Abstract
Recognition of viral dsRNA by endosomal toll-like receptor 3 (TLR3) activates innate immune response during virus infection. Trafficking of TLR3 to the endo-lysosomal (EL) compartment arising from fusion of late endosome (LE) with lysosome is required for recognition and detection of Pathogen Associated Molecular Patterns (PAMP). PAMP detection results in activation of TLR3-dependent signaling cascade. Existing knowledge about the mechanism(s) and cellular factor(s) governing TLR3 trafficking is limited. In the current study we have identified intracellular S100A9 protein as a critical regulator of TLR3 trafficking. S100A9 was required for maturation of TLR3 containing early endosome (EE) into LE, the compartment that fuses with lysosome to form EL compartment. Drastic reduction in cytokine production was observed in S100A9 knockout (KO) primary macrophages following RNA virus infection and treatment of cells with polyIC (a dsRNA mimetic that acts as a TLR3 agonist). Mechanistic studies revealed co-localization and interaction of S100A9 with TLR3 following polyIC treatment. S100A9-TLR3 interaction was critical for maturation of TLR3 containing EE into LE since TLR3 could not be detected in the LE of polyIC treated S100A9 KO macrophages. Subsequently, TLR3 failed to co-localize with its agonist (i.e. biotin-labeled polyIC) in S100A9 deficient macrophages. The in vivo physiological role of S100A9 was evident from loss of cytokine production in polyIC treated S100A9 KO mice. Thus, we have identified intracellular S100A9 as a regulator of TLR3 signaling and demonstrated that S100A9 functions during pre-TLR3 activation stages by facilitating maturation of TLR3 containing EE into LE.
Introduction
Innate antiviral immunity comprised of type-I interferon (IFN-α/β) mediated antiviral response and pro-inflammatory response by cytokines (like TNF, IL-6, IL-12) is required to initiate an adaptive immune response for virus clearance (1–10). Virus associated Pathogen Associated Molecular Patterns (PAMPs) are recognized by Pattern Recognition Receptors (PRRs) to induce innate immunity in response to virus infection (11, 12). Toll-like receptor 3 (TLR3) is a crucial PRR that plays a vital role in innate antiviral immunity (13, 14, 15). TLR3, TLR9, and TLR7 are endosomal PRRs recognizing viral-specific PAMPs (e.g. viral RNA) that trigger activation of IRF3 and NF-κB to produce IFN-β and cytokines (e.g. IL-12) from infected cells. An essential role of TLR3 in immunity, infection and pathogenesis has been demonstrated for a wide spectrum of viruses including Coxsackievirus B3 and B4 (CVB3, CVB4), respiratory syncytial virus (RSV), influenza A virus (IAV), West Nile virus (WNV), Rhinovirus (RV), Hepatitis C and B viruses (HCV and HBV), Herpes Simplex Virus 1 (HSV-1), HIV-1, Chikungunya virus (CGV), Vesicular Stomatitis virus (VSV), and Punta Toro virus (PTV) (14–24). Immune cells like macrophages and DCs (like pDCs) express TLR3 and TLR7, and these PRRs contribute to innate immune response following RNA virus infection (25–28). In these cells, it is postulated that upon virus infection, TLR3 undergoes intracellular trafficking from ER-golgi to endo-lysosomal (EL) compartment, which is generated after fusion of late endosome (LE) with lysosome (25–33). Once in the lumen of EL compartment, TLR3 senses its agonist/PAMP. Subsequently, TLR3 is activated following recruitment of the adaptor protein TRIF to its cytosolic tail and assembly of key signaling complexes which activates IRF3 and NF-κB (25–33).
Although the cellular and molecular apparatus required for endosomal TLR (TLR3, TLR7, TLR9) activation is well established, limited knowledge exists regarding the cellular mechanism(s)/factor(s) that regulates “proper” intracellular sorting of TLR3 to the endosomal compartment for PAMP sensing/detection. For example, it is still unclear whether TLR3 undergoes a “classical” sorting pathway constituting of TLR3 targeting from ER-golgi to early endosome (EE) followed by sorting to LE (from EE) and subsequent fusion with lysosome to form EL compartment. The latter is the subcellular site for PAMP sensing/detection by TLR3. In addition, there is very limited knowledge about specific cellular factor(s) that can act as a “chaperone” to facilitate targeting of TLR3 from ER-golgi to EL for PAMP sensing/detection
Spatial regulation of endosomal TLR compartmentalization is critical for their activation and innate immune function. Multiple endosomal TLRs can be activated in one cell (for example, TLR7 and TLR3 could be activated simultaneously in virus infected cells) and therefore, a differential/distinct cellular-factor driven mechanism may exist to ensure proper compartmentalization of TLR3 and TLR7 during such event, although a TLR3 specific “chaperone” that may facilitate intracellular trafficking and proper compartmentalization of TLR3 is still not known. Very limited knowledge exist regarding regulation of TLR3 trafficking in macrophages since the majority of studies were performed with DCs (25–28) or epithelial cells (including over-expression studies by using HEK293 and CHO cells) (34–37). TLR3 could reach EL compartment (following stimulation of resting cells with TLR3 agonist/PAMP) possibly via three routes – vesicle (endosome) arising from ER-golgi can directly transport TLR3 to the lysosomal/EL compartment, TLR3 from ER-golgi can be targeted directly to LE (which will fuse with lysosome to form EL compartment), thus bypassing EE, and lastly, TLR3 from ER-golgi can traffic via “classical sorting” pathway involving ER-golgi-EE-LE-EL. In addition, it is still unknown whether EE and LE play any “functional” role during TLR3 trafficking to the EL compartment for agonist/PAMP sensing/detection.
Recent studies have identified extracellular S100A9 as an important pro-inflammatory factor in influenza A virus infection (38). S100A9 is a small calcium-binding protein heavily expressed by myeloid cells (38, 39, 40). In the current study, we have identified intracellular S100A9 protein as an essential cellular factor regulating TLR3 trafficking and activation. S100A9 was required for maturation of TLR3 containing EE to LE, the compartment that fuses with lysosome to form EL compartment where TLR3 detects PAMP for its activation. Our studies have delineated the involvement of a TLR3-specific factor/chaperone (i.e. S100A9) that ensures proper localization of TLR3 in the “correct” intracellular compartment (i.e. in the LE and EL compartment which arise from fusion of TLR3 containing LE with lysosome) for PAMP detection and activation.
Materials and Methods
Virus, cell culture, mice
RSV (A2 strain) and VSV-IND (Mudd–Summers strain) was propagated in CV-1 and baby hamster kidney (BHK) cells as described previously (7, 8, 9, 10, 38, 41, 42). RSV and VSV were purified by centrifugation (two times) on discontinuous sucrose gradients as described previously (7, 8, 9, 10, 38, 41, 42). Bone-marrow-derived macrophages (BMDMs) were obtained from femurs and tibias of wild-type (WT) and indicated knock-out mice and were cultured for 6–8 days as described earlier (9, 10, 38, 41). Cells were plated on 12-well plates containing RPMI 1640, 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 20 ng/ml GM-CSF. THP-1 cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, 1 mM sodium pyruvate, and 100 nM HEPES. RAW 264.7 cells were maintained in DMEM, 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin. S100A9 KO mice were generated at Laval University, Quebec, Canada. TLR3 KO mice were obtained from Jackson Laboratory, Bar Harbor, ME.
Antibodies and reagents
DMEM, RPMI 1640, FBS and penicillin-streptomycin were purchased from Life Technologies (Grand Island, NY). PolyIC, CpG DNA, and imiquimod were purchased from Invivogen. Primary antibodies: anti-TLR3 (rabbit anti-TLR3 from Abcam and mouse anti-TLR3 from Thermo Scientific), anti-Rab4 (Abcam), anti-Rab9 (Abcam), anti-S100A9 (R&D), anti-GFP (Santa Cruz), anti-FLAG (Sigma), and anti-Myc (Sigma). Control siRNA and S100A9 siRNA were purchased from Santa Cruz. ELISA kits: IFN-β (PBL Assay Science), IL-12-p70 (eBioscience), TNF (eBioscience). Control IgG (purified rabbit IgG) was purchased from Innovative Research, Novi, MI. Biotinylated polyIC and FITC-Avidin was purchased from Invivogen and Invitrogen, respectively.
S100A9 neutralizing antibody and purified S100A9 protein
Murine S100A9 neutralizing IgG antibody (S100A9 Ab) was purified from the serum of S100A9 immunized rabbits as described previously (38). Recombinant mouse S100A9 proteins were generated as described previously (38). Recombinant proteins were prepared in Hank’s buffered salt solution (HBSS) buffer. The absence of endotoxin contamination in antibody and protein preparations was confirmed using the limulus amebocyte assay (Cambrex). The neutralizing activity of S100A9 Ab was tested by ELISA analysis of IL-6 levels in the medium supernatant of WT BMDMs treated with recombinant mouse S100A9 protein (1 ng/ml) in the presence of either control IgG or S100A9 Ab (3 ng/ml) for 8h.
Reverse transcription-PCR (RT-PCR)
Total RNA was extracted using TriZOL Reagent (Invitrogen). cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). PCR was performed using 0.25 units of Taq polymerase, 10 pmol of each oligonucleotide primer, 1 mM MgCl2, and 100 μM deoxynucleotide triphosphates in a final reaction volume of 25 μl. Following amplification, the PCR products were analyzed on 1% agarose gel. Equal loading in each well was confirmed by analyzing expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers used to detect the indicated genes by RT-PCR were:
Human GAPDH forward, 5′-GTCAGTGGTGGACCTGACCT
Human GAPDH reverse, 5′-AGGGGTCTACATGGCAACT
Mouse GAPDH forward, 5′-GCCAAGGTCATCCATGACAACTTTGG
Mouse GAPDH reverse, 5′-GCCTGCTTCACCACCTTCTTGATGTC
Human S100A9 forward, 5′-CAGCTGGAACGCAACATAGA
Human S100A9 reverse, 5′-TCAGCTGCTTGTCTGCATTT
Human IFN-β forward, 5′-TCCACGACAGCTCTTTCCAT
Human IFN-β reverse, 5′-TCATAGATGGTCACTGCGGC
Mouse IFN-β forward, 5′-CAGCTCCAGCTCCAAGAAAGGACGAACATTCG
Mouse IFN-β reverse, 5′-CCACCACTCATTCTGAGGCATCAACTGACAGG
Viral infection of cells
BMDM was infected with purified RSV or VSV [1 multiplicity of infection (MOI) for RSV and 0.01 MOI for VSV] in serum-free, antibiotic-free OPTI-MEM medium (Gibco). Virus adsorption was done for 1.5 h at 37°C, after which cells were washed twice with PBS. Infection was continued in the presence of serum containing RPMI medium for the specified time points.
PolyIC, CpG and imiquimod treatment
BMDM or THP-1 cells were treated with polyIC (10μg/mL) as indicated. BMDMs were also treated with CpG (5μM) or imiquimod (5μg/mL). In some experiments, BMDMs were treated with polyIC in the presence of 5 ng/ml control IgG or 5 ng/ml anti-S100A9 blocking antibody. In addition, in some experiments polyIC treatment of S100A9 KO BMDMs was done in the presence of purified S100A9 protein or HBSS buffer (vehicle control). Purified S100A9 protein [200 pg S100A9 per 100,000 cells (for 8h polyIC treatment) and 900 pg of S100A9 per 100,000 cells (for 12h polyIC treatment] was added to S100A9 KO BMDMs at the same time as polyIC addition. Purified protein was present during polyIC treatment.
siRNA
Control siRNA and human S100A9 siRNA were purchased from Santa Cruz Biotechnology. PMA differentiated THP-1 cells were transfected with 40 pmol of siRNAs using Lipofectamine 2000 (Invitrogen). At 48 h post-transfection, the cells were treated with polyIC.
ELISA assays
Medium supernatants from BMDMs were analyzed for IFN-β and IL-12 (p70) levels by using a cytokine-specific ELISA kit. For S100A9 ELISA, Costar High-Binding 96-well plates (Corning, NY) were coated overnight at 4°C with 800 ng/well of purified rabbit IgG against mouse S100A9 (Abcam) diluted in 0.1 M carbonate buffer, pH 9.6. Wells were blocked with PBST + 1% BSA for 1 h at room temperature. Samples were added and incubated overnight at 4°C. The plates were washed three times with PBST and incubated with goat anti-mouse IgG (300 ng/well) (R&D) in PBST + 0.1% BSA for 2 h at room temperature. The plates were then washed three times in PBST. To detect mouse S100A9, rabbit anti-goat HRP (Bio-Rad) was added to the plates. After 1 h incubation at room temperature, the plates were washed three times with PBST. TMB-S substrate (100 μl/well) (Sigma-Aldrich) was added to the plates. The ODs were detected at 450 nm, using a Modulas micro-plate reader.
Cell transfection
HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen) at 80% confluency with expression plasmids encoding myc-S100A9 (4μg/well), FLAG-TLR3 (4μg/well) or empty pcDNA6.1 (4μg/well) vector expressing FLAG or, myc tags alone. At 24 h post-transfection, cell lysate was collected for co-immunoprecipitation analysis. RAW 264.7 cells were transfected with Lipofectamine 2000 (Invitrogen) at 80% confluency with expression plasmids encoding GFP and GFP tagged wild type (WT) and dominant negative (DN) Rab7 (0.8μg/well). At 24h post-transfection, cells were treated with polyIC (50μg/mL) and following 8h polyIC treatment, cell lysate and RNA was collected for Western blotting and RT-PCR, respectively. The medium supernatants collected from these cells were also subjected to IFN-β specific ELISA analysis.
Mice treatment
For the in vivo experiment, eight week old female C57BL/6 mice (WT and S100A9 KO) were injected intraperitoneally (IP) with polyIC (100μg/mouse) in PBS for 3h. Mice were then bled through retro-orbital method and sacrificed. Cytokine levels in serum were detected by ELISA. All animal experiments were approved by the Institutional Animal Care and Use Committee.
S100A9 KO mice were generated for us by Knockout Mouse Project (KOMP, Davis, CA). S100A9 KO mice were generated by eliminating exons from the S100A9 gene of C57BL/6 embryonic stem cells. Loss of S100A9 expression was confirmed by genotyping S100A9 by PCR using tail snip samples from WT and S100A9 KO mice. Other groups have reported generation of S100A9 KO mice. S100A9 mice are fertile, viable and devoid of any obvious phenotype (43). We have recently utilized S100A9 KO mice to demonstrate the role of S100A9 in inflammatory response during influenza A virus infection (38).
Immunofluorescence
For immunofluorescence, 1 × 105 BMDMs were seeded onto 12mm #1.5 glass cover slips (Fisher) and cultured for 48 h. BMDMs were treated (with either polyIC or biotinylated-polyIC) following the protocol above. BMDMs were then fixed with 10% stock formaldehyde in PBS for 15 min at RT, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked with 1% BSA in PBS for 1 h, probed with the indicated primary antibodies overnight at 4°C, washed 3x (5 min per wash) with PBS, probed with corresponding fluorescently labeled (FITC or Texas red) secondary antibodies or FITC-avidin for1h, washed 3x and finally mounted onto glass slides using 5μL SlowFade Gold antifade reagent with DAPI (Invitrogen) and clear nail polish. Slides were then analyzed on an Olympus FV-1000 Confocal MultiPhoton Spectral Laser Scanning Microscope.
Immunoprecipitation and Western blotting
For immunoprecipitation, HEK293 lysate was harvested using 1% Triton-X 100 in PBS with 5mM TRIS pH 7.4 and 1x Roche complete Mini EDTA-free protease inhibitor cocktail. Lysates were sonicated 3 times for 5 s in cold conditions. Lysates were then centrifuged at 13k rpm for 10 min and clear lysate supernatant was transferred to a new tube. Lysates were mixed with Mouse Anti-FLAG Agarose (Clontech) for 12h at 4°C. Samples were centrifuged at 1k rpm for 1 min, and pellets were washed 4 times with PBS/5mM TRIS pH 7.4 and protease inhibitor. For each sample, protein was eluted from the final pellet with 100mM glycine pH 2.9. Eluted proteins were precipitated with 20% TCA overnight at 4°C. The precipitated protein samples were pelleted by centrifugation at 13k rpm for 10 min, washed twice with ice cold acetone, resuspended in SDS dissolving buffer and separated on SDS-PAGE. Protein samples were transferred onto 0.2μm nitrocellulose membranes (BIO-RAD) for Western blot analysis with anti-myc or anti FLAG antibodies.
For immuno-precipitation with BMDMs, cell lysate was mixed with anti-mouse S100A9 antibody for 1 h, followed by addition of protein G Agarose (Thermo Scientific) for 12h at 4°C. Samples were centrifuged at 1k rpm for 1 min and the pellet was washed 4 times with PBS 5mM TRIS pH 7.4 and protease inhibitor. For each sample, protein was eluted from the final pellet with 100mM glycine pH 2.9. Eluted protein was precipitated with 20% TCA overnight at 4°C. Precipitated protein was pelleted by centrifugation at 13k rpm for 10 min, washed twice with ice cold acetone, resuspended in SDS dissolving buffer and separated on a SDS-PAGE. Protein was transferred onto a .2μm nitrocellulose membrane (BIO-RAD) for Western blot analysis with anti-TLR3 or anti-S100A9 antibody.
Results
S100A9 is required for TLR3-dependent IFN-β induction
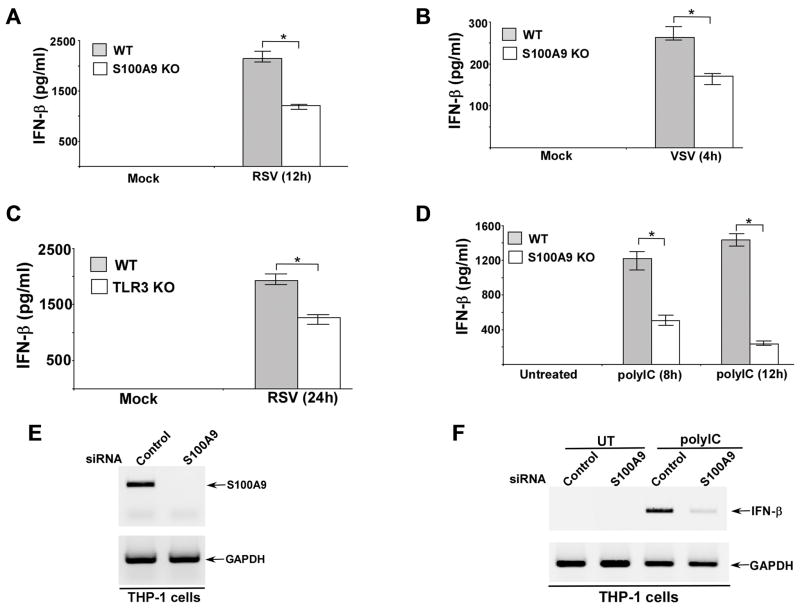
In order to study the role of S100A9 during IFN-β response we infected wild type (WT) and S100A9 knockout (KO) bone marrow derived macrophages (BMDMs) with two RNA viruses RSV and VSV. The levels of IFN-β in the medium supernatants of infected cells were analyzed by ELISA. We detected significantly reduced levels of IFN-β in the medium supernatant of S100A9 KO cells infected with RSV and VSV (Fig. 1A, B). RT-PCR analysis revealed diminished IFN-β expression in VSV (Supplemental Fig. 1A) and RSV (data not shown) infected S100A9 KO BMDMs. In accordance with reduced IFN-β production (and resultant loss of IFN-β mediated antiviral activity) from infected S100A9 deficient cells, we observed increased RSV (Supplemental Fig. 1B) and VSV (data not shown) infectivity in S100A9 KO BMDMs compared to WT cells. Various TLRs have been implicated in activation of the innate response during RNA virus infection. To evaluate the role of TLR3 activation in IFN-β production following RNA virus infection, we infected WT and TLR3 KO BMDMs with RSV. TLR3 is required for IFN-β induction, since significant loss of IFN-β production was noted in RSV infected TLR3 KO cells (Fig. 1C). Similar results were obtained when WT and TLR3 KO cells were infected with VSV (data not shown). These results suggested an important role of S100A9 in triggering TLR3-dependent IFN-β response following RNA virus infection.
FIGURE 1. S100A9 is required for optimal TLR3-dependent interferon-beta (IFN-β) induction.
Primary bone marrow derived macrophages (BMDM) isolated from wild-type (WT) and S100A9 knockout (KO) mice were infected with either human respiratory syncytial virus (RSV) or vesicular stomatitis virus (VSV). The medium supernatant from RSV (A) and VSV (B) infected cells were collected to assess levels of mouse IFN-β by ELISA. (C) WT and TLR3 KO BMDM were infected with RSV. At 24h post-infection, medium supernatant was collected to assess levels of mouse IFN-β by ELISA. (D) WT and S100A9 KO BMDM were treated with polyIC for 8h and 12h. At each time-point, medium supernatant was collected to assess levels of mouse IFN-β by ELISA. (E) Human macrophage cell-line THP-1 was transfected with either control siRNA or S100A9 specific siRNA. At 48h post-transfection, RT-PCR analysis was performed to examine expression of S100A9 in control and S100A9 silenced cells. (F) THP-1 cells transfected with either control siRNA or S100A9 specific siRNA were treated with polyIC. At 8h post-treatment, RT-PCR analysis was performed to examine IFN-β induction. UT; untreated cells. The values shown in (A), (B), (C) and (D) represent the mean ± standard deviation from three independent experiments performed in triplicate. *p <0.05 using a Student’s t test.
The above results were validated using TLR3 agonist polyIC. WT and S100A9 KO BMDMs were treated with polyIC and IFN-β levels were measured in the medium supernatant at 8h and 12h post-treatment. In accordance with the results obtained with RSV and VSV, drastic reduction in IFN-β production was observed in polyIC treated S100A9 KO cells (Fig. 1D). Loss of IFN-β was due to reduced IFN-β gene expression in S100A9 KO cells (Supplemental Fig. 1C). Moreover, TLR3 expression was similar in polyIC treated WT and S100A9 KO BMDMs (Supplemental Fig. 1D). We also did not find any difference in S100A8 protein expression in WT vs. S100A9 KO BMDMs (Supplemental Fig. 1E). In fact, S100A8 expression was enhanced in S100A9 deficient BMDMs. In addition, we noted similar levels of IFN-β production from polyIC treated WT and S100A8 KO BMDMs (data not shown). These results suggested S100A9-dependent (S100A8-independent) TLR3 activation in BMDMs. Furthermore, S100A9 was not required for activation of cytosolic PRRs like RIG-I and MDA5. Specificity of S100A9 in regulating TLR3 activity was borne out by our result showing similar IFN-β production from WT and S100A9 KO cells following transfection of these cells with RIG-I/MDA5 activator LyoVec-polyIC and LyoVec-5′ppp-dsRNA (Supplemental Fig. 1F, 1G).
To assess S100A9 function in human macrophages, we silenced (via siRNA) S100A9 expression in human macrophage cell-line THP-1. Efficient silencing was evident in THP-1 cells treated with S100A9-specific siRNA (Fig. 1E). Human S100A9 also functioned as a positive regulator of TLR3-mediated IFN-β response since IFN-β expression was diminished in S100A9 silenced THP-1 cells (Fig. 1F) following stimulation with polyIC. These results demonstrated that S100A9 is a critical cellular factor regulating activated TLR3-dependent IFN-β response.
Intracellular S100A9 regulates TLR3 activation
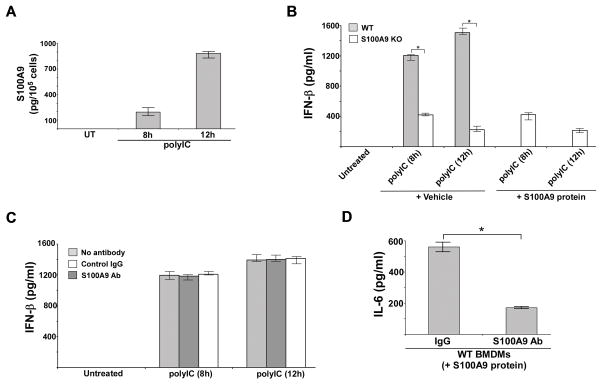
We recently demonstrated a role of extracellular (secreted) S100A9 in regulating pro-inflammatory response during influenza A virus infection (38). Therefore, it was important to dissect the role of extracellular vs. intracellular S100A9 (38, 39, 40, 44) in regulating TLR3 activation. We initiated our studies by measuring levels of extracelluar S100A9 in polyIC treated WT BMDMs. ELISA analysis of medium supernatant revealed release of S100A9 protein following polyIC treatment. We detected ~200 pg of S100A9 protein per 100,000 cells and ~900 pg of S100A9 protein per 100,000 cells after 8h and 12h polyIC treatment, respectively (Fig. 2A). The role of intracellular vs. extracellular S100A9 was further examined by adding purified S100A9 protein to polyIC treated S100A9 KO BMDMs. S100A9 KO BMDMs were treated with polyIC and purified S100A9 protein for 8h and 12h. IFN-β levels in the medium supernatant was analyzed by ELISA. We added 200 pg of S100A9 protein per 100,000 cells (for 8h polyIC treatment) and 900 pg of S100A9 protein per 100,000 cells (for 12h polyIC treatment), reflecting the amount of endogenous S100A9 protein detected in the medium supernatant of polyIC treated WT cells at 8h and 12h post-polyIC treatment (please see Fig. 2A). Addition of purified S100A9 protein did not result in recovery of IFN-β production from polyIC treated S100A9 KO cells (Fig. 2B). This result suggested that intracellular S100A9 is involved in regulating TLR3 activation. Involvement of intracellular S100A9 during this process was further confirmed by using S100A9 blocking (neutralizing) antibody, which inhibits the extracellular S100A9 activity (38, 40–42, 44–48). Extracellular S100A9 does not play a role during TLR3 mediated IFN-β response since S100A9 blocking antibody did not inhibit IFN-β production from polyIC treated cells (Fig. 2C). The neutralizing function of this antibody is evident from loss of IL-6 production from cells treated with purified S100A9 protein in the presence of S100A9 blocking antibody (Fig. 2D). These results demonstrated a role of intracellular S100A9 in positively regulating TLR3 activity.
FIGURE 2. Intracellular S100A9 regulates TLR3 activation.
(A) Primary bone marrow derived macrophages (BMDM) from wild type (WT) mice were treated with polyIC. At indicated post-treatment time-periods the medium supernatant was collected to assess levels of S100A9 protein by ELISA. UT; untreated cells. The value represents the mean ± standard deviation from three independent experiments performed in triplicate. (B) S100A9 knockout (KO) BMDMs was treated with polyIC in the absence or presence of purified recombinant mouse S100A9 protein (200 pg/100,000 cells for 8h polyIC treatment and 900 pg/100,000 cells for 12h polyIC treatment). Medium supernatant was collected from polyIC treated cells to assess levels of IFN-β by ELISA. Vehicle control cells (vehicle) were incubated with HBSS buffer. (C) WT BMDMs were treated with polyIC in the presence of either control IgG or anti-S100A9 blocking (neutralizing) antibody (S100A9 Ab). At indicated post-treatment time-periods the medium supernatant was collected to assess levels of mouse IFN-β by ELISA. (D) WT BMDMs were treated with purified recombinant mouse S100A9 protein (1 ng/ml) in the presence of either control IgG or S100A9 Ab (3 ng/ml). At 8h post-treatment the medium supernatant was collected to assess levels of mouse IL-6 by ELISA. The ELISA values shown in (B), (C) and (D) represent the mean ± standard deviation from three independent experiments performed in triplicate. *p <0.05 using a Student’s t test.
Positive regulatory function of intracellular S100A9 is specific for TLR3
TLR3 activation occurs in the endosome, where it can detect its PAMP. Likewise, TLR7 and TLR9 also detect their corresponding PAMPs in the endosomal compartment. Therefore we investigated whether the requirement of S100A9 is specific for TLR3 or if it has a more general function in positively regulating other endosomal TLRs (i.e. TLR9 and TLR7). TLR9 and TLR7 activation occurs in macrophages (BMDMs) following treatment with their corresponding agonists, imiquimod and CpG DNA, respectively. Unlike TLR3, activation of TLR7 and TLR9 does not result in production of discernible levels of IFN-β. Since TLR3, TLR7 and TLR9 activation in macrophages results in production of IL-12 (p70); we measured IL-12 (p70) in the medium supernatants to study the role of S100A9 in regulating endosomal TLR (i.e. TLR3, TLR7 and TLR9) activation.
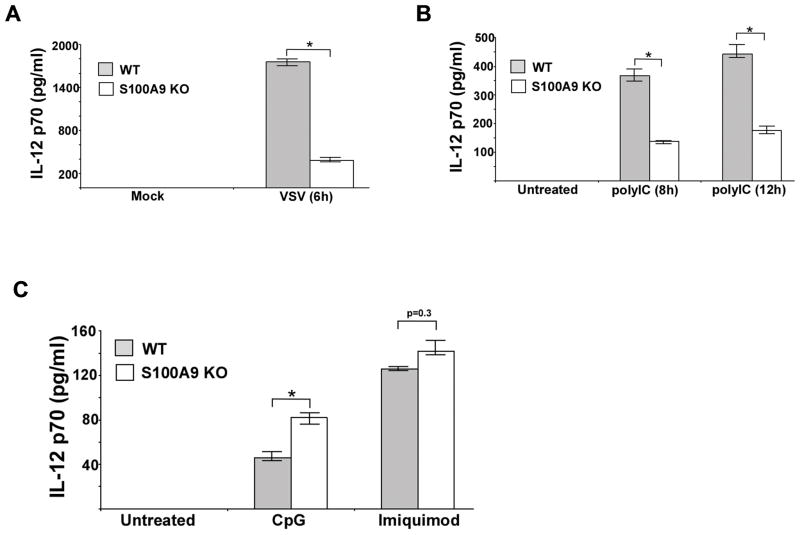
RNA viruses like VSV require S100A9 for IFN-β production (Fig. 1B). In addition to IFN-β, S100A9 also plays a role in production of IL-12 from VSV infected cells. Drastic loss of IL-12 production was observed in VSV infected S100A9 KO BMDMs (Fig. 3A). PolyIC treatment of S100A9 KO BMDMs led to diminished production of IL-12 (Fig. 3B). FACS analysis of FITC-polyIC treated BMDMs revealed similar level of polyIC uptake by WT and S100A9 KO cells (Supplemental Fig. 2). Thus, loss of TLR3 activity in S100A9 KO cells was not due to diminished polyIC internalization.
FIGURE 3. Positive-regulatory function of S100A9 is TLR3-specific.
(A) Primary bone marrow derived macrophages (BMDM) isolated from wild-type (WT) and S100A9 knockout (KO) mice were infected with vesicular stomatitis virus (VSV). The medium supernatant from VSV infected cells was collected to assess levels of IL-12 (p70) by ELISA. (B) WT and S100A9 KO BMDM were treated with polyIC for 8h and 12h. At each time-point, medium supernatant was collected to assess levels of mouse IL-12 (p70) by ELISA. (C) WT and S100A9 KO BMDM were treated with either CpG DNA (CpG) or imiquimod. After treatment, medium supernatant was collected to assess levels of mouse IL-12 (p70) by ELISA. The values represent the mean ± standard deviation from three independent experiments performed in triplicate. *p <0.05 using a Student’s t test.
The role of S100A9 during TLR7 and TLR9 response was next investigated by treating WT and S100A9 KO BMDMs with imiquimod (TLR7 agonist) and CpG DNA (TLR9 agonist). S100A9 is not required for TLR7 activation since IL-12 production was similar in imiquimod treated WT and S100A9 KO BMDMs (Fig. 3C). Surprisingly, we observed enhanced IL-12 production from CpG DNA treated S100A9 KO cells (Fig. 3C), suggesting an involvement of S100A9 in negative-regulation of TLR9 signaling. These observations suggested differential function (positive-regulation of TLR3 signaling and negative-regulation of TLR9 signaling) of S100A9 during activation of two endosomal TLRs, TLR3 and TLR9. Nevertheless, our studies demonstrated that positive regulatory function of S100A9 is specific for TLR3 activation.
Interaction of S100A9 with TLR3
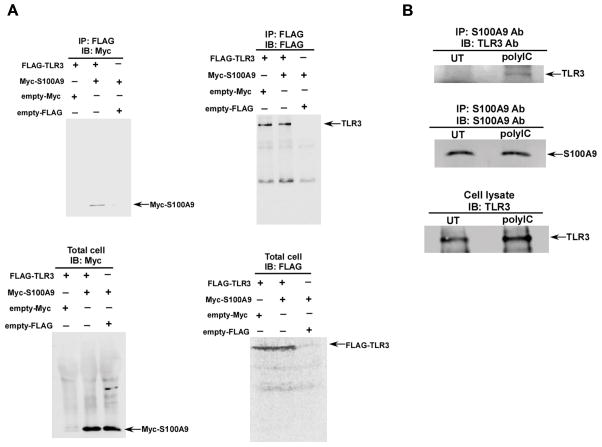
In order to elucidate the mechanistic aspect of TLR3 activation by S100A9, we first examined possible interaction of TLR3 with S100A9. We co-expressed FLAG-TLR3 and Myc-S100A9 in HEK293 cells. Cell lysate collected from these cells were immuno-precipitated (IP) with anti-FLAG antibody and subsequently blotted with anti-Myc antibody. Co-IP analysis revealed interaction of S100A9 with TLR3, since Myc-S100A9 could be immuno-precipitated with FLAG-TLR3 (Fig. 4A, upper left). Following co-IP, Myc-S100A9 was not detected in control cells, (i.e. in cells co-expressing either FLAG-TLR3 and empty-Myc or Myc-S100A9 and empty-FLAG). Amount of FLAG-TLR3 protein pulled down following immuno-precipitation with anti-FLAG antibody were similar in control cells (i.e. in cells expressing FLAG-TLR3 and empty-Myc) and in cells co-expressing FLAG-TLR3 and Myc-S100A9 (Fig. 4A, upper right). Furthermore expression of FLAG-TLR3 and Myc-S100A9 were similar in control cells and cells co-expressing FLAG-TLR3 and Myc-S100A9 as deduced by Western blotting of cell lysate with anti-FLAG and anti-Myc antibodies (Fig. 4A, lower right and lower left). Interaction of S100A9 with TLR3 was specific, since we failed to observe interaction of S100A9 with RIG-I following co-IP analysis with HEK293 cells co-expressing Myc-S100A9 and FLAG-RIG-I (Supplemental Fig. 3A). These studies suggested that interaction of S100A9 with TLR3 occurs in the cellular milieu.
FIGURE 4. Interaction of S100A9 with TLR3.
(A) Cell lysate collected from HEK293 cells expressing FLAG-TLR3 and Myc-S100A9 were immuno-precipitated (IP) with FLAG antibody, followed by immuno-blotting (IB) with either Myc antibody (upper panel, left) or FLAG antibody (upper panel, right). Total lysate was also blotted with Myc (lower panel, left) and FLAG (lower panel, right) antibodies. (B) Primary bone marrow derived macrophages (BMDM) isolated from wild-type (WT) mice were treated with polyIC for 8h. The cell lysate was subjected to co-IP [IP with S100A9 antibody (Ab) and IB with TLR3 Ab] (upper panel). The co-IP samples were also IP and IB with S100A9 Ab (middle panel). Total cell lysate was subjected to immuno-blotting with TLR3 Ab (lower panel).
We further extended our studies to investigate the interaction of endogenous S100A9 with TLR3 in WT BMDMs. WT BMDMs were treated with poly-IC and at 6h post-polyIC treatment, cell lysate was subjected to IP with anti-S100A9 antibody and immuno-blotting with anti-TLR3 antibody. Although interaction of S100A9 with TLR3 was not detected in untreated cells, polyIC treatment prompted interaction of S100A9 with TLR3 (Fig. 4B). These results demonstrated that TLR3 activation (e.g. polyIC treatment) triggers interaction of S100A9 with TLR3.
Co-localization of TLR3 with S100A9
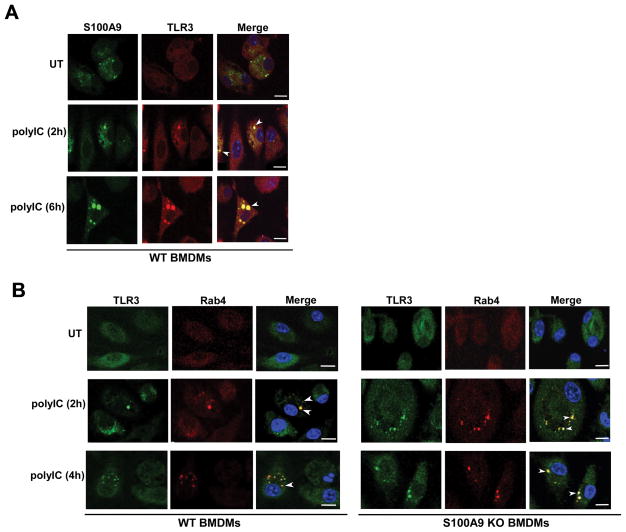
Co-IFA was conducted to examine intracellular localization of S100A9 and TLR3 in untreated and polyIC-treated cells. WT BMDMs treated with polyIC (or vehicle control) were fixed and labeled with anti-TLR3 (red-Texas red) and anti-S100A9 (green-FITC) antibodies. Co-localization of S100A9 (green) with TLR3 (red) was not observed in untreated cells (Fig. 5A). PolyIC treatment triggered co-localization of TLR3 with S100A9 (Fig. 5A). Interestingly, S100A9 co-localized with TLR3 in intracellular specks, which may represent vesicular and/or endosomal compartments. We failed to visualize co-localization by using an irrelevant control antibody (i.e. isotype matched control IgG) (Supplemental Fig. 3B). This result suggested targeting of S100A9 and TLR3 to the same intracellular compartment following priming of cells with TLR3 agonist polyIC.
FIGURE 5. S100A9 co-localizes with TLR3 upon polyIC stimulation and S100A9 is not required for TLR3 trafficking to early endosome.
(A) Co-immunofluorescence (co-IFA) analysis was performed by treating primary bone marrow derived macrophages (BMDM) isolated from wild-type (WT) mice with polyIC. Following treatment, cells were fixed and labeled with S100A9 (FITC- green) and TLR3 (Texas red - red) antibodies. The images of polyIC treated cells represent 70% of cells (i.e. 70 cells out of 100 cells) with S100A9 and TLR3 co-localization. (B) BMDM isolated from WT and S100A9 knockout (KO) mice were treated with polyIC for co-IFA analysis. Following polyIC treatment, cells were fixed and labeled with antibodies specific for TLR3 (FITC- green) and early endosome marker Rab4 (Texas red - red). The images of polyIC treated WT and S100A9 KO cells represent 68% and 72% of cells (i.e. 68 cells or 72 cells out of 100 cells) with Rab4 and TLR3 co-localization, respectively. Merged images in (A) and (B) are represented as yellow. The images are representative of thirty viewing fields from two independent experiments with similar results. White arrow heads in merged images represents co-localization. The white bar represents 10 μm. UT; untreated cells.
S100A9 is not required for TLR3 targeting to the early endosome following polyIC treatment
Interaction of TLR3 with S100A9 following polyIC treatment suggested possible involvement of S100A9 in either promoting TLR3 activation by facilitating TLR3 adaptor protein (e.g. TRIF) recruitment or S100A9 playing a role during pre-activation stages of TLR3 activation by ensuring “correct” intracellular sorting of TLR3 from ER-golgi to LE (and EL) for PAMP recognition. S100A9 was not involved in recruitment of TLR3 adaptors since S100A9 did not interact with TRIF and S100A9 expression in HEK293 cells did not alter interaction kinetics of TRIF with TLR3 (data not shown). Based on these results, our studies focused on the role of S100A9 at pre-activation stages of TLR3 activation. In order to explore the role of S100A9 in TLR3 trafficking, we first investigated whether S100A9 is required for sorting of TLR3 from the ER-golgi compartment to EE. WT and S100A9 KO BMDMs were treated with polyIC and TLR3 localization in the EE was monitored by labeling cells with anti-TLR3 (green-FITC) and anti-Rab4 (red-Texas red) antibodies. S100A9 is dispensable for vesicular trafficking of TLR3 from ER-golgi to EE, since in both WT and S100A9 KO BMDMs we observed localization of TLR3 in Rab4 [Rab4 is a marker of early and recycling endosomes (49)] positive intracellular compartment (Fig. 5B). These results indicated that S100A9 is not required for TLR3 targeting to EE following cell stimulation with TLR3 agonist.
S100A9 is essential for maturation of TLR3 containing late endosome
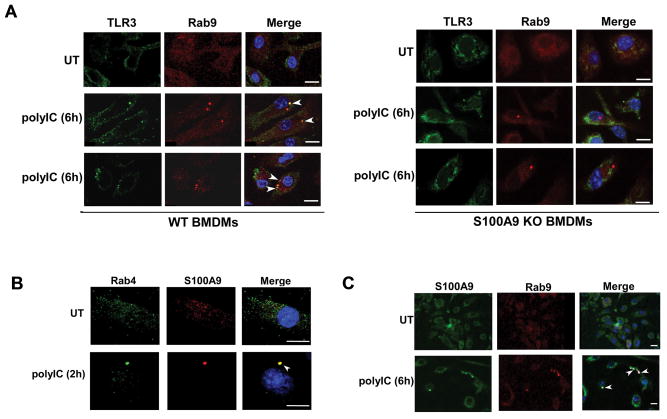
Our study suggested a role of S100A9 in post-EE stages of TLR3 activation since TLR3 was detected in EE of S100A9 deficient cells (Fig. 5B). We next investigated whether S100A9 play a role in maturation of TLR3 containing EE into LE. PolyIC treated WT and S100A9 KO BMDMs were labeled with anti-TLR3 (green-FITC) and anti-Rab9 (red-Texas red) antibodies. Confocal microscopic analysis revealed localization of TLR3 in Rab9 [Rab9 is a marker of late endosomes (50)] positive intracellular compartment of polyIC treated WT cells (Fig. 6A). In contrast, such localization was lacking in S100A9 KO cells (Fig. 6A). Increasing polyIC treatment time (10h–24h) did not lead to co-localization of TLR3 with Rab9 positive compartment (data not shown). Interestingly, we could visualize Rab9 positive punctuate structures in polyIC treated S100A9 KO cells. Thus ours results suggested a critical role of S100A9 in promoting maturation of TLR3 containing EE into LE following stimulation of cells with TLR3 agonist.
FIGURE 6. S100A9 regulates TLR3 sorting to late endosome.
(A) Co-immunofluorescence analysis was performed by treating primary bone marrow derived macrophages (BMDM) isolated from wild-type (WT) and S100A9 knockout (KO) mice with polyIC. Following treatment, cells were fixed and labeled with antibodies specific for TLR3 (FITC- green) and late endosome marker Rab9 (Texas red - red). The images of polyIC treated WT cells represent 71% of cells (i.e. 71 cells out of 100 cells) with Rab9 and TLR3 co-localization. We failed to detect Rab9 and TLR3 co-localization in S100A9 KO cells. Two representative images from polyIC treated (6h treatment) cells are shown. (B) WT BMDM treated with polyIC for 2h were fixed and labeled with antibodies specific for S100A9 (Texas red - red) and early endosome marker Rab4 (FITC- green). The images of polyIC treated cells represent 65% of cells (i.e. 65 cells out of 100 cells) with S100A9 and Rab4 co-localization. (C) WT BMDM treated with polyIC for 6h were fixed and labeled with antibodies specific for S100A9 (FITC- green) and late endosome marker Rab9 (Texas red - red). The images of polyIC treated cells represent 58% of cells (i.e. 58 cells out of 100 cells) with S100A9 and Rab9 co-localization. Merged images in (A), (B) and (C) are represented as yellow. The images are representative of thirty viewing fields from two independent experiments with similar results. The white bar represents 10 μm. White arrow heads in merged images represents co-localization. UT; untreated cells.
Our results suggested polyIC treatment triggering intracellular trafficking of S100A9 to TLR3-postive intracellular compartment. Since TLR3 is detected in EE (Fig. 5B) and LE (Fig. 6A) following polyIC treatment, we next performed co-IFA studies to examine whether S100A9 is targeted to EE and LE after polyIC stimulation. Our studies revealed that indeed polyIC alters intracellular dynamics and distribution of S100A9. While S100A9 could not be detected in the endosomal compartment of untreated cells, polyIC treatment for 2h led to sorting of S100A9 to EE (Fig. 6B). Longer polyIC exposure (i.e. 6h polyIC treatment) resulted in sorting of S100A9 to LE (Fig. 6C). PolyIC mediated intra-cellular sorting of S100A9 and TLR3 to EE and LE illuminated a critical role of S100A9 in facilitating TLR3 trafficking to LE (and EL) for PAMP recognition. Overall our results suggested that S100A9 is involved in maturation of TLR3 containing EE into LE and therefore in cells lacking S100A9, TLR3 was not localized in LE. As a consequence, TLR3 fails to detect its agonist/PAMP for activation since agonist/PAMP detection and activation occurs in the EL compartment arising from fusion of TLR3 containing LE with lysosome.
S100A9 deficiency prevents TLR3 targeting to the polyIC positive subcellular compartment
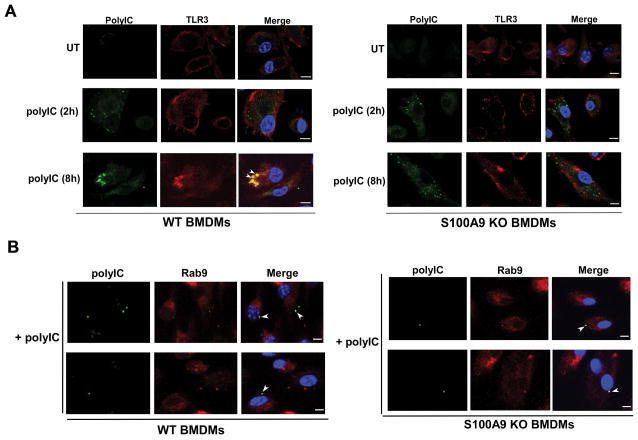
Since TLR3 was not targeted to LE in S100A9 lacking cells (Fig. 6A) and TLR3 targeting to LE is a pre-requisite for PAMP/agonist detection, we envisioned that TLR3 will not be recruited to polyIC positive subcellular compartment of S100A9 lacking cells. In order to study intracellular localization of TLR3 and polyIC, we treated WT and S100A9 KO BMDMs with biotinylated-polyIC (biotin-polyIC). At 2h and 8h post-polyIC treatment, cells were labeled with anti-TLR3 antibody (red-TexasRed) and FITC-avidin (green-FITC) to detect biotin-polyIC. As expected, TLR3 colocalized with polyIC in WT cells (Fig. 7A). However, in the absence of S100A9 (i.e. in S100A9 KO cells) TLR3 and polyIC could not be detected in the same compartment (Fig. 7A). Increasing polyIC treatment time (10h–24h) did not lead to co-localization of TLR3 with biotin-polyIC (data not shown). It is interesting to note that although polyIC could be visualized in intracellular vesicular structures (represented by punctuate staining) of S100A9 deficient cells, it was not co-localizing with TLR3 (Fig. 7A).
FIGURE 7. S100A9 is essential for TLR3 targeting to polyIC positive intracellular compartment.
(A) Co-immunofluorescence analysis was performed by treating primary bone marrow derived macrophages (BMDM) isolated from wild-type (WT) and S100A9 knockout (KO) mice with biotinylated-polyIC (polyIC). Following treatment, cells were fixed and labeled with Avidin-FITC (green) and TLR3 (Texas red - red) antibody. The images of polyIC (8h treatment) treated WT cells represent 69% of cells (i.e. 69 cells out of 100 cells) with polyIC and TLR3 co-localization. We failed to detect polyIC and TLR3 co-localization in S100A9 KO cells. (B) BMDM isolated from WT and S100A9 KO mice were treated with biotinylated-polyIC (polyIC) for co-IFA analysis. Following polyIC treatment, cells were fixed and labeled with Avidin-FITC (green) and antibody specific for late endosome marker Rab9 (Texas red - red). The images of polyIC treated WT and S100A9 KO cells represent 67% and 59% of cells (i.e. 67 cells or 59 cells out of 100 cells) with polyIC and Rab9 co-localization, respectively. Merged images in (A) and (B) are represented as yellow. The images are representative of thirty viewing fields from two independent experiments with similar results. The white bar represents 10 μm. White arrow heads in merged images represents co-localization. UT; untreated cells.
Since lack of S100A9 prevented TLR3 sorting to LE (Fig. 6A), we next asked the question whether S100A9 deficiency may also prevent targeting of TLR3 agonist (i.e. polyIC) to LE. For these studies, we treated WT and S100A9 KO BMDMs with biotin-polyIC. At 6h post-polyIC treatment we performed co-IFA analysis to visualize co-localization of polyIC with LE marker Rab9. Surprisingly, we detected polyIC in LE of WT and S100A9 KO cells (Fig. 7B). Thus, unlike TLR3, polyIC targeting to the LE was not compromised in S100A9 deficient cells. This result along with our polyIC uptake studies (Supplemental Fig. 2) suggested that S100A9 is not involved in internalization and intracellular trafficking of polyIC to LE. These results demonstrated that S100A9 plays an important role in TLR3 biology at pre-activation stage by ensuring proper spatial distribution (and targeting) of TLR3 to LE.
Late endosome (LE) is required for TLR3 activation
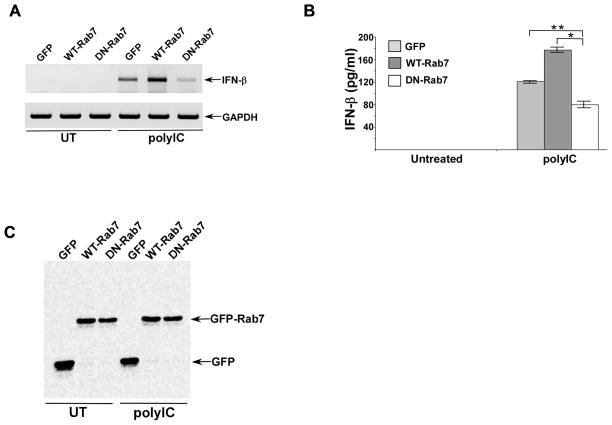
Our studies suggested an important role of LE in TLR3 activation since absence of TLR3 in the LE of S100A9 KO cells resulted in diminished TLR3-dependent response (Figs. 1, 6A, 7A). To date the role of LE in TLR3 trafficking has not been assessed. In order to examine the involvement of LE in TLR3 activation, we expressed GFP tagged dominant-negative Rab7 (DN-Rab7) in mouse macrophage cell-line RAW 264.7. Expression of DN-Rab7 blocks vesicular traffic to LE and therefore has been widely utilized to study function of LE during various physiological/biological processes (51). As a control we expressed GFP and GFP tagged wild type Rab7 (WT-Rab7) in RAW 264.7 cells. Cells expressing GFP, WT-Rab7 and DN-Rab7 were treated with polyIC and at 8h post-treatment we assessed TLR3 activation by analyzing IFN-β expression by RT-PCR (Fig. 8A). We also analyzed IFN-β production from these cells (Fig. 8B). LE plays a critical role in TLR3 activation since we observed loss of IFN-β expression (Fig. 8A) and production (Fig. 8B) following expression of DN-Rab7. Expression of GFP, WT-Rab7 and DN-Rab7 is shown in Fig. 8C. These results demonstrated that LE play an essential role in TLR3 activation, by virtue of ensuring proper subcellular compartmentalization of TLR3 for PAMP detection and subsequent activation of TLR3-signalling cascade.
FIGURE 8. Late endosome play an important role in TLR3 activation.
(A) RAW 264.7 cells were transfected with plasmids expressing either GFP, GFP tagged wild-type Rab7 (WT-Rab7) or GFP tagged dominant-negative Rab7 (DN-Rab7). Following polyIC treatment of these cells, interferon-β (IFN-β) expression was examined by RT-PCR analysis. UT; untreated cells. (B) Medium supernatant collected from untreated and polyIC treated RAW 264.7 cells expressing either GFP, GFP-WT-Rab7 or GFP-DN-Rab7 were subjected to ELISA analysis to detect IFN-β levels. The ELISA values represent the mean ± standard deviation from three independent experiments performed in triplicate. *p and **p <0.05 using a Student’s t test. (C) Cell lysates from GFP and GFP tagged Rab7 (WT-Rab7 and DN-Rab7) expressing RAW 264.7 cells were subjected to Western blotting with anti-GFP antibody.
S100A9 is essential for in vivo TLR3 signaling
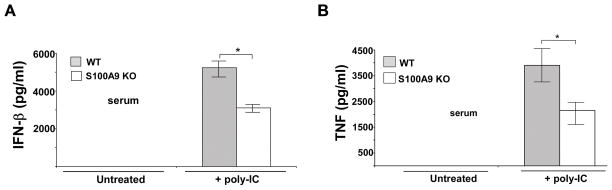
The physiological role of S100A9 during TLR3 activation was then investigated by administering polyIC to WT and S100A9 KO mice. PolyIC has been previously administered to mice via intra-peritoneal (i.p.) route to analyze TLR3 activation in vivo (52). PolyIC was administered to WT and S100A9 KO mice (via i.p. route). At 3h post poly-IC treatment, serum was collected to analyze levels of IFN-β and TNF-α. As shown in Fig, 9, both IFN-β and TNF-α levels were significantly diminished in polyIC treated S100A9 KO mice compared to WT animals. These studies demonstrated an in vivo role of S100A9 in positively regulating TLR3 activation.
FIGURE 9. S100A9 is required in vivo for TLR3 activation.
Wild-type (WT) and S100A9 knockout (KO) mice [n=5 per group] were injected (via intraperitoneal or i.p. route) with polyIC (100 μg/mouse). Serum collected from the mice at 3h post-polyIC treatment was analyzed for interferon-β or IFN-β (A) and TNF-α or TNF (B) by ELISA. The values represent the mean ± standard deviation. *p <0.05 using Student’s t test.
Discussion
In this study, we examined the interplay between S100A9 and TLR3 activation. Reduction in IFN-β and IL-12 production was observed in S100A9 deficient (S100A9 knockout or KO cells) primary macrophages following RNA virus [respiratory syncytial virus (RSV) and vesicular stomatitis virus (VSV)] infection and treatment of cells with polyIC (a dsRNA mimetic that acts as a TLR3 agonist). Interestingly, the positive regulatory function of S100A9 was specific for TLR3 since it did not play a similar role during TLR7 and TLR9 activation. Mechanistic studies revealed co-localization and interaction of S100A9 with TLR3 following polyIC treatment. We also observed that the S100A9-TLR3 interaction was critical for maturation of TLR3 containing EE into LE. Although we observed localization of TLR3 in the EE of both WT and S100A9 KO cells following polyIC treatment, TLR3 could not be detected in the LE of polyIC treated S100A9 KO macrophages. Subsequently, TLR3 failed to co-localize with its agonist (i.e. biotin-labeled polyIC) in S100A9 deficient macrophages. The importance of TLR3 trafficking to the LE was further borne out by our studies demonstrating loss of TLR3 activation in cells with defective LE traffic due to expression of dominant negative Rab7. The in vivo physiological role of S100A9 was evident from loss of IFN-β and TNF levels in the serum of S100A9 KO mice treated with TLR3 agonist polyIC. Thus, we have identified intracellular S100A9 as a regulator of TLR3 signaling and demonstrated that S100A9 functions during pre-TLR3 activation stages by facilitating maturation of TLR3 containing EE to LE.
TLR3 is an endosomal PRR involved in sensing viral dsRNA (13, 14, 15). TLR3 is also expressed on the plasma membrane (53). TLR3 activation regulates immunity, infection and pathogenesis of a wide range of viruses [e.g. Coxsackievirus B3 and B4 (CVB3, CVB4), respiratory syncytial virus (RSV), influenza A virus (IAV), West Nile virus (WNV), Rhinovirus (RV), Hepatitis C and B viruses (HCV and HBV), Herpes Simplex Virus 1 (HSV-1), HIV-1, Vesicular Stomatitis virus (VSV), Chikungunya virus (CGV), Punta Toro virus (PTV)] (14–24). Upon infection or stimulation with synthetic dsRNA mimic (e.g. polyIC), TLR3 is targeted to endosome which fuses with lysosome to form the endolysosomal (EL) compartment. It is in this compartment that TLR3 is presumed to sense/detect its PAMP and undergo proteolytic cleavage by lysosomal enzymes for activation (25–34, 54, 55, 56). Thus, correct compartmentalization of TLR3 is critical for its activity and resulting innate immune response. PAMP mediated TLR3 activation by recruitment of “singnalosome” complex (i.e. complex comprising of TRIF:TRAF3:TBK1:IKK-ε) to the cytosolic face of endosome is well studied. In contrast, existing knowledge about the cellular mechanism(s) [and cellular factor(s)] regulating intracellular sorting of TLR3 to the endosomal (and EL) compartment for PAMP sensing/detection and activation is limited. Moreover, information on how TLR3 trafficking is regulated in macrophages is lacking since majority of studies with myeloid cells were performed with DCs (25–28)?
In the current study, we uncovered a role of intracellular S100A9 in regulating TLR3 trafficking in macrophages. We demonstrate that lack of S100A9 diminishes TLR3 activation and subsequent TLR3-mediated IFN-β and IL-12 production. S100A9 co-localized and interacted with TLR3 following treatment with TLR3 agonist polyIC. Biological significance of such interaction was evident from S100A9’s role in facilitating intracellular trafficking of TLR3 to LE, the compartment which fuses with lysosome to form the endolysomal (EL) body where TLR3 senses/detect its PAMP/agonist for activation. Interestingly, S100A9 was not required for polyIC uptake/internalization and trafficking to the LE. The physiological role of S100A9 was further validated following polyIC treatment of S100A9 KO mice. Significant reduction in serum cytokine levels were observed in polyIC treated S100A9 KO mice. Thus, we have identified S100A9 as a critical regulator of TLR3 activation by virtue of it acting as a “chaperone” like cellular factor to aid intracellular trafficking of TLR3 to the correct subcellular compartment for activation.
Our result suggested that positive regulatory activity of S100A9 is specific for TLR3, since we did not observe such activity following activation of TLR9 and TLR7. It is important to mention that for our studies we exogenously added 10 μg/ml of polyIC to BMDMs which triggers type-I interferon production and pro-inflammatory response primarily via TLR3 activation (57). However, exogenous addition of higher concentrations (more than 20 μg/ml) of polyIC to BMDMs was shown to activate MAVS pathway (58). Thus, it is conceivable that the result obtained with BMDMs were mediated by TLR3 since we used 10 μg/ml of exogenous polyIC for our experiments. This dosage does not induce MAVS in BMDMs. Concomitantly we have utilized activators of MAVS signaling to demonstrate that S100A9 is dispensable for MAVS activation.
The cellular endosomal system is involved in proper sorting of cellular proteins to correct subcellular compartment for its activity or degradation via the lysosomal enzymes (59). EE and LE are the major endosomal compartments achieving this function. EE receives cellular proteins (cargo) from the ER-golgi and plasma membrane (directly or via recycling endosome). EE represents a major sorting compartment in the endocytic pathway, where stringent selection is made whether to transport part of the EE cargo to lysosomes for degradation or proteolytic cleavage by lysosomal enzymes for activation (e.g. endosomal TLRs). EE possess a unique morphology comprising of tubular extensions and vacuolar domains. Cargo destined for lysosome are segregated in the EE vacuole, which matures into LE. Formation of nascent LE occurs when the EE vacuoles (containing selected subset of cargo destined for lysosome) are disengaged from the EE. LE maturation occurs as it fuses with other LE (e.g. intraluminal vesicles or ILV) en route to lysosome. Finally, LE fuses with lysosome to form endolysosome which contain the lysosomal hydrolases for degradation and cleavage (e.g. endosomal TLRs). Endocytic pathway for trafficking of endosomal TLRs to endosome is well studied for TLR7 and TLR9. Chaperones like PRAT4A facilitates TLR7 and TLR9 sorting from ER-golgi to the endosome (31). PRAT4A is required for TLR9 and TLR7 trafficking (and activity), but dispensable for TLR3 activation. Thus, discrete endosomal TLR specific sorting mechanism may exist to facilitate proper targeting to the correct subcellular compartment for PAMP sensing/detection. Cells may have evolved to stringently regulate individual endosomal TLR-specific response by distinct cellular factor(s). However, such examples are few, especially a cellular factor that has TLR3-specific positive regulatory role was yet unknown. In that regard, our studies have identified S100A9 as a specific regulator of TLR3 activation.
Comprehensive knowledge about molecular and cellular mechanism regulating endosomal TLR trafficking is not well-defined. Especially the role of endosomal sorting pathway in TLR3 activation is unclear. Although TLR3 has been detected in EE and LE, it is not known what role these compartments play in TLR3 activation. In the current study we show that indeed “classical” sorting mechanism via LE is required for TLR3 activation since inhibiting TLR3 targeting to LE (by expressing DN-Rab7) abrogated TLR3 activity.
S100A9 (also known as MRP14 and calgranulin-B) belongs to the family of S100 proteins characterized by the presence of EF hand (39, 40). Members of the S100 family of proteins regulate a wide spectrum of cellular activities. S100 proteins are primarily localized in the cytoplasm. However, stress responses trigger secretion of S100A9 via active process. Recently, we demonstrated that extracellular S100A9 (secreted form) triggers pro-inflammatory response and apoptosis during influenza A virus infection (38). In the current study, we did not observe a role of extracellular S100A9 in regulation of TLR3 activity. In contrast, our studies revealed the involvement of intracellular S100A9 in controlling TLR3 activity by promoting maturation of TLR3 containing LE. It is important to note that extracellular S100 proteins were previously implicated in modulating immune and inflammatory response as a Damage Associated Molecular Pattern (DAMP) molecule by activating PRRs (38, 39, 40, 44). In contrast, whether intracellular S100A9 can act as an immune regulator was not known. Our current studies suggested a role of intracellular S100A9 in regulating innate immunity.
How does S100A9 may specifically facilitate maturation of TLR3 specific LE? LE formation from the EE occurs due to a mechanism known as “rab switching” (59–65). Rabs are GTPases that are involved in various cellular processes including intracellular trafficking (59–65). A hallmark of EE is the recruitment of activated Rab5 to its membrane. During formation of LE, Rab5 is displaced by Rab7. This Rab5/Rab7 exchange initiates a chain of events that culminates in formation of LE from the vacuolar body of EE. Since following polyIC treatment we detected S100A9 in both EE and LE and S100A9 co-localized (and interacted) with TLR3, it is possible that once TLR3 is localized in the vacuolar body of the EE, S100A9 interacts with the cytosolic tail of TLR. This event may trigger displacement of EE-specific Rab5 and recruitment of LE promoting Rab7. This process may occur due to S100A9 altering Rab5 folding for displacement and/or facilitating uploading of Rab7 to the vacuolar body of EE that is destined to form a LE. It is an intriguing idea that individual endosomal TLRs may possess specific sets of cellular factors for regulating formation/maturation of endosomal compartment that contain TLRs as cargo. Cells may have evolved such a ploy to prevent aberrant endosomal TLR activation and to ensure that upon stimulation with multiple endosomal TLR ligands, each TLR is sorted to distinct LE (and EL) for activation. This could only be achieved if specific regulatory cellular factors exist for individual endosomal TLR. Further studies will be performed in the future to dissect the exact mechanism governing S100A9 mediated regulation of maturation of TLR3 containing LE.
In summary, our studies have identified and delineated the involvement of S100A9 as a TLR3-specific factor/chaperone that ensures proper localization of TLR3 in the “correct” intracellular compartment for PAMP detection and activation.
Supplementary Material
Acknowledgments
We would like to thank Dr. William C. Davis, Ph.D. (Director, Flow Cytometry Laboratory, College of Veterinary Medicine, Washington State University) for assistance with the FACS analysis. We would like to thank Stephanie Baker for technical assistance.
This work was supported by National Institutes of Health (NIH) grants AI083387 (to S.B) and U19AI070412 (to J.B.B) and a grant from the Canadian Institutes of Health Research (to P.A.T). J.A.S. was supported by NIH training grant 5T32AI007271.
Abbreviations
- BMDMs
bone marrow-derived macrophages
- WT
wild type
- KO
knockout
- TLR
toll-like receptor
- PAMP
Pathogen Associated Molecular Pattern
- PRR
Pattern Recognition Receptor
- DC
dendritic cell
- IFN
interferon
- RSV
respiratory syncytial virus
- VSV
vesicular stomatitis virus
- ER
endoplasmic reticulum
- EE
early endosome
- LE
late endosome
- EL
endolysosomal
- i.p
intraperitoneal
- Ab
antibody
- DN
dominant negative
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Bose S, Banerjee AK. Innate immune response against nonsegmented negative strand RNA viruses. J Interferon Cytokine Res. 2003;23:401–412. doi: 10.1089/107999003322277810. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, I, Kerr M, Williams BRG, Silverman RH, Schreiber RD. How cells respond to inteferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Uematsu S, Akira S. Toll-like Receptors and Type I Interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 4.Versteeg GA, García-Sastre A. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol. 2010;13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmolke M, García-Sastre A. Evasion of innate and adaptive immune responses by influenza A virus. Cell Microbiol. 2010;12:873–880. doi: 10.1111/j.1462-5822.2010.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bose S, Kar N, Maitra R, DiDonato JA, Banerjee AK. Temporal activation of NF-κB regulates an interferon-independent innate antiviral response against cytoplasmic RNA viruses. Proc Natl Acad Sci USA. 2003;100:10890–10895. doi: 10.1073/pnas.1832775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kota S, Sabbah A, Chang TH, Harnack R, Xiang Y, Meng X, Bose S. Role of Human β-Defensin-2 during Tumor Necrosis Factor-α/NF-κB-mediated Innate Antiviral Response against Human Respiratory Syncytial Virus. J Biol Chem. 2008;283:22417–22429. doi: 10.1074/jbc.M710415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segovia J, Sabbah A, Mgbemena V, Tsai S, Chang T, Berton MT, Morris IR, Allen IC, Ting JP, Bose S. TLR2/MyD88/NF-κB Pathway, Reactive Oxygen Species, Potassium Efflux Activates NLRP3/ASC Inflammasome during Respiratory Syncytial Virus Infection. PLoS ONE. 2012;7:e29695. doi: 10.1371/journal.pone.0029695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 12.Brencicova E, Diebold SS. Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Front Cell Infect Microbiol. 2013;3:37. doi: 10.3389/fcimb.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto M, Seya T. TLR3: Interferon induction by double-stranded RNA including poly(I:C) Adv Drug Deliv Rev. 2008;60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Herman M, Ciancanelli MJ, Pérez de Diego R, Sancho-Shimizu V, Abel L, Casanova J. TLR3 immunity to infection in mice and humans. Curr Opin Immunol. 2013;25:19–33. doi: 10.1016/j.coi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perales-Linares R, Navas-Martin S. Toll-like receptor 3 in viral pathogenesis: friend or foe? Immunology. 2013;140:153–167. doi: 10.1111/imm.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Miller DJ, Bowman ER, Nagarkar DR, Schneider D, Zhao Y, Linn MJ, Goldsmith AM, Bentley JK, Sajjan US, Hershenson MB. MDA5 and TLR3 Initiate Pro-Inflammatory Signaling Pathways Leading to Rhinovirus-Induced Airways Inflammation and Hyperresponsiveness. PLoS Pathogens. 2011;7:e1002070. doi: 10.1371/journal.ppat.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goffic RL, Balloy V, Lagranderie M, Alexopoulou L, Escriou N, Flavell R, Chignard M, Si-Tahar M. Detrimental Contribution of the Toll-Like Receptor (TLR)3 to Influenza A Virus-Induced Acute Pneumonia. PLoS Pathog. 2006;2:e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groskreutz DJ, Monick MM, Powers LS, Yarovinsky TO, Look DC, Hunninghake GW. Respiratory Syncytial Virus Induces TLR3 Protein and Protein Kinase R, Leading to Increased Double-Stranded RNA Responsiveness in Airway Epithelial Cells. J Immunol. 2006;176:1733–1740. doi: 10.4049/jimmunol.176.3.1733. [DOI] [PubMed] [Google Scholar]
- 19.Aeffner F, Traylor ZP, Yu ENZ, Davis IC. Double-stranded RNA induces similar pulmonary dysfunction to respiratory syncytial virus in BALB/c mice. Am J Physiol Lung Cell Mol Physiol. 2011;301:L99–L109. doi: 10.1152/ajplung.00398.2010. [DOI] [PubMed] [Google Scholar]
- 20.Rudd BD, Smit JJ, Flavell RA, Alexopoulou L, Schaller MA, Gruber A, Berlin AA, Lukacs NW. Deletion of TLR3 Alters the Pulmonary Immune Environment and Mucus Production during Respiratory Syncytial Virus Infection. J Immunol. 2006;176:1937–1942. doi: 10.4049/jimmunol.176.3.1937. [DOI] [PubMed] [Google Scholar]
- 21.Her Z, Teng T, Tan JJ, Teo T, Kam Y, Lum F, Lee WW, Gabriel C, Melchiotti R, Andiappan AK, Lulla V, Lulla A, Win MK, Chow A, Biswas SK, Leo Y, Lecuit M, Merits A, Rénia L, Ng LF. Loss of TLR3 aggravates CHIKV replication and pathology due to an altered virus-specific neutralizing antibody response. EMBO Mol Med. 2014;7:24–41. doi: 10.15252/emmm.201404459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-Like Receptor 3 Mediates Establishment of an Antiviral State against Hepatitis C Virus in Hepatoma Cells. J Virol. 2009;83:9824–9834. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Huang S, Zhao X, Chen M, Lin Y, Xia Y, Sun C, Yang X, Wang J, Guo Y, Song J, Zhang E, Wang B, Zheng X, Schlaak JF, Lu M, Yang D. Poly(I:C) Treatment Leads to Interferon-Dependent Clearance of Hepatitis B Virus in a Hydrodynamic Injection Mouse Model. J Virol. 2014;88:10421–10431. doi: 10.1128/JVI.00996-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karimi-Googheri M, Arababadi M. TLR3 plays significant roles against hepatitis B virus. Mol Biol Rep. 2014;41:3279–3286. doi: 10.1007/s11033-014-3190-x. [DOI] [PubMed] [Google Scholar]
- 25.Jelinek I, Leonard JN, Price GE, Brown KN, Meyer-Manlapat A, Goldsmith PK, Wang Y, Venzon D, Epstein SL, Segal DM. TLR3-Specific Double-Stranded RNA Oligonucleotide Adjuvants Induce Dendritic Cell Cross-Presentation, CTL Responses, and Antiviral Protection. J Immunol. 2011;186:2422–2429. doi: 10.4049/jimmunol.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. Subcellular Localization of Toll-Like Receptor 3 in Human Dendritic Cells. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 27.Crozat K, Beutler B. TLR7: A new sensor of viral infection. Proc Natl Acad Sci USA. 2004;101:6835–6836. doi: 10.1073/pnas.0401347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muzio M, Bosisio D, Polentarutti N, D’amico G, Stoppacciaro A, Mancinelli R, van’t Veer C, Penton-Rol G, Ruco LP, Allavena P, Mantovani A. Differential Expression and Regulation of Toll-Like Receptors (TLR) in Human Leukocytes: Selective Expression of TLR3 in Dendritic Cells. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- 29.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BL, Barton GM. Trafficking of endosomal Toll-like receptors. Trends Cell Biol. 2014;24:360–369. doi: 10.1016/j.tcb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Shibata T, Akashi-Takamura S, Kiyokawa T, Wakabayashi Y, Tanimura N, Kobayashi T, Matsumoto F, Fukui R, Kouro T, Nagai Y, Takatsu K, Saitoh S, Miyake K. A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses. J Exp Med. 2007;204:2963–2976. doi: 10.1084/jem.20071132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGettrick AF, O’Neill LA. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr Opin Immunol. 2010;22:20–27. doi: 10.1016/j.coi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Johnsen IB, Nguyen TT, Ringdal M, Tryggestad AM, Bakke O, Lien E, Espevik T, Anthonsen MW. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 2006;25:3335–3346. doi: 10.1038/sj.emboj.7601222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Dunzendorfer S, Soldau K, Tobias PS. Double-Stranded RNA-Mediated TLR3 Activation Is Enhanced by CD14. Immunity. 2006;24:153–163. doi: 10.1016/j.immuni.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Niimi K, Asano K, Shiraishi Y, Nakajima T, Wakaki M, Kagyo J, Takihara T, Suzuki Y, Fukunaga K, Shiomi T, Oguma T, Sayama K, Yamaguchi K, Natori Y, Matsumoto M, Seya T, Yamaya M, Ishizaka A. TLR3-Mediated Synthesis and Release of Eotaxin-1/CCL11 from Human Bronchial Smooth Muscle Cells Stimulated with Double-Stranded RNA. J Immunol. 2007;178:489–495. doi: 10.4049/jimmunol.178.1.489. [DOI] [PubMed] [Google Scholar]
- 36.Funami K, Sasai M, Ohba Y, Oshiumi H, Seya T, Matsumoto M. Spatiotemporal Mobilization of Toll/IL-1 Receptor Domain-Containing Adaptor Molecule-1 in Response to dsRNA. J Immunol. 2007;179:6867–6872. doi: 10.4049/jimmunol.179.10.6867. [DOI] [PubMed] [Google Scholar]
- 37.Qi R, Singh D, Kao CC. Proteolytic Processing Regulates Toll-like Receptor 3 Stability and Endosomal Localization. J Biol Chem. 2012;287:32617–32629. doi: 10.1074/jbc.M112.387803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai S, Segovia JA, Chang T, Morris IR, Berton MT, Tessier PA, Tardif MR, Cesaro A, Bose S. DAMP Molecule S100A9 Acts as a Molecular Pattern to Enhance Inflammation during Influenza A Virus Infection: Role of DDX21-TRIF-TLR4-MyD88 Pathway. PLoS Pathog. 2014;10:e1003848. doi: 10.1371/journal.ppat.1003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donato R, Cannon B, Sorci G, Riuzzi F, Hsu K, Weber D, Geczy C. Functions of S100 Proteins. Curr Mol Med. 2013;13:24–57. [PMC free article] [PubMed] [Google Scholar]
- 40.Hermann A, Donato R, Weiger TM, Chazin WJ. S100 Calcium Binding Proteins and Ion Channels. Front Pharmacol. 2012;3:67. doi: 10.3389/fphar.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segovia JA, Tsai S, Chang T, Shil NK, Weintraub ST, Short JD, Bose S. Nedd8 Regulates Inflammasome-Dependent Caspase-1 Activation. Mol Cell Biol. 2015;35:582–597. doi: 10.1128/MCB.00775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bose S, Mathur M, Bates P, Joshi N, Banerjee AK. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J Gen Virol. 2003;84:1687–1699. doi: 10.1099/vir.0.19074-0. [DOI] [PubMed] [Google Scholar]
- 43.Manitz MP, Horst B, Seeliger S, Strey A, Skryabin BV, Gunzer M, Frings W, Schönlau F, Roth J, Sorg C, Nacken W. Loss of S100A9 (MRP14) results in reduced interleukin-8-induced CD11b surface expression, a polarized microfilament system, and diminished responsiveness to chemoattractants in vitro. Mol Cell Biol. 2003;23:1034–1043. doi: 10.1128/MCB.23.3.1034-1043.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halayko AJ, Ghavami S. S100A8/A9: a mediator of severe asthma pathogenesis and morbidity? Can J Physiol Pharmacol. 2009;87:743–755. doi: 10.1139/Y09-054. [DOI] [PubMed] [Google Scholar]
- 45.Simard J, Girard D, Tessier PA. Induction of neutrophil degranulation by S100A9 via a MAPK-dependent mechanism. J Leukoc Biol. 2010;87:905–914. doi: 10.1189/jlb.1009676. [DOI] [PubMed] [Google Scholar]
- 46.Simard J, Simon M, Tessier PA, Girard D. Damage-Associated Molecular Pattern S100A9 Increases Bactericidal Activity of Human Neutrophils by Enhancing Phagocytosis. J Immunol. 2011;186:3622–3631. doi: 10.4049/jimmunol.1002956. [DOI] [PubMed] [Google Scholar]
- 47.Raquil M, Anceriz N, Rouleau P, Tessier PA. Blockade of Antimicrobial Proteins S100A8 and S100A9 Inhibits Phagocyte Migration to the Alveoli in Streptococcal Pneumonia. J Immunol. 2008;180:3366–3374. doi: 10.4049/jimmunol.180.5.3366. [DOI] [PubMed] [Google Scholar]
- 48.Vandal K, Rouleau P, Boivin A, Ryckman C, Talbot M, Tessier PA. Blockade of S100A8 and S100A9 Suppresses Neutrophil Migration in Response to Lipopolysaccharide. J Immunol. 2003;171:2602–2609. doi: 10.4049/jimmunol.171.5.2602. [DOI] [PubMed] [Google Scholar]
- 49.Kalia M, Kumari S, Chadda R, Hill MM, Parton RG, Mayor S. Arf6-independent GPI-anchored Protein-enriched Early Endosomal Compartments Fuse with Sorting Endosomes via a Rab5/Phosphatidylinositol-3′-Kinase–dependent Machinery. Mol Biol Cell. 2006;17:3689–3704. doi: 10.1091/mbc.E05-10-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugimoto Y, Ninomiya H, Ohsaki Y, Higaki K, Davies JP, Ioannou YA, Ohno K. Accumulation of cholera toxin and GM1 ganglioside in the early endosome of Niemann–Pick C1-deficient cells. Proc Natl Acad Sci USA. 2001;98:12391–12396. doi: 10.1073/pnas.221181998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastin G, Heximer SP. Rab Family Proteins Regulate the Endosomal Trafficking and Function of RGS4. J Biol Chem. 2013;288:21836–21849. doi: 10.1074/jbc.M113.466888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar H, Koyama S, Ishii KJ, Kawai T, Akira S. Cutting Edge: Cooperation of IPS-1- and TRIF-Dependent Pathways in Poly IC-Enhanced Antibody Production and Cytotoxic T Cell Responses. J Immunol. 2008;180:683–687. doi: 10.4049/jimmunol.180.2.683. [DOI] [PubMed] [Google Scholar]
- 53.Murakami Y, Fukui R, Motoi Y, Kanno A, Shibata T, Tanimura N, Saitoh S, Miyake K. Roles of the cleaved N-terminal TLR3 fragment and cell surface TLR3 in double-stranded RNA sensing. J Immunol. 2014;193:5208–5217. doi: 10.4049/jimmunol.1400386. [DOI] [PubMed] [Google Scholar]
- 54.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe A, Tatematsu M, Saeki K, Shibata S, Shime H, Yoshimura A, Obuse C, Seya T, Matsumoto M. Raftlin Is Involved in the Nucleocapture Complex to Induce Poly(I:C)-mediated TLR3 Activation. J Biol Chem. 2011;286:10702–10711. doi: 10.1074/jbc.M110.185793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Cattaneo A, Gobert F, Müller M, Toscano F, Flores M, Lescure A, Del Nery E, Benaroch P. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc Natl Acad Sci USA. 2012;109:9053–9058. doi: 10.1073/pnas.1115091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- 61.Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–160. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- 62.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab Conversion as a Mechanism of Progression from Early to Late Endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 63.Meresse S, Gorvel JP, Chavrier P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J Cell Sci. 1995;108:3349–3358. doi: 10.1242/jcs.108.11.3349. [DOI] [PubMed] [Google Scholar]
- 64.Vonderheit A, Helenius A. Rab7 Associates with Early Endosomes to Mediate Sorting and Transport of Semliki Forest Virus to Late Endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Del Conte-Zerial P, Brusch L, Rink JC, Collinet C, Kalaidzidis Y, Zerial M, Deutsch A. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol Syst Biol. 2008;4:206–206. doi: 10.1038/msb.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.