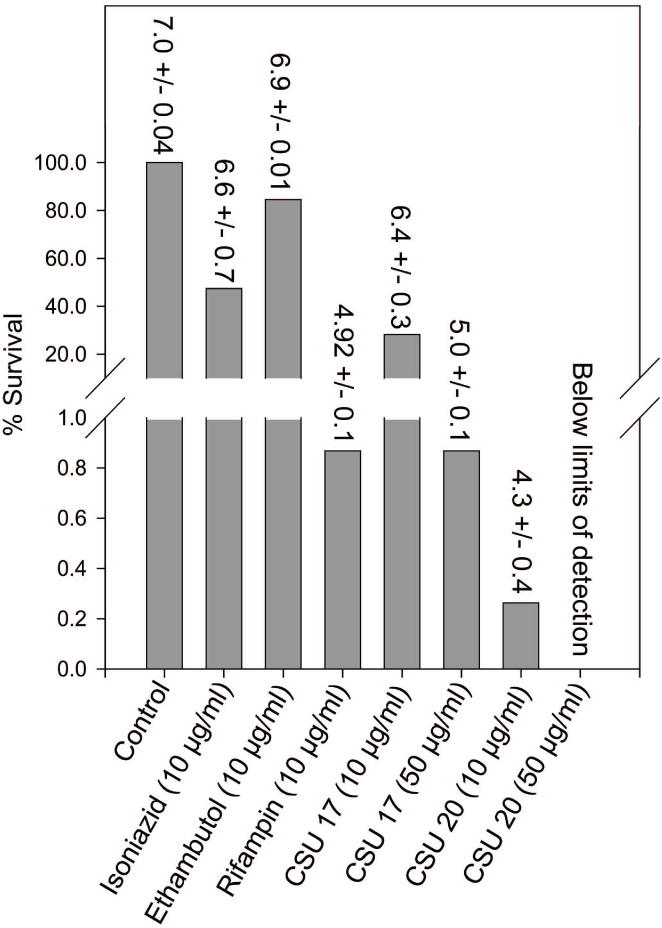
Cultures were exposed to gradual oxygen depletion for 24 days (NRP phase 2 as previously defined (Wayne and Sohaskey, 2001) and then treated with compounds at the indicated concentrations for 96 h. Subsequently, the bacterial suspension was diluted, plated and colonies were counted. The lower limit of detection is 100 bacilli using this protocol. The numbers above the bars are the average Log10 CFU/ml +/− SEM.
Discussion
One of the goals of TB drug development is to identify lead compounds with activity against latent TB infections since a large number of tuberculosis cases are suspected to arise from reactivation. Initially, transcript detection via hybridization was utilized to define the metabolic tendencies of NRPB. This analysis indicated that genes encoding components of metabolism relating to energy and reducing potential, including the electron transport chain and ATP synthesis, are abundantly represented in the transcriptome after prolonged exposure to microaerophilic conditions in the Wayne model of NRP. In fact, the expression indices for all genes encoding cytochrome C reductase, aa3 cytochrome C oxidase and F1F0-ATP synthase were > 1, suggesting that oxidative phosphorylation and related metabolic functions are required to maintain viability in this persistent state and, perhaps, latent infections in agreement with the recent observation that ATP homeostasis and proton motive force is important for the survival of M. tuberculosis in NRP (Koul et al., 2008;Rao et al., 2008). Notably, all of these genes were also predicted to encode essential enzymes in M. tuberculosis (Sassetti et al., 2003;Sassetti and Rubin, 2003). Thus, the ability to maintain ATP synthesis appears to be vital to replicating and non-replicating M. tuberculosis and this, in turn, appears to be dependent on electron transport. In addition, the terminal acceptor for the cytochrome C reductase/aa3 cytochrome C oxidase supercomplex is dioxygen suggesting that the presence of oxygen may be required to maintain NRP.
Even though MenA was not predicted to be essential based on random transposon mutagenesis experiments (Sassetti et al., 2003;Sassetti and Rubin, 2003), enzymes upstream and downstream in the pathway [MenC, MenD, MenE and UbiE, which likely encodes the methyltransferase (MenG) in M. tuberculosis] are predicted to be essential, suggesting that MenA may also be essential. The facts that treatment with Ro 48-8071 inhibits MenA activity, reduces resipiration and bacterial levels of menaquinone and that addition of vitamins K1 and K2 to the medium rescues the growth and oxygen consumption of mycobacteria in the presence of Ro 48-8071 strongly implies that this compound inhibits the synthesis of menaquinone, generating menaquinone auxotrophy.
Attempts to generate resistant strains of mycobacteria by transformation of bacilli with plasmids carrying menA were unsuccessful; likely, in part, due to the fact that Mycobacterium strains transformed with plasmids carrying menA did not express significant levels of recombinant MenA as judged by SDS-PAGE and Western blot. Although it is well known that overexpression of genes encoding drug targets within sensitive cells can lead to decreased sensitivity, overexpression does not always result in a significant change in MIC (Koul et al., 2007). In addition, attempts to generate spontaneous resistant mutants were also unsuccessful. This is often thought to indicate that a compound inhibits multiple targets. However, since the K vitamins rescue both growth and oxygen consumption at Ro 48-8071 concentrations several fold higher than its MIC (supplemental Fig. S1) there does not appear to be a secondary target in this case.
It has previously been shown that Gram-positive bacilli are capable of utilizing exogenous DHNA to satisfy nutritional requirements in organisms with mutations in genes upstream of MenA (Taber et al., 1981;Meganathan et al., 1981); thus, the observation that addition of DHNA to the culture medium does not rescue bacterial growth in the presence of Ro 48-8071 indicated that the inhibition likely occurs at a step in menaquinone biosynthesis downstream of the action of MenB, a hypothesis supported by the observation that Ro 48-8071 specifically inhibits MenA activity in vitro.
Of the compounds designed to be MenA inhibitors CSU-20 demonstrated somewhat greater inhibitory activity against NRPB than rifampin, a clinically available RNA polymerase inhibitor that maintains efficacy in aerobic and microaerophilic conditions (Cho et al., 2007). In addition, both CSU-20 and CSU-17 demonstrated greater inhibitory activity against NRPB than isoniazid or ethambutol; a result that could be predicted as both of these clinically available drugs are cell-wall synthesis inhibitors, which show significantly reduced activity under anaerobic or microaerophilic conditions in vitro (Cho et al., 2007;Lenaerts et al., 2005), presumably due to decreased cell-wall synthesis in NRP. There are a number of compounds known to be active against NRPB, and many, including rifampin, rifabutin, RU66252, amikacin, streptomycin, capreomycin, minocycline, fusidic acid, moxifloxacin, ciprofoxacin and ofloxacin, inhibit DNA replication, transcription or translation (Cho et al., 2007). A series of rhodanine compounds, which target DlaT (dihhydrolipoamide acyltransferase), also has activity against NRP bacilli (Bryk et al., 2008). In addition, phenothiazines, which inhibit NADH:menaquinone oxidoreductase activity (Weinstein et al., 2005) are bactericidal in a starvation model of NRP (Xie et al., 2005). Two reports, published during the preparation of this manuscript, indicate that de novo ATP synthesis is indeed required for viability of NRP M. tuberculosis (Rao et al., 2008;Koul et al., 2008) as predicted by the transcriptional analysis reported here. Thus, the data reported here and elsewhere strongly indicate that electron transport and oxidative phosphorylation in M. tuberculosis are critical for survival in NRP.
The MenA inhibitors prevent aerobic bacterial growth and respiration without inducing a dormancy response in M. tuberculosis. This is in agreement with data from mycobacteria treated with cyanide (Voskuil et al., 2003;Boshoff et al., 2004), phenothiazines (Weinstein et al., 2005) or blocked in the bc1-aa3 respiratory pathway (Matsoso et al., 2005). Thus, inhibitors of menaquinone synthesis, electron transport and oxidative phosphorylation may have significant advantages in treating rapidly growing bacilli as well as NRPB.
The observation that interruption of the electron transport chain in microaerophilic or anaerobic conditions is lethal to the bacilli is somewhat counterintuitive, as many organisms can survive using substrate level phosphorylation to produce ATP. However, an explanation for this apparent aberration lies in the observation that Mycobacterium smegmatis requires the F1F0-ATP synthase for growth on both fermentable and nonfermentable carbon sources (Tran and Cook, 2005). Thus, it has been hypothesized that ATP production from substrate level phosphorylation alone may be insufficient to sustain growth of mycobacteria, or that mycobacteria may not support uncoupled respiration (Tran and Cook, 2005). The results presented here support this view, in that, menaquinone synthesis appears to be required for mycobacterial survival.
Overall, the results presented indicate that NRP M. tuberculosis in the Wayne model are not “metabolically dormant”, requiring ongoing menaquinone synthesis to support electron transport and oxidative phosphorylation for survival. It is not yet clear if survival of NRPB is based on a requirement for ATP for energy, nucleic acid synthesis or, more probably, some combination of these. The requirement for ATP per se is suggested by the fact that several nucleic acid synthesis inhibitors are active against both actively growing and NRP M. tuberculosis (Cho et al., 2007). In addition, the expression analysis, menaquinone depletion, growth and oxygen consumption inhibition and rescue, and in vitro MenA assays indicate that menaquinone synthesis presents a viable drug target in both actively growing and NRP M. tuberculosis.
Experimental Procedures
M. tuberculosis (H37Rv) genomic DNA and M. tuberculosis whole genome DNA-microarray slides were obtained from the TB Vaccine Testing and Research Material Contract, NIH/NIAID contract NO1-A1-75320. Each microarray contains probes representing the open reading frames of M. tuberculosis strain H37Rv and strain CDC1551. All PCR product and plasmid purifications were performed using Qiagen kits. All antibiotics and other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
RNA Isolation
For expression profiling using the Wayne model of gradual oxygen depletion in M. tuberculosis (H37Rv) cultures, an aerobic preculture was diluted 100-fold (from an OD600 = 0.5) in tubes closed with rubber septa to ensure gradual oxygen depletion. Cultures were incubated on stirring platforms at 150 rpm for six months prior to harvesting by centrifugation. Bacilli from three biological replicates were resuspended in 1 ml of Trizol reagent and total RNA was liberated from the Trizol suspended cells by intermittent bead beating, in the presence of 0.5 ml of 0.1 mm dia. zirconium beads, for a total of three minutes. RNA was partitioned from other cellular products by addition of chloroform and subsequent centrifugation at 13,000 × g for 20 minutes at 4°C. The resulting aqueous phase was transferred to a fresh tube and an equal volume of 70% ethanol was added. RNA was purified using an RNeasy miniprep kit (Qiagen) following manufacturer’s instructions and total RNA was estimated spectrophotometrically.
cDNA labeling, microarray hybridization and analysis
Labeled cDNA was prepared as follows: 5 μg of total RNA (extracted from exponentially growing or NRP M. tuberculosis) and 4.5 μg of random oligonucleotide hexamers (Invitrogen) were incubated 2 min at 98°C in RNase free water and cooled on ice; subsequently 10X Stratascript RTase buffer, 1.8 μl Stratascript RTase (Stratagene), 0.5 mM dATP, dGTP, dCTP, 0.02 mM dTTP and 5.8 μM Cy3-dUTP or Cy5-dUTP (GE Healthcare) were added and brought to a final volume of 25.6 μl with RNase free water. The resulting mixture was incubated for 10 min at 25°C and 90 min at 42°C. cDNA was then purified by filtration using a microcon-10 (Amicon) and applied to whole genome M. tuberculosis oligonucleotide microarrays, which had been prehybridized for 1 h in 5X SSC, 1% BSA, and 0.1% SDS. The arrays were then washed with water and isopropanol and hybridization solution containing labeled cDNA, 5 μg yeast tRNA, 2X SCC, 25% formamide and 0.1%SDS was added and incubated overnight at 42°C.
A two color, competitive hybridization format was used for microarray analysis. RNA extracted from exponentially growing M. tuberculosis was used to generate Cy3-labeled labeled reference cDNA for comparison to Cy5-labeled cDNA synthesized from RNA extracted from bacilli in a state of NRP. The relative intensities of the two dyes at each spot on the hybridized microarray slides were visualized and analyzed using a Genepix 4000B scanner (Molecular Devices Co.). The scanned images of the microarray slides were also analyzed using GeneSifter analysis software at the Rocky Mountain Regional Center of Excellence Genomics/Proteomics Core.
The data were normalized relative to the mean channel intensity of the transcriptionally active genes, determined using GeneSifter, seen in each growth condition and are presented as expression indices. Expression index is defined as (the expression intensity of RvXXXX in NRP/mean channel expression intensity in NRP)(the expression intensity of RvXXXX in exponentially growing cultures under aerobic conditions/mean channel intensity in exponentially growing cultures under aerobic conditions)−1. Results presented are averages of three biological replicates +/− variance. Significance was considered to be > a 1.5 fold alteration in expression having a variance of <0.05.
Whole cell labeling
Whole cell labeling was done using [1-14C]isopentenyl diphosphate (50 mCi/mmol, Amersham Biosciences), and unlabeled geranyl diphosphate and farnesyl diphosphate, or L-[methyl-14C]methionine (55 mCi/mmol, Amersham Biosciences). M. bovis BCG cells were grown to mid log-phase (OD600 of 0.6–0.8) in Sauton’s media. Cells were centrifuged and resuspended in fresh media at 1/10 of the original volume, and subsets were incubated with Ro 48-8071 at 200 μM for 15 min at room temperature. Both control and treated cells were transferred to a 24 well plate containing radiolabeled precursor, 200 μM Ro 48-8071 (treated cells only) and incubated at 37° C for two hours. The reaction mixtures were transferred to glass tubes and labeling was terminated by addition 6 ml of chloroform/methanol (2:1, v/v) and the mixture was gently rocked for 2 hrs. The resulting biphasic solution was centrifuged and the upper aqueous layer was removed. The organic, lower, phase was washed with water:methanol:chloroform (48:47:3, v/v/v) and transferred to new tubes. The solvent was evaporated under a stream of N2 and the sample dissolved in chloroform. Polar lipids were removed from the sample by silicic acid column chromatography using chloroform to elute the apolar lipids. The chloroform was removed from the resulting material under a N2 stream and the sample was dissolved in hexane and applied to silica gel TLC plates, which were developed in hexane:diethyl ether (95:5 v/v).
Identification of menaquinone
For unambiguous identification of the neutral lipids of interest 2 L cultures of M. bovis BCG were grown to mid log-phase, harvested and the bacilli were then extracted and the apolar lipids were prepared as described above. The resulting material was subjected to preparative TLC, side by side with radioactive material derived from a whole cell labeling experiment, in hexane:diethyl ether (95:5 v/v). Material co-migrating with the radioactive compound of interest was scraped from the TLC plate and extracted from the silica gel with chloroform. This material was then subjected to HPLC/MS on a HP1100 Series HPLC connected to a 2000 Finnigan LCQ-DUO ion-trap mass spectrometer with APCI interface. HPLC separation was achieved using a reverse-phase hypersil ODS column (Agilent) and a gradient running from 100% methanol to methanol/isopropanol (1:1, v/v) over 50 minutes at 0.4 ml/min and 40 °C. Eluted molecules were subjected to positive ion MS using APCI as the ionization interface. Capillary temperature was 150 °C and APCI vaporizer temperature was 450 °C. Source voltage and current were 6 kV and 5 μA, respectively. Sheath gas flow was maintained at 40 units.
Quantitation of menaquinone in bacilli treated with Ro 48-8071
M. smegmatis was grown in Sauton’s medium containing 0.05% Tween 80 (40 ml cultures) to mid-log phase (0.6 OD600), at which time Ro 48-8071 was added to a final concentration of 40 μM to some of the cultures. The cultures were incubated at 37°C for 8 hours, OD600 was determined and the cells were harvested by centrifugation. The cell pellet was washed with water, vitamin K2 was added as an internal standard and the bacteria were extracted with chloroform/methanol (2:1 v/v). The organic solvent was transferred to a clean tube and evaporated under nitrogen. The lipids were dissolved in an aliquot of chloroform and applied to a silica gel column. Nonpolar lipids in the chloroform flow through were subjected to HPLC-MS on a Agilent Technologies 1200 HPLC connected to a Agilent Technologies 6210 TOF-MS. HPLC separation was achieved using Waters XBridge C18 column and a solvent gradient running from 100% methanol to methanol/isopropanol (3:1, v/v) over 59 minutes at 0.4 ml/min and 40 °C. Eluted molecules were subjected to positive ion MS using APPI as the ionization interface. Gas temperature was 350 °C, vaporizer temperature was 300 °C and nebulizer pressure was 45 psig. Results were normalized to the amount of recovered vitamin K2 and the OD600 at the time of harvest, all values presented are averages of triplicate experiments +/− standard deviation.
Oxygen consumption
M. smegmatis cells were grown in Sauton’s medium to mid log-phase [OD600nm 0.5–0.8] and 1.5 ml aliquots of the culture were treated with the indicated concentration of Ro 48-8071 or trifluoperazine (Sigma) and incubated at 37° C for 2h in the presence of 0.01% methylene blue. In rescue experiments, the indicated concentrations of vitamin K2 or K1 were added prior to addition of the inhibitor.
In the case of M. tuberculosis, cells were grown in 7H9 medium supplemented with OADC and 0.05% Tween 80 to mid log-phase. Cultures were treated as described above with the indicated concentrations of Ro 48-8071 in the presence and absence of vitamin K2 and 0.03% methylene blue and incubated at 37° C for 8h.
MIC determinations
The MIC values of all compounds were determined by a microtiter plate based spectrophotometric method (Gruppo et al., 2006) or by a colorimetric microtiter plate based method using Alamar blue/visual inspection (Yajko et al., 1996).
Cloning and expression of Rv0534c
Based on the nucleotide sequence of the open reading frame Rv0534c, the following primers were designed and synthesized (Invitrogen): F, 5′-AATGATCATATGGCCAGTTTCGCACAGTGGGTC-3′, and R, 5′-AACAAGCTTAGCTCAACTGACCAAACGCCAATGC-3′. NdeI and HindIII restriction sites (underlined) were engineered in the N-terminal and C-terminal primers, respectively. Rv0534c was amplified from M. tuberculosis chromosomal DNA using a Perkin-Elmer GeneAmp 2400 PCR system and polymerase (PE Biosystems). The PCR product was digested with appropriate enzymes and cloned into the multiple cloning site of pET28a(+) (EMD Biosciences, Inc.). Plasmids were purified, analyzed by restriction endonuclease digestion and sequenced (Macromolecular Resources, Colorado State University).
Membrane isolation and MenA Assays
The MenA activity was characterized using membrane fractions prepared from M. tuberculosis. Briefly, M. tuberculosis (H37Rv) was grown to mid-log phase in glycerol-alanine-salts medium, washed with saline and harvested by centrifugation. The resulting pellet was irradiated for 18 h at 2,315 Rads/min using a JL Shepard instrument with a 137Cs source. The washed cell pellet was resuspended in homogenization buffer containing 50 mM potassium phosphate (pH 7.2), 10% glycerol, 5 mM MgCl2 and 5 mM DL-dithiothreitol and disrupted by probe sonication on ice with a Sanyo Soniprep 150 (10 cycles of 60 sec on and 90 sec off). The resulting suspension was centrifuged at 27,000 × g for 15 min. The pellet was discarded and the supernatant was centrifuged at 100,000 × g for 1hr in a Beckman Ti 70.1 rotor. The pellet (membranes) was resuspended in homogenization buffer, divided into aliquots and frozen at −70°C. The protein concentration of the membrane preparation was estimated using a BCA protein assay kit (Pierce, Rockford, IL).
Reaction mixtures contained 500 μM DHNA, 10 μM [3H]farnesyl diphosphate (American Radiolabeled Chemicals), 5 mM MgCl2 and 0.1% CHAPs in 100 mM Tris-HCl (pH 8.0) and 50–100 μg membrane protein. Reactions were stopped by the addition of 0.1 M acetic acid in methanol. The resulting mixture was extracted twice with hexane, the combined extracts were evaporated to dryness under N2 and dissolved in chloroform/methanol (2:1, v/v). An aliquot was taken for liquid scintillation counting and the remaining material was subjected to TLC on C18 reverse-phase plates, which were developed in acetone/water (95:5 v/v). Radioactive spots on the thin layer plated were located and the relative abundance of each radiolabeled compound was determined using a System 200 Imaging Scanner (Bioscan Inc). The relative abundance was then used to calculate the portion of the total radioactivity, determined by liquid scintillation counting, in the sample which could be attributed to newly synthesized menaquinone.
Testing compounds against M. tuberculosis grown under low oxygen conditions
The effectiveness of two representative compounds (CSU-17 and CSU-20) was evaluated against NRPB using an in vitro assay of M. tuberculosis grown under low oxygen conditions using the Wayne model (Wayne and Hayes, 1996) with minor modifications (Lenaerts et al., 2005). Briefly, an aerobic preculture was diluted 100-fold (from an OD600 = 0.5) in tubes closed with rubber septa to ensure gradual oxygen depletion. Cultures were incubated on stirring platforms at 150 rpm for 24 days (well into NRP phase 2 as defined by the Wayne model). Compounds in solution were deoxygenated by purging with nitrogen and added by injection through the septa. Drug exposure lasted for 96 h, after which the bacterial suspension was diluted and plated. Bacilli that recover (are culturable) are then grown aerobically at 37 °C and colonies counted.
Other procedures
Restriction digests, ligations and electroporations were done as described by Sambrook et al. (Sambrook and Russell, 2001) unless otherwise noted. BLAST searches were done on the National Center for Biotechnology Information website or the Mycobacterium tuberculosis Structural Genomics Consortium website using standard protein-protein BLAST (blastp). Alignments were done using multiple sequence alignments with hierarchical clustering (Corpet, 1988) using the ‘Multalin’ interface at the Institut National de la Recherche Agronomique (Toulouse, France) website.
Supplementary Material
Acknowledgments
Funding for this research was provided through the National Institutes of Health grants AI049151, AI06357, AI055298 and AI057836.
Footnotes
Online Supplemental Material
Online supplemental material includes a table of expression indices for genes in M. tuberculosis during non-replicating persistence (Table S1), a table of the structures of selected compounds discussed in the text (Table S2), results of growth rescue experiments (Fig. S1), the effects of Ro 48-8071 on prenyl diphosphate synthase activity (Fig. S2) and quantitative real-time PCR analysis of selected genes in response to treatment with Ro 48-8071 (Figure S3).
The authors have no conflicting financial interests.
Reference List
- Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Anonymous. Tuberculosis. Nat Med. 2007;13:263. [Google Scholar]
- Boshoff HI, Barry CE., III Tuberculosis – metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., III The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- Brauer L, Brandt W, Wessjohann LA. Modeling the E. coli 4-hydroxybenzoic acid oligoprenyltransferase (ubiA transferase) and characterization of potential active sites. J Mol Model. 2004;10:317–327. doi: 10.1007/s00894-004-0197-6. [DOI] [PubMed] [Google Scholar]
- Bryk R, Gold B, Venugopal A, Singh J, Samy R, Pupek K, et al. Selective killing of nonreplicating mycobacteria. Cell Host & Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Warit S, Wan B, Hwang CH, Pauli GF, Franzblau SG. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh A, Ray A, Gupta JB. Squalene epoxidase as hypocholesterolemic drug target revisited. Prog Lipid Res. 2003;42:37–50. doi: 10.1016/s0163-7827(02)00029-2. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence [published erratum appears in Nature 1998 Nov 12;396(6707):190] Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma CL, Sherman DR, Ramakrishnan L. The secret lives of the pathogenic mycobacteria. Annu Rev Microbiol. 2003;57:641–676. doi: 10.1146/annurev.micro.57.030502.091033. [DOI] [PubMed] [Google Scholar]
- Embley TM, Stackebrandt E. The molecular phylogeny and systematics of the Actinomycetes. Annu Rev Microbiol. 1994;48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- Gennis RB, Stewart V. Respiration. In: Neidhardt FC, editor. Eschericia coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- Georgellis D, Kwon O, Lin EC. Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Gruppo V, Johnson CM, Marietta KS, Scherman H, Zink EE, Crick DC, et al. Rapid microbiologic and pharmacologic evaluation of experimental compounds against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:1245–1250. doi: 10.1128/AAC.50.4.1245-1250.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, Mizrahi V. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J Bacteriol. 2001;183:7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol. 2007;3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem. 2008;283:25273–25280. doi: 10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- Kurosu M, Narayanasamy P, Biswas K, Dhiman R, Crick DC. Discovery of 1,4-dihydroxy-2-naphthoate prenyltransferase inhibitors: New drug leads for multidrug-resistant gram-positive pathogens. J Med Chem. 2007;50:3973–3975. doi: 10.1021/jm070638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts AJ, Gruppo V, Marietta KS, Johnson CM, Driscoll DK, Tompkins NM, et al. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob Agents Chemother. 2005;49:2294–2301. doi: 10.1128/AAC.49.6.2294-2301.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsoso LG, Kana BD, Crellin PK, Lea-Smith DJ, Pelosi A, Powell D, et al. Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J Bacteriol. 2005;187:6300–6308. doi: 10.1128/JB.187.18.6300-6308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan R. Biosynthesis of vitamin K (menaquinone) and ubiquinone (coenzyme Q) In: Neihardt FC, editor. Escherichia coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 642–656. [Google Scholar]
- Meganathan R, Bentley R, Taber H. Identification of Bacillus subtilis men mutants which lack ortho-succinylbenzoyl-coenzyme-A synthetase and dihydroxynaphthoate synthase. J Bacteriol. 1981;145:328–332. doi: 10.1128/jb.145.1.328-332.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE. Lipids: complex lipids, their chemistry, biosynthesis and roles. In: Ratledge C, Stanford J, editors. The Biology of Mycobacteria. London: Academic Press; 1982. pp. 95–184. [Google Scholar]
- Morand OH, Aebi JD, Dehmlow H, Ji YH, Gains N, Lengsfeld H, Himber J. Ro 48-8.071, a new 2,3-oxidosqualene:lanosterol cyclase inhibitor lowering plasma cholesterol in hamsters, squirrel monkeys, and minipigs: comparison to simvastatin. J Lipid Res. 1997;38:373–390. [PubMed] [Google Scholar]
- Niebisch A, Bott M. Purification of a cytochrome bc(1)-aa(3) supercomplex with quinol oxidase activity from Corynebacterium glutamicum – Identification of a fourth subunit of cytochrome aa(3) oxidase and mutational analysis of diheme cytochrome c(1) J Biol Chem. 2003;278:4339–4346. doi: 10.1074/jbc.M210499200. [DOI] [PubMed] [Google Scholar]
- Orme IM. The latent tuberculosis bacillus (I’ll let you know if I ever meet one) Int J Tuberc Lung Dis. 2001;5:589–593. [PubMed] [Google Scholar]
- Pandya KP, King HK. Ubiquinone and menaquinone in bacteria – a comparative study of some bacterial respiratory systems. Arch Biochem Biophys. 1966;114:154–&. doi: 10.1016/0003-9861(66)90316-x. [DOI] [PubMed] [Google Scholar]
- Rao SPS, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual Cold. Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MM, Petersen LA, Poulter CD. Isolation and characterization of isoprene mutants of Escherichia coli. J Bacteriol. 1989;171:3619–3628. doi: 10.1128/jb.171.7.3619-3628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LB, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci U S A. 2005;102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GR, Robertson BD, Young DB. Tuberculosis: A problem with persistence. Nat Rev Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- Taber HW, Dellers EA, Lombardo LR. Menaquinone biosynthesis in Bacillus subtilis – Isolation of men mutants and evidence for clustering of men genes. J Bacteriol. 1981;145:321–327. doi: 10.1128/jb.145.1.321-327.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran SL, Cook GM. The F1F0-ATP synthase of Mycobacterium smegmatis is essential for growth. J Bacteriol. 2005;187:5023–5028. doi: 10.1128/JB.187.14.5023-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrichs T, Kaufmann SHE. Mycobacterial persistence and immunity. Front Biosci. 2002;7:D458–D469. doi: 10.2741/A788. [DOI] [PubMed] [Google Scholar]
- Ulrichs T, Kaufmann SHE. New insights into the function of granulomas in human tuberculosis. J Path. 2006;208:261–269. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- Weinstein EA, Yano T, Li LS, Avarbock D, Avarbock A, Helm D, et al. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc Natl Acad Sci U S A. 2005;102:4548–4553. doi: 10.1073/pnas.0500469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler PD, Blanchard JS. Metabolism and biochemical pathways. In: Cole S, Eisenach KD, McMurray DN, Jacobs WR, editors. Tuberculosis and the tubercle bacillus. Washington, DC: ASM Press; 2005. pp. 309–339. [Google Scholar]
- Xie ZF, Siddiqi N, Rubin EJ. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2005;49:4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajko DM, Sanders CA, Madej JJ, Cawthon VL, Hadley WK. In vitro activities of rifabutin, azithromycin, ciprofloxacin, clarithromycin, clofazimine, ethambutol, and amikacin in combinations of two, three, and four drugs against Mycobacterium avium. Antimicrob Agents Chemother. 1996;40:743–749. doi: 10.1128/aac.40.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


