Abstract
MicroRNAs are known to regulate critical developmental stages during embryogenesis. Here, we defined an Etv2-miR-130a cascade that regulates mesodermal specification and determination. Ablation of Dicer in the Etv2-expressing precursors resulted in altered mesodermal lineages and embryonic lethality. We identified miR-130a as a direct target of Etv2 and demonstrated its role in the segregation of bipotent hemato-endothelial progenitors towards the endothelial lineage. Gain-of-function experiments demonstrated that miR-130a promoted the endothelial program at the expense of the cardiac program without impacting the hematopoietic lineages. In contrast, CRISPR/Cas9-mediated knockout of miR-130a demonstrated a reduction of the endothelial program without affecting hematopoiesis. Mechanistically, miR-130a directly suppresses Pdgfra expression and promotes the endothelial program by blocking Pdgfra signaling. Inhibition or activation of Pdgfra signaling phenocopied the miR-130a over-expression and knockout, phenotypes, respectively. In summary, we report the function of a miRNA that specifically promotes the divergence of the hemato-endothelial progenitor to the endothelial lineage.
Keywords: Lineage specification, Dicer, microRNA, Etv2, miR-130a
Graphical Abstract
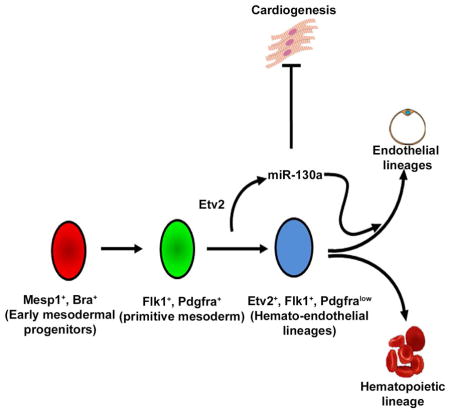
Introduction
During embryogenesis mesodermal progenitors give rise to multiple lineages including hemato-endothelial and cardiac lineages. For example, Flk1−/Pdgfra+ (paraxial mesoderm) and Flk1+/Pdgfra− (lateral plate mesoderm) cells arise from the Flk1+/Pdgfra+ unpatterned mesoderm (Kataoka et al., 2011; Sakurai et al., 2006). These lineages respond to distinct transcriptional networks and signaling cues. Precise control of the specification of these lineages is necessary for proper development and embryogenesis. The transcriptional regulators and signaling pathways that govern these mesodermal progenitors are incompletely defined (Kattman et al., 2006; Loebel et al., 2003).
Studies have established a hierarchy of transcriptional regulators including Mesp1, Vegf/Flk1 and Etv2 as modulators of hemato-endothelial development (Shalaby et al., 1995; Saga et al., 1999; Ferdous et al., 2009). Ablation of Etv2 results in embryonic lethality by E9.5 with complete absence of the hemato-endothelial lineages (Ferdous et al., 2009; Koyano-Nakagawa et al., 2012). Etv2 serves as a key regulator of hemato-endothelial lineages through its interaction with multiple factors including: Tie2, Lmo2, Gata2 and FoxC2 (De Val et al., 2008; Rasmussen et al., 2011; Koyano-Nakagawa et al., 2012; Shi et al., 2014). While the transcriptional hierarchy in hemato-endothelial development has been well described, the role of miRNAs in these progenitors are unknown.
MicroRNAs (miRNAs) govern the molecular switch by suppressing gene expression, thereby modulating and fine-tuning cell fate decisions (Ivey and Srivastava, 2010). Although global deletion as well as hypomorphic mutants of Dicer (miRNA processing enzyme) results in embryonic lethality (Bernstein et al., 2003; Yang et al., 2005), it is unclear whether Dicer and miRNAs play any role in the hemato-endothelial segregation and vascular develoment.
In the present study, we deciphered the requirement of miRNA biogenesis in early mesodermal precursors. We discovered a Etv2-miR-130a-Pdgfra network that directs the hemato-endothelial progenitors towards the endothelial fate without affecting the hematopoietic lineage. These findings define a factor that directs endothelial development without affecting the hematopoietic lineage.
Results
Etv2-Cre-mediated Dicer deletion results in altered mesodermal lineages and embryonic lethality
Analysis of mesodermal transcripts during embryonic stem cell/embryoid body (ES/EB) differentiation indicated that Mesp1 was transiently, but robustly expressed at day (d) 3, of differentiation with subsequent expression of both Flk1 and Etv2 at d4, marking the appearance of the mesodermal lineages (Figure S1A). To evaluate the functional role of Dicer (miRNA-dependent and -independent) in the mesodermal progenitors, we conditionally deleted floxed-Dicer using Cre recombinase under the control of either Mesp1, Flk1 and Etv2 promoter elements. qPCR analysis revealed efficient deletion of the Dicer allele from the respective FACS-sorted cells (Figure S1B). Whole mount analysis revealed embryonic lethality in Mesp1-Cre;DicerL/L embryos by E10.5; and the Flk1-Cre;DicerL/L and the Etv2-Cre;DicerL/L embryos were lethal by E12.5 due to vascular defects (Figure S1C, Table S1). These results indicated an essential requirement for miRNAs in the endothelial lineage. Importantly, Tie2-Cre;DicerL/L or VE-Cad-Cre;DicerL/L conditional mutants were viable with no obvious vascular defects (Suarez et al., 2007). Based on our findings and these reports, we hypothesized that miRNAs induced during Flk1 and Etv2 expression are essential for endothelial development.
Previously, we reported that Etv2-mutants have altered mesodermal populations (Rasmussen et al., 2011). To evaluate whether mesodermal derivatives were affected in Etv2-Cre;DicerL/L mutants, we crossed Etv2-Cre;DicerL/+ and Etv2-EYFP;DicerL/L lines and undertook FACS analysis at E7.5. The Etv2-EYFP reporter was used to label the hemato-endothelial lineages (Rasmussen et al., 2011). We did not observe any change in the number of EYFP+ cells between DicerL/L and Etv2-Cre;DicerL/L embryos at E7.5 (Figure S1D). Further, our analysis showed a significant increase in the number of the Flk1−/Pdgfra+ cells in the EYFP+ compartment of the Dicer conditional mutants (Figures 1A–C and Figure S1E) without any change in the Flk1+/Pdgfra+ and Flk1+/Pdgfra− populations (Figures S1F and S1G). In contrast, in the EYFP− fractions, no changes in the Flk1−/Pdgfra+ or Flk1+/Pdgfra+ populations were observed (Figures 1D–F). These results demonstrated that miRNAs expressed in the Etv2+ progenitors are critical for segregation of the mesodermal lineages.
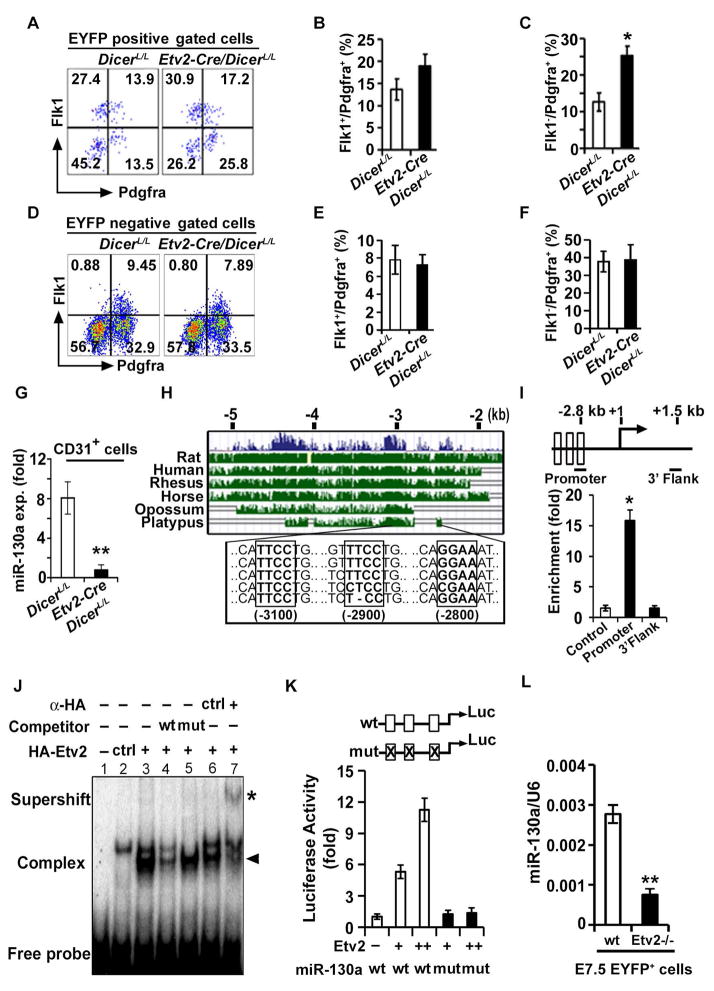
Figure 1. Etv2 modulates miR-130a expression in the endothelial progenitors.
(A–F) Representative FACS profiles (A, D) and quantification (B, C, E, F) of mesodermal populations in Etv2-EYFP:Etv2-Cre;DicerL/L embryos at E7.5 from EYFP+ and EYFP− compartments. (G) qPCR analysis of miR-130a in CD31+ cells sorted from DicerL/L and Etv2-Cre;DicerL/L embryos at E9.5. (H) Evolutionary conservation of the 5.0 kb upstream fragment of the miR-130a gene. (I) Top: Schematic of the 2.8 kb upstream region of the miR-130a promoter. Bottom: ChIP analysis of d4 Dox-inducible HA-Etv2 EBs using an HA antibody. ChIP assay for the Gapdh promoter (Control) and a non-specific locus (miR-130a 3′ UTR region: 3′ Flank) are shown as controls. (J) EMSA showing Etv2 bound to the Ets binding site in the miR-130a promoter region. (K) Luciferase reporter constructs using the miR-130a promoter (-1.0 kb) harboring wild-type (wt; open box) or mutant (mut; crossed box) Etv2 binding sites. (L) qPCR analysis of miR-130a using EYFP+ sorted cells from Etv2 wild-type and mutant embryos at E7.5. Error bars indicate SEM (n = 4; *p<0.05; **p < 0.005) (see also Figure S1).
To decipher the role of specific miRNA in the endothelial progenitors, we analyzed miRNAs that were enriched in endothelial lineages. qPCR analysis from sorted CD31+ cells vs. CD31− cells in E9.5 embryos revealed robust expression of miR-126, miR-126*, miR-221, miR-27b, miR-130a and miR-130b (Figure S1H). We focused on miR-130a as relatively little is known regarding its role during embryogenesis and lineage determination. To evaluate the role of miR-130a in the regulation of hemato-endothelial lineages, we FACS sorted Flk1+ cells from d4 EBs and observed significant enrichment of miR-130a in Flk1+ cells relative to Flk1− cells (Figure S1I). Next, to assess the lineage specific expression of miR-130a in vivo, we sorted CD31+, CD41+, and cardiac progenitor cells (CPCs; Nkx-2.5-EYFP+ cells) from E9.5 embryos and detected robust expression of miR-130a in CD31+ cells relative to other lineages (Figure S1J). These results indicated that miR-130a is enriched in the endothelial progenitors. FACS-sorted CD31+ cells from the Etv2-Cre;DicerL/L embryos demonstrated reduced expression of mature miR-130a, indicating that Dicer is required for miR-130a biogenesis in the Etv2+ lineage (Figure 1G). To define the regulatory network for miR-130a, we analyzed the 5.0kb upstream region of the miR-130a locus and found three highly conserved binding motifs for Etv2 (Figure 1H). Using ChIP assays and doxycycline (Dox) inducible HA-Etv2 cell lysates at d4 of differentiation observed a 15-fold enrichment of Etv2 at the cis-regulatory region of the miR-130a promoter, but not in the non-specific region of the miR-130a locus (Figure 1I). Gel-shift assays further confirmed that Etv2 could bind to the miR-130a promoter containing the Etv2 recognition sequence (Figure 1J). Transcriptional assays using the 1.0kb miR-130a promoter-reporter construct revealed that Etv2 potently transactivated the miR-130a promoter in a dose-dependent fashion and mutagenesis of the Etv2 binding motifs resulted in abolishment of the transcriptional activity (Figure 1K). To monitor whether Etv2 could regulate miR-130a expression in vivo, we FACS-sorted the Etv2-EYFP+ cells from the Etv2-wildtype (Etv2+/+) and null (Etv2−/−) embryos at E7.5. qPCR analysis for miR-130a showed reduced expression of miR-130a in Etv2−/− embryos compared to controls (Figure 1L). These results indicated that miR-130a was expressed in early endothelial progenitors and that Etv2 is a direct upstream regulator of miR-130a.
miR-130a modulates the endothelial lineage during embryogenesis
qPCR analysis revealed robust (~8-fold) and significant expression of mature miR-130a transcripts by d4 (hemato-endothelial specification stage) and remained high at d8 (endothelial maturation stage) of EB differentiation (Figure S2A). To define the role miR-130a in endothelial progenitors, we generated an inducible ESC line which over-expresses miR-130a in response to Dox (miR-130a iES) (Figure 2A). Induction of miR-130a from d2-d6 resulted in significantly increased expression of endothelial transcripts including: CD31, Tie2, and Flk1 with no effect in the key hematopoietic transcripts including CD41, c-kit, gata1 and Lmo2 (Figure 2B). Further, FACS analysis showed that the over-expression of miR-130a resulted in an increase of the endothelial program (CD31+/Tie2+ and CD31+/VE-cadherin− cells) by 3-fold and 2-fold, respectively (Figures 2C and 2D; Figures S2B and S2C) without any significant changes in the hematopoietic lineage (CD41+/CD45+ cells) at d6 of EB differentiation (Figures 2E and 2F). Colony forming (CFC) assays revealed that the definitive hematopoietic colonies (Ery-D, GEMM, GM and Mac) were not significantly changed between uninduced and Dox-induced ES/EBs (Figure S2D). We next evaluated whether the hemogenic endothelial lineages were affected in response to miR-130a induction. Interestingly, we did not observe a significant change in the percentages of hemogenic endothelial lineages (CD41+/Tie2+), whereas the endothelial lineages (CD41−/Tie2+) were robustly increased in the presence of Dox (Figures S2E and S2F). Altogether these data demonstrated that miR-130a promoted the endothelial lineage with no effect on hematopoiesis. To examine the differential effect of miR-130a, we performed miR-130a antagomir-mediated knockdown assays using ES/EBs (Figure S2G). Knockdown of miR-130a resulted in a robust increase in Meox2 [a known target of miR-130a (Chen and Gorski, 2008)] expression (Figure S2H). FACS analysis revealed that knockdown of miR-130a resulted in a significant reduction of the endothelial lineage (Figures S2I and S2J), whereas the hematopoietic lineages were unaffected (Figures S2K and S2L). To validate these findings, we generated miR-130a knockout ESC lines (miR-130a−/−) using CRISPR/Cas9 technology (Figures S2M–O). Consistent with the knockdown studies, the deletion of miR-130a resulted in reduced endothelial lineage (CD31+/Tie2+ cells) development (Figures 2G and 2H) without affecting the hematopoietic lineages (CD41+/CD45+ cells) (Figures 2I and 2J). These results confirmed the preferential effect of miR-130a on endothelial development.
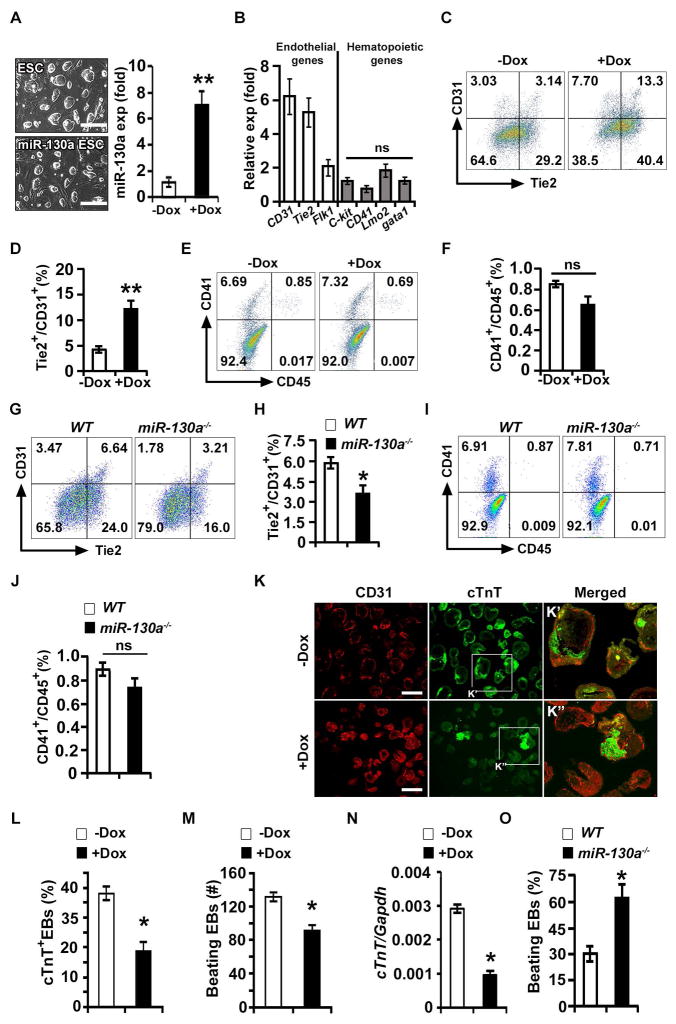
Figure 2. miR-130a promotes the endothelial lineage.
(A) Left panel: phase contrast images of wild-type E14 (ESCs) and Dox-inducible miR-130a ES cell (miR-130a iES) colonies. Right panel: qPCR analysis of miR-130a in the absence (−Dox) and presence of Dox (+Dox). (B) qPCR analyses for endothelial and hematopoietic markers using −Dox and +Dox EBs at d6 of differentiation (ratio shown as +Dox/−Dox). (C–F) FACS profiles (C, E) and quantification (D, F) of endothelial [Tie2 and CD31 (C, D)]; and hematopoietic [CD41 and CD45 (E, F)] markers in −Dox and +Dox conditions. (G–J) Representative FACS profiles (G, I) and quantification (H, J) of endothelial (G, H) and hematopoietic (I, J) markers in WT and miR130a−/− EBs. (K) Sections of −Dox and +Dox miR-130a iES/EBs immunostained with CD31 (red) and cTnT (green) at d10 of EB differentiation. Boxed regions are shown in panel K′ and K″. (L) Quantitative analysis of cTnT+ EBs in −Dox (n = 123) and +Dox (n = 142) EBs. (M) Contractility assay using −Dox (n = 146) and +Dox (n = 135) EBs. (N) qPCR analyses of a cardiac marker (cTnT) from −Dox and +Dox miR-130a iES/EBs. (O) Cardiogenic assay using WT and miR-130a−/− embryoid bodies. Error bars indicate SEM (n = 5; *p < 0.05). Scale bar: 200 μm, (see also Figure S2).
We next examined whether miR-130a induction affects other mesodermal-derived lineages. Induction of miR-130a from d2-d6 resulted in a significant reduction of cardiac Troponin (cTnT) expression in d10 EBs with a corresponding increase of CD31 expression (Figure 2K). Quantification of cTnT+ EBs and beating EBs revealed a significant decrease in the cardiogenic program upon induction of miR-130a (Figures 2L and 2M). Furthermore, qPCR analysis revealed decreased expression of cTnT, concurrent with reduced cardiac contractility in the Dox-induced EBs (Figure 2N). In contrast, miR-130a−/− ES/EBs demonstrated increased cardiogenic potential as we observed increased percentage of beating embryoid bodies (Figure 2O). These data suggested that miR-130a promoted the endothelial lineage at the expense of the cardiac program.
miR-130a targets Pdgfra expression and modulates mesodermal lineage development
To explore the mechanism by which miR-130a modulates endothelial lineage development, we employed three miRNA target prediction tools including TargetScan 6.2, PicTar and miRANDA to mine common predicted targets. Among the multiple targets, we identified Pdgfra as a highly conserved target of miR-130a with a high percentile score using TargetScan 6.2 in both mouse and zebrafish genomes (data not shown). Multiple sequence alignment revealed a highly conserved miR-130a seed-sequence in the Pdgfra-3′-UTR region (Figure 3A). To examine whether miR-130a targets Pdgfra mRNA, we performed luciferase assays using a PGK promoter-driven luciferase reporter (PGK-Luc-Pdgfra-3′-UTR) construct. Co-transfection of the PGK-Luc-Pdgfra-3′-UTR reporter with a miR-130a mimic resulted in a statistically significant reduction (~40%) of the luciferase activity whereas co-transfection with miR-130a antagomir resulted in enhancement of the luciferase activity, indicating that endogenous miR-130a could target the Pdgfra mRNA (Figure 3B). To assess whether miR-130a could target Pdgfra transcripts in vivo, we injected morpholinos against miR-130a (miR-130a-MO) at the one-two cell stage into fertilized zebrafish eggs and performed in situ hybridization for pdgfra mRNA. Our data demonstrated increased expression of pdgfra in the miR-130a morphants, indicating that miR-130a could suppress pdgfra expression in vivo (Figure 3C, Figures S3A and S3B). To further examine its effect on the endothelial lineage, we utilized an endothelial specific transgenic reporter [Tg(fli1-EGFP)] line and injected control MO or miR-130a MO into the fertilized eggs. qPCR analysis from FACS sorted fli1-EGFP+ cells revealed enrichment of pdgfra transcripts in the miR-130a morphants relative to the control (Figure 3D). These results indicated that miR-130a could target Pdgfra transcripts in endothelial cells in vitro and in vivo. We next evaluated whether injection of miR-130a morpholinos could affect the hemato-endothelial lineage specification at early somitogenesis stages. Initially, we performed FACS analysis using the fli1-EGFP reporter to quantify the angioblasts at the 8 somite stage (Shoji et al., 2003). Our analysis revealed a reduced percentage of EGFP+ populations in the miR-130a MO relative to control at the 8 somite stage (Figure S3C and S3D). Next, we examined kdrl expression using in situ hybridization in control and miR130a morphants. Our analysis revealed reduced expression of kdrl transcripts (brackets) at 14–16 hpf hours post fertilization (hpf) (Figure S3E). These results indicated that miR-130a modulates endothelial precursors. Furthermore, qPCR analysis at 14 hpf and 24 hpf revealed reduction in the levels of the endothelial program (kdrl) without affecting the hematopoietic program (gata1) (Figures S3F and S3G). Similar to the observation in miR-130a−/− embryoid bodies, we found increased expression of the cardiogenic marker, myl2f, at 24 hpf (Figure S3H). Together, these results indicated that miR-130a modulates endothelial lineages both in vitro and in vivo.
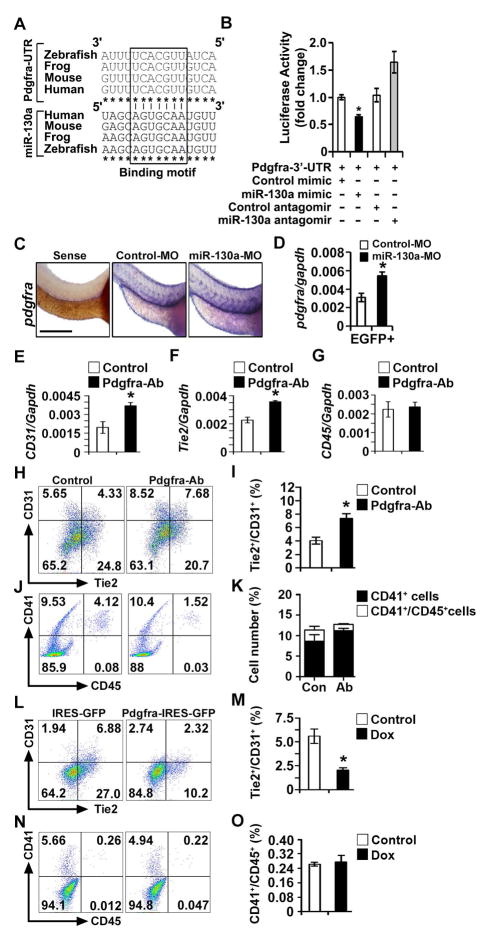
Figure 3. miR-130a targets Pdgfra and miR-130a-Pdgfra pathway modulates mesodermal progenitors.
(A) ClustalW multiple sequence alignment of Pdgfra 3′ UTR and miR-130a. (B) Luciferase activity of Luc-Pdgfra-3′-UTR reporter constructs in the presence of miR-130a mimic and miR-130a antagomir. (C) Whole-mount in situ hybridization images of control and miR-130a morphants using pdgfra probes at 48 hpf. (D) qPCR analysis of pdgfra transcripts using fli1-EGFP+ sorted cells from control and miR-130a morphants at 48 hpf. (E–G) qPCR analysis of CD31, Tie2 and CD45 transcripts from control and Pdgfra neutralizing antibody-treated EBs. (H–K) Representative FACS profile (H, J) and quantification (I, K) of endothelial lineages (H, I) and hematopoietic lineages (J, K) in control and Pdgfra neutralizing antibody-treated EBs. (L–O) Representative FACS profile (L, N) and quantification (M, O) of endothelial lineages (L, M) and hematopoietic lineages (N, O) using d6 EB in control and Pdgfra over-expression conditions. Error bars indicate SEM (n = 5; *p < 0.05). Scale bar: 200 μm (see also Figure S3).
Next, we examined whether down regulation of Pdgfra is the mechanism by which miR-130a regulates endothelial lineages. Antibody-mediated inhibition of Pdgfra signaling resulted in a significant induction of CD31 and Tie2 transcripts with no effect on CD45 transcripts (Figures 3E–G). Similarly, FACS analysis showed that blocking Pdgfra signaling led to a significant increase (~ 2-fold) in the endothelial lineage (Tie2+/CD31+ cells) with no effect on the hematopoietic lineage (Figures 3H–K). To validate these findings, we generated a Dox-inducible lentiviral vector expressing Pdgfra (Pdgfra-IRES-GFP). In contrast to the inhibition studies, Dox-mediated overexpression of Pdgfra repressed the endothelial program (Tie2+/CD31+ cells) from 6.90%±1.5% to 2.5%±0.75% without affecting the hematopoietic lineage (Figures 3L–O). To further investigate the interaction between miR-130a and Pdgfra signaling, we engineered an inducible Pdgfra mouse ES cell line using miR-130a iES cells, which over-expressed both miR-130a and Pdgfra in response to doxycycline (miR-130a+Pdgfra-IRES-GFP iESC) (Figure S3I). As expected, induction of miR-130a alone resulted in an induction of the endothelial lineage. Forced overexpression of miR-130a and Pdgfra resulted in a significant reduction (~2.5 fold) of Tie2+/CD31+ cells with no effect on the hematopoietic populations (Figures S3J–M). These results demonstrated that miR-130a regulates the endothelial lineage via down-regulation of Pdgfra signaling.
miR-130a modulates Flk1+/Pdgfra+ mesodermal progenitors towards lateral plate mesodermal lineage
Our results above demonstrated that miR-130a regulates the differentiation of the endothelial lineage in vivo. We next examined whether miR-130a has a role during the mesodermal specification stage. Studies have shown that Flk1−/Pdgfra+ (paraxial mesoderm) and Flk1+/Pdgfra− (lateral plate mesoderm) cells arise from the Flk1+/Pdgfra+ (unpatterned mesoderm) by modulating Pdgfra levels at early stages of ES/EB differentiation (Kataoka et al., 2011; Sakurai et al., 2006). Therefore, we performed a shorter pulse (48h) and examined whether miR-130a promoted the endothelial lineage from mesodermal progenitors. A 48h Dox pulse between d2-d4 using miR-130a iES/EBs resulted in a markedly reduced percentage (54%±4% to 35%±2%) of the Flk1−/Pdgfra+, but not Flk1+/Pdgfra+ populations (Figures 4A–C). qPCR analyses showed reduction of Pdgfra transcripts in Dox-pulsed miR-130a EBs, confirming that miR-130a targets Pdgfra mRNA (Figure 4D). Notably, similar to a long pulse (d2-6) of miR-130a (Figure 2), a shorter pulse (d2-4) resulted in an equivalent promotion of the endothelial lineage (Figures 4E and 4F).
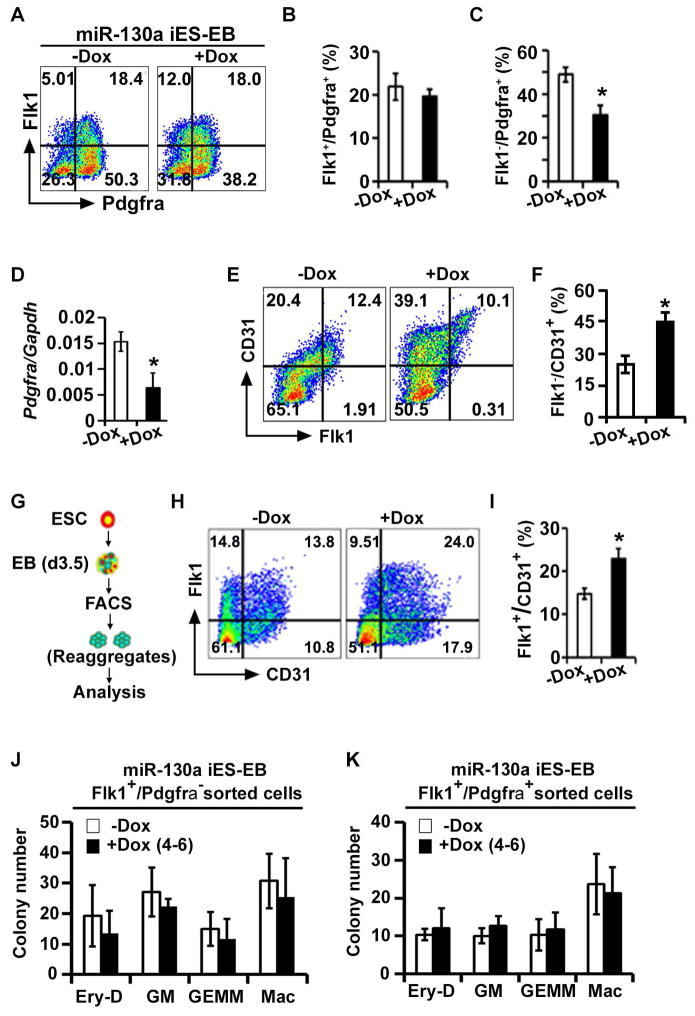
Figure 4. miR-130a targets Pdgfra in vivo and promotes lateral plate mesodermal lineage.
(A–C) FACS profile (A) and quantification (B, C) of mesodermal populations in miR-130a iES/EB differentiation in −Dox and +Dox conditions. (D) qPCR analysis of Pdgfra in −Dox and +Dox conditions using d4 EBs. (E, F) FACS profile and quantification (F) of endothelial (Flk1 and CD31) markers during miR-130a iES/EB differentiation in −Dox and +Dox conditions. (G) Schematic of the experiment to determine the ability of miR-130a to re-specify mesoderm. (H, I) FACS profiles (H) and quantification (I) of endothelial markers at d6. (J, K) Hematopoietic colony forming assay from Flk1+/Pdgfra− (J) and Flk+/Pdgfra+ (K) sorted cells using d3.5 miR-130a iES/EB system. Error bars indicate SEM (n = 3; *p < 0.05).
We next performed aggregation-reaggregation assays to test the ability of miR-130a to modulate mesodermal precursors. Uninduced FACS sorted Flk1+/Pdgfra− and Flk1+/Pdgfra+ cells from miR-130a iES/EBs (d3.5) were reaggregated for an additional 48h in the absence or presence of Dox and were analyzed for mesodermal-derivatives at d6 using cell surface markers (Figure 4G). We did not detect significant differences in the number of endothelial cells derived from the Flk1+/Pdgfra− population, demonstrating that over-expression of miR-130a did not influence the fate of lineage-committed endothelial populations (data not shown). FACS analyses of Flk1+/Pdgfra+ reaggregates revealed an emergence of the endothelial lineages (Figure 4H), supporting the existence of early unpatterned mesodermal cells within this population (Sakurai et al., 2006). Importantly, a Dox-pulse (d4-6) in the double-positive reaggregates resulted in a significant increase (12% ± 3% to 22% ± 2%) of endothelial cells (Figures 4H and 4I). In contrast, the blood colony forming activity from either Flk1+/Pdgfra− or Flk1+/Pdgfra+ cell populations did not show significant changes (Figures 4J and 4K), demonstrating that the effect of miR-130a is specific to the promotion of the endothelial lineage. These data indicated that miR-130a directed the unpatterned mesodermal lineage towards lateral plate mesoderm by suppression of Pdgfra expression.
Discussion
Here, we defined the essential role of the Etv2-miR-130a-Pdgfra network in the divergence of hemato-endothelial progenitors. Mechanistically, miR-130a promotes endothelial lineage development by modulating Pdgfra signaling. Together, we have uncovered an important role for a miRNA in the specific promotion of the endothelial lineage without affecting the hematopoietic lineage.
miRNAs are known to be important regulators of transcript expression, however, only a limited number of miRNAs have specific developmental roles. For example, the miR-430/427/302 family controls mesendodermal fate specification and miR-1/miR-133 (muscle-specific miRNAs) can promote mesodermal formation during embryogenesis (Ivey et al., 2008; Rosa et al., 2009). Our study reports a functional role for miR-130a that specifically promotes differentiation of the endothelial lineage.
Previous reports suggest that Etv2 is essential for the progression of Flk1+/Pdgfra+ progenitors (primitive mesoderm) to the Flk1+/Pdgfra− (vascular mesoderm) state (Kataoka et al., 2011). Based on our observations, we propose that the Etv2-miR-130a cascade down-regulates Pdgfra expression and promotes the transition of Flk1+/Pdgfra+ to the Flk1+/Pdgfra− lineages. We further propose that miR-130a facilitates the segregation of bipotent progenitors towards the endothelial lineage without affecting the hematopoietic lineage.
Since mutation of Etv2 affects both hematopoietic and endothelial lineages (Koyano-Nakagawa et al., 2012), it was unexpected to discover that miR-130a targets Pdgfra and promotes only the endothelial lineage. However, these results are supported by the finding that fetal liver hematopoiesis was not affected in the Pdgfra conditional knockout embryos (Ding et al., 2013). Our data together with others (Ding et al., 2013) support the notion that the endothelial, but not the hematopoietic lineage is sensitive to the level of Pdgfra. We propose that as Flk1+/Pdgfra+ common mesodermal progenitors progress to the Flk1+/Pdgfra− hemato-endothelial progenitors, miR-130a further down-regulates Pdgfra expression to direct cells towards the endothelial lineage (Figure S4). This is in agreement with a previous report highlighting that newly-generated Flk1 single positive cells are still plastic in nature and could switch between the Pdgfra single positive state (and vice versa) before lineage commitment (Sakurai et al., 2006). We propose that the Etv2-miR-130a-Pdgfra pathway may function to stabilize this plastic intermediate state towards an endothelial fate by suppressing the fluctuating levels of Pdgfra. Despite several reports emphasizing the existence of a common progenitor for endothelial, cardiac and smooth muscle lineages (Kattman et al., 2006; Moretti et al., 2006), it is unclear whether Flk1+/Pdgfra+ (unpatterned mesoderm) cells have equivalent cardiac and vascular potential. Therefore, it is possible that miR-130a induces the endothelial program by promoting a pre-specified lineage from a heterogeneous mix of cells. Our findings, together with others, illustrate the critical role of signaling pathways, transcription factors and miRNAs in the regulation of cell fate decisions, thereby forming an integral part of the regulatory network in the emergence of mesodermal lineages. While several factors including HoxA3, Runx1, Notch1, Sox17 and miR-142-3p have been shown to have a critical role in hematopoietic lineage segregation and hemangioblast specification (Iacovino et al., 2011; Nimmo et al., 2013; Lizama et al., 2015), endothelial specific regulators have not been described. In summary, miR-130a is an important regulator of endothelial development from the hemato-endothelial progenitors (Figure S4).
Materials and Methods
Detailed experimental procedures can be found in the supplemental data section.
Generation of Dicer conditional null mice and morphological analysis
All animal studies were approved by the IACUC, University of Minnesota. DicerL/L mice (strain: B6.Cg-Dicertm1Bdh/J) (Harfe et al., 2005) were intercrossed with Mesp1-Cre (Saga et al., 1999), Flk1-Cre (Shalaby et al., 1995) and Etv2-Cre (Rasmussen et al., 2011) mice to generate Mesp1-Cre;DicerL/+, Flk1-Cre;DicerL/+ and Etv2-Cre;DicerL/+ mice and subsequently crossed with DicerL/L mice to generate conditional Dicer knockouts.
Generation of inducible miR-130a mouse ESC line, inducible Pdgfra ESC lines, ES/EB culture conditions and FACS analysis
ESC culture conditions, ES/EB differentiation and the generation of the Dox-inducible miR-130a ESCs were performed as previously reported (Iacovino et al., 2011).
Generation of miR130a−/− ES cells
Targeted deletion of miR-130a locus was achieved using the CRISPR/Cas9 technology. For the detailed description please see the supplemental data section.
Statistical Analysis
All experiments were repeated at least three times and the data represent the mean ± SEM. Statistical significance was determined using the Student’s t-test and a p-value ≤ 0.05 was considered as a significant change.
Supplementary Material
Highlights.
Etv2 transactivates miR-130a in the endothelial progenitors
miR-130a specifically promotes the endothelial fate and without affecting hematopoiesis
miR-130a regulates Pdgfra expression
Acknowledgments
Funding support was obtained from NIH (U01HL100407 and R01HL122576 to DJG and R01AR064195 to YK). The authors thank Naoyuki Tahara for his technical assistance with the zebrafish studies.
Footnotes
Author Contributions:
B.N.S., X.S., R.A., S.D., T.L.R., M.G.G., W.G., Y.K., N.K.N.: Conception, experimental design, collection, assembly, data analysis, and manuscript writing. D.J.G.: Experimental design, data interpretation, manuscript writing, and financial support.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Tanaka Y, Hayashi M, Nishikawa S, Kataoka H. PDGF receptor alpha+ mesoderm contributes to endothelial and hematopoietic cells in mice. Dev Dyn. 2013;242:254–268. doi: 10.1002/dvdy.23923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson EN, et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci USA. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovino M, Chong D, Szatmari I, Hartweck L, Rux D, Caprioli A, Cleaver O, Kyba M. HoxA3 is an apical regulator of haemogenic endothelium. Nat Cell Biol. 2011;13:72–78. doi: 10.1038/ncb2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood. 2011;118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kweon J, Iacovino M, Shi X, Rasmussen TL, Borges L, Zirbes KM, Li T, Perlingeiro RC, Kyba M, et al. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells. 2012;30:1611–1623. doi: 10.1002/stem.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel DA, Watson CM, De Young RA, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- Lizama CO, Hawkins JS, Schmitt CE, Bos FL, Zape JP, Cautivo KM, Borges Pinto H, Rhyner AM, Yu H, Donohoe ME, Wythe JD, Zovein AC. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat Commun. 2015;6:7739. doi: 10.1038/ncomms8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Nimmo R, Ciau-Uitz A, Ruiz-Herguido C, Soneji S, Bigas A, Patient R, Enver T. MiR-142-3p controls the specification of definitive hemangioblasts during ontogeny. Dev Cell. 2013;26:237–249. doi: 10.1016/j.devcel.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, Shi X, Ferdous A, Li T, Kyba M, et al. ER71 directs mesodermal fate decisions during embryogenesis. Development. 2011;138:4801–4812. doi: 10.1242/dev.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki Ji, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Era T, Jakt LM, Okada M, Nakai S, Nishikawa S, Nishikawa S. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586. doi: 10.1634/stemcells.2005-0256. [DOI] [PubMed] [Google Scholar]
- Shi X, Richard J, Zirbes KM, Gong W, Lin G, Kyba M, Thomson JA, Koyano-Nakagawa N, Garry DJ. Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev Biol. 2014;389:208–218. doi: 10.1016/j.ydbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji W, Isogai S, Sato-Maeda M, Obinata M, Kuwada JY. Semaphorin3a1 regulates angioblast migration and vascular development in zebrafish embryos. Development. 2003;130:3227–3236. doi: 10.1242/dev.00516. [DOI] [PubMed] [Google Scholar]
- Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.