Abstract
The acid-sensing ion channels (ASICs) are a family of ion channels expressed throughout the mammalian nervous system. The principal activator of ASICs is extracellular protons, and ASICs have been demonstrated to play a significant role in many physiologic and pathophysiologic processes, including synaptic transmission, nociception, and fear. However, not all ASICs are proton-sensitive: ASIC2a is activated by acid, whereas its splice variant ASIC2b is not. We made a series of chimeric ASIC2 proteins, and using whole-cell electrophysiology we have identified the minimal region of the ASIC2a extracellular domain that is required for ASIC2 proton activation: the first 87 amino acids after transmembrane domain 1. We next examined the function of different domains within the ASIC2b N-terminus and identified a region proximal to the first transmembrane domain that confers tachyphylaxis upon ASIC2a. We have thus identified domains of ASIC2 that are crucial to channel function and may be important for the function of other members of the ASIC family.
Introduction
The acid-sensing ion channel (ASIC) family of ion channels comprises six subunits that are encoded by four genes: ASIC1a/b, ASIC2a/b, ASIC3, and ASIC4, the ASIC1 and ASIC2 genes having splice variants a and b (Sherwood et al., 2012; Kellenberger and Schild, 2015). Recombinant expression of ASICs has demonstrated that they are activated by extracellular protons (Waldmann et al., 1997), and therefore, situations in which lowering of extracellular pH occurs are liable to activate ASICs. In humans, acid-evoked pain is largely mediated by ASICs (Ugawa et al., 2002; Jones et al., 2004), and it has recently been shown that the acidification occurring upon synaptic vesicle release is sufficient to drive ASIC-mediated excitatory postsynaptic potentials (Du et al., 2014). The ASIC crystal structure identified that functional ASICs form trimers (Jasti et al., 2007; Gonzales et al., 2009), a finding that has been corroborated by atomic force microscopy (Carnally et al., 2008). Moreover, functional analysis has demonstrated that ASICs can form both homo- and heterotrimeric complexes (Hesselager et al., 2004; Chen et al., 2006; Smith et al., 2007), a phenomenon that adds to the functional diversity of acid-mediated currents.
Although it is clear that ASICs are activated by acid and thereby contribute to numerous physiologic and pathologic conditions (Lingueglia, 2007; Sherwood et al., 2012; Wemmie et al., 2013; Kellenberger and Schild, 2015), it remains unclear how they are actually gated by acid. The crystal structure of chicken ASIC1a (cASIC1a) identified a region containing many acidic residues, which has been termed the acidic pocket and contains three carboxylate pairs (D238–D350, E239–D346, and E220–D408; cASIC1a numbering), which were suggested to be the primary sites for proton sensing (Jasti et al., 2007). However, subsequent mutagenesis has shown that mutation of these residues, while decreasing pH sensitivity, does not fully abolish the ability of protons to activate ASIC1a (Paukert et al., 2008; Li et al., 2009). Interestingly, ASIC2a contains all of the acidic pocket carboxylates apart from D350, which may in part explain its decreased proton sensitivity compared with ASIC1a (EC50 pH 4.53 compared with pH 6.27); however, ASIC2b, which is not activated by protons, also contains all carboxylates, with the exception of D350 (Smith et al., 2007). These results suggest that sites outside of the acidic pocket are important for ASIC proton sensing and channel gating, and several studies have identified further amino acids that are required for normal proton sensing by ASIC1a (Paukert et al., 2008; Li et al., 2009; Liechti et al., 2010; Della Vecchia et al., 2013). Moreover, in a study comparing the extracellular domains (EC domain) of the proton-sensitive ASIC2a and the proton-insensitive ASIC2b, we and others identified 5 amino acids in ASIC2a, which are absent in ASIC2b, and that when mutated in ASIC2a produced a proton-insensitive ion channel that trafficked normally to the plasma membrane (H72, D77, E78, H109, and H180) (Baron et al., 2001; Smith et al., 2007). Mutation of both D77 and E78 also modulates, but does not abolish, the proton sensitivity of ASIC1a (Paukert et al., 2008); the post-transmembrane domain 1 (TM1) location of these residues suggests that this region is critical in determining ASIC proton sensitivity.
The post-TM1 domain also includes the β1-β2 linker segment that connects the β1 and β2 strands, which is essential in conferring proton sensitivity to the proton-insensitive lamprey ASIC1 (Li et al., 2010b). This region also includes the nonprotonatable leucine residue L85, mutation of which has a significant impact upon ASIC proton sensitivity, and variation of the residue at position 85 explains some species variation in ASIC1 proton sensitivity (Li et al., 2010a). However, L85 is conserved in all ASICs apart from ASIC4, and thus, although this residue might contribute to the proton insensitivity of ASIC4, it cannot account for the proton insensitivity of ASIC2b.
Finally, it is also unnecessary for all three subunits of an ASIC trimer to be proton-sensitive in order for functional ASICs to form: ASIC2b forms heteromers with ASIC2a that shows decreased proton sensitivity compared with ASIC2a homomers (Lingueglia et al., 1997), and the proton-insensitive ASIC2aE78R mutant forms heteromers with ASIC1a that show decreased proton sensitivity compared with ASIC1a homomers (Smith et al., 2007).
In the current study, we sought to exploit the difference in proton sensitivity of the splice variants ASIC2a (proton-sensitive) and ASIC2b (proton-insensitive) by constructing chimeric ion channels that would enable the identification of a minimum region necessary for ASIC2a activation and a full examination of ASIC2 N-terminus function.
Materials and Methods
Chinese Hamster Ovary Cell Culture.
Chinese hamster ovary (CHO) cells (Sigma-Aldrich, St. Louis, MO) were grown using standard procedures, as previously described (Smith et al., 2011; Brand et al., 2012).
Transfection of CHO Cells.
Twenty-four hours before transfecting, 35-mm dishes (Fisher Scientific, Waltham, MA) were coated with 100 µg/ml poly-l-lysine (Sigma-Aldrich); cells from a 70–80% confluent flask were trypsinized and resuspended in 5 ml CHO medium; and a volume was taken to seed cells at a 1:10 dilution, 2 ml/dish. For transfections, an enhanced green fluorescent protein expression vector was used to enable identification of transfected cells, and, apart from where otherwise stated, DNA was transfected at a ratio of 10:1, ASICx:GFP using 0.9 µg ASICx DNA and 0.09 µg enhanced green fluorescent protein DNA; the transfection reagent Lipofectamine LTX (Life Technologies, Carlsbad, CA) was used according to the manufacturer’s protocol.
Chimera Construction.
Cloning was performed according to the FastCloning protocol (Li et al., 2011) using primers detailed in Table 1 with a Phusion polymerase chain reaction kit, according to the manufacturer’s protocol (Thermo Scientific, Waltham, MA), and chimeras were constructed from rat ASIC2a cDNA in a pCI expression plasmid and rat ASIC2b cDNA in a pIRES expression plasmid that we have previously characterized (Smith et al., 2007). The plasmid DNA was isolated using PureLink Plasmid Miniprep Kit (Life Technologies), and Sanger sequencing was used to verify sequences (Department of Biochemistry, University of Cambridge).
TABLE 1.
Primers used for chimera construction
List of primers used to design the chimeras analyzed in this study.
| Chimera Number | Forward Primer | Reverse Primer |
|---|---|---|
| 1 Insert | 5′-ACGCTCGGTGCACTGGCACAGGAAGGCACCCAA-3′ | 5′-TGCAGTCGACGGTACCATGCCCATCCGGATCTTCTGCTC-3′ |
| 1 Backbone | 5′-TGCACCGAGCGTGTGCAGTACTA-3′ | 5′-GGTACCGTCGACTGCAGAATTCG-3′ |
| 2 Insert | 5′-TTGAGATTGCACAGGGTGACTG CTGGGAAAGTGAGCT-3′ | 5′-TGCAGTCGACGGTACCATGCCCATCCGGATCTTCTGCTC-3′ |
| 2 Backbone | 5′-CTGTGCAATCTCAATGAGTTCCGCTT-3′ | 5′-GGTACCGTCGACTGCAGAATTCG-3′ |
| 3 Insert | 5′-GAGCTGCTGGCCCGGCCAGGTACAGCAAGTCAGGGTA-3′ | 5′-TGCAGTCGACGGTACCATGCCCATCCGGATCTTCTGCTC-3′ |
| 3 Backbone | 5′-GAGCTGCTGGCCCTGCTCAACAA-3′ | 5′-GGTACCGTCGACTGCAGAATTCG-3′ |
| 4 Insert | 5′-CAATATCTCCAGCTGTCTTGCTCCACCTGGGCCAGAG-3′ | 5′-TGCAGTCGACGGTACCATGCCCATCCGGATCTTCTGCTC-3′ |
| 4 Backbone | 5′-CAGCTGGAGATATTGCAGGACAA-3′ | 5′-GGTACCGTCGACTGCAGAATTCG-3′ |
| 5 Insert | 5′-ATAGAATTCACGCATAATCGCTCCTGCCACCGACTG-3′ | 5′-TGCAGTCGACGGTACCATGCCCATCCGGATCTTCTGCTC-3′ |
| 5 Backbone | 5′-ATGCGTGAATTCTATGACCGTGCA-3′ | 5′-GGTACCGTCGACTGCAGAATTCG-3′ |
| 5-N Insert | 5′-TGCGGCCGCCACCATGATCCGGCGCTGCAGGGGCCA-3′ | 5′-AAGGCCACTGCCCAAAGCGCCCGTCGCTGGAAAGAGCCT-3′ |
| 5-N Backbone | 5′-CATGGTGGCGGCCGCAGATATCGAT-3′ | 5′-CTTTGGGCAGTGGCCTTCGTCGGAT-3′ |
| 6 Insert | 5′-TATTTTGTAAACACTGTGGTGAAGTCTTGATGCCCACACT-3′ | 5′-CTGCAGGGGCCAATGGACCTCAAGGAGAGCCCCAGT-3′ |
| 6 Backbone | 5′-GTGTTTACAAAATACGGGAAGTGTTACAT-3′ | 5′-TGGCCCCTGCAGCGCCGGAT-3′ |
| 7 Insert | 5′-AGGGACGTGCAGAAGGCGATGGTCAGCGGCCCATACACGAA-3′ | 5′-GGGGAACGGCAGCACCTTGGTAACATGCTGATAAGA-3′ |
| 7 Backbone | 5′-GCCTTCTGCACGTCCCTCGGCTT-3′ | 5′-GGACCAGGACAGCAGCAAGCCGA-3′ |
| 8 Insert | 5′-TCGGCTTGCTGCTGTCCTGGTCCTCGGAGAGGGTGTCCTACTAT-3′ | 5′-ACAGCAGCAAGCCGAGGGACGTGCAGAAGGCCAGCACCCA-3′ |
| 8 Backbone | 5′-CGCCGCTCCGGTCTCCTCCGG-3′ | 5′-CTGCCGTTCCCCGCCGTCA-3′ |
| 9 Insert | 5′-TGCGGAACCACTGGCGGTCTGCCAGATGCGGGTCGGGAA-3′ | 5′-TATCCTCCAGCTGGTGGCCTACCCGGTGCAGGAACTCCA-3′ |
| 9 Backbone | 5′-CGCCAGTGGTTCCGCAAACT-3′ | 5′-CACCAGCTGGAGGATATGCTGCT-3′ |
| 10 | 5′-TCTCTTATCAGCATGTTACCAAGGTGGATGAAGTGGTGCGCCAGCTGCCGTTCCCCG-3′ | 5′-ATGCTGATAAGAGAAATAGTAGGACACCCTCTCCGAGCTCTCCACCAGCAGCAGGCCCA-3′ |
| 10-1 | 5′-CTGGGAAGACCAGGCTCTGGGCCACCACTTCATCCACCTTGGTAA-3′ | 5′-CCTGGTCTTCCCAGCTGTGACCTGCAACAACAACCCCCTGCGCTT-3′ |
| 10-2 | 5′-GATGCGGGTCGGGAATCTGTAGGCGGTTGGGAAGCAGCAGCCCTA-3′ | 5′-TCCCGACCCGCATCTGGCAGACCCCACGGTGCTGGAGGCC-3′ |
| 11 | 5′-TCGAGGGCATCAGCGCTGCCTTCATGGACCGTTTGGGCCACCAGCTGGAGGATATGCTGCT-3′ | 5′-AGCGCTGATGCCCTCGAAGTGGCGCGGCGGCAGGAAGAGGCGCTTCTGTCGGAGGGCCTCCAGCA-3′ |
| 11-1 | 5′-ATTGAGGTTGCAGAGGGTCACAGCTGGGAAGACCAGGCT-3′ | 5′-CTCTGCAACCTCAATCTGCGCTTCCCGCGCCTCTCC-3′ |
| 11-2 | 5′-GGAAGCCATTGAGGTTGCAGAGGGTCACAGCTGGGAAGACCAGGCTCT-3′ | 5′-CCTCAATGGCTTCCGGTTCTCCCTCTCCAAGGGGGACCTCTACTA-3′ |
| 11-3 | 5′-CCAGCAGGGCCAGCAACTCCCCGCCCGCGTAGTAGAGGTCCCCCTT-3′ | 5′-GCTGGCCCTGCTGGATGTCAACCTACAGATTCCCGACCCGCATCT-3′ |
| 11-4 | 5′-TAGTAGAGGTCCCCCTTGGAGAGGGAGAACCGGAAGCCATTGAGGTT-3′ | 5′-AGGGGGACCTCTACTACGCGGGCGGGGAGTTGCTGGCCCTGCT-3′ |
| 11-5 | 5′-AAGGGGGACGACTTGTACCACGCTGGGGAGTTGCTGGCCCTGCTGGAT-3′ | 5′-TGGTACAAGTCGTCCCCCTTGGAGAGGGAGAACCGGAAGCCATTGAGGTT-3′ |
| 11-6 | 5′-ACCACCAACCTCTACTACGCGGGCGGGGAGTTGCTGGCCCTGCTGGAT-3′ | 5′-TAGTAGAGGTTGGTGGTAAGCCTGGAGAACCGGAAGCCATTGAGGTT-3′ |
Sequence Alignment.
Sequences of rat ASIC2a and 2b were obtained by sequencing of cDNA clones. Rat ASIC1a (ENSRNOT00000025476), ASIC1b (ENSRNOT00000047887), ASIC3 (ENSRNOT00000011300), and ASIC4 (ENSRNOT00000027135) sequences were downloaded from Ensembl genome browser release 76. Alignment was made using MAFFT version 7 with default settings and unalignment factor 0.8 and visualized in Jalview 2.8. Colors indicate percentage identity.
Whole-Cell Electrophysiology.
Whole-cell patch-clamp recordings from CHO cells were performed at room temperature 24 hours after transfection, unless otherwise stated. The intracellular solution contained (in mM): 110 KCl, 10 NaCl, 1 MgCl2, 1 EGTA, 10 HEPES, 2 Na2ATP, and 0.5 Na2GTP in MilliQ water; pH was adjusted to pH 7.3 by adding KOH, and the osmolarity was adjusted to 310–315 mOsm with sucrose. The extracellular solution contained (in mM): 140 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES (solutions >pH 6) or MES (solutions <pH 6), and 4 glucose in MilliQ water; osmolarity was adjusted to 300–310 mOsm with sucrose, and pH was adjusted as required using NaOH and HCl. Patch pipettes were pulled (Model P-97, Flaming/Brown puller; Sutter Instruments, Novato, CA) from borosilicate glass capillaries (Hilgenberg GmbH, Malsfeld, Germany) and had a resistance of 3–6 MΩ. Data were acquired using an EPC10 amplifier (HEKA, Bellmore, NY) and Patchmaster software (HEKA). Whole-cell currents were recorded at 20 kHz, pipette and membrane capacitance were compensated using Patchmaster macros, and series resistance was compensated by >60%. For measurement of current amplitude and inactivation time constant, the following protocol was used: 5 seconds, pH 7.4; 5-second test, pH; and 5 seconds, pH 7.4, the holding potential of −60 mV. For tachyphlaxis experiments, the test pH was applied five times with a 1-minute wash period between stimuli. To measure the current-voltage relationship, recordings were made at different holding potentials (−60 to +60 mV in 30-mV steps) with stimuli being applied at 1-minute intervals. For pH-response curves, 2.5-second stimuli between pH 3 and pH 6, at 0.5 pH intervals, were administered in random order with 30 seconds at pH 7.4 in between stimuli. To measure steady-state inactivation (SSI), the response to pH 4 was measured as usual and then the pH of the bath solution was lowered stepwise in 0.3 pH steps from pH 6.5 to pH 5, each step being 60-second duration, before applying a 2.5-second pH 4 stimulus.
Biotinylation.
Biotinylation assays were performed, as conducted previously (Balasuriya et al., 2012). Briefly, transfected CHO cells were incubated with biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinimide ester sodium salt (0.2 mg/ml) for 60 minutes at 4°C. Biotin was then quenched with 10 mM Tris-HCl and solubilized in 1% Triton solution. To remove any insoluble material, the solubilized mix was centrifuged at 61,740g for 60 minutes at 4°C. The supernatant containing the biotinylated proteins was isolated using streptavidin beads, and ASIC2 proteins were detected with an anti-ACCN1 antibody (1:100; Abcam, Cambridge, MA).
Data Analysis.
Electrophysiological data were analyzed and plotted using Fitmaster software (HEKA) and Prism (GraphPad Software, La Jolla, CA). Peak current density was determined by subtracting baseline current (average current amplitude in the 5- to 10-second preceding stimulation) from the peak current amplitude using Fitmaster software (HEKA) and then dividing by cell capacitance (pA/pF). The inactivation time constant τ was measured using a built-in function of Fitmaster. Peak current density data distribution was skewed, and therefore every datapoint x was transformed using yi = log10(xi), as advised for log-normally distributed data (http://www.graphpad.com/guides/prism/6/statistics/stat_the_lognormal_distribution.htm). Results are expressed as the S.E.M., unless otherwise stated; this might, however, not necessarily represent the statistical differences for peak current density data, which were transformed as above. Statistical analysis was performed in GraphPad Prism using ordinary one-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons, comparing data from each chimera with ASIC2a for transformed peak current density data and inactivation data. For concentration-response and SSI curves, a nonlinear regression curve was fitted and differences were analyzed using a sum-of-squares F test; data points were normalized to the largest response in each cell, and curves were constrained between 0 and 1 because of this normalization. Tachyphylaxis data were fitted with an exponential curve, and the slope was compared using a sum-of-squares F test. Current-voltage curves were fitted with a linear regression line and analyzed using a built-in comparison function of Prism equivalent to analysis of covariance. Figures were made using Prism, Adobe CS6 Photoshop, and Illustrator.
Results
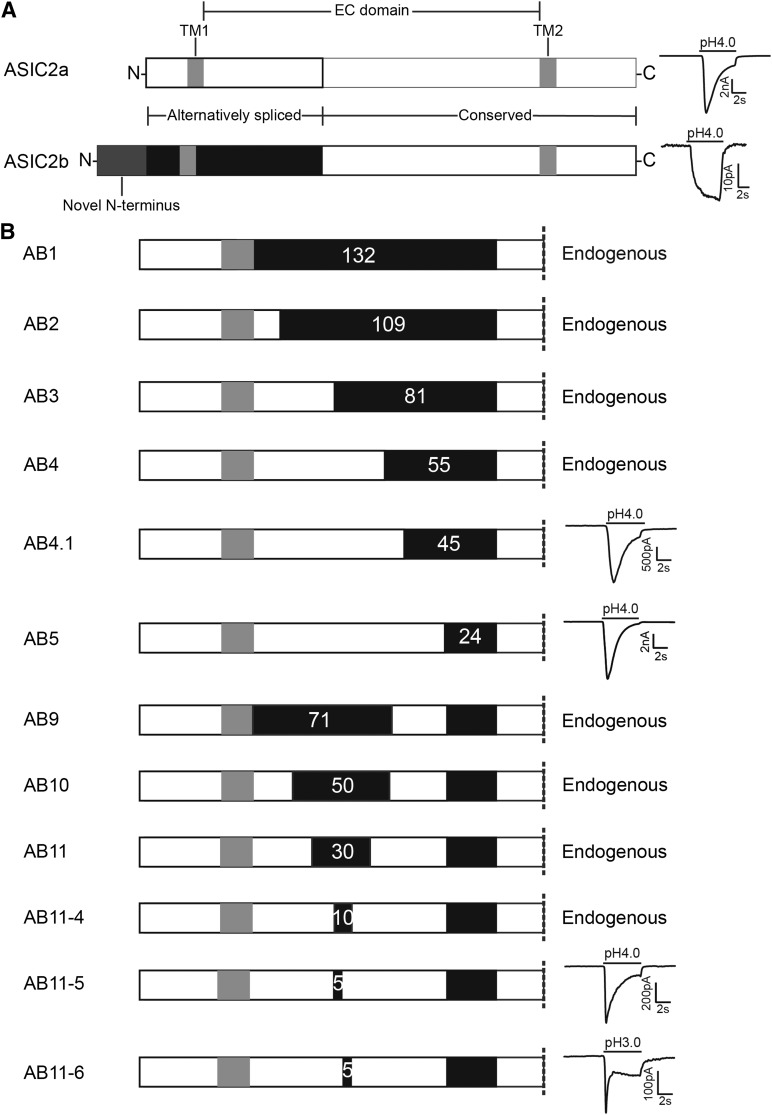
ASIC2a and ASIC2b are splice variants of the same gene, which differ only in the first exon, but whereas ASIC2a is activated by low pH, ASIC2b is not (Fig. 1A). As characterized previously, the small, slowly activating, and sustained response evoked in cells transfected with ASIC2b is not significantly different from the endogenous CHO cell response to acidic solutions (Smith et al., 2007). ASIC2a is a 512-amino-acid protein, and the first 185 amino acids are different in ASIC2b, which also has an added 51 amino acids at the N-terminus. Because extracellular protons are necessary for ASIC activation, we sought to determine the minimum sequence of the ASIC2a EC domain that is necessary to render ASIC2b proton-sensitive. We did this by making a series of chimeras in which increasing sections of ASIC2a were inserted into the extracellular domain of ASIC2b.
Fig. 1.
Identification of the minimum extracellular sequence required for ASIC2 acid sensitivity. (A) The ASIC2 protein has two splice variants, ASIC2a and ASIC2b, and the alternatively spliced region consists of one third of the EC domain, TM1, and the intracellular domain with ASIC2b having an extended, novel N-terminus region; ASIC2a is activated by a pH 4 stimulus, whereas ASIC2b is not. (B) Chimeras were constructed in which evermore of the ASIC2b EC domain (black) was replaced with the equivalent ASIC2a sequence (white), and pH sensitivity was tested. Chimeras AB1–AB4 produced a response that was indistinguishable from the endogenous CHO cell current, like that observed in ASIC2b-transfected cells. Chimeras with 45 (AB4.1) and 24 (AB5) amino acids of ASIC2b both responded to pH 4; see text for details. AB9 containing the 31 amino acids that distinguish AB4 (pH insensitive) from AB5 (pH sensitive) was pH insensitive, as were AB10 and AB11 that included evermore ASIC2a sequence. Chimeras AB11-5 and AB11-6 contained the 24 ASIC2b amino acids of AB5 and an additional 5 amino acids (102–106) and (107–111) of ASIC2b, and both responded to a pH 4 stimulus.
The first chimera (AB1) consisted of the EC domain of ASIC2b and both the intracellular N-terminus and first TM of ASIC2a (Fig. 1B), and, unsurprisingly, considering that ASICs are activated by extracellular protons and that ASIC2b is proton-insensitive, this construct did not produce ASIC-like responses when transfected into CHO cells (data not shown for this or subsequent proton-insensitive chimeras). AB2 contained the first 16 amino acids of the ASIC2a EC domain, which does not include either D77 or E78, mutation of which has previously been shown to inactivate ASIC2a (Smith et al., 2007), but does include E62, which is required for normal pH sensitivity of ASIC1a (Paukert et al., 2008), and H72, which is necessary for normal acid sensing by both ASIC1a and ASIC2a (Baron et al., 2001; Smith et al., 2007; Paukert et al., 2008); however, this chimera also failed to respond to stimulation by a pH 4 solution. AB3 contained 44 amino acids of ASIC2b with the ASIC2a sequence, thus including both D77 and E78, but also failed to respond, as did AB4, which contained 70 amino acids of ASIC2a (Fig. 1B). However, AB5, which contained 101 amino acids of ASIC2a, produced an ASIC-like transient response to a pH 4 stimulus, thus demonstrating that the remaining 24 amino acids in the ASIC2a EC domain are not necessary for proton sensitivity.
The proton insensitivity of AB4 could be due to a functional loss of the ability of protons to bind to and activate the channel, or due to an inability of the protein to traffic to the cell membrane. We therefore performed a biotinylation assay and observed that, like the proton-sensitive ASIC2a and the proton-insensitive ASIC2b, AB4 trafficked to the cell membrane (Fig. 2, A and B). We thus conclude that the lack of response of AB4 to protons is a result of an inability of protons to gate the chimeric ion channel, rather than it being unable to be processed and trafficked normally within the cell.
Fig. 2.
Characterization of functional EC domain chimeras. (A) Protein isolation using cell surface biotinylation in intact cells and analysis by SDS-PAGE, immunoblots of protein prior to isolation of biotinylated proteins, and (B) purified biotinylated proteins using streptavidin agarose beads. (C) Currents evoked by a pH 4 stimulus were of significantly larger magnitude in AB5-transfected cells than those transfected by ASIC2a; both AB4.1 and AB11-5 produced significantly smaller currents than ASIC2a. (D) The inactivation time constant was not significantly different between ASIC2a, AB5, AB4.1, and AB11-5. (E and F) pH-response curves were evoked by applying pH stimuli in a random order, as shown by the example ASIC2a trace; the EC50 for AB4.1 was significantly shifted to the right compared with ASIC2a. (G and H) SSI curves were evoked by stimulating with a pH 4 stimulus every minute, with the bath solution becoming ever more acidic between stimuli, as indicated in the example ASIC2a trace; the AB5 SSI curve was significantly shifted to the left compared with ASIC2a. *P < 0.05, **P < 0.005, ****P < 0.0001.
We next characterized AB5 to determine whether it fully replicates the function of ASIC2a (summary data for these and all other experiments are given in Table 2). The peak current densities at pH 6 and 5 were not significantly different between ASIC2a and AB5, but at pH 4 the response of AB5 was greater than ASIC2a: 924 ± 156 pA/pF (n = 26) versus 421 ± 57 pA/pF (n = 50, P = 0.0129; ANOVA with Dunnett’s multiple comparisons test) (Fig. 2C). Because the distribution of peak current density data was highly skewed and followed a log-normal distribution, we transformed the data (see above) to test significance. By contrast, there was no significant difference in the inactivation time constant between AB5 (1471 ± 150 milliseconds) and ASIC2a (1275 ± 111 milliseconds, P = 0.383, ANOVA with Dunnett’s multiple comparisons test) (Fig. 2D). We next measured currents activated by pH 4 at different holding potentials and calculated the reversal potential as 31.4 mV (n = 9) for ASIC2a and 33.5 mV (n = 7) for AB5, which is similar to values reported by others for ASIC2a using similar solutions (Lingueglia et al., 1997) and demonstrates that the ion selectivity of AB5 is not significantly different from that of ASIC2a (P = 0.342, analysis of covariance). We next calculated the EC50 from concentration-response curves and identified the proton sensitivity of ASIC2a and AB5 to be virtually identical (pH 4.44 and pH 4.48, respectively, P = 0.599, F test), which further suggests that AB5 fully recapitulates the function of ASIC2a (Fig. 2, E and F). However, when we examined SSI, the inactivation curve for AB5 was slightly, yet significantly shifted to the left, suggesting that some element in the remaining 24 amino acids of ASIC2a might play a role in determining SSI (ASIC2a IC50, pH 6.13 versus AB5 IC50, 6.34, P = 0.034, F test; Fig. 2, G and H).
TABLE 2.
Summary data
Summary electrophysiological data for ion channels analyzed in this study. Brackets in numbers represent the number of cells tested for a particular measurement. Statistically significant differences are indicated in comparison with ASIC2a.
| Chimera | Mean Peak Current Density + S.E.M. at pH 4 | Mean τ + S.E.M. at pH 4 | EC50 | IC50 | Hill Coefficient EC/IC | Reversal + S.E.M. |
|---|---|---|---|---|---|---|
| pA/pF (n) | ms (n) | pH (n) | mV (n) | |||
| ASIC2a | 421 ± 57 (50) | 1275 ± 111 (17) | 4.44 (19) | 6.13 (9) | 0.88/1.46 | 31.4 (9) |
| AB5 | 924 ± 156 (26)* | 1471 ± 150 (12) | 4.48 (14) | 6.34 (7)* | 0.83/1.73 | 33.5 (7) |
| AB4.1 | 43 ± 14 (11)**** | 1336 ± 141 (9) | 4.13 (6)** | — | 0.84 | — |
| AB11-5 | 23 ± 7 (8)**** | 1150 ± 89 (5) | — | — | — | — |
| AB6 | 560 ± 160 (20) | 1637 ± 150 (9)* | 4.1 (13)**** | 6.44 (6)**** | 0.8/1.86 | 39.2 (6) |
| AB5-N | 499 ± 203 (15)* | 561 ± 74 (8)** | — | — | — | — |
—, data not collected.
P < 0.05; ** P < 0.005; **** P < 0.0001.
AB5 contains 31 extra amino acids compared with AB4, and we therefore made a set of chimeras to determine which regions, in addition to the crucial 31 amino acids, are necessary for ASIC2a function. We predicted that a chimera containing the 31 amino acids that are novel to AB5 would alone be insufficient to make the EC domain of ASIC2b proton-sensitive because it lacks amino acids that in the post-TM1 domain that we and others have been shown to be critical for proton sensitivity (Baron et al., 2001; Smith et al., 2007; Paukert et al., 2008), and indeed AB9 was nonfunctional (Fig. 1A). The next chimera used AB9 as a template with the addition of 20 amino acids of ASIC2a in the post-TM1 region (AB10), which contains the critical amino acids H72, D77, and E78, but this chimera also lacked proton sensitivity (Fig. 1A). We produced further chimeras, which narrowed in on the critical region by further addition of ASIC2 amino acids from both the post-TM1 region and the 31 amino acids of AB5. Surprisingly, only when we created chimeras with 5 amino acids left of ASIC2b between ASIC2a regions were we able to record proton-gated currents, either amino acids L102–D106, or amino acids L107–G111, AB11-5, and AB11-6, respectively. AB11-4, in which all 10 amino acids between L102 and G111 were from ASIC2b, was also insensitive to protons (Fig. 1A).
Both 11-5 and 11-6 produced characteristic ASIC-like currents at pH 4, but they were significantly smaller than ASIC2a (23 ± 7 pA/pF, n = 8 and 19 ± 12 pA/pF, n = 3, P < 0.001 and 0.05, respectively, compared with ASIC2a, ANOVA with Dunnett’s multiple comparisons test; Figs. 1A and 2C). Because of the small current amplitude at pH ≥ 4, it was not possible to generate a pH-response or SSI curve. We tried to circumvent this problem by increasing the amount of DNA transfected from 0.9 μg to 1.8 μg and waiting for 48 hours after transfection, but neither modification resulted in any measureable difference. We also hypothesized that the small response at pH 4 represented a large shift in the pH-response relationship; however, at pH lower than pH 3, the cells failed to survive stimulation, and we were thus unable to identify if the small response at pH 4 represents a significant shift in the pH-response relationship. The inactivation time constant for AB11-5 was not significantly different from ASIC2a 1150 ± 89 milliseconds (n = 5, P = 0.994; Fig. 2D), but we were unable to calculate inactivation time constants for AB11-6 due to the small nature of the response, which was often heavily mixed with the endogenous current.
We next sought to determine whether a chimera containing more ASIC2b amino acids than AB5 could be proton-sensitive. AB4.1 contains an extra 10 amino acids of ASIC2a, compared with the proton-insensitive AB4, and thus has a total of 45 amino acids from ASIC2b in the EC domain and responds to protons (Fig. 1A). However, the peak current density of AB4.1 at pH 4 (43 ± 14 pA/pF, n = 11) was significantly smaller compared with ASIC2a (P < 0.0001; Fig. 2C), although the inactivation time constant was not significantly different from ASIC2a (1336 ± 141 ms, n = 9, P = 0.959; Fig. 2D). Unlike AB11-5 and AB11-6, AB4.1 currents were consistently larger than the endogenous current across the pH range used, which enabled us to construct a pH-response curve. In contrast to AB5, which was almost identical to ASIC2a, the pH-response curve for AB4.1 is significantly shifted to the right compared with ASIC2a, EC50 = 4.13 (n = 6, P < 0.01, F test; Fig. 2F). This suggests that, although proton-sensitive, AB4.1, unlike AB5, does not display ASIC2a-like proton sensitivity. Because AB4.1 currents were very small, we were unable to calculate SSI because the currents measured were negligible after the first pH step; increasing the amount of DNA transfected and increasing the time post-transfection for making recordings did not produce any significant increase in current amplitude.
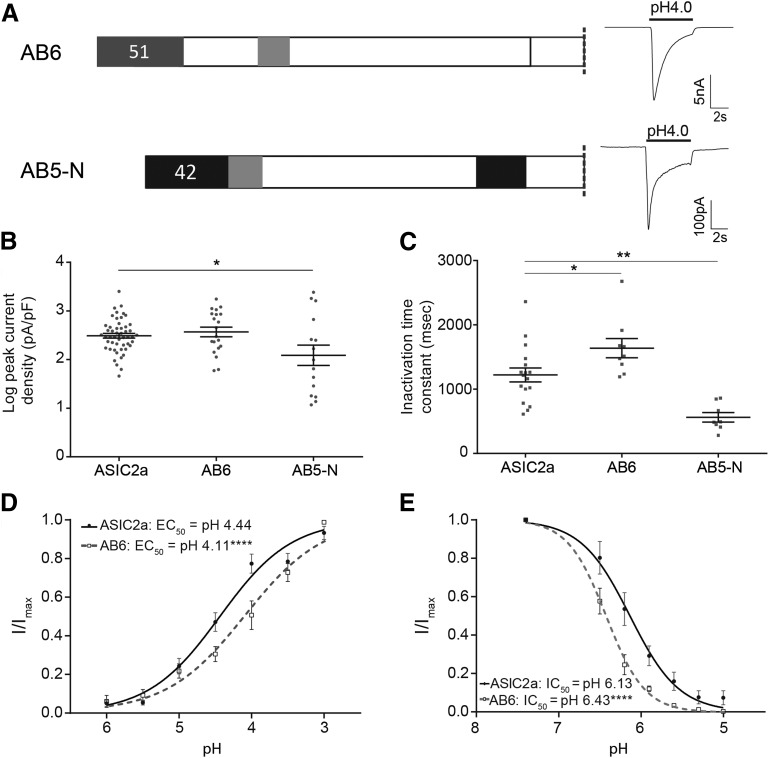
We next constructed chimeras that enabled us to examine the function of the ASIC2b N-terminus, part of which has an equivalent sequence in ASIC2a (44 amino acids), but which also contains an extended region that has no equivalent structure in ASIC2a (51 amino acids). AB6 contains the novel 51 amino acids of ASIC2b attached to the N-terminus of ASIC2a, and AB5-N is a chimera in which the homologous regions have been swapped (Fig. 3A). Initial recordings showed that AB6 had similar characteristics to ASIC2a, transient inward currents were evoked by low pH (Fig. 3A), and peak current density was not significantly different from ASIC2a (at pH 4, 560 ± 160 pA/pF, n = 20, P = 0.966; Fig. 3B). The inactivation time constant was, however, significantly longer than ASIC2a (1637 ± 150 milliseconds, n = 9, P = 0.036; Fig. 3C); the reversal potential was also similar to that of ASIC2a, 39.2 mV (n = 6, P = 0.343). However, the pH-response and SSI curves indicate that this channel is significantly less proton-sensitive compared with ASIC2a: the pH-response curve was shifted to the right (EC50, pH 4.1, P < 0.001, F test; Fig. 3D), and the SSI curve was shifted to the left (IC50, pH 6.44, P < 0.001, F test; Fig. 3E). These data illustrate that AB6 is activated less efficiently than ASIC2a, needing more protons for activation, but that SSI occurs at higher pH, meaning that fewer protons are required to inactivate the channel.
Fig. 3.
Characterization of functional chimeras with changes in the N-terminus. (A) AB6 consisted of ASIC2a with the addition of the 51-amino-acid–long novel N-terminus of ASIC2b, and AB5-N was constructed from AB5 by exchanging the 42 amino acids prior to the first TM domain. Both chimeras responded to pH 4 with ASIC-like currents. (B) AB5-N produced significantly smaller currents than ASIC2a at pH 4, whereas currents of AB6 did not differ from ASIC2a. (C) Inactivation time constant was significantly slower in AB6, but faster for AB5-N compared with ASIC2a. (D) pH-response curve of AB6 was significantly shifted to the right compared with ASIC2a. (E) SSI curve of AB6 was significantly shifted to the left compared with ASIC2a. *P < 0.05, **P < 0.005, ****P < 0.0001.
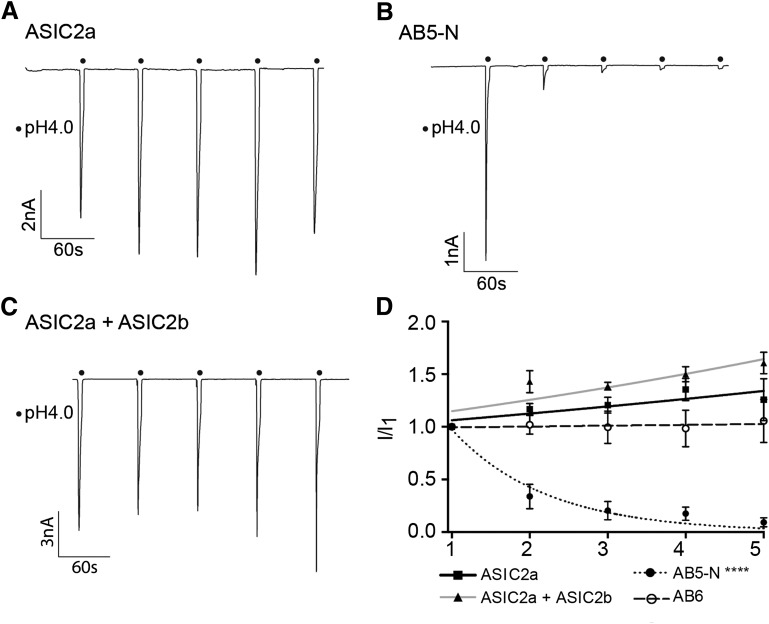
AB5-N also produced ASIC-like currents; however, the peak current density was significantly smaller than that of ASIC2a (499 ± 203 pA/pF, n = 15, P = 0.016, ANOVA with Dunnett’s multiple comparison test; Fig. 3, A and B), and the inactivation time constant was significantly faster (561 ± 74 milliseconds, n = 8, P < 0.01, one-way ANOVA and Dunnett’s post hoc test; Fig. 3C). Moreover, it was immediately apparent that AB5-N underwent strong tachyphylaxis in response to multiple stimulations, which made it thus impossible to obtain pH-response or SSI curves using our standard protocol. Among ASICs, tachyphylaxis only occurs in ASIC1a (Chen and Grunder, 2007), and therefore, we examined further the function of AB5-N. Repetitive stimulation of ASIC2a for 5 seconds every minute produces currents with approximately the same amplitude each time (Fig. 4A), whereas repetitive stimulation of AB5-N produces rapid tachyphylaxis (Fig. 4B). The data could be fitted with a single exponential function, and the slope of AB5-N (n = 7) was significantly different from that of ASIC2a (n = 13), respectively (P < 0.001, F test; Fig. 4D). To determine whether any alteration to the ASIC2a N-terminus could induce tachyphylaxis, we tested AB6 using the same protocol, but, like ASIC2a, no tachyphlaxis was observed (n = 7, P = 0.334; Fig. 4D).
Fig. 4.
Tachyphylaxis in N-terminal chimeras. (A) Representative current trace for ASIC2a elicited by five stimulations with pH 4 (tachyphylaxis protocol). (B) Representative current elicited by tachyphylaxis protocol in AB5-N. (C) Representative current elicited by tachyphylaxis protocol in ASIC2a-ASIC2b heteromers. (D) Graph comparing tachyphylaxis in ASIC2a, AB6, AB5-N, and ASIC2a + ASIC2b heteromers, currents normalized to the first current. AB5-N currents, but none of the others, were significantly different from ASIC2a. ****P < 0.0001.
Because ASIC2b is not activated by protons, but can modulate the function of other ASICs, we hypothesized that one function of ASIC2b might be to confer tachyphylaxis onto ASIC2b-containing heteromers through the N-terminus domain that we have identified in chimera AB5-N. We therefore cotransfected ASIC2a and ASIC2b at various ratios and measured the responses to repetitive stimulation. However, even at a ratio of 3:1, we did not observe any tachyphylaxis (n = 4, P = 0.494, F test; Fig. 4, C and D), which suggests that other domains of ASIC2b counteract any tachyphylaxis-inducing effect that the AB5-N portion of ASIC2b may have.
Discussion
In this study, we sought to identify the minimum ASIC2a sequence that when exchanged for the equivalent sequence of ASIC2b would produce a proton-sensitive ion channel and thereby identify regions of the ASIC EC domain that play important roles in proton sensing. Chimera AB5 lacks only the last 24 amino acids of the alternatively spliced region and displays properties that are almost identical to ASIC2a. Comparison of ASIC2 sequences in this area shows that there is a high sequence similarity in the part that was exchanged in AB5 (Fig. 5A). The 24 amino acids that were exchanged in AB5 contain four protonatable residues that are not shared with ASIC2b, D163, E177, H180, and D182 (Fig. 5A), which thus appear to be unnecessary for proton sensing. Of those four, only D163 and D182 are conserved between ASIC2a and ASIC1a even though the overall sequence similarity is very high between the two subunits, and only D163 is conserved between all six subunits. The other residues are not conserved, which might be a further indication that protonation of these residues does not play a substantial role in channel opening. Interestingly, we previously reported that point mutation of H180 to alanine abolishes acid sensing in ASIC2a (Smith et al., 2007), but AB5, which lacks H180, is proton-sensitive. This could be explained by the fact that the ASIC2b sequence has a histidine in position H181, which would be available in AB5, but missing altogether in mutant H180A. Therefore, a histidine in this area is important for acid sensing by ASIC2a, but is not alone sufficient to confer proton activation of an ion channel and does not contribute to the proton insensitivity of ASIC2b. In the same study, mutations of D163, E177, and D182, in agreement with our current results, do not affect proton sensing and react like ASIC2a (Smith et al., 2007).
Fig. 5.
Sequence and structure of chimeras. (A) Sequence comparison between ASIC2a, ASIC2b, ASIC1a, ASIC1b, ASIC3, and ASIC4 of the region exchanged in AB5; greyness indicates percentage identity. (B) Sequence comparison between ASIC2a, ASIC2b, AB11-4, AB11-5, AB11-6, and ASIC1a; greyness indicates percentage identity. (C) Overview of ASIC2 structure indicating regions swapped in chimera experiments.
Unlike AB5, AB4.1, although being proton-sensitive, produced dramatically smaller pH 4 gated currents compared with ASIC2a and demonstrated a large shift in its proton sensitivity. AB4.1 contains an additional 21 amino acids of ASIC2b compared with AB5, and lacks the following potentially protonatable sites that are expressed by the equivalent ASIC2a sequence: H145, E155, and H158. In the domain exchanged in AB4.1, there is little sequence conservation between ASIC subunits, with the exception of residues shared between ASIC2a and ASIC1a. Residues H145 and E155 are conserved between ASIC2a and ASIC1a, but not with other ASIC subunits. Residue 158 is highly variable among the mammalian ASICs, and thus, lack of this histidine is unlikely to account for ASIC2b insensitivity, and we have previously shown that mutation H158 in ASIC2a (as well as H145 and E155) did not have any significant impact upon ASIC2a proton sensitivity (Smith et al., 2007). Within the 21 ASIC2b amino acids that differentiate AB4.1 from AB5 is a double-proline motif P145–P146, which might influence the structure of the protein and therefore change its properties; recent evidence from a study using voltage-clamp fluorometry shows that many conformational changes occur during ASIC gating (Bonifacio et al., 2014), and thus, residues that limit or alter how these changes can occur would be expected to affect channel gating. In the proton-insensitive AB4, an additional 10 amino acids were exchanged. These are highly divergent and contain only one protonatable site, E135, which has previously been mutated without effect upon proton sensitivity (Smith et al., 2007).
As expected from previous studies, AB9, which contains several amino acids that are critical for ASIC proton sensitivity, did not respond to acid. In constructing chimeras that built on AB9 and the critical amino acids identified from comparing AB4 (proton-insensitive, but normal membrane trafficking) and AB5 (proton-sensitive), we identified that amino acids 102–111 also play a role in proton sensitivity. AB11-4, which contains L102–G111 from ASIC2b, did not respond to protons, but AB11-5 and AB11-6, which contained L102–D106 and L107–G111 from ASIC2b, respectively, both responded to protons, albeit not as robustly as ASIC2a or AB5. A sequence alignment of this region illustrates that the changes in activation are most likely due to a combination of several amino acids (Fig. 5B). Residues 104 and 105 are important in determining the difference in proton sensitivity between ASIC1a and ASIC1b (Babini et al., 2002), and therefore, differences in this region might contribute to the lack of proton sensitivity in ASIC2b. It is unlikely, however, that S103, K104, and G105 influence the activation of ASIC2b negatively, because S103 and K104 can be found in ASIC1a, which responds strongly to protons, and all three are present in AB11-5, which could be activated by protons. The 107–111 region only contains one residue difference: 109 is a histidine in ASIC2a, but a tyrosine in ASIC2b. Mutation of H109 inactivates ASIC2a (Smith et al., 2007), but AB11-6, which lacks H109, is proton-sensitive. An explanation for this difference in the role of H109 might be that in the current study we observed very small currents in cells expressing AB11-6 and used a more efficient transfection method (Lipofectamine LTX) compared with our previous study (calcium phosphate) in which data from only four cells were measured, and thus, our previous conclusion on the role of H109 may have been overstated. We can conclude from our data on AB11-6 that, although H109 seems to play an important role in proton sensitivity, it is not necessary for activation of the channel. The observation that most of the residues of the ASIC2a extracellular domain are needed for activation of the channel suggests that there are complex structural interactions that are disrupted in all chimeras up to chimera 11-5 and 11-6.
The ASIC2b N-terminus contains both an equivalent and novel domain compared with ASIC2a. Adding the novel domain to ASIC2a (AB6) produced a proton-sensitive ion channel, but one that was less sensitive to protons for activation compared with ASIC2a and also underwent SSI at more alkaline pH. Although extracellular protons activate ASICs, mutations at the N-terminus of ASIC2a have been previously shown to affect proton sensitivity (Coscoy et al., 1999). However, the most striking phenotype that we observed in this study was that swapping the equivalent N-terminus domains between ASIC2a and ASIC2b (AB5-N) resulted in an ion channel that demonstrated profound tachyphylaxis and a significantly faster time constant of inactivation. Previous work has demonstrated that, of the wild-type ASICs, only ASIC1a undergoes tachyphylaxis (Chen and Grunder, 2007), and we hypothesized that the tachyphylaxis we observed with AB5-N may have revealed a potential modulatory role for ASIC2b. However, coexpression of ASIC2b with ASIC2a did not produce ion channels displaying tachyphylaxis, and thus, our initial results rule out the possibility that ASIC2b negatively regulates other ASICs by inducing tachyphylaxis.
In summary, our chimeric study has identified that the last 24 amino acids of the ASIC2a-spliced region are not required for normal proton activation (AB5), and that a proton-sensitive ASIC2 ion channel could still be produced when the last 45 amino acids of ASIC2a were exchanged for ASIC2b, although this ion channel was less sensitive to proton activation (AB4.1; Fig. 5C). These results illustrate that large regions of the ASIC EC domain are necessary for protons to activate ASICs and that the carboxylates of the acidic pocket are insufficient (ASIC2a and ASIC2b contain the same number of acidic pocket carboxylates). We have also shown novel roles for the modulation of ASIC function by the intracellular N-terminus, which supports previous studies that have shown how intracellular domains are also involved in ASIC function (Bassler et al., 2001; Babini et al., 2002; Chen and Grunder, 2007; Salinas et al., 2009).
Acknowledgments
The authors thank Jonathan Raby, Leanne Young, Victoria Ball, Helena Sivaloganathan, Nicholas Gianaris, and Ines Serra for assistance in the laboratory, and Marie Brunet for assistance with the biotinylation assay.
Abbreviations
- ANOVA
analysis of variance
- ASIC
acid-sensing ion channel
- CHO
Chinese hamster ovary
- EC domain
extracellular domain
- SSI
steady-state inactivation
- TM
transmembrane domain
Authorship Contributions
Participated in research design: Schuhmacher, Srivats, Smith.
Conducted experiments: Schuhmacher, Srivats.
Performed data analysis: Schuhmacher, Srivats, Smith.
Wrote or contributed to the writing of the manuscript: Schuhmacher, Srivats, Smith.
Footnotes
L.-N.S. is a student in the Biotechnology and Biological Sciences Research Council Doctoral Training Programme [Grant BB/J014540/1] and a recipient of the David James Pharmacology Award. S.S. is supported by the Cambridge International and European Trust. E.St.J.S. was supported by the Isaac Newton Trust/Wellcome Trust Institutional Strategic Support Fund/University of Cambridge Joint Research Grants Scheme.
References
- Babini E, Paukert M, Geisler HS, Grunder S. (2002) Alternative splicing and interaction with di- and polyvalent cations control the dynamic range of acid-sensing ion channel 1 (ASIC1). J Biol Chem 277:41597–41603. [DOI] [PubMed] [Google Scholar]
- Balasuriya D, Stewart AP, Crottès D, Borgese F, Soriani O, Edwardson JM. (2012) The sigma-1 receptor binds to the Nav1.5 voltage-gated Na+ channel with 4-fold symmetry. J Biol Chem 287:37021–37029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron A, Schaefer L, Lingueglia E, Champigny G, Lazdunski M. (2001) Zn2+ and H+ are coactivators of acid-sensing ion channels. J Biol Chem 276:35361–35367. [DOI] [PubMed] [Google Scholar]
- Bässler EL, Ngo-Anh TJ, Geisler HS, Ruppersberg JP, Gründer S. (2001) Molecular and functional characterization of acid-sensing ion channel (ASIC) 1b. J Biol Chem 276:33782–33787. [DOI] [PubMed] [Google Scholar]
- Bonifacio G, Lelli CIS, Kellenberger S. (2014) Protonation controls ASIC1a activity via coordinated movements in multiple domains. J Gen Physiol 143:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand J, Smith ESJ, Schwefel D, Lapatsina L, Poole K, Omerbašić D, Kozlenkov A, Behlke J, Lewin GR, Daumke O. (2012) A stomatin dimer modulates the activity of acid-sensing ion channels. EMBO J 31:3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnally SM, Dev HS, Stewart AP, Barrera NP, Van Bemmelen MX, Schild L, Henderson RM, Edwardson JM. (2008) Direct visualization of the trimeric structure of the ASIC1a channel, using AFM imaging. Biochem Biophys Res Commun 372:752–755. [DOI] [PubMed] [Google Scholar]
- Chen X, Gründer S. (2007) Permeating protons contribute to tachyphylaxis of the acid-sensing ion channel (ASIC) 1a. J Physiol 579:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Paukert M, Kadurin I, Pusch M, Gründer S. (2006) Strong modulation by RFamide neuropeptides of the ASIC1b/3 heteromer in competition with extracellular calcium. Neuropharmacology 50:964–974. [DOI] [PubMed] [Google Scholar]
- Coscoy S, de Weille JR, Lingueglia E, Lazdunski M. (1999) The pre-transmembrane 1 domain of acid-sensing ion channels participates in the ion pore. J Biol Chem 274:10129–10132. [DOI] [PubMed] [Google Scholar]
- Della Vecchia MC, Rued AC, Carattino MD. (2013) Gating transitions in the palm domain of ASIC1a. J Biol Chem 288:5487–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Reznikov LR, Price MP, Zha X-M, Lu Y, Moninger TO, Wemmie JA, Welsh MJ. (2014) Protons are a neurotransmitter that regulates synaptic plasticity in the lateral amygdala. Proc Natl Acad Sci USA 111:8961–8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. (2009) Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselager M, Timmermann DB, Ahring PK. (2004) pH dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem 279:11006–11015. [DOI] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449:316–323. [DOI] [PubMed] [Google Scholar]
- Jones NG, Slater R, Cadiou H, McNaughton P, McMahon SB. (2004) Acid-induced pain and its modulation in humans. J Neurosci 24:10974–10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Schild L. (2015) International union of basic and clinical pharmacology. XCI. Structure, function, and pharmacology of acid-sensing ion channels and the epithelial Na+ channel. Pharmacol Rev 67:1–35. [DOI] [PubMed] [Google Scholar]
- Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. (2011) FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol 11:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. (2009) Interaction of the aromatics Tyr-72/Trp-288 in the interface of the extracellular and transmembrane domains is essential for proton gating of acid-sensing ion channels. J Biol Chem 284:4689–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. (2010a) Two residues in the extracellular domain convert a non-functional ASIC1 into a proton-activated channel. Am J Physiol Cell Physiol, 299:C66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang Y, Canessa CM. (2010b) Leu85 in the β1-β2 Linker of ASIC1 Slows Activation and Decreases the Apparent Proton Affinity by Stabilizing a Closed Conformation. J Biol Chem 285:22706–22712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti LA, Bernèche S, Bargeton B, Iwaszkiewicz J, Roy S, Michielin O, Kellenberger S. (2010) A combined computational and functional approach identifies new residues involved in pH-dependent gating of ASIC1a. J Biol Chem 285:16315–16329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E. (2007) Acid-sensing ion channels in sensory perception. J Biol Chem 282:17325–17329. [DOI] [PubMed] [Google Scholar]
- Lingueglia E, de Weille JR, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. (1997) A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem 272:29778–29783. [DOI] [PubMed] [Google Scholar]
- Paukert M, Chen X, Polleichtner G, Schindelin H, Gründer S. (2008) Candidate amino acids involved in H+ gating of acid-sensing ion channel 1a. J Biol Chem 283:572–581. [DOI] [PubMed] [Google Scholar]
- Salinas M, Lazdunski M, Lingueglia E. (2009) Structural elements for the generation of sustained currents by the acid pain sensor ASIC3. J Biol Chem 284:31851–31859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood TW, Frey EN, Askwith CC. (2012) Structure and activity of the acid-sensing ion channels. Am J Physiol Cell Physiol 303:C699–C710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ES, Zhang X, Cadiou H, McNaughton PA. (2007) Proton binding sites involved in the activation of acid-sensing ion channel ASIC2a. Neurosci Lett 426:12–17. [DOI] [PubMed] [Google Scholar]
- Smith ESJ, Omerbašić D, Lechner SG, Anirudhan G, Lapatsina L, Lewin GR. (2011) The molecular basis of acid insensitivity in the African naked mole-rat. Science 334:1557–1560. [DOI] [PubMed] [Google Scholar]
- Ugawa S, Ueda T, Ishida Y, Nishigaki M, Shibata Y, Shimada S. (2002) Amiloride-blockable acid-sensing ion channels are leading acid sensors expressed in human nociceptors. J Clin Invest 110:1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. (1997) A proton-gated cation channel involved in acid-sensing. Nature 386:173–177. [DOI] [PubMed] [Google Scholar]
- Wemmie JA, Taugher RJ, Kreple CJ. (2013) Acid-sensing ion channels in pain and disease. Nat Rev Neurosci 14:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]