Abstract
Hydrogenases are enzymes of great biotechnological relevance because they catalyse the interconversion of H2, water (protons) and electricity using non-precious metal catalytic active sites. Electrochemical studies into the reactivity of NiFe membrane-bound hydrogenases (MBH) have provided a particularly detailed insight into the reactivity and mechanism of this group of enzymes. Significantly, the control centre for enabling O2 tolerance has been revealed as the electron-transfer relay of FeS clusters, rather than the NiFe bimetallic active site. The present review paper will discuss how electrochemistry results have complemented those obtained from structural and spectroscopic studies, to present a complete picture of our current understanding of NiFe MBH.
Keywords: iron–sulfur cluster relay, membrane-bound hydrogenase, NiFe hydrogenase, oxygen tolerance, protein film electrochemistry
Introduction
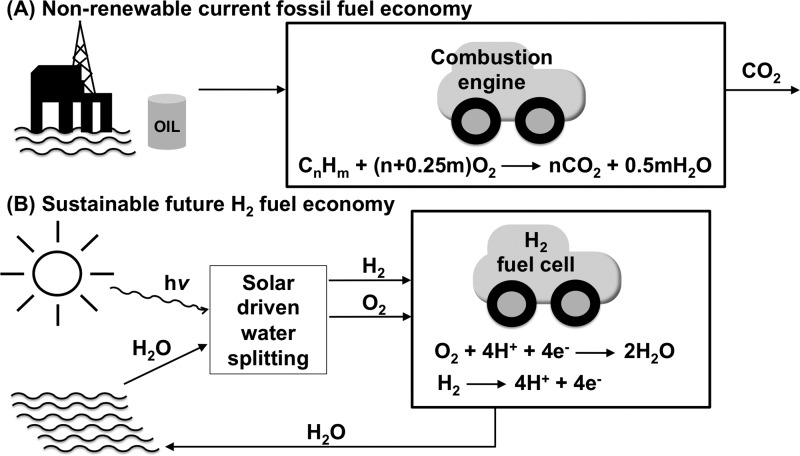
Humankind's current fossil fuel economy is unsustainable: it is non-renewable, generates the greenhouse gas carbon dioxide and relies on finite resources which are not evenly distributed across the globe, creating geo- and political- access issues [1]. As depicted in Figure 1, in comparison with the use of fossil fuels, a renewable H2 fuel economy presents many advantages: generating H2 from water is a cyclical, sustainable process, and vehicular H2 technology is a commercial reality [2]. The challenge lies in finding redox catalysts for the half cell processes of proton reduction (2H++2e− → H2) and water oxidation (H2O → ½O2+2H++2e−) to achieve the overall reaction of solar driven water splitting (H2O+hν → H2+½O2) [3]. There is a requirement for highly efficient catalysts built from commonly available elements that combine protons and electrons to produce H2.
Figure 1. Cartoon depicting the contrast between (A) the non-renewable, current fossil fuel economy, and (B) a sustainable, future H2 fuel economy.
Hydrogenases are H2 enzymes that are produced by a wide variety of microbes to catalyse either H2 splitting or the reverse reaction, H2 production [4,5]. As biological catalysts, hydrogenases are stable in water, built from earth-abundant elements and have high substrate affinities and fast turnover rates [6], and these combined factors have fuelled an interest into how hydrogenases can be utilized within a future H2 economy. Understanding how hydrogenases function as molecular H2 catalysts is of fundamental importance to any biological-H2 technology development and this paper will highlight how electrochemistry, in conjunction with other techniques, has played a vital role in deconvoluting the mechanism of NiFe hydrogenases.
Hydrogenases
There are three main types of hydrogenases, named for the composition of their active site as the NiFe, FeFe and Fe only hydrogenases [7]. The majority of biotechnological hydrogenase enzyme-devices, whether fuel cells or H2 producing devices, have made use of one subclass of NiFe hydrogenases, the Group 1 membrane-bound hydrogenases (MBH) [8,9]. These enzymes are biotechnologically useful because they react reversibly with O2, whereas the FeFe and Fe only hydrogenases sustain permanent damage after reaction with O2. Many MBH also adsorb onto carbon surfaces in an electrocatalytic configuration, generating a heterogeneous catalyst of “wired” enzyme molecules without the need for complex surface modification.
The NiFe MBH will be the focus of this paper, although reference will be made to soluble periplasmic enzymes in order to clarify aspects of the reactivity. Membrane-bound hydrogenases are periplasmically located enzymes which are embedded in the inner membrane of a bacterial cell [10,11], as shown in Figure 2. The physiological function of NiFe MBH is H2 uptake, the conversion of H2 into protons and electrons (H2 → 2H++2e−). The electrons are transferred from the hydrogenase into the quinone pool, with the NiFe MBH therefore forming part of the bacterial respiratory chain [12]. NiFe MBH are found in bacteria from a diverse range of ecological niches, from human pathogens such as Salmonella enterica, in which hydrogenase activity is linked to virulence [13], to soil bacteria like Ralstonia eutropha, which can survive by using H2 as their sole energy source [14], and Allochromatium vinosum, a photosynthetic purple sulfur bacteria which uses light energy to oxidize hydrogen sulfide to elemental sulfur [15].
Figure 2. The orientation of a NiFe MBH within a bacterial cell.
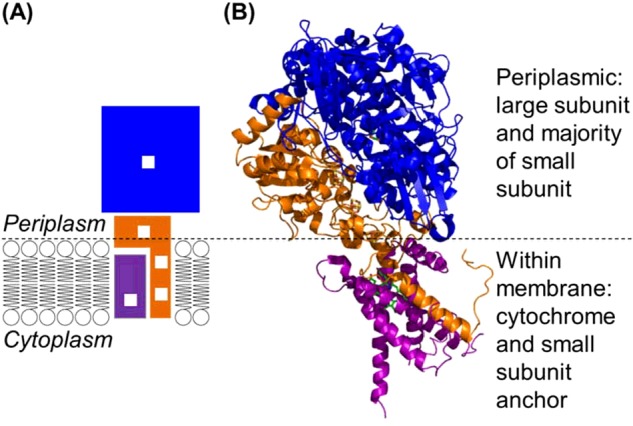
(A) Cartoon depiction of how a NiFe MBH is located within the cytoplasmic membrane, with white boxes representing the redox active metal centres and blue, orange and purple blocks indicating the large, small and cytochrome subunits, respectively. (B) Crystallographic insight into how the E. coli hydrogenase-1 large (blue ribbon), small (orange ribbon) and cytochrome (purple ribbon) subunits can interact. Figure generated from PDB 4GD3 [68].
The NiFe MBH are sub-categorized into enzymes which are able to function in O2 and enzymes which do not, with the former classed as O2 tolerant while the latter are known as O2 sensitive MBH [16]. The O2 reactivity of hydrogenases is considered important because most water splitting technology requires an O2 insensitive H2-catalyst [17]. Additionally, O2 tolerant NiFe MBH have been used to develop membrane-free H2 fuel cells, powered by non-explosive H2/O2 mixes [9]. Such devices are amenable for miniaturization because of their simple design.
In addition to highlighting the utility of electrochemistry as a technique for studying NiFe MBH, the present paper also will review our current mechanistic understanding of what controls the reaction of a hydrogenase with O2.
Hydrogenase film electrochemistry
Protein film electrochemistry applied to hydrogenases
Protein film electrochemistry, a technique in which enzyme is adsorbed to the surface of an electrode, has been a particularly useful tool for interrogating the reactivity of NiFe MBH. The Armstrong Group at the University of Oxford has played a major role in pioneering this field of research [18,19]. In these studies, the hydrogenase molecules under interrogation are commonly the “minimal functional unit” of an MBH, i.e. the active site (large) and electron-transfer (small) subunits, since these often purify separately from other subunits [20]. The possible impact of this is returned to later in the present paper (Section ‘The cytochrome: beyond the dimeric unit of a NiFe membrane-bound hydrogenase’). Although the enzyme molecules are assumed to be randomly orientated on the electrode surface, the electroactive orientations–where redox cofactors are in close enough proximity to the electrode to facilitate rapid electron transfer [21]–are thought analogous to the “wiring” of the hydrogenase to the cytoplasmic membrane.
The electrochemical cell, shown in Figure 3, is designed to facilitate temperature control via the water jacket, and pH control is determined by the buffer composition of the experimental solution within the main body of the cell. There are three separate electrodes, the working electrode, onto which the hydrogenase is adsorbed; the reference electrode, which is the reference point against which the voltage of the working electrode is set; and the counter/auxiliary electrode, which completes the electrical circuit by “countering” the electron flow at the working electrode. The working electrode is capable of rotation, which allows the investigation and minimization of diffusion effects, such as product inhibition. Gas flow controllers upstream of the electrochemical cell allow precise control of headspace gas composition, and mixtures of H2, N2, and the inhibitors O2 and CO can be made.
Figure 3. Diagram of the electrochemical cell.
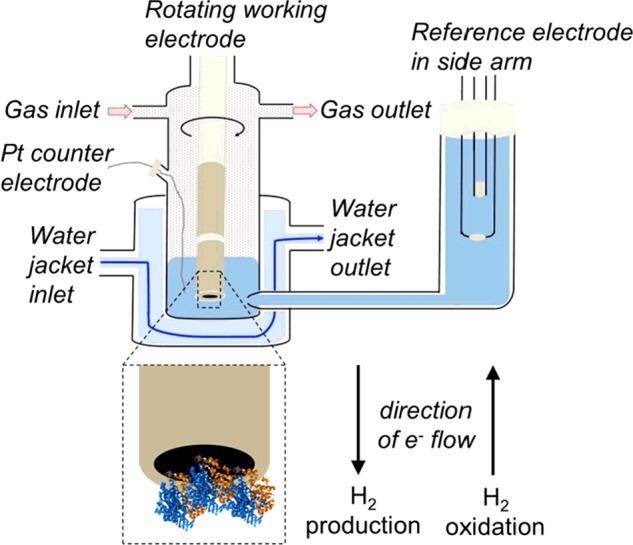
with detail of how the direction of electron flow in/out of the working electrode changes depending whether H2 production (H+ reduction, an electron uptake reaction) or H2 oxidation (an electron producing reaction) is catalysed by the hydrogenase adsorbed on the electrode surface.
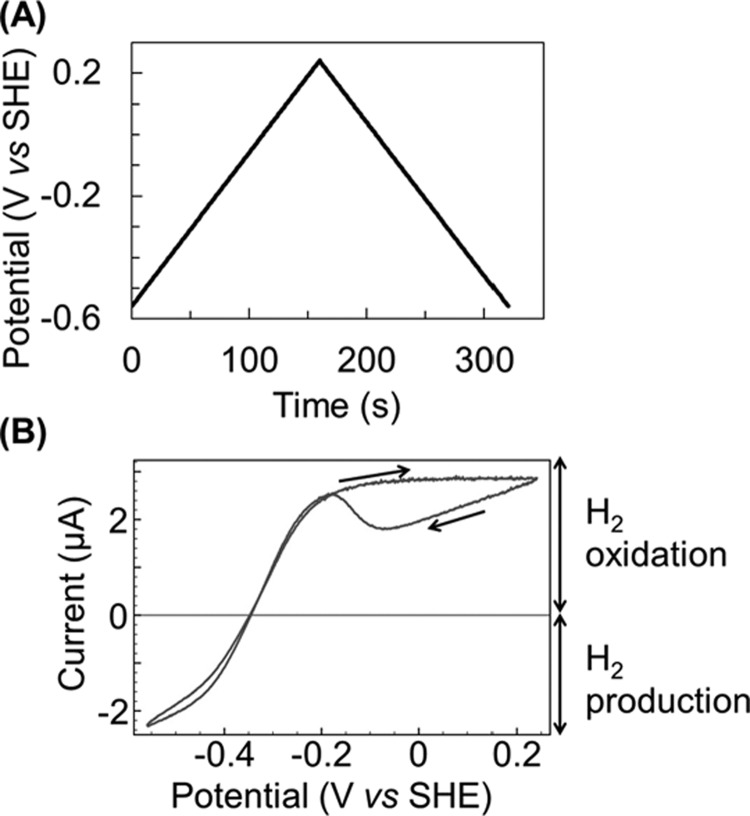
The bidirectional catalytic activity of NiFe hydrogenases is clearly revealed in cyclic voltammetry experiments such as that shown in Figure 4. The electrochemical potential of the hydrogenase-coated electrode is raised from negative (reducing) potentials to positive (oxidizing) potentials and the resultant current is monitored. Electrical current corresponds to enzymatic activity, with negative current providing a direct measure of H2 production (when electrons are transferred from the electrode into the enzyme as in Figure 3) and with positive current measuring H2 oxidation (where the direction of electron flow is reversed, also shown in Figure 3). If the number of enzyme molecules adsorbed onto the surface of the electrode is known (the product of the electrode surface area, A, and the surface density of enzyme on the electrode, Γ) then the catalytic current, icat, can be converted into a turnover rate, kcat, via eqn (1), where n denotes the number of electrons, 2 in the case of H2 catalysis, and F is Faraday's constant [18].
Figure 4. How cyclic voltammetry can be used to measure the bidirectional H2 catalytic activity of a hydrogenase under a H2-containing gas atmosphere.
(A) The potential–time linear sweep applied to the hydrogenase-coated working electrode, and (B) the resultant current–potential response. The enzyme used in this example is E. coli Hyd-2.
| 1 |
In most studies it has not been possible to measure Γ as the number of enzyme molecules on the electrode surface is often too low to produce the necessary non-catalytic signals needed to determine accurate coverage [22]. Therefore, electrochemistry is not commonly used to measure hydrogenase turnover rates, instead dye assays are used to quantify hydrogenase rate constants [23]. For enzyme-catalysed H2 oxidation, the concomitant reduction in either methylene blue or benzyl viologen can be followed spectrophotometrically [24]. Alternatively, the rate of H2 production/H+ reduction can be followed by measuring the rate of oxidation of reduced methyl viologen. A significant limitation of the dye assays is that they do not work in the presence of O2 and electrochemistry has provided an unparalleled comparison of the reactions of hydrogenases under anaerobic compared with aerobic conditions.
Electrochemical definition of hydrogenase O2 tolerance
Electrochemistry is particularly effective at categorizing a hydrogenase as either O2 sensitive or O2 tolerant [25]. Figure 5(A) shows a chronoamperometry experiment where the potential of the electrode is poised at a constant value and the current is measured as a function of time. The phenotypic behaviour of an O2 tolerant hydrogenase is that upon exposure to a mixture of H2 and O2, a substantial amount of the H2 oxidation activity (positive current) observed in the absence of O2 is maintained [26]. Upon removal of the O2, the electrical current returns to almost 100% of the initial activity, indicating full re-activation of the enzyme. This is shown by the “Hyd-1” data in Figure 5(A), measured for Escherichia coli hydrogenase-1 (Hyd-1). Conversely, O2 sensitive enzymes such as E. coli hydrogenase-2, “Hyd-2” in Figure 5(A), are fully inhibited (current tends towards zero) under O2-containing gas mixtures [18,26]. When O2 is removed from the experimental setup, not all of the enzyme activity is recovered within a short timeframe (the experiment shown in Figure 5A allows a reactivation period of 30 min).
Figure 5. Contrasting the electrochemical signature of an O2 tolerant NiFe MBH (black line, E. coli Hyd-1) with that of an O2 sensitive isozyme (grey line, E. coli Hyd-2) under (A) O2 inhibition conditions and (B) O2-free conditions.
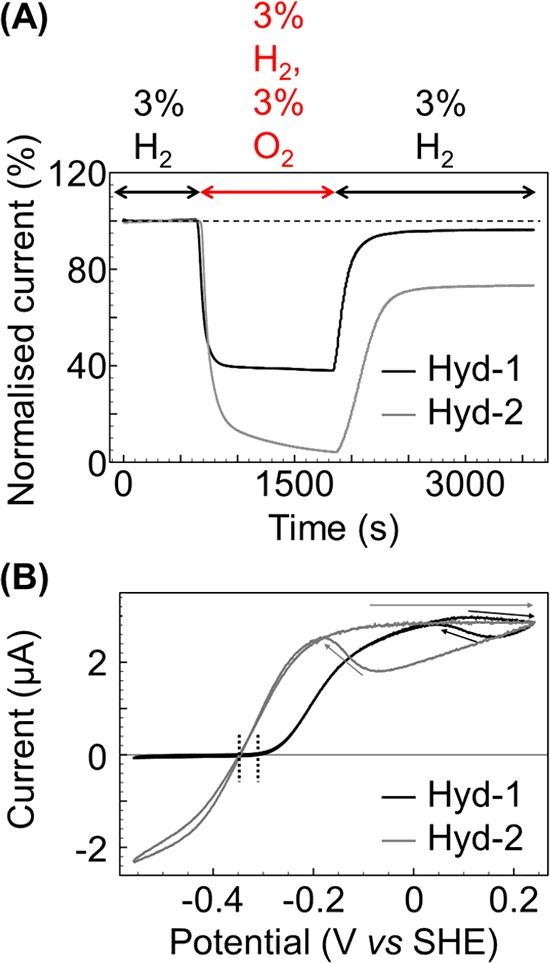
(A) Constant potential experiments measuring the percentage of initial H2 oxidation activity which is sustained when the enzyme is exposed to O2, and also quantifying the reversibility of the inhibition. For Hyd-1 a voltage of +0.06 V versus SHE is used, for Hyd-2 -0.16 V versus SHE. (B) 5 mV s−1 cyclic voltammetry experiments highlighting the difference in catalytic activity of O2 tolerant and sensitive NiFe MBH under conditions of 3% H2. The dotted vertical bars mark the potential onset of H2 oxidation and the arrows indicate regions of Ni-B formation and reactivation. Other experimental conditions: pH 6, 37°C, rotation rate 3000 rpm and total gas flow rate of 100 scc min−1 with N2 as carrier gas.
As well as the characteristic differences in O2 reactivity, electrochemistry also reveals notable heterogeneity between O2 tolerant and O2 sensitive hydrogenases in the absence of O2, as shown by Figure 5(B). Firstly, measurement of the catalytic activity via cyclic voltammetry shows that whereas enzymes which are tolerant to O2 produce very little H2 at pH levels above 6 (there is negligible negative current for Hyd-1 in Figure 5B), the ratio of maximum negative current to maximum positive current is almost 1:1 for O2 sensitive hydrogenases at near-neutral pH [26]. We can summarize this observation by stating that the catalytic “bias” is different for O2 tolerant and sensitive MBH; the O2 tolerant enzymes are essentially unidirectional H2-uptake enzymes at pH >6 while the O2 sensitive hydrogenases are bidirectional H2 catalysts over a wide pH range (5–8) [10]. The crucial role of pH in controlling the H2 production activity of O2 tolerant hydrogenases will be described later on.
A second observation related to catalysis is that O2 sensitive hydrogenases are “ideal” H2-catalyts; the potential of zero current (the voltage at which there is no net H2 oxidation or H2 production activity) equates to the reduction potential for the 2H+/H2 couple under the experimental conditions, E(2H+/H2) [26] (the value of E(2H+/H2) is calculated using the Nernst equation and the standard reduction potential value EΘ(2H+/H2)=0 V versus SHE). In contrast with this ideal behaviour, O2 tolerant hydrogenases such as Hyd-1 manifest an “overpotential requirement” at pH >5, meaning that the potential at which H2 oxidation catalysis commences is more positive than for Hyd-2 [27]. This is highlighted by the vertical dotted lines in Figure 5(B), which show that the onset of H2 oxidation is ∼50 mV higher for Hyd-1 than for Hyd-2.
It is helpful to correlate the differences in reactivity measured by electrochemistry with structural insights gained by spectroscopic and crystallographic studies, in order to build a mechanistic understanding of the difference between O2 tolerant and O2 sensitive MBH. The next sections will describe how all these data can be combined to build a picture of hydrogenase structure–function properties.
Active site states
States formed in the absence of O2
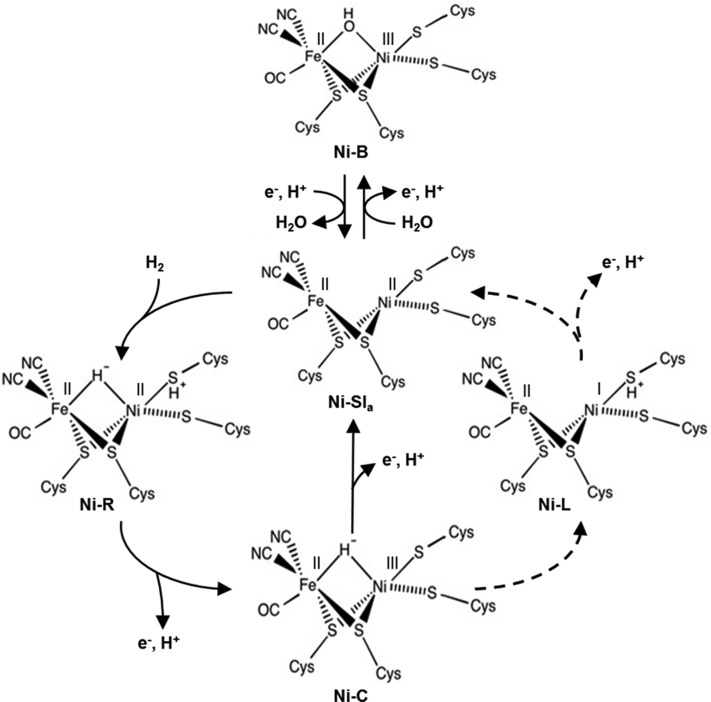
Detailed structural information about the active site of NiFe MBH has been obtained from spectroscopic measurements using a variety of techniques including EPR [28] and IR [29–32]. A summary of the proposed key states in the catalytic cycle is given by Figure 6; for simplicity this figure shows uni-directional H2 oxidation, although the same states are proposed to occur for H2 production/H+ reduction.
Figure 6. Catalytic and inhibited active site redox states accessed by NiFe hydrogenases under O2-free conditions.
The solid arrows indicate the reaction mechanism for O2 sensitive NiFe hydrogenases. The dashed arrows indicate how the Ni-L state may participate as a reaction intermediate in the catalytic cycle of O2 tolerant MBH.
When catalysing H2 oxidation, NiFe hydrogenases operate via a heterolytic cleavage mechanism whereby the H2 initially binds to the Ni-Sia (NiII–FeII) state as a proton and a hydride to form the most reduced Ni-R (NiII–H−–FeII) state of the active site (Figure 6) [33]. Loss of a proton and an electron then generates the Ni-C (NiIII–H−–FeII) state. At this stage in the H2 uptake reaction the proposed mechanisms for O2 tolerant and O2 sensitive enzymes differ, with EPR and FTIR studies showing that different active site states predominate within each hydrogenase subgroup. For O2 sensitive NiFe hydrogenases the Ni-L (NiI–FeII) state is not thought to be a true catalytic intermediate because it is only formed under light exposure at very low temperatures [34–36]. As denoted by the solid arrows in Figure 6, catalysis by O2 sensitive NiFe hydrogenases is thought to proceed via loss of an electron and proton from the Ni-C state to give direct regeneration of Ni-SIa. In contrast, for O2 tolerant hydrogenases the Ni-L state is readily detectable at room temperature, even when the enzyme has not been illuminated [31], so this state is considered an important catalytic intermediate. Figure 6 shows a putative Ni-C to Ni-L conversion that involves electron and proton rearrangement; NiIII is reduced to NiI with concomitant movement of the bridging hydrogen to the sulfur of a coordinating cysteine.
Under conditions of oxidative stress, but in the absence of O2, all NiFe MBH form the inactive Ni-B state, of which the structure is well established [29,30], as shown in Figure 6. Formation of the Ni-B state is also thought to be observable in cyclic voltammetry electrochemistry experiments, such as those shown in Figures 4 and 5(B). Under O2-free conditions, when the potential is steadily increased above -0.2 V for Hyd-2 or 0 V for Hyd-1, a current plateau is observed. This means that the H2 oxidation activity levels off despite the application of more oxidizing potentials. This is denoted by the arrows pointing from left to right in Figure 5(B). Upon reversal of the potential sweep, so that the voltage of the electrode is now being made progressively more negative, the MBH is activated, as shown by the sharp rise in current from -0.1 to -0.2 V for Hyd-2 and +0.15 to +0.05 V for Hyd-1, and highlighted by the diagonal arrows in Figure 5(B). Comparison of electrochemically measured inactivation and reactivation kinetics with spectroscopic studies has led to attribution of the current plateau and recovery to formation and reactivation of the Ni-B state, respectively [25,37]. The observation that all O2 tolerant hydrogenases apparently require an additional oxidative driving force to form the Ni-B state in cyclic voltammetry experiments has been used as the basis for a model for understanding how these enzymes remain catalytically active in the presence of O2 [38]. However, there is nothing about the active site structures which explains why formation of the Ni-B state should require a different electrochemical potential for O2 tolerant compared with O2 sensitive enzymes [29,38]. Modern research has focused on understanding how the movement of electrons within the hydrogenase controls the active site chemistry, and this is described later on.
States formed by reaction with O2
The Ni-B state is also formed when NiFe MBH are exposed to O2. The mechanism of this conversion is still controversial. One proposed reaction [39] involves O2 binding at the active site followed by reduction by four electrons and three protons to form water and a hydroxide molecule (O2+3H++4e− → H2O+OH−). The water molecule formed from O2 is then proposed to diffuse out of the enzyme via a solvent channel, while the hydroxide remains bound to the oxidized active site. Envisaging Ni-B formation as arising from O2 binding at the active site has been used to reconcile the observation that O2 tolerant hydrogenases have decreased sensitivity to CO and very low Michaelis constants for H2; it has been interpreted that these enzymes have evolved to optimize H2 binding at the active site in order to minimize competitive inhibitor ligation at the NiFe centre [26,40].
However, the notion that aerobic Ni-B formation involves a long lived active site species formed from O2 has been challenged by isotope studies showing evidence that the bridging OH− ligand in Ni-B is solvent derived [41], and oxygenic species originating from O2 are not observable in the first coordination sphere of the nickel [42]. Based on these studies, alternative mechanisms which do not involve the binding of O2 to the active site have been proposed. For example, O2 could prompt oxidation of the active site by acting as an electron acceptor, as proposed by Hamdan et al. in their work which proves that “aerobic” inactivation can occur under O2-free oxidizing conditions [43].
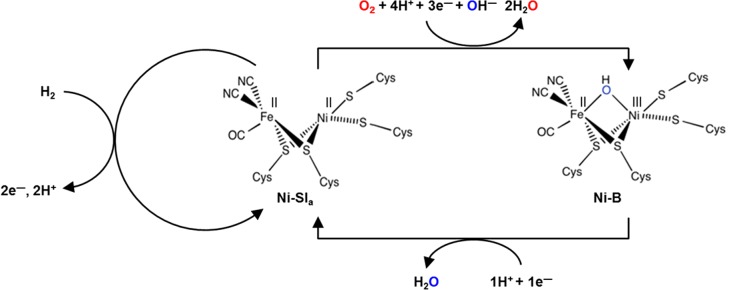
Regardless of the precise site of O2 binding, it is known that an O2 tolerant NiFe hydrogenase exposed to O2 can catalyse H2 oxidation and also act as an O2-reductase, converting O2 into two molecules of H2O (Figure 7) [44]. The rapid reactivation of Ni-B explains why O2 tolerant hydrogenases sustain H2 catalytic activity in the presence of O2; any enzyme molecule which forms the Ni-B state upon inactivation by O2 is quickly reactivated by addition of one electron, as shown in Figure 7. Electrochemical experiments reveal that the “Eswitch” potential, a complex voltammetric parameter used to characterize the reactivation window of a hydrogenase, is positive for an O2 tolerant MBH, reflecting the ease of Ni-B reduction [45]. For O2 sensitive enzymes, Eswitch has a negative potential, but slower Ni-B activation is not the only reason that these enzymes are inactivated by O2 exposure.
Figure 7. Reaction of an O2 tolerant NiFe MBH with O2 to generate the Ni-B state.
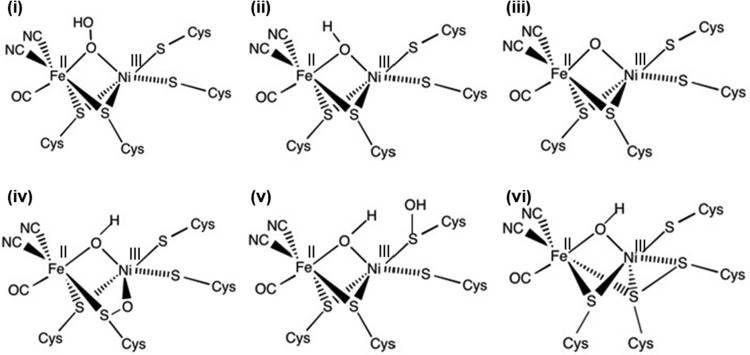
In the case of O2 sensitive NiFe hydrogenases, spectroscopic experiments using a variety of techniques all conclude that reaction with O2 generates both the Ni-B state and a very kinetically inert “Unready”, Ni-A, state [46–50]. This correlates with the electrochemical observation that following O2 exposure, O2 sensitive enzymes such as Hyd-2 rapidly recover some activity as soon as the O2 is removed from the experiment, but a substantial proportion of the enzyme remains inactivated (Figure 5A). The rapid reactivation is attributed to recovery of molecules which formed the Ni-B state, while slow reactivation is assigned to enzyme which formed the Ni-A state [43,49]. A significant challenge in discovering the precise mechanism for Ni-A formation originates from the difficulty in reconciling the spectroscopic and electrochemical experiments with the fact that crystallographic experiments have isolated a myriad of different structures, all denoted as possible representatives of the Ni-A state (Figure 8) [48,51–54]. Due to the observation of extended electron density about the bridging ligand, early structural studies proposed that the Ni-A state contains an O2 derived peroxide group bridging the Ni and Fe [48] (Figure 8i). This model has been revised in recent years to consider the possibility of oxidation of the S group of cysteine ligands, as in several structures proposed by Volbeda et al. [53,54] and shown in Figures 8(iv)–8(vi). A bridging oxo group, O2−, was also postulated, as shown in Figure 8(iii). Carepo et al. [41] used 17O labelled water, H217O, to confirm that there is a bridging ligand but it is solvent derived. The isotope labelling study instead proposed that the Ni-A state could contain a bridging hydroxide in a different orientation to that seen in Ni-B (Figure 8ii). This is supported by recent work by Barilone et al. [55] who used single crystal ENDOR spectroscopy to confirm the Ni-A bridging ligand as a hydroxide, again suggesting that the difference in the structure of Ni-B and Ni-A is due to rotation at the nickel, specifically identifying a cysteine side chain as the mobile element. Altogether, the issue of the structural identity of Ni-A remains contentious, rendering the task of explaining why O2 sensitive NiFe hydrogenases are slow to reactivate very difficult.
Figure 8. Different postulated structures for the Ni-A inhibited active site state.
(i) Bridging peroxo structure [48]; (ii) stereoisomer of the Ni-B, hydroxide bridged, active site [41]; (iii) bridging oxo structure [41]; (iv) Ni and S bridging oxo species [53]; (v) S–OH and bridging hydroxide form [54] and (vi) disulfide bond containing hydroxide-bridging structure [54].
The lack of consensus in determining exactly what active site structure is generated when O2 sensitive NiFe hydrogenases react with O2 may be in part attributable to the fact that O2 inactivation can also generate “dead” states of permanently inactivated enzyme [19]. In electrochemistry it is difficult to separate an estimate of dead phase formation from “film loss”, the normal steady drop in enzyme activity which is observed over long experiments and is probably due to desorption of enzyme from the electrode [56]. If the hydrogenase dead phase is non-paramagnetic then it will also be rendered spectroscopically silent via EPR, and small numbers of molecules are hard to detect using FTIR. However, at least some of the structural electron density maps may reflect dead phase enzyme and so reconciling X-ray data and spectroscopic information could remain challenging.
The essential role of the electron-transfer relay
Description of the iron–sulfur “wire”
The minimal functional unit of a NiFe MBH is a heterodimer comprising two protein chains designated the “large” and “small” subunits [57]. As shown by Figure 9(A), over the past 10 years substantial progress has been made in the determination of NiFe hydrogenase X-ray structures. Enzymes from both aerobic (Hydrogenovibrio marinus [58] and R. eutropha [59,60]), anaerobic (Desulfovibrio gigas [48,51], Desulfovibrio fructosovorans [53,54,61], Desulfovibrio vulgaris [52,62–64], Desulfomicrobium baculatum [65,66] and A. vinosum [67]) and facultative anaerobic organisms (E. coli [68,69] and S. enterica [70]) have been crystallized (Figure 9B) [71], resulting in the elucidation of the heterodimer structures of several O2 sensitive and O2 tolerant NiFe hydrogenases. The overall structure of all these enzymes is remarkably similar (Figure 10). In addition to the NiFe active site centre (contained within the ∼60 kDa large subunit), three iron–sulfur (FeS) clusters are ligated by the ∼35 kDa small subunit [58,59]. This “wire” of FeS clusters is thought to function as an electron-transfer relay, shuttling electrons from the buried active site to the surface of the protein, thus allowing the passage of electrons between the active site and the inner membrane [72]. The three clusters are commonly denoted as “proximal”, “distal” or “medial” in reference to their distance from the active site.
Figure 9. Graphs relating number of NiFe hydrogenase PDB structures to (A) publication year and (B) bacterial species.
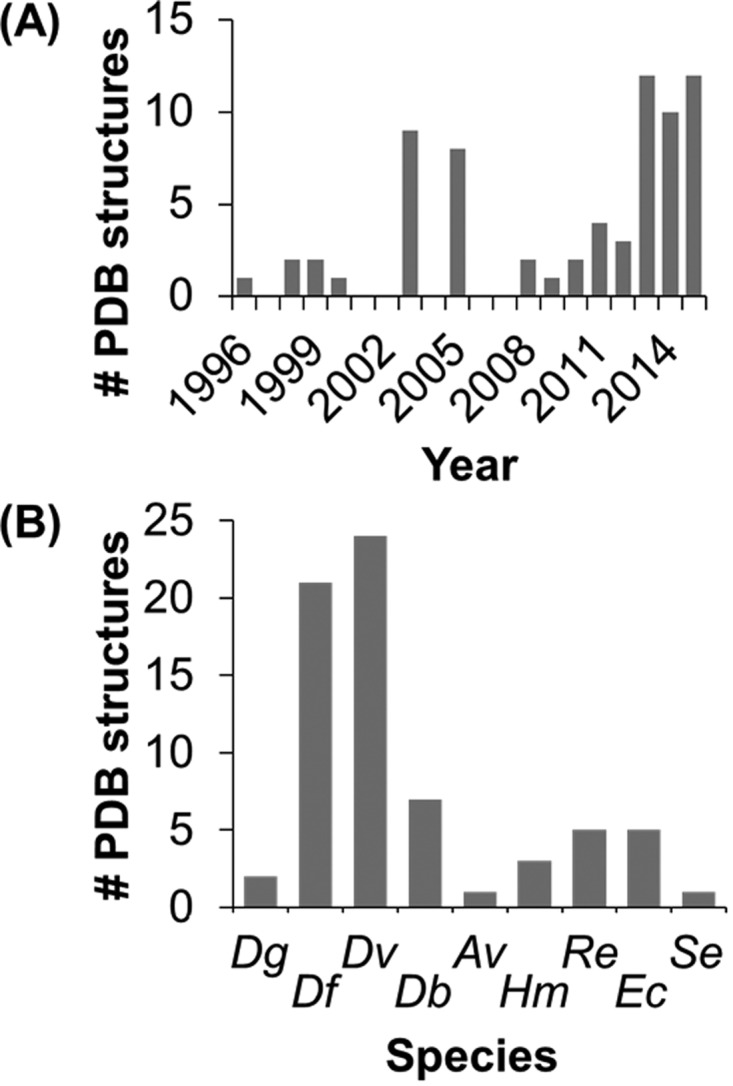
Abbreviations: Dg, D. gigas; Dv, D. vulgaris; Av, A. vinosum; Re, R. eutropha; Se, S. enterica; Df, D. fructosovorans; Db, D. baculatum; Hm, H. marinus; Ec, E. coli. Generated from data in the Protein Data Bank [71].
Figure 10. (A and B) The minimal functional unit of a NiFe hydrogenase.
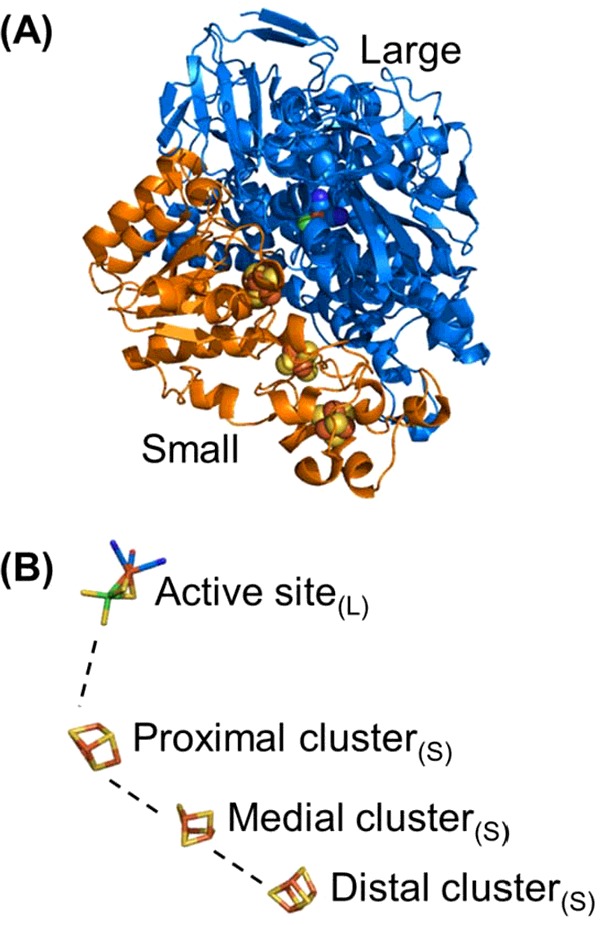
(PDB 3RGW [60]) comprising a NiFe active site co-ordinated by the large subunit (blue ribbon) and three FeS clusters co-ordinated by the small subunit (orange ribbon).
As discussed above, electrochemistry reveals clear differences in the reactivity of O2 tolerant and O2 sensitive MBH. The overpotential-requirement and uni-directional activity of the O2 tolerant hydrogenases may be linked to their propensity to form the Ni-L state as part of the catalytic cycle (Figure 6) but nothing about the active site architecture suggests why the Ni-L state might be more accessible. Instead, the active site binding pocket is highly conserved for both O2 tolerant and sensitive MBH. Recent studies have revealed that it is the electron-transfer clusters which play a critical role in tuning the reaction mechanism of the hydrogenases, and this will now be explained in detail.
Proximal cluster
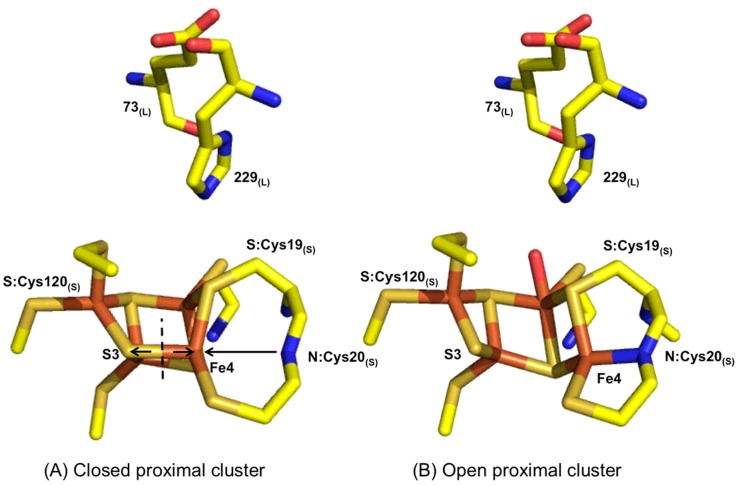
Experiments on a number of different O2 tolerant hydrogenases have revealed that it is a uniquely structured FeS cluster that confers O2 tolerance upon NiFe MBH [59,72,73]. Crystallography has shown that while O2 sensitive NiFe hydrogenases contain a standard 4Fe4S cluster proximal to the active site, all O2 tolerant MBH contain a novel 4Fe3S cluster in this position [58,59]. As highlighted in the sequence alignment in Figure 11, additional cysteines are contained within O2 tolerant MBH, and these assist in the stabilization of the unusual proximal cluster. These “supernumerary” cysteines support a structural transition which occurs when the cluster is oxidized, for example when the enzyme is exposed to O2. During this transition, the backbone N of cysteine 20 ligates Fe4 of the cluster, and the bond between Fe4 and S3 of the cluster is broken, thus creating an “open” structure [60], as seen in Figure 12. This “over oxidation” can also be observed in EPR studies which show that the 4Fe3S cluster can undergo two redox transformations, i.e. it is stable in three different oxidation states: fully reduced, [4Fe3S]3+; oxidized, [4Fe3S]4+; and over oxidized, [4Fe3S]5+ [60,69,72,74].
Figure 11. Sequence alignment highlighting how O2 tolerant and O2 sensitive NiFe hydrogenases differ in the small and large subunit amino acids located near to the proximal cluster.
Ec (E. coli) Hyd-1 numbering is used, this is the same as Se Hyd-1/Hyd-5. Other abbreviations are: Se, S. enterica; Re, R. eutropha; Aa, A. aeolicus; Av, A. vinosum; Dg, D. gigas; Dv, D. vulgaris; Db, D. baculatum.
Figure 12. Structural insight into the oxidative “opening” of the proximal cluster obtained from R. eutropha NiFe MBH crystallography studies (PDB reduced=3RGW and oxidized=4IUB) [60].
The cluster “opening” is ascribed to the breaking of the bond between S3 and Fe4, indicated by the dashed line and outward arrows in (A), and formation of a bond between Fe4 and the backbone N of cysteine 20, indicated by the arrow pointing from N:Cys20 to Fe4 in (A). Colours: orange=iron, ochre=sulfur, yellow=carbon, blue=nitrogen, red=oxygen. E. coli hydrogenase-1 / S. enterica Hyd-5 residue numbering is used.
Electrochemical studies on hydrogenase variants show that using glycine to replace the supernumerary cysteine which supports the cluster opening mechanism (E. coli Hyd-1 numbering: C19G) turns an O2 tolerant hydrogenase into an enzyme which is highly sensitive to O2 [75]. This suggests that formation of the over oxidized state of the proximal cluster plays an important role in O2 tolerance. Returning to Figure 7, the idea is that because the proximal cluster can access three oxidation states it is therefore able to rapidly deliver two of the electrons which are needed to ensure that inhibitory O2 is rapidly neutralized to H2O and OH− [44], ensuring that the Ni-B state is the only product of reaction between an O2-tolerant hydrogenase and O2.
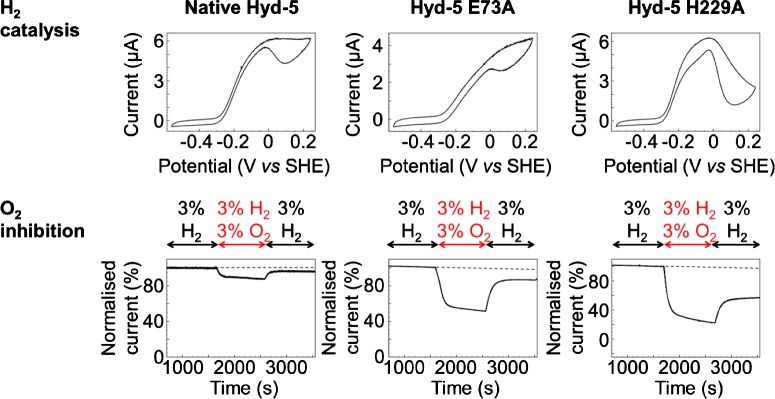
More recent work has shown that large subunit residues also play a vital role in the proximal cluster chemistry [60,70]. The sequence alignment shown in Figure 11 demonstrates that all O2 sensitive hydrogenases possess a glutamine in the large subunit, while the majority of O2 tolerant MBH encode a glutamate at this site. Structural analysis indicates that this residue is too removed from the proximal cluster to play a direct role in stabilizing the 3Fe4S redox transitions. However, in conjunction with a conserved histidine as an acid/base pair, a possible proton transfer mechanism was hypothesized and tested for variants of the O2 tolerant MBH S. enterica hydrogenase-5 (Hyd-5) [70]. While exchange of S. enterica Hyd-5 glutamate 73 for alanine had a minimal effect on catalysis in the absence of O2, in the presence of O2 the enzyme exhibited decreased O2 tolerance relative to the native enzyme (Figure 13). The essential role of E73 in tuning O2 tolerance was therefore confirmed. Replacing histidine 229 with alanine had a substantial effect on the H2 oxidizing ability of the enzyme under anaerobic and aerobic conditions. The increase in O2 sensitivity in large subunit histidine variants was also observed in R. eutropha MBH [60] and crystallographic data from that study provided evidence for a direct proton transfer route from the large subunit to the proximal cluster via H229, which points direct at one of the cluster's Fe centres, as shown in Figure 12. The role of this proton transfer pathway in triggering the structural and redox transition of the proximal cluster into the over oxidized state has been elaborated through recent density functional theory studies. Dance [76] suggests that O2 binding at the active site induces a change in protonation which is communicated to the proximal cluster via a proton transfer relay which ends at H229. Because the change in histidine coordination induces the proximal cluster to open up, the proton transfer pathway therefore ensures that O2 binding at the active site is rapidly followed by electron delivery from the proximal cluster.
Figure 13. Electrochemical characterization of the catalytic (top) and O2 inhibition properties (bottom) of different proximal cluster variants (left to right) of S. enterica Hyd-5.
The 5 mV s−1 cyclic voltammetry H2 catalysis experiments were measured under a gas atmosphere of 10% H2 while the chronoamperometry O2 inhibition experiments were conducted at a potential of +0.06 V versus SHE with changing gas atmosphere as indicated. Other experimental conditions: pH 6, 37°C, rotation rate 4000 rpm and total gas flow rate of 100 scc min−1 with N2 as carrier gas.
Interestingly, in all proximal cluster variants to date, although it has been possible to turn an O2 tolerant hydrogenase into an O2 sensitive enzyme, the bi-directional catalytic activity of a true O2 sensitive NiFe MBH was not conferred upon any of the single site variants [60,70,75]. Instead, it is the distal cluster which is thought to hold the key to understanding the origins of the O2 tolerant over-potential requirement [77], while the medial cluster is thought to play a role in supporting the O2 tolerance mechanism [78].
Medial cluster
Studies of the proximal cluster highlighted that this site was capable of providing two electrons to reduce inhibitory O2 [69,74], however if O2 attacks a NiII oxidation state of the enzyme, then generation of the Ni-B state requires a total of three electrons from the FeS relay [44] (and one from the oxidation of the active site, forming NiIII) (see Figure 7). Given that electrons one and two can be supplied by the proximal cluster, the most obvious source of the third FeS relay electron is the medial cluster. In both O2 tolerant and sensitive MBH the medial cluster is a 3Fe4S centre, ligated by three cysteines. Evans et al. [78] generated and electrochemically studied a variant of E. coli Hyd-1 which contained a cysteine in position 242 instead of proline. The amino acid exchange caused the medial cluster to become a 4Fe4S centre with a concomitant increase in O2 sensitivity. The loss of O2 tolerance probably arises because the medial redox potential has become more negative as a result of the amino acid exchange. This will cause the medial cluster to lose electrons to the distal cluster too readily, rather than storing them in order to reduce the proximal cluster.
A combined proximal and medial cluster variant, E. coli Hyd-1 C19G/C120G/P242C, showed a complete and irreversible loss of H2 oxidation activity following O2 exposure, supporting the proposed mechanism that three electrons are needed from the medial and proximal cluster in order to achieve O2 tolerance [78].
Distal cluster
The distal cluster of the NiFe MBH remains the most elusive FeS centre. Although midpoint redox potentials have been quoted for Aquifex aeolicus [72], R. eutropha and Ralstonia metallidurans [79], it is not possible to determine the same parameter from analogous EPR experiments on E. coli Hyd-1. Indeed, the very careful study on Hyd-1 instead concluded that the distal cluster does not form an S=½ state even under reducing conditions [74,75]. Measuring the reduction potential of the Hyd-1 distal cluster using electrochemistry instead of EPR has so far proved impossible because of our inability to obtain “non-turnover” signals for O2-tolerant NiFe MBH. However, simulation of hydrogenase electrocatalytic cyclic voltammetry data supports the notion of the distal cluster controlling catalytic bidirectionality via its “gateway” role in mediating intermolecular electron transfers [77].
At low pH, substantial H2 production has been observed for the O2 tolerant hydrogenase E. coli Hyd-1, and there is a loss of the overpotential requirement for catalysis [27]. This result is interpreted in terms of the electrocatalysis voltammetric simulation model [77]; because the redox potential of the 2H+/H2 couple is strongly pH dependent while the distal cluster reduction potential is thought to be pH independent, there is a threshold pH below which it is thermodynamically favourable for electrons to transfer from the distal cluster into the active site thus facilitating H+ reduction to generate H2 [27,77].
The only study of a distal cluster NiFe hydrogenase variant was conducted on an O2 sensitive enzyme at high pH [80]. That study shows that NiFe hydrogenases are non-functional without a correctly ligated distal cluster because when the normal Cys3His coordination was converted to Cys4 or Cys3Gly a substantial drop (≈ 95%) in H2 oxidation rates was observed for D. fructosovorans variants. Imidazole was observed to recover the activity of the variants, emphasizing the vital role of the histidine ligand at the distal cluster.
The cytochrome: beyond the dimeric unit of a NiFe membrane-bound hydrogenase
In a bacterial cell, a NiFe MBH is anchored on the periplasmic face of the inner membrane by a trans-membrane helix on the C-terminus of the small subunit (Figure 2) [10]. This helix is believed to associate with a hydrogenase cytochrome which embeds into the membrane [81]. The physiological electron-transfer chain of a hydrogenase does not, therefore, end at the distal cluster and it may be dangerous to interpret the in vivo role of a NiFe MBH without considering the mediating role which the cytochrome and quinone pool will play. There is, at the date of writing, only one MBH crystal structure which incorporates the cytochrome subunit (PDB 4GD3 [68]). This E. coli Hyd-1 structure contained a 2:1 ratio of large–small dimer:cytochrome, which is likely an experimental artefact since the operon of all NiFe MBH contain a single gene for each of the large, small and cytochrome subunits, all under control of the same promoter. Figure 2 therefore shows a fragment of the X-ray structure with one small, large and cytochrome subunit, this combination of subunits is classed as the heterotrimeric unit.
All the electrochemical experiments described above were conducted on dimers of the large and small subunits only, because the cytochrome easily dissociates during enzyme purification. However, it has been possible to isolate the O2 tolerant MBH from R. eutropha as a unit of three heterotrimers [81]. Electrochemical experiments have been conducted to analyse the behaviour of the trimeric complex embedded within a membrane-coated electrode, with a ubiquinone pool acting to shuttle electrons between the electrode and the hydrogenase [82]. In this configuration, the enzyme's electrochemical response is dramatically different from that of an isolated dimer on a membrane-free electrode. When the electrode wiring was mediated via the quinone pool, the hydrogenase trimer showed little to no anaerobic inactivation and a higher level of activity was sustained in the presence of O2. It has been proposed that “short-circuiting” multiple hydrogenases by forming multimers of trimers should enhance O2 tolerance because an active site which has H2 bound can provide the electrons to reduce a Ni-B inactivated neighbouring active site. There may therefore be a physiological benefit for hydrogenase molecules associating with one another within the cytoplasmic membrane. It is important to consider that while biotechnological applications of isolated dimeric enzymes require an understanding of the catalytic features of the same molecules, knowing how to harness hydrogenase activity within whole cell applications (such as cyanobacterial photosynthetic H2 production) may require an alternative approach such as the study of MBH within a membrane. Spectroscopic methods have also been developed to permit analysis of MBH within cell membranes [30], but there is a requirement for hydrogenase over-expression which is not always feasible because of the complex biosynthesis of these enzymes.
Conclusion
In conclusion, when used in combination with spectroscopy and crystallography, electrochemistry has been proved an extremely useful tool for investigating NiFe MBH reactivity. Enormous progress has been made in recent years to understand the origin of O2 tolerance in MBH, and the essential roles of redox centres and residues remote from the NiFe active site have been elucidated and understood. There remain some core aspects of NiFe MBH biochemistry, such as the origin of catalytic bias and the structure of the Ni-A state, formed when O2 sensitive enzymes react with O2, which remain elusive. Beyond the membrane-bound Group 1 NiFe hydrogenases, there are four further categories of NiFe hydrogenases which are woefully under-studied but now becoming accessible due to modern molecular biology techniques. It is hoped that hydrogenase research will continue to contribute to the vital field of sustainable H2 production, inspiring the development of stable non-precious metal catalysts with high turnover frequency. It is noted that some synthetic catalysts can already outperform the enzymes under certain conditions, and purifying hydrogenases is a costly process. However, the biological synthesis of these highly active NiFe catalysts under conditions of ambient temperature, pressure and in an aqueous solvent remains an inspirational goal, and it may be that whole cell technology is where microbial catalysts are best deployed.
Abbreviations
- FTIR
Fourier transformed infrared
- Hyd-1
hydrogenase-1
- Hyd-2
hydrogenase-2
- Hyd-5
Hydrogenase-5
- MBH
membrane-bound hydrogenase(s)
Footnotes
Bioenergetics in mitochondria, bacteria and chloroplasts: Held at Schloss Rauischholzhausen, Ebsdorfergrund, Germany, 10–13 April 2013.
Funding
This work was supported by a Junior Research Fellowship from Merton College, Oxford and laboratory start-up funds from the University of York (to A. Parkin); and the Wellcome Trust 4-year PhD programme [grant number WT095024MA (to L. Flanagan)] ‘Combating infectious disease: computational approaches in translational science’.
References
- 1.King D.A. Climate change science: adapt, mitigate, or ignore? Science. 2004;303:176–177. doi: 10.1126/science.1094329. [DOI] [PubMed] [Google Scholar]
- 2.Momirlan M., Veziroglu T. The properties of hydrogen as fuel tomorrow in sustainable energy system for a cleaner planet. Int. J. Hydrogen Energy. 2005;30:795–802. doi: 10.1016/j.ijhydene.2004.10.011. [DOI] [Google Scholar]
- 3.RSC. Solar fuels and artificial photosynthesis. 2012. www.rsc.org/images/Solar-fuels_tcm18-221433.pdf.
- 4.Greening C., Biswas A., Carere C.R., Jackson C.J., Taylor M.C., Stott M.B., Cook G.M., Morales S.E. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2015 doi: 10.1038/ismej.2015.153. doi:10.1038/ismej.2015.153. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubitz W., Ogata H., Rüdiger O., Reijerse E. Hydrogenases. Chem. Rev. 2014;114:4081–4148. doi: 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- 6.Shafaat H.S., Rüdiger O., Ogata H., Lubitz W. [NiFe] hydrogenases: a common active site for hydrogen metabolism under diverse conditions. Biochim. Biophys. Acta. 2013;1827:986–1002. doi: 10.1016/j.bbabio.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Vignais P.M., Billoud B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 8.Cracknell J.A., Vincent K.A., Armstrong F.A. Enzymes as working or inspirational electrocatalysts for fuel cells and electrolysis. Chem. Rev. 2008;108:2439–2461. doi: 10.1021/cr0680639. [DOI] [PubMed] [Google Scholar]
- 9.Vincent K.A., Cracknell J.A., Lenz O., Zebger I., Friedrich B., Armstrong F.A. Electrocatalytic hydrogen oxidation by an enzyme at high carbon monoxide or oxygen levels. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16951–16954. doi: 10.1073/pnas.0504499102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballantine S.P., Boxer D.H. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur. J. Biochem. 1986;156:277–284. doi: 10.1111/j.1432-1033.1986.tb09578.x. [DOI] [PubMed] [Google Scholar]
- 11.Sawers R.G., Boxer D.H. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K12. Eur. J. Biochem. 1986;156:265–275. doi: 10.1111/j.1432-1033.1986.tb09577.x. [DOI] [PubMed] [Google Scholar]
- 12.Jones R.W. The role of the membrane-bound hydrogenase in the energy-conserving oxidation of molecular hydrogen by Escherichia coli. Biochem. J. 1980;188:345–350. doi: 10.1042/bj1880345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier R.J., Olczak A., Maier S., Soni S., Gunn J. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect. Immun. 2004;72:6294–6299. doi: 10.1128/IAI.72.11.6294-6299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenz O., Ludwig M., Schubert T., Bürstel I., Ganskow S., Goris T., Schwarze A., Friedrich B. H2 conversion in the presence of O2 as performed by the membrane-bound [NiFe]-hydrogenase of Ralstonia eutropha. ChemPhysChem. 2010;11:1107–1119. doi: 10.1002/cphc.200901002. [DOI] [PubMed] [Google Scholar]
- 15.Weissgerber T., Zigann R., Bruce D., Chang Y., Detter J.C., Han C., Hauser L., Jeffries C.D., Land M., Munk A.C., et al. Complete genome sequence of Allochromatium vinosum DSM 180T. Stand. Genomic Sci. 2011;5:311–330. doi: 10.4056/sigs.2335270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandelia M.-E., Lubitz W., Nitschke W. Evolution and diversification of Group 1 [NiFe] hydrogenases. Is there a phylogenetic marker for O2-tolerance? Biochim. Biophys. Acta. 2012;1817:1565–1575. doi: 10.1016/j.bbabio.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 17.United States. Congress. House. Committee on Science. Subcommittee on Energy. Fueling the future: on the road to the hydrogen economy. 2006. Available from: http://purl.access.gpo.gov/GPO/LPS67640.
- 18.Lamle S.E., Vincent K.A., Halliwell L.M., Albracht S.P.J., Armstrong F.A. Hydrogenase on an electrode: a remarkable heterogeneous catalyst. Dalton Trans. 2003;21:4152–4157. doi: 10.1039/b306234c. [DOI] [Google Scholar]
- 19.Vincent K.A., Parkin A., Armstrong F.A. Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem. Rev. 2007;107:4366–4413. doi: 10.1021/cr050191u. [DOI] [PubMed] [Google Scholar]
- 20.Dubini A., Pye R.L., Jack R.L., Palmer T., Sargent F. How bacteria get energy from hydrogen: a genetic analysis of periplasmic hydrogen oxidation in Escherichia coli. Int. J. Hydrogen Energy. 2002;27:1413–1420. doi: 10.1016/S0360-3199(02)00112-X. [DOI] [Google Scholar]
- 21.Hirst J. Elucidating the mechanisms of coupled electron transfer and catalytic reactions by protein film voltammetry. Biochim. Biophys. Acta. 2006;1757:225–239. doi: 10.1016/j.bbabio.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Léger C., Bertrand P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem. Rev. 2008;108:2379–2438. doi: 10.1021/cr0680742. [DOI] [PubMed] [Google Scholar]
- 23.Gitlitz P.H., Krasna A.I. Structural and catalytic properties of hydrogenase from Chromatium. Biochemistry. 1975;14:2561–2568. doi: 10.1021/bi00683a001. [DOI] [PubMed] [Google Scholar]
- 24.Whiteley H.R., Ordal E.J. The reduction of methylene blue by hydrogenase. J. Bacteriol. 1955;70:608–613. doi: 10.1128/jb.70.5.608-613.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vincent K.A., Parkin A., Lenz O., Albracht S.P.J., Fontecilla-Camps J.C., Cammack R., Friedrich B., Armstrong F.A. Electrochemical definitions of O2 sensitivity and oxidative inactivation in hydrogenases. J. Am. Chem. Soc. 2005;127:18179–18189. doi: 10.1021/ja055160v. [DOI] [PubMed] [Google Scholar]
- 26.Lukey M.J., Parkin A., Roessler M.M., Murphy B.J., Harmer J., Palmer T., Sargent F., Armstrong F.A. How Escherichia coli is equipped to oxidize hydrogen under different redox conditions. J. Biol. Chem. 2010;285:3928–3938. doi: 10.1074/jbc.M109.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy B.J., Sargent F., Armstrong F.A. Transforming an oxygen-tolerant [NiFe] uptake hydrogenase into a proficient, reversible hydrogen producer. Energy Environ. Sci. 2014;7:1426. doi: 10.1039/c3ee43652g. [DOI] [Google Scholar]
- 28.Pandelia M.-E., Infossi P., Stein M., Giudici-Orticoni M.-T., Lubitz W. Spectroscopic characterization of the key catalytic intermediate Ni–C in the O2-tolerant [NiFe] hydrogenase I from Aquifex aeolicus: evidence of a weakly bound hydride. Chem. Commun. 2011;48:823–825. doi: 10.1039/C1CC16109A. [DOI] [PubMed] [Google Scholar]
- 29.Pandelia M.-E., Fourmond V., Tron-Infossi P., Lojou E., Bertrand P., Léger C., Giudici-Orticoni M.-T., Lubitz W. Membrane-bound hydrogenase I from the hyperthermophilic bacterium Aquifex aeolicus: enzyme activation, redox intermediates and oxygen tolerance. J. Am. Chem. Soc. 2010;132:6991–7004. doi: 10.1021/ja910838d. [DOI] [PubMed] [Google Scholar]
- 30.Saggu M., Zebger I., Ludwig M., Lenz O., Friedrich B., Hildebrandt P., Lendzian F. Spectroscopic insights into the oxygen-tolerant membrane-associated [NiFe] hydrogenase of Ralstonia eutropha H16. J. Biol. Chem. 2009;284:16264–16276. doi: 10.1074/jbc.M805690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hidalgo R., Ash P.A., Healy A.J., Vincent K.A. Infrared spectroscopy during electrocatalytic turnover reveals the Ni-L active site state during H2 oxidation by a NiFe hydrogenase. Angew. Chem. Int. Ed. 2015;54:7110–7113. doi: 10.1002/anie.201502338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisitruangsakul N., Lenz O., Ludwig M., Friedrich B., Lendzian F., Hildebrandt P., Zebger I. Monitoring catalysis of the membrane-bound hydrogenase from Ralstonia eutropha H16 by surface-enhanced IR absorption spectroscopy. Angew. Chem. Int. Ed. 2009;48:611–613. doi: 10.1002/anie.200802633. [DOI] [PubMed] [Google Scholar]
- 33.Ogata H., Nishikawa K., Lubitz W. Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase. Nature. 2015;520:571–574. doi: 10.1038/nature14110. [DOI] [PubMed] [Google Scholar]
- 34.Tai H., Nishikawa K., Inoue S., Higuchi Y., Hirota S. FT-IR characterization of the light-induced Ni-L2 and Ni-L3 states of [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F. J. Phys. Chem. B. 2015;119:13668–13674. doi: 10.1021/acs.jpcb.5b03075. [DOI] [PubMed] [Google Scholar]
- 35.Kellers P., Pandelia M.-E., Currell L.J., Görner H., Lubitz W. FTIR study on the light sensitivity of the [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F: Ni–C to Ni–L photoconversion, kinetics of proton rebinding and H/D isotope effect. Phys. Chem. Chem. Phys. 2009;11:8680–8683. doi: 10.1039/b913635e. [DOI] [PubMed] [Google Scholar]
- 36.Medina M., Williams R., Cammack R., Hatchikian E.C. Studies of light-induced nickel EPR signals in Desulfovibrio gigas hydrogenase. J. Chem. Soc. Faraday Trans. 1994;90:2921–2924. doi: 10.1039/ft9949002921. [DOI] [Google Scholar]
- 37.Jones A.K., Lamle S.E., Pershad H.R., Vincent K.A., Albracht S.P.J., Armstrong F.A. Enzyme electrokinetics: electrochemical studies of the anaerobic interconversions between active and inactive states of Allochromatium vinosum [NiFe]-hydrogenase. J. Am. Chem. Soc. 2003;125:8505–8514. doi: 10.1021/ja035296y. [DOI] [PubMed] [Google Scholar]
- 38.Hamdan A.A., Liebgott P.-P., Fourmond V., Gutiérrez-Sanz O., Lacey A.L.D., Infossi P., Rousset M., Dementin S., Léger C. Relation between anaerobic inactivation and oxygen tolerance in a large series of NiFe hydrogenase mutants. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19916–19921. doi: 10.1073/pnas.1212258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkin A., Sargent F. The hows and whys of aerobic H2 metabolism. Curr. Opin. Chem. Biol. 2012;16:26–34. doi: 10.1016/j.cbpa.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Cracknell J.A., Wait A.F., Lenz O., Friedrich B., Armstrong F.A. A kinetic and thermodynamic understanding of O2 tolerance in [NiFe]-hydrogenases. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20681–20686. doi: 10.1073/pnas.0905959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carepo M., Tierney D.L., Brondino C.D., Yang T.C., Pamplona A., Telser J., Moura I., Moura J.J.G., Hoffman B.M. 17O ENDOR detection of a solvent-derived Ni−(OHx)−Fe bridge that is lost upon activation of the hydrogenase from Desulfovibrio gigas. J. Am. Chem. Soc. 2002;124:281–286. doi: 10.1021/ja010204v. [DOI] [PubMed] [Google Scholar]
- 42.van der Zwaan J.W., Coremans J.M., Bouwens E.C., Albracht S.P. Effect of 17O2 and 13CO on EPR spectra of nickel in hydrogenase from Chromatium vinosum. Biochim. Biophys. Acta. 1990;1041:101–110. doi: 10.1016/0167-4838(90)90051-G. [DOI] [PubMed] [Google Scholar]
- 43.Abou Hamdan A., Burlat B., Gutiérrez-Sanz O., Liebgott P.-P., Baffert C., De Lacey A.L., Rousset M., Guigliarelli B., Léger C., Dementin S. O2-independent formation of the inactive states of NiFe hydrogenase. Nat. Chem. Biol. 2013;9:15–17. doi: 10.1038/nchembio.1110. [DOI] [PubMed] [Google Scholar]
- 44.Wulff P., Day C.C., Sargent F., Armstrong F.A. How oxygen reacts with oxygen-tolerant respiratory [NiFe]-hydrogenases. Proc. Natl. Acad. Sci. 2014;111:6606–6611. doi: 10.1073/pnas.1322393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fourmond V., Infossi P., Giudici-Orticoni M.-T., Bertrand P., Léger C. “Two-step” chronoamperometric method for studying the anaerobic inactivation of an oxygen tolerant NiFe hydrogenase. J. Am. Chem. Soc. 2010;132:4848–4857. doi: 10.1021/ja910685j. [DOI] [PubMed] [Google Scholar]
- 46.de Lacey A.L., Hatchikian E.C., Volbeda A., Frey M., Fontecilla-Camps J.C., Fernandez V.M. Infrared-spectroelectrochemical characterization of the [NiFe] hydrogenase of Desulfovibrio gigas. J. Am. Chem. Soc. 1997;119:7181–7189. doi: 10.1021/ja963802w. [DOI] [Google Scholar]
- 47.Chapman A., Cammack R., Hatchikian C.E., McCracken J., Peisach J. A pulsed EPR study of redox-dependent hyperfine interactions for the nickel centre of Desulfovibrio gigas hydrogenase. FEBS Lett. 1988;242:134–138. doi: 10.1016/0014-5793(88)81001-9. [DOI] [PubMed] [Google Scholar]
- 48.Volbeda A., Martin L., Cavazza C., Matho M., Faber B.W., Roseboom W., Albracht S.P.J., Garcin E., Rousset M., Fontecilla-Camps J.C. Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases. J. Biol. Inorg. Chem. 2005;10:239–249. doi: 10.1007/s00775-005-0632-x. [DOI] [PubMed] [Google Scholar]
- 49.Lamle S.E., Albracht S.P.J., Armstrong F.A. Electrochemical potential-step investigations of the aerobic interconversions of [NiFe]-hydrogenase from Allochromatium vinosum: insights into the puzzling difference between unready and ready oxidized inactive states. J. Am. Chem. Soc. 2004;126:14899–14909. doi: 10.1021/ja047939v. [DOI] [PubMed] [Google Scholar]
- 50.Volbeda A., Garcin E., Piras C., de Lacey A.L., Fernandez V.M., Hatchikian E.C., Frey M., Fontecilla-Camps J.C. Structure of the [NiFe] hydrogenase active site: evidence for biologically uncommon Fe ligands. J. Am. Chem. Soc. 1996;118:12989–12996. doi: 10.1021/ja962270g. [DOI] [Google Scholar]
- 51.Volbeda A., Charon M.H., Piras C., Hatchikian E.C., Frey M., Fontecilla-Camps J.C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 52.Ogata H., Hirota S., Nakahara A., Komori H., Shibata N., Kato T., Kano K., Higuchi Y. Activation process of [NiFe] hydrogenase elucidated by high-resolution X-ray analyses: conversion of the ready to the unready state. Structure. 2005;13:1635–1642. doi: 10.1016/j.str.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 53.Volbeda A., Martin L., Barbier E., Gutiérrez-Sanz O., Lacey A.L.D., Liebgott P.-P., Dementin S., Rousset M., Fontecilla-Camps J.C. Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states. J. Biol. Inorg. Chem. 2014;20:11–22. doi: 10.1007/s00775-014-1203-9. [DOI] [PubMed] [Google Scholar]
- 54.Volbeda A., Martin L., Liebgott P.-P., Lacey A.L.D., Fontecilla-Camps J.C. [NiFe]-hydrogenases revisited: nickel–carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states. Metallomics. 2015;7:710–718. doi: 10.1039/C4MT00309H. [DOI] [PubMed] [Google Scholar]
- 55.Barilone J.L., Ogata H., Lubitz W., van Gastel M. Structural differences between the active sites of the Ni-A and Ni-B states of the [NiFe] hydrogenase: an approach by quantum chemistry and single crystal ENDOR spectroscopy. Phys. Chem. Chem. Phys. 2015;17:16204–16212. doi: 10.1039/C5CP01322D. [DOI] [PubMed] [Google Scholar]
- 56.Butt J.N., Gates A.J., Marritt S.J., Richardson D.J. Enzyme film electrochemistry. In: Lewenstam A., Gorton L., editors. Electrochemical Processes in Biological Systems. Hoboken, NJ: John Wiley & Sons; 2015. [Google Scholar]
- 57.Pinske C., Krüger S., Soboh B., Ihling C., Kuhns M., Braussemann M., Jaroschinsky M., Sauer C., Sargent F., Sinz A., et al. Efficient electron transfer from hydrogen to benzyl viologen by the [NiFe]-hydrogenases of Escherichia coli is dependent on the coexpression of the iron–sulfur cluster-containing small subunit. Arch. Microbiol. 2011;193:893–903. doi: 10.1007/s00203-011-0726-5. [DOI] [PubMed] [Google Scholar]
- 58.Shomura Y., Yoon K.-S., Nishihara H., Higuchi Y. Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature. 2011;479:253–256. doi: 10.1038/nature10504. [DOI] [PubMed] [Google Scholar]
- 59.Fritsch J., Scheerer P., Frielingsdorf S., Kroschinsky S., Friedrich B., Lenz O., Spahn C.M.T. The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature. 2011;479:249–252. doi: 10.1038/nature10505. [DOI] [PubMed] [Google Scholar]
- 60.Frielingsdorf S., Fritsch J., Schmidt A., Hammer M., Löwenstein J., Siebert E., Pelmenschikov V., Jaenicke T., Kalms J., Rippers Y., et al. Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase. Nat. Chem. Biol. 2014;10:378–385. doi: 10.1038/nchembio.1500. [DOI] [PubMed] [Google Scholar]
- 61.Abou-Hamdan A., Ceccaldi P., Lebrette H., Gutiérrez-Sanz O., Richaud P., Cournac L., Guigliarelli B., Lacey A.L.D., Léger C., Volbeda A., et al. A threonine stabilizes the NiC and NiR catalytic intermediates of [NiFe]-hydrogenase. J. Biol. Chem. 2015;290:8550–8558. doi: 10.1074/jbc.M114.630491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogata H., Mizoguchi Y., Mizuno N., Miki K., Adachi S., Yasuoka N., Yagi T., Yamauchi O., Hirota S., Higuchi Y. Structural studies of the carbon monoxide complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: suggestion for the initial activation site for dihydrogen. J. Am. Chem. Soc. 2002;124:11628–11635. doi: 10.1021/ja012645k. [DOI] [PubMed] [Google Scholar]
- 63.Higuchi Y., Ogata H., Miki K., Yasuoka N., Yagi T. Removal of the bridging ligand atom at the Ni-Fe active site of [NiFe] hydrogenase upon reduction with H2, as revealed by X-ray structure analysis at 1.4 Å resolution. Structure. 1999;7:549–556. doi: 10.1016/S0969-2126(99)80071-9. [DOI] [PubMed] [Google Scholar]
- 64.Higuchi Y., Yagi T., Yasuoka N. Unusual ligand structure in Ni–Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis. Structure. 1997;5:1671–1680. doi: 10.1016/S0969-2126(97)00313-4. [DOI] [PubMed] [Google Scholar]
- 65.Volbeda A., Amara P., Iannello M., Lacey A.L.D., Cavazza C., Fontecilla-Camps J.C. Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase. Chem. Commun. 2013;49:7061–7063. doi: 10.1039/c3cc43619e. [DOI] [PubMed] [Google Scholar]
- 66.Garcin E., Vernede X., Hatchikian E.C., Volbeda A., Frey M., Fontecilla-Camps J.C. The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center. Structure. 1999;7:557–566. doi: 10.1016/S0969-2126(99)80072-0. [DOI] [PubMed] [Google Scholar]
- 67.Ogata H., Kellers P., Lubitz W. The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni-A State) J. Mol. Biol. 2010;402:428–444. doi: 10.1016/j.jmb.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 68.Volbeda A., Darnault C., Parkin A., Sargent F., Armstrong F.A., Fontecilla-Camps J.C. Crystal structure of the O2-tolerant membrane-bound hydrogenase 1 from Escherichia coli in complex with its cognate cytochrome b. Structure. 2013;21:184–190. doi: 10.1016/j.str.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Volbeda A., Amara P., Darnault C., Mouesca J.-M., Parkin A., Roessler M.M., Armstrong F.A., Fontecilla-Camps J.C. X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5305–5310. doi: 10.1073/pnas.1119806109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bowman L., Flanagan L., Fyfe P.K., Parkin A., Hunter W.N., Sargent F. How the structure of the large subunit controls function in an oxygen-tolerant [NiFe]-hydrogenase. Biochem. J. 2014;458:449–458. doi: 10.1042/BJ20131520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pandelia M.-E., Nitschke W., Infossi P., Giudici-Orticoni M.-T., Bill E., Lubitz W. Characterization of a unique [FeS] cluster in the electron transfer chain of the oxygen tolerant [NiFe] hydrogenase from Aquifex aeolicus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6097–6102. doi: 10.1073/pnas.1100610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goris T., Wait A.F., Saggu M., Fritsch J., Heidary N., Stein M., Zebger I., Lendzian F., Armstrong F.A., Friedrich B., et al. A unique iron-sulfur cluster is crucial for oxygen tolerance of a [NiFe]-hydrogenase. Nat. Chem. Biol. 2011;7:310–318. doi: 10.1038/nchembio.555. [DOI] [PubMed] [Google Scholar]
- 74.Roessler M.M., Evans R.M., Davies R.A., Harmer J., Armstrong F.A. EPR spectroscopic studies of the Fe–S clusters in the O2-tolerant [NiFe]-hydrogenase Hyd-1 from Escherichia coli and characterization of the unique [4Fe–3S] cluster by HYSCORE. J. Am. Chem. Soc. 2012;134:15581–15594. doi: 10.1021/ja307117y. [DOI] [PubMed] [Google Scholar]
- 75.Lukey M.J., Roessler M.M., Parkin A., Evans R.M., Davies R.A., Lenz O., Friedrich B., Sargent F., Armstrong F.A. Oxygen-tolerant [NiFe]-hydrogenases: the individual and collective importance of supernumerary cysteines at the proximal Fe-S cluster. J. Am. Chem. Soc. 2011;133:16881–16892. doi: 10.1021/ja205393w. [DOI] [PubMed] [Google Scholar]
- 76.Dance I. What is the trigger mechanism for the reversal of electron flow in oxygen-tolerant [NiFe] hydrogenases? Chem. Sci. 2015;6:1433–1443. doi: 10.1039/C4SC03223C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hexter S.V., Grey F., Happe T., Climent V., Armstrong F.A. Electrocatalytic mechanism of reversible hydrogen cycling by enzymes and distinctions between the major classes of hydrogenases. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11516–11521. doi: 10.1073/pnas.1204770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Evans R.M., Parkin A., Roessler M.M., Murphy B.J., Adamson H., Lukey M.J., Sargent F., Volbeda A., Fontecilla-Camps J.C., Armstrong F.A. Principles of sustained enzymatic hydrogen oxidation in the presence of oxygen–the crucial influence of high potential Fe–S clusters in the electron relay of [NiFe]-hydrogenases. J. Am. Chem. Soc. 2013;135:2694–2707. doi: 10.1021/ja311055d. [DOI] [PubMed] [Google Scholar]
- 79.Knuettel K., Schneider K., Erkens A., Plass W., Mueller A., Bill E., Trautwein A. Redox properties of the metal centers in the membrane-bound hydrogenase from Alcaligenes eutrophus CH34. Bull. Pol. Acad. Sci. Chem. 1994;42:495. [Google Scholar]
- 80.Dementin S., Belle V., Bertrand P., Guigliarelli B., Adryanczyk-Perrier G., De Lacey A.L., Fernandez V.M., Rousset M., Léger C. Changing the ligation of the distal [4Fe4S] cluster in NiFe hydrogenase impairs inter- and intramolecular electron transfers. J. Am. Chem. Soc. 2006;128:5209–5218. doi: 10.1021/ja060233b. [DOI] [PubMed] [Google Scholar]
- 81.Frielingsdorf S., Schubert T., Pohlmann A., Lenz O., Friedrich B. A trimeric supercomplex of the oxygen-tolerant membrane-bound [NiFe]-hydrogenase from Ralstonia eutropha H16. Biochemistry. 2011;50:10836–10843. doi: 10.1021/bi201594m. [DOI] [PubMed] [Google Scholar]
- 82.Radu V., Frielingsdorf S., Evans S.D., Lenz O., Jeuken L.J.C. Enhanced oxygen-tolerance of the full heterotrimeric membrane-bound [NiFe]-hydrogenase of Ralstonia eutropha. J. Am. Chem. Soc. 2014;136:8512–8515. doi: 10.1021/ja503138p. [DOI] [PMC free article] [PubMed] [Google Scholar]