Summary
Autophagy is originally described as the main catabolic pathway responsible for maintaining intracellular nutritional homeosta-sis that involves the formation of a unique vacuole, the autophago-some, and the interaction with the endosome-lysosome pathways. This conserved machinery plays a key role in immune-protection against different invaders, including pathogenic bacteria, intracellular parasites, and some viruses like herpes simplex and hepatitis C virus. Importantly, autophagy is linked to a number of human diseases and disorders including neurodegenerative disease, Crohn’s disease, type II diabetes, tumorigenesis, cardiomyopathy, and fatty liver disease. On the other hand, inflammasomes are multiprotein platforms stimulated upon several environmental conditions and microbial infection. Once assembled, the inflammasomes mediate the maturation of pro-inflammatory cytokines and promote phagosome-lysosome fusion to sustain an innate immune response. The intersections between autophagy and inflammasome have been observed in various diseases and microbial infections. This review highlights the molecular aspects involved in autophagy and inflammasome interactions during different medical conditions and microbial infections.
Keywords: monocytes/macrophages, bacteria, autophagy, Toll-like receptors/pattern recognition receptors, cell trafficking
Introduction
Inflammasome and autophagy are essential elements of the innate immune system, and their disruption have been implicated in specific infections and disease conditions. Inflammasomes are cytosolic multiprotein complexes that govern the maturation and secretion of select pro-inflammatory cytokines, such as IL-1β, IL-18, and IL-33 (1). Within this complex, cytosolic receptors of the NOD-like receptor (NLR) family (i.e. NLRP3 and NLRP1) interact with an adapter protein, apoptosis-associated speck like protein containing CARD (ASC), which recruits and activates the procaspase-1 by proteolytic cleavage (1). In turn, active cas-pase-1 activates interleukin-1β (IL-1β) and IL-18 that are subsequently released (2). The inflammasome induces and is induced by autophagy through direct interactions with autophagy proteins or through the effects of secondary molecules, such as mitochondrial reactive oxygen species and mitochondrial DNA.
Autophagy is a biological process characterized by self-digestion and involves induction of autophagosome formation, leading to degradation of autophagic cargo, and reuse of building blocks such as amino acids. Autophagy cooperates with apoptosis, inflammation, and adaptive immune system to orchestrate the cellular homeostasis and the appropriate immune response against endogenous or exogenous danger signals (3). Autophagy controls inflammation through interactions with innate immune pathways, by removing endogenous inflammasome components, and affects the secretion of immune mediators. Moreover, autophagy contributes to antigen presentation and T-cell homeostasis (4, 5). Besides maintaining cellular homeostasis, autophagy plays important roles in multiple biological processes including development, aging, and degeneration (6). The delicate interplay between autophagy and inflammasome to orchestrate the appropriate immune response and the impact of cross-talk between these pathways will be the focus of this review.
Autophagy: the devouring machinery and sometimes the packaging carrier
The formation of the autophagosome membrane requires a major regulatory complex that includes beclin-1 and Vps34, a Class III phosphatidylinositol-3-kinase (PIK3C3) (7). Subsequently, elongation of autophagosomes requires the activation of two ubiquitin-like conjugation systems (8). The ubiquitin-like protein Atg12 is conjugated to Atg5 by Atg7 and Atg10 enzymes. Then, the Atg5–Atg12 complex associates with Atg16L (the mammalian homolog of yeast Atg16). The resulting multimeric complex promotes the elongation of the autophagic membrane (8). A second conjugation system requires the ubiquitin-like protein, microtubule-associated protein-1 light chain 3 (LC3) (the mammalian homolog of yeast Atg8) (9). Then, Atg4B catalyzes the proteolytic processing the LC3 pro-form to generate the cleaved form LC3-1. Conjugation of LC3-I with PE is catalyzed by Atg7 and Atg3 activities (9). The conversion of LC3-I to LC3-II is considered the key regulatory step and indicator of autophagosome formation (10). Finally, the autophagosome matures and fuses with the lysosome, where enclosed cargoes are digested (11). Autophagy is a selective process since specific adapters such as p62 and neighbor of BRCA1 gene (NBR1), target ubiquitinated substrates for selective degradation (12). Additional proteins such as Rubicon, UV-RAG, and Bcl-2 family proteins interact with and influence the early steps of autophagosome formation (13). Interestingly, Dupont et al. (14) demonstrated that autophagy acts as a novel secretory pathway for IL-1β. Together, these findings demonstrate that autophagy is not only a devouring machinery but it also enables a subset of cytosolic proteins devoid of signal peptide sequences to exit from the cell (14) (Fig. 1).
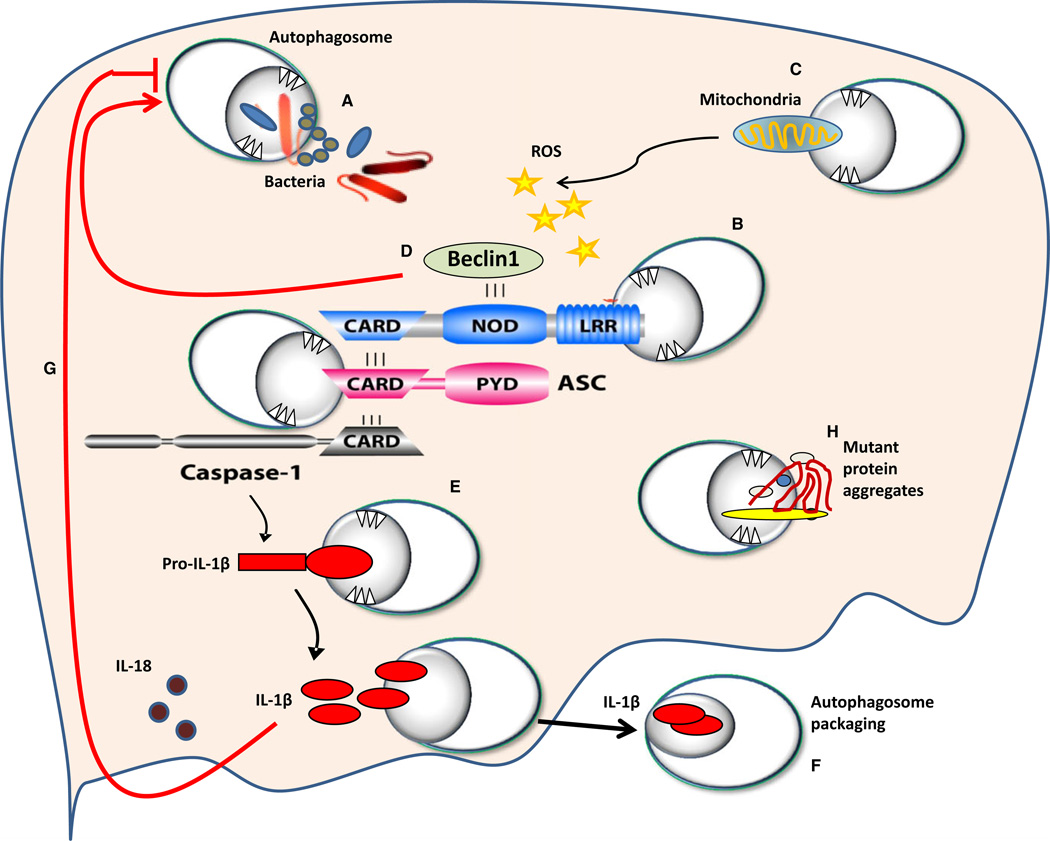
Fig. 1. Schematic representation of molecular interaction between autophagosomes and members of the inflammasome.
NOD-like receptors (NLRs) sense bacterial or viral molecules leading to the assembly of the inflammasome complex. (A) Autophagy can uptake certain invading organisms. The inflammasome complex is composed of the NLR, the adapter molecule ASC, and caspase-1 (B). Members of the inflammasome are ubiquitinated and then targeted by autophagosomes (B). The inflammasome can be activated by reactive oxygen species (ROS) released from mitochondria that were not cleared by autophagosomes (C). NLRs bind beclin-1, and upon their recruitment to the inflammasome complex, beclin-1 is released and contributes to the formation of autophagosomes (D). Autophagosomes also target pro-IL-1β to maintain its level under control (E). The inflammasome complex cleaves pro-IL-1β leading to active IL-1β. Active IL-1β is escorted outside the cell within autophagosomes (F). IL-1β inhibits autophagosome formation via unknown mechanisms (G). Mutant protein aggregates are targeted by autophagy to clear them from the cytosol; however, if they accumulate, autophagy molecules will be sequestered reducing their availability to form autophagosomes (H). Red arrows depict pathways by which the inflammasome contributes to the modulation of autophagy.
Cell sensors and autophagy
Toll-like receptors (TLRs) are cellular sensors for pathogen associated molecular patterns (PAMPs), and can regulate autophagy through the activation of signaling processes in macrophages and other cell types (15, 16). The TLRs reside either within the cell surface membrane (TLR1, TLR2, TLR4, TLR5, and TLR6) or in endosomal compartments (TLR3, TLR7, TLR8, and TLR9). Two different ligands of TLR7, single-stranded RNA (ssRNA) and the chemical compound imiquimod, induce autophagosome formation, characterized by LC3 puncta formation in murine macrophages (17). TLR2 stimulation by various pathogens such as Listeria monocytogenes, induces autophagy (18). Another study showed that Staphylococcus aureus-mediated stimulation of TLR2 in RAW 264.7 mouse macrophages induces phagocytosis and autophagy. In addition, the bacterial CpG motifs is recognized by TLR9, which can activate autophagy via the MAPK signaling pathway (19). Although still a matter of debate, reports suggest that bacterial LPS, a TLR4 ligand, could be implicated in stimulation of autophagy in cultured macrophage cell lines (20). NLRs are cytoplasmic members of the pattern recognition receptor family, and more than 20 NLRs have been identified in mammals. Many NLRs, such as NLRP1, NLRP3, and NLRC4, employ ASC to recruit procas-pase-1, but this does not apply to all inflammasomes. In mouse embryonic fibroblasts, NOD2 recruits Atg16L1 to the plasma membrane at the site of bacterial entry, in turn facilitating bacterial trafficking to the autophagosomes and fusion of the autophagosomes with lysosomes to promote bacterial clearance and antigen presentation via major histocompatibility complex class II (MHCII) (21). Another study using human DCs demonstrated that stimulation of NOD2 with muramyl dipeptide induces autophagosome formation and consequently enhances MHCII-associated antigen presentation (22). A recent study indicated that autophagy induced by inflammatory signals targets ubiquitinated inflammasomes, thereby limiting IL-1β production through inflammasome destruction (23, 24). Induction of absent in melanoma 2 (AIM2) or NLRP3 inflammasomes triggers nucleotide exchange on Ras-like small G protein, RalB, and autophagosome assembly to form isolation membranes (25). During autophagy, ubiquitinated assembled inflammasomes are engulfed through autophagic adaptors such as p62 (24). Thus, activation of inflammasomes can stimulate autophagosome formation. Conversely, NLRs also negatively regulate autophagy. NLRC4, NLRP3, NLRP4, and NLRP10 interact with beclin-1, and NLRP4 in particular has a strong affinity for beclin-1 (Fig. 1). Upon infection, NLRP4 transiently dissociates from beclin-1, enabling the initiation of beclin-1-mediated autophagy. Moreover, NLRP4 physically interacts with the class C vacuolar protein-sorting complex, resulting in inhibition of autophagosome and endosome maturation (26). Taken together, the available data indicate that homeostasis is maintained through reciprocal regulation of NLR activation and autophagy.
The interaction between the inflammasome and the autophagy machinery: a case of mutual regulation
The inhibition of inflammasome activity by autophagy was reported for the first time in 2008. A pioneer study by Saitoh et al. (27) revealed that ablation of Atg16L1, an autophagy protein that is fundamental for the autophagosome formation, potentiates the LPS-induced secretion of IL-1b and IL-18. Then several biologic and genetic studies on both human and mouse cells consistently reported the negative regulation of inflammasome by autophagy. Depletion of be-clin-1 and LC3B in mouse macrophages upregulates caspase-1 activation and secretion of mature IL-1b and IL-18 in response to LPS and ATP in an NLRP3-dependent manner (28). In addition, pharmacological inhibition of autophagy, with the PI3K inhibitors 3-methyl adenine (3-MA) or wort-mannin, augments LPS-induced inflammasome activation in mouse macrophages, dendritic cells, and human peripheral blood monocyte-derived macrophages (29). Induction of autophagy by IL-1α and IL-1β suggests a potential negative feedback loop for the control of inflammasome by inflammation (4, 30). Interestingly, very recent work demonstrated that blocking IL-1 receptor restores autophagy in autophagy-defective chronic granulomatous disease macrophages (31). Together, these data suggest that the autophagy machinery regulates the amount of IL-1β, which in turn inhibits autophagosome formation by a yet unresolved mechanism.
Induction of AIM2 or NLRP3 inflammasomes triggers nucleotide exchange on RalB and autophagosome assembly to form isolation membranes (32). During autophagy, ubiquitinated assembled inflammasomes are engulfed through autophagic adapters such as p62. Reciprocally, several NLRs could attenuate autophagy through binding to key autophagy proteins (22). Not only NOD2 has affinity to Atg16L (33) but also NLRC4, and NLRP4 could interact with Atg6 (be-clin-1) (26). Consequently, the detection of PAMPs with the NLRs results in release of free beclin-1, which could initiate autophagy. Autophagy contains the inflammatory response; the major example is that AIM2 and NLRP3 inflammasomes are sequestered in the autophagosome for degradation. The inflammasome components are delivered into the autophagosome in P62-dependent manner. It has also been reported that ASC aggregates are subjected to K63-linked ubiquitination, which is targeted by p62 into the autophagy pathway (32). Another study has reported that IL-1β but not caspase-1, was co-localized with LC3II in macrophages and dendritic cells, denoting that autophagosomes do not act as a platform for inflammasome recruitment and processing of IL-1β, but rather separate pro-IL-1β from caspase-1. This sequestration of pro-IL-1β by autophagosomes limits the available pro-IL-1β in the cytosol for activation by caspase-1, and this might explain why inhibition of autophagy leads to increased secretion of IL-1β (5). On the other hand, a recent study has reported that in human blood monocytes-derived macrophages, autophagy controls the expression of IL-1β on the transcriptional level but not on activation step (29, 34). This divergence could be explained partially by the different cell populations used, i.e., primary human monocytes in the studies by Crisan et al. and Plantinga et al. versus mouse macrophages in most other studies (29, 34). Collectively, autophagy is activated in response to inflammasome induction to contain the inflammation by physical destruction of inflammasome components.
Moreover, recent studies have shown that activation of NOD2 by muramyl dipeptide (MDP) induces autophago-some formation, which in turn enhances bacterial clearance (35). NOD2 stimulated with MDP induces autophagosome formation, which promotes MHCII-associated antigen presentation. Atg5, Atg7, Atg16L1, and receptor-interacting serine-threonine kinase-2 (RIPK2), the latter being one of the downstream effectors of the NOD2 signaling pathway, are required for autophagosome formation and antigen presentation by MDP (21, 36). Another study also showed that stimulation of NOD1 and NOD2 by bacterial peptidoglycans activates the autophagy pathway in mouse embryonic fibro-blasts (21). Upon bacterial invasion, NOD2 recruits Atg16L1 to the bacterial entry sites, facilitating bacterial trafficking to the autophagosomes. This, in turn, induces the fusion of the autophagosomes with the lysosomes to form the autophago-lysosomes and promotes antigen presentation. In mouse embryonic fibroblasts, this process does not require the adapter RIP2 nor the transcription factor NF-κB.
Bad house-keeping by autophagy leads to stimulation of the inflammasome: the role of mitochondrial reactive oxygen species and mitochondrial DNA
One of the fundamental house-keeping duties of autophagy pathway is to remove the damaged mitochondria (mitophagy) (37). Cells with genetically defective autophagy are characterized by abundance of damaged mitochondria and subsequently excessive amount of the mitochondrial reactive oxygen species (ROS) (28, 37) (Fig. 1). In addition, autophagy inhibitor (3-MA) induces the accumulation of damaged mitochondria and increased concentrations of mitochondrial ROS in monocytic cells in dose dependent manner (38, 39). On the other hand, robust mitochondrial ROS generation due to blocking of key enzymes in respiratory chain, is accompanied by increase in the NLRP3 inflam-masome-dependent mature IL-1β secretion (39). In turn, elevated levels of ROS are prerequisite for NLRP3 inflammasome activation with subsequent secretion of mature IL-1β and IL-18. Supporting this notion, the increase in caspase-1-mediated inflammatory response in the autophagy-defective macrophages was abolished by treatment with specific scavenger for mitochondrial ROS (40, 41). ROS are short-lived molecules and work only for very short distance, this requires the proximity of NLRP3 to mitochondria to interact with mitochondrial ROS. Interestingly, despite localization of NLRP3 to the endoplasmic reticulum in resting cells, the activation of inflammasome shifts the localization of NLRP3 to the mitochondria and mitochondria-associated ER membranes. The re-location of NLRP3 upon activation may be related to the redistribution of thioredoxin-interacting protein, which has been reported to be shuttled to the mitochondria upon oxidative stress (42).
ROS-induced NLRP3 activation was diminished by the knocking-down of voltage-dependent anion channel-1, which is tightly regulated by Bcl-2 (43). Intriguingly, mito-chondrial ROS have been reported to be necessary for induction of mitochondrial membrane permeability transition that make the mitochondrial inner membrane abruptly permeable to high molecular weight molecules such as mitochondrial DNA (44). Mitochondrial DNA (mtDNA) represents one of the damage-associated molecular patterns that could be detected by innate immune system using NLRs that alternatively sense pathogens producing inflammatory response. The induction of mitochondrial membrane permeability transition and subsequent release of mtDNA into cytosol has been found to be NLRP3-dependent and has been reported to be partially implicated in activation of cas-pase-1. Recently, NLRP3 has been reported to bind mtDNA causing its stabilization in the cytosol (45). Collectively, the disruption of mitochondrial quality control by defective autophagy is responsible for the induction of caspase-1 inflammatory response in NLRP3-dependent manner.
Cross-talk between autophagy and inflammasome in inflammatory disease conditions
Crohn’s disease
Crohn’s disease (CD) is a chronic inflammatory bowel disease that develops predominantly at terminal ileum and colon, where commensal bacteria intensively increase in mass. It is believed that CD results from robust increase in challenge by microbiota with an anomalous innate immune response (46). Three mutations (R702W, G908R, and L1007insC) within or near the leucine-rich repeats (LRR) domain of nucleotide oligomerization domain-containing protein 2 (NOD2) are associated with susceptibility to Crohn’s disease (47). NOD2 is an intracellular recognition receptor for bacterial peptidoglycan and is expressed in macrophages, dendritic cells, and intestinal epithelial cells. Mechanistic in vitro studies in cell lines and primary mono-cytes showed that the CD-associated NOD2 variants have a reduced capability of NF-κB activation and cytokine production in response to the bacterial cell wall molecule MDP (21, 34, 48). The role played by NOD2 in modulation of TLRs inflammatory signaling in intestinal phagocytic cells is conflicting because studies using human and mouse cells gave controversial results (49). Despite that mouse macrophages harbor L1007InsC NOD2 variant, displayed high IL-1β in response to MDP stimulation (50). The peripheral blood monocyte-derived macrophages isolated from patients with the same mutation exhibit defective IL-1β secretion in response to the same stressor (51). Interestingly, genome-wide association studies in CD have revealed the association of certain polymorphisms in two autophagy related genes, Atg16L1 (T300A) and IRGM, with the disease (52), several biochemical and genetic studies have investigated the mechanism lying behind the association between Atg16L1 and CD. A landmark study by Saitoh’s group has revealed that transgenic mice in which Atg16L1 gene deleted for the CCD (the Coiled Coil Domain) die within 1 day of birth, a phenomenon previously observed with the Atg5 knockout mice. Exposure of Atg16L1 ΔCCD macrophages to Escherichia coli elicited dramatically high IL-1β which is reminiscent of CD NOD2 variants, that also exhibit higher IL-1β in mouse models (53). Consistent with this, monocytes isolated from patients bearing the ATG16L1 Thr300Ala risk variant, which is shown to decrease ATG16L1 protein expression, display augmented secretion of IL-1β and IL-6, specifically in response to NOD2 ligands (54). A study by Travassos et al. (21) has provided a functional link between NOD2 and ATG16L1. The intracellular recognition receptor NOD2 directly interacts with ATG16L1 at the site of bacterial entry. In cells homozygous for the mutant NOD2, ATG16L1 fails to reach the plasma membrane, and consequently, the sequestration of invading bacteria by autophagosomes is compromised. Accordingly, the balance between the two actions employed by the NOD2, the recruitment of ATG16L1 to induce autophagy and induction of pro-inflammatory response via activation of NF-κB pathway, will be deviated in favor of NF-κB activation and IL-1β production in patients bearing the risk variant of ATG16L1 (55). One effect of the increased IL-1β is enhancement of the epithelial barrier permeability, which may increase the microbial products translocation (56). Polymorphisms in another autophagy gene, ULK1, are also associated with CD (57). This genetic evidence and other studies implicate autophagy in chronic inflammatory disease disorders.
Alzheimer’s disease
Alzheimer’s disease (AD) is the most common neurodegen-erative disease that causes long-term disruptions in the cognitive and intellectual capabilities. The histopathological indicators of AD are the accumulation of amyloid-β-containing neuritic plaques and intracellular tau protein tangles (58). A solid body of evidence has shown that neuronal autophagosome formation and lysosomal degradation is impaired in AD. It has been reported that the expression of beclin-1, a key autophagy protein, was markedly decreased in the brains of AD patients. In addition, the depletion of beclin-1 in cultured cells and transgenic mice exaggerates the deposition of amyloid-β peptides whereas its over expression diminishes the accumulation of amyloid-β (58).
The level of beclin-1 reduction was more prominent in the brains of AD compared with the patients suffering from mild cognitive impairment (59). The reduction in beclin-1 was localized into brain regions, which were most vulnerable to AD pathology. Several mechanisms may be implicated in decline of beclin-1 level in AD. The transcription and translation of beclin-1 have been shown to be decreased either via DNA methylation or microRNAs (miRNAs) (miR30a, miR376b) that target beclin-1 mRNA. However, the entire role of miRNAs in AD is still elusive (60–62). However, there is a mounting evidence that proteolysis of beclin-1 by caspases (caspase-3, −6, −8) is a key player in decreasing its level in AD. One of the caspases implicated in beclin-1 cleavage is caspase-6, an interesting non-executioner caspase, which can cleave the tau protein leading to the formation of neurofiberilly tangles, a major hallmark of AD (63). Of note, it is reported that caspase-1, the major inflammatory caspase, is upstream activator for caspase-6 in human neurons (64). This represents an interesting link between the inflammasome and autophagy in AD pathogen-esis. In addition, the formation of inhibitory complexes between beclin-1 and inflammasome proteins may be one of the mechanisms implicated in impaired autophagy in AD (Fig. 1). Many NLRs (NLRP3, NLRP4, NLRP10, NLRC4) could sequester beclin-1 and inhibit autophagocytosis. In particular, NLRP4 has a high affinity for beclin-1. The binding is mediated through the NACHT domain of NLR proteins to the evolutionarily conserved domain of beclin-1. The depletion of NLRC4 and NLRP4 markedly enhances the autophagocytosis in cell culture (26).
It has been demonstrated that the NLRP3 inflammasome is activated in response to the fibillary amyloid-β and consequently activates a caspase-1-mediated inflammatory response in microglia (65). Concurrently, it has been clearly shown that the expression of caspase-1 and IL-18 was increased in the brains of AD patients (66). Intriguingly, the knocking-down of Nlrp3 in transgenic mouse model of AD (APP/PS1) significantly reduced the deposition of amyloid-β aggregates and improved their memory (65). Remarkably, the phagocytosis capacity of the microglia from APP/PS1 mice deficient in Nlrp3 or Casp1 was significantly enhanced as compared to their wildtype. Collectively, this denotes that the recruitment of NLRP3 inflammasomes with subsequent secretion of inflammatory cytokines suppress the microglial capacity to phagocytose amyloid-β fibrils (58, 67). These findings represent models of interplay between inflammasome and autophagy in AD pathogenesis.
Atherosclerosis
Atherosclerosis is characterized by chronic inflammation within the atherosclerotic plaques. Autophagy becomes dysfunctional in atherosclerosis and its deficiency promotes atherosclerosis in part through inflammasome hyperactivation. Atherosclerotic blood vessels have elevated levels of p62, suggesting that dysfunctional autophagy is characteristic of plaques. Atg5 knockout mice had increased plaques, suggesting an essential role for basal levels of autophagy in athero-protection. Defective autophagy is associated with proatherogenic inflammasome activation. Classic inflammasome markers were robustly induced in Atg5-null animals, in response to cholesterol crystals. Conversely, cholesterol crystals are increased in Atg5-null plaques, suggesting a potentially vicious cycle of crystal formation and inflammasome activation in autophagy-deficient plaques (68).
Cystic fibrosis
Cystic fibrosis (CF) is the most common hereditary lethal disease in Caucasians (69). CF patients are characterized by mutations in the gene encoding the cystic fibrosis trans-membrane conductance regulator (CFTR), which leads to the accumulation of misfolded proteins in the macrophages and airway epithelial cells (70). Autophagy contributes to the selective clearance of aggregated and denatured protein, a process termed ‘aggrephagy’. Macrophages and epithelial cells from CF patients, which bear the mutation in the CFTR gene, have an impaired autophagic response, reduced autophagosome formation, and accumulation of p62 (71). Cells with CFTR mutation also displayed an abnormal accumulation of polyubiquitinated protein aggregate. Mutations in CFTR were associated with elevated ROS production and increased tissue transglutaminase 2 (TG2) levels (72). Defective autophagy associated with CF led to overproduction of inflammatory cytokines and failure to clear specific infections such as Burkholderia cenocepacia, Pseudomonas aeruginosa, and Staphylococcus aureus (71, 73–75). Notably, these organisms that are cleared by autophagy in healthy macrophages.
Sepsis
Sepsis remains a leading cause of mortality in intensive care units. This condition arises as a consequence of systemic responses to inflammation caused by acquired bacterial, fungal, parasitic, or viral infections and may lead to multiple organ failure (76). Marked autophagosome accumulation has been observed in the livers of patients who die from sepsis (77). However, it currently remains unclear whether this observation represents increased autophagic activity (flux) in sepsis patients or inhibition of autophagic processing which leads to the inappropriate accumulation of autophagosomes. Genetic deletion of critical autophagic proteins has recently been shown to increase sepsis-induced inflammatory responses in mice subjected to the cecal-ligation and puncture model of polymicrobial sepsis and LPS (28). Furthermore, BECN1+/− mice and LC3B−/− mice were found to be susceptible to the lethal effects septic shock in mice, and to express higher levels of IL-18 in the plasma, one of the inflammasome-associated cytokines (28). Collectively, these studies suggest a potential link between autophagy and inflammatory responses during the pathogenesis of sepsis.
The interplay between autophagy and the inflammasome during infection
Mycobacteria tuberculosis
Immune response to M. tuberculosis is Th1-biased and is characterized by release of interferon-γ (IFN-γ), tumor necrosis factor-a (TNF-α), and IL-12, while IL-4, a Th2 cytokine, is linked to the disease severity (4, 5). Autophagy denotes the divergence of Th1/Th2 T-cell response, as it is induced by Th1 cytokines, and repressed by Th2 cytokines (78). Autophagy plays pivotal roles in the host defense against M. tuberculosis. Notably, autophagy induction is associated with diminished M. tuberculosis replication in several models (79). IFN-γ, a key player in immune response to M. tuberculosis, exerts its protective effect partially by induction of autophagic pathway (79–81). In mouse macrophages, the immunological induction of autophagy, by IFN-γ, is mediated by the p47 guanosine triphosphatase IRGM-1 (82). Diminished levels of IRGM1 have been linked to disrupted autophagy (83). Yet, the role of IRGM-1 in autophagy is not fully understood, but recently, IRGM-1 has been reported to translocate to mitochondria, where it controls autophagy together with mitochondrial fission (84). Interestingly, single nucleotide polymorphism in IRGM-1 has been reported to be associated with susceptibility to M. tuberculosis infection, confirming the crucial role of autophagy in containing M. tuberculosis infection (79). In addition, M. tuberculosis peptidoglycan efficiently activates NOD2 intracellular recognition receptor with subsequent NF-κB activation (85). NOD2 knockout mice exert low production of pro-inflammatory cytokines and nitric oxide in response to M. tuberculosis infection (86). Notably, NF-κB activation with subsequent cytokine production is not the sole effect of NOD2 activation, as it also has the ability to recruit ATG16L1 to plasma membrane at the site of bacterial entry causing autophagosome formation representing another mechanism for autophagy induction in response to M. tuberculosis infection (87). Besides being an antibacterial mechanism for the eradication of M. tuberculosis intracellular infection, autophagy can also limit the secretion of pro-inflammatory cytokine to avoid excessive inflammation (88). On the one hand, infection of Atg5-deficient mice with M. tuberculosis was associated with increased bacterial count and excessive pulmonary inflammation compared with the wildtype animals. This effect was independent of the inflammasome (89). On the other hand, in human peripheral monocyte-derived macrophages, autophagy stimulation by starvation or IFN-γ has exerted differential effect on cytokine secretion in response to M. tuberculosis infection. It increases TNF-α and decreases IL-1β levels and this effect occurs on transcriptional level and is inflammasome-independent (90). Collectively, during M. tuberculosis infection, autophagy acts as an anti-mycobacterial mechanism for the eradication of intracellular bacteria, besides controlling the secretion of proinflammatory cytokine such as IL-1β, to avoid excessive inflammation.
Shigella flexneri
Shigella are highly adapted intracellular pathogens that cause bacillary dysentery (shigellosis). The distinct pathogenic character of Shigella is their ability to infect a variety of host cells, including epithelial cells, and macrophages, leading to excessive inflammation in intestinal tissues. Shigella-infected macrophages undergo caspase-1-mediated cell death, termed pyroptosis (91). Suzuki et al. (91) have reported that infection of mouse bone marrow-derived macrophages with Shi-gella induces caspase-1 activation in Nlrc4 and Asc-dependent manner with subsequent IL-1β processing and pyroptosis. Strikingly, unlike Salmonella and Legionella, Shigella-induced Nlrc4 activation is flagellin-independent (92–95). During multiplication, Shigella can escape from phagosomes into the cytoplasm, disseminating into neighboring cells. Fragments of phagosomes ruptured by Shigella in epithelial cells are studded by components of both the autophagy and inflammasome machineries (96). In epithelial cells, Shigella can avoid autophagy by secreting IcsB, which competitively inhibits the interaction of host Atg5 with bacterial surface protein VirG. Besides being necessary for intracellular actin-based motility, VirG also acts as molecular target of host autophagy (97). Contrasting to that observed in epithelial cells, Suzuki et al. have reported that VirG is dispensable for Shigella-induced autophagy in bone marrow macrophages. In addition, autophagy triggered by Shigella is markedly enhanced in caspase-1-deficient and Nlrc4-deficient but not Asc-deficient bone marrow-derived macrophages with marked resistance to pyroptotic cell death (98). In their study, the exact mechanism by which Nlrc4 and caspase-1 inhibit autophagy was not resolved. However, they proposed that active caspase-1 degrades some bacterial factors that are essential for the induction of autophagic pathway. Their rationale was that caspase-1 and Nlrc4 knockout cells showed activation of autophagy comparable to wildtype in response to rapamycin and amino acid starvation, suggesting that the autophagic machinery is intact and efficiently responds to physiological or pharmacological stresses (98). Interestingly, a recent report demonstrated that NLRC4 has inhibitory effects on autophagy as it interacts with beclin-1 via NACHT domain and consequently inhibits the autopha-gic initiation (26) (Fig. 1). This may partially explain the above-mentioned findings by Suzuki and coworkers.
Legionella pneumophila
Legionella pneumophila is an aerobic Gram-negative intracellular opportunistic organism. It is the causative agent of a specific kind of pneumonia called Legionnaires’ disease. Although human phagocytic cells are susceptible to Legionella infection, wildtype mouse macrophages are resistant (99–101). Interestingly, wildtype mouse (C57BL/6J) macrophages detect the ubiquitinated Legionella vacuole bound to the adapter molecule p62 (102). The bacterial protein LegA9 seems to promote the recognition of the Legionella-containing vacuole by the ubiquitination machinery (102). Thus, once tagged by ubiquitin, the Legionella vacuole is targeted by autophagy for delivery to the lysosome for degradation. On the other hand, bacterial flagellin monomers which leak into the cytoplasm through the bacterial type IV secretion system are recognized by Nlrc4 (100, 103). During physiological level of infection, bacterial flagellin detection results in the assembly of Nlrc4 inflammasome inducing caspase-1 activation with subsequent lysosome-phagosome fusion (95). If caspase-1 activation is excessive, pyroptosis occurs and the cell containing several Legionellae succumbs to death, preventing further propagation of infection. Given that in resting macrophages Nlrc4 is linked to beclin-1 repressing auto-phagy, the infection of wildtype mouse macrophages with Legionella infection activates the Nlrc4 and consequently alleviates its repression on beclin-1 and this induces autophagy. The autophagosome then sequesters and disposes of the pathogen by fusion with lysosomes (6). Recent reports have revealed that Nlrc4 and procaspase-1 but not caspase-1 activity are crucial for modulation of autophagy in bone marrow-derived macrophages. Pro-caspase-1 was shown to play a role in robust autophagy induction in response not only to Legionella infection but also to amino acid starvation and potassium efflux. However, the exact mechanism by which procaspase-1 modulates autophagy is unknown. The induction of autophagy protects macrophages against the pyroptosis. However, when the infection is exceeding the capacity of autophagy for eliminating the pathogen, the inflammatory pathway proceeds and the cells commit pyroptosis to eliminate the pathogen and recruit more leukocytes (104).
Influenza
The innate immune system is able to elicit an inflammatory response to influenza A virus (IAV) infection; however, the efficient clearance of infection necessitates the coordination of both innate and adaptive immunity. Several NLRs have been reported to be implicated in the innate immune response to influenza A virus infection. Primarily, N LRP3 has been reported to be activated by IAV with consequent caspase-1 and IL-1β activation. Yet the exact mechanism by which IAV activates NLRP3 is a matter of debate. The leakage of ROS from mitochondria has been reported to be crucial for the IAV mediated NLRP3 activation, which is reminiscent with the role of mitophagy in inflammasome activation. Interestingly, a recent study has shown that NOD2 was activated by IAV resulting in binding to the adapter RIPK2. The interaction between NOD2 and RIPK2 induces autophagy, specifically mitophagy, instead of activation of NF-κB pathway as in bacterial infection. Thus, NOD2 activation by IAV may strikingly mitigate the inflammation indirectly by controlling the ROS available for NLRP3 activation (105, 106).
Acknowledgements
Studies in Dr. Amer’s laboratory are supported by The Cystic Fibrosis Foundation, The Ohio State University Center for Clinical and Translational Science Longitudinal Pilot Award (CCTS) and Public Health Preparedness for Infectious Diseases (PHPID). HK is supported by Egyptian Science and Technology Development Fund (STDF) through project ID 6117.
References
- 1.Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 2.Fantuzzi G, Dinarello CA. Interleukin-18 and interleukin-1 beta: two cytokine substrates for ICE (caspase-1) J Clin Immunol. 1999;19:1–11. doi: 10.1023/a:1020506300324. [DOI] [PubMed] [Google Scholar]
- 3.Choi AJ, Ryter SW. Autophagy in inflammatory diseases. Int J Cell Biol. 2011;2011:732798. doi: 10.1155/2011/732798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris J. Autophagy and IL-1 family cytokines. Front Immunol. 2013;4:83. doi: 10.3389/fimmu.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris J, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 9.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 13.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 14.Dupont N, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 17.De Meyer I, et al. Toll-like receptor 7 stimulation by imiquimod induces macrophage autophagy and inflammation in atherosclerotic plaques. Basic Res Cardiol. 2012;107:269. doi: 10.1007/s00395-012-0269-1. [DOI] [PubMed] [Google Scholar]
- 18.Into T, Inomata M, Takayama E, Takigawa T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal. 2012;24:1150–1162. doi: 10.1016/j.cellsig.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Oka T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delgado M, et al. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev. 2009;227:189–202. doi: 10.1111/j.1600-065X.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 22.Oh JE, Lee HK. Pattern recognition receptors and autophagy. Front Immunol. 2014;5:300. doi: 10.3389/fimmu.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont N, Temime-Smaali N, Lafont F. How ubiquitination and autophagy participate in the regulation of the cell response to bacterial infection. Biol Cell. 2010;102:621–634. doi: 10.1042/BC20100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang LJ, et al. The microtubule-associated protein, EB1, links AIM2 inflammasomes with autophagy-dependent secretion. J Biol Chem. 2014;289:29322–29333. doi: 10.1074/jbc.M114.559153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jounai N, et al. NLRP4 negatively regulates autophagic processes through an association with beclin-1. J Immunol. 2011;186:1646–1655. doi: 10.4049/jimmunol.1001654. [DOI] [PubMed] [Google Scholar]
- 27.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crisan TO, et al. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PLoS ONE. 2011;6:e18666. doi: 10.1371/journal.pone.0018666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi CS, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy. 2010;6:986–987. doi: 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Luca A, et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci USA. 2014;111:3526–3531. doi: 10.1073/pnas.1322831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodemann BO, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 34.Plantinga TS, Joosten LA, van der Meer JW, Netea MG. Modulation of inflammation by autophagy: consequences for Crohn’s disease. Curr Opin Pharmacol. 2012;12:497–502. doi: 10.1016/j.coph.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Yano T, Kurata S. Induction of autophagy via innate bacterial recognition. Autophagy. 2008;4:958–960. doi: 10.4161/auto.6802. [DOI] [PubMed] [Google Scholar]
- 36.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 37.Tal MC, et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Q, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 40.Jiang B, et al. ATP-binding domain of heat shock protein 70 is essential for its effects on the inhibition of the release of the second mitochondria-derived activator of caspase and apoptosis in C2C12 cells. FEBS J. 2009;276:2615–2624. doi: 10.1111/j.1742-4658.2009.06989.x. [DOI] [PubMed] [Google Scholar]
- 41.Trnka J, Blaikie FH, Logan A, Smith RA, Murphy MP. Antioxidant properties of MitoTEMPOL and its hydroxylamine. Free Radic Res. 2009;43:4–12. doi: 10.1080/10715760802582183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxena G, Chen J, Shalev A. Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. J Biol Chem. 2010;285:3997–4005. doi: 10.1074/jbc.M109.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colombini M. VDAC: the channel at the interface between mitochondria and the cytosol. Mol Cell Biochem. 2004;256–257:107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 44.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 45.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 48.Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. 2011;32:171–179. doi: 10.1016/j.it.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe T, et al. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity. 2006;25:473–485. doi: 10.1016/j.immuni.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 50.Maeda S, et al. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–738. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 51.Netea MG, et al. NOD2 3020insC mutation and the pathogenesis of Crohn’s disease: impaired IL-1beta production points to a loss-of-function phenotype. Neth J Med. 2005;63:305–308. [PubMed] [Google Scholar]
- 52.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 53.Tsujikawa T, et al. Novel single-balloon enteroscopy for diagnosis and treatment of the small intestine: preliminary experiences. Endoscopy. 2008;40:11–15. doi: 10.1055/s-2007-966976. [DOI] [PubMed] [Google Scholar]
- 54.Plantinga TS, et al. Crohn’s disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–1235. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 55.Plantinga TS, Joosten LA, Netea MG. ATG16L1 polymorphisms are associated with NOD2-induced hyperinflammation. Autophagy. 2011;7:1074–1075. doi: 10.4161/auto.7.9.15867. [DOI] [PubMed] [Google Scholar]
- 56.Al-Sadi R, Ye D, Dokladny K, Ma TY. Mechanism of IL-1beta-induced increase in intestinal epithelial tight junction permeability. J Immunol. 2008;180:5653–5661. doi: 10.4049/jimmunol.180.8.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henckaerts L, et al. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm Bowel Dis. 2011;17:1392–1397. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 58.Salminen A, et al. Impaired autophagy and APP processing in Alzheimer’s disease: the potential role of Beclin 1 interactome. Prog Neurobiol. 2013;106–107:33–54. doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, et al. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer. 2010;10:98. doi: 10.1186/1471-2407-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou Z, et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287:4148–4156. doi: 10.1074/jbc.M111.307405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8:165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 63.Horowitz PM, et al. Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer’s disease. J Neurosci. 2004;24:7895–7902. doi: 10.1523/JNEUROSCI.1988-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo H, et al. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13:285–292. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- 65.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ojala J, et al. Expression of interleukin-18 is increased in the brains of Alzheimer’s disease patients. Neurobiol Aging. 2009;30:198–209. doi: 10.1016/j.neurobiolaging.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Salminen A, Kauppinen A, Suuronen T, Kaarniranta K, Ojala J. ER stress in Alzheimer’s disease: a novel neuronal trigger for inflammation and Alzheimer’s pathology. J Neuroinflammation. 2009;6:41. doi: 10.1186/1742-2094-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Razani B, et al. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15:534–544. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kerem E, et al. The relation between genotype and phenotype in cystic fibrosis-analysis of the most common mutation (delta F508) N Engl J Med. 1990;323:1517–1522. doi: 10.1056/NEJM199011293232203. [DOI] [PubMed] [Google Scholar]
- 70.Luciani A, et al. Cystic fibrosis: a disorder with defective autophagy. Autophagy. 2011;7:104–106. doi: 10.4161/auto.7.1.13987. [DOI] [PubMed] [Google Scholar]
- 71.Abdulrahman BA, et al. Depletion of the ubiquitin-binding adaptor molecule SQSTM1/ p62 from macrophages harboring cftr DeltaF508 mutation improves the delivery of Burkholderia cenocepacia to the autophagic machinery. J Biol Chem. 2013;288:2049–2058. doi: 10.1074/jbc.M112.411728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luciani A, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12:863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 73.Abdulrahman BA, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy. 2011;7:1359–1370. doi: 10.4161/auto.7.11.17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Assani K, Tazi MF, Amer AO, Kopp BT. IFN-gamma stimulates autophagy-mediated clearance of Burkholderia cenocepacia in human cystic fibrosis macrophages. PLoS ONE. 2014;9:e96681. doi: 10.1371/journal.pone.0096681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kopp BT, et al. Exaggerated inflammatory responses mediated by Burkholderia cenocepacia in human macrophages derived from Cystic fibrosis patients. Biochem Biophys Res Commun. 2012;424:221–227. doi: 10.1016/j.bbrc.2012.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balk RA. Severe sepsis septic shock. Definitions, epidemiology, and clinical manifestations. Crit Care Clin. 2000;16:179–192. doi: 10.1016/s0749-0704(05)70106-8. [DOI] [PubMed] [Google Scholar]
- 77.Watanabe E, et al. Sepsis induces extensive autophagic vacuolization in hepatocytes: a clinical and laboratory-based study. Lab Invest. 2009;89:549–561. doi: 10.1038/labinvest.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 80.Ponpuak M, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ponpuak M, Delgado MA, Elmaoued RA, Deretic V. Monitoring autophagy during Mycobacterium tuberculosis infection. Methods Enzymol. 2009;452:345–361. doi: 10.1016/S0076-6879(08)03621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 83.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 84.Singh SB, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coulombe F, et al. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J Exp Med. 2009;206:1709–1716. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jo EK. Innate immunity to mycobacteria: vitamin D and autophagy. Cell Microbiol. 2010;12:1026–1035. doi: 10.1111/j.1462-5822.2010.01491.x. [DOI] [PubMed] [Google Scholar]
- 87.Cheallaigh CN, et al. Interferon gamma release assays for the diagnosis of latent TB infection in HIV-infected individuals in a low TB burden country. PLoS ONE. 2013;8:e53330. doi: 10.1371/journal.pone.0053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li JC, et al. HIV-1 trans-activator protein dysregulates IFN-gamma signaling and contributes to the suppression of autophagy induction. AIDS. 2011;25:15–25. doi: 10.1097/QAD.0b013e328340fd61. [DOI] [PubMed] [Google Scholar]
- 89.Castillo EF, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA. 2012;109:E3168–E3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kleinnijenhuis J, et al. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response. Immunology. 2011;134:341–348. doi: 10.1111/j.1365-2567.2011.03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pereira MS, et al. Activation of NLRC4 by flagellated bacteria triggers caspase-1-dependent and -independent responses to restrict Legionella pneumophila replication in macrophages and in vivo. J Immunol. 2011;187:6447–6455. doi: 10.4049/jimmunol.1003784. [DOI] [PubMed] [Google Scholar]
- 93.Sauer JD, et al. Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci USA. 2011;108:12419–12424. doi: 10.1073/pnas.1019041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245 . doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abdelaziz DH, Amr K, Amer AO. Nlrc4/Ipaf/ CLAN/CARD12: more than a flagellin sensor. Int J Biochem Cell Biol. 2010;42:789–791. doi: 10.1016/j.biocel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dupont N, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 97.Ogawa M, Sasakawa C. Intracellular survival of Shigella. Cell Microbiol. 2006;8:177–184. doi: 10.1111/j.1462-5822.2005.00652.x. [DOI] [PubMed] [Google Scholar]
- 98.Suzuki T, Nunez G. A role for Nod-like receptors in autophagy induced by Shigella infection. Autophagy. 2008;4:73–75. doi: 10.4161/auto.5101. [DOI] [PubMed] [Google Scholar]
- 99.Khweek AA, Amer A. Replication of legionella pneumophila in human cells: why are we susceptible? Front Microbiol. 2010;1:133. doi: 10.3389/fmicb.2010.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 101.Abdelaziz DH, et al. Apoptosis-associated specklike protein (ASC) controls Legionella pneumophila infection in human monocytes. J Biol Chem. 2011;286:3203–3208. doi: 10.1074/jbc.M110.197681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khweek AA, et al. A bacterial protein promotes the recognition of the Legionella pneumophila vacuole by autophagy. Eur J Immunol. 2013;43:1333–1344. doi: 10.1002/eji.201242835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 104.Amer AO. Modulation of caspases and their non-apoptotic functions by Legionella pneumophila. Cell Microbiol. 2010;12:140–147. doi: 10.1111/j.1462-5822.2009.01401.x. [DOI] [PubMed] [Google Scholar]
- 105.Lupfer C, Kanneganti TD. The expanding role of NLRs in antiviral immunity. Immunol Rev. 2013;255:13–24. doi: 10.1111/imr.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lupfer C, Thomas PG, Kanneganti TD. Nucleotide oligomerization and binding domain 2-dependent dendritic cell activation is necessary for innate immunity and optimal CD8+ T Cell responses to influenza A virus infection. J Virol. 2014;88:8946–8955. doi: 10.1128/JVI.01110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]