Abstract
Recent advances in cell reprogramming via employing different sets of factors, which allows generation of various cell types that are beyond the downstream developmental lineages from the starting cell type, provide significant opportunities to study fundamental biology and hold enormous promise in regenerative medicine. Small molecules have been identified to enhance and enable reprogramming by regulating various mechanisms, and provide a highly temporal and tunable approach to modulate cellular fate and functions. Here, we review the latest development in cell reprogramming from the perspective of small molecule modulation.
Introduction
During development and in tissue homeostasis, cell identities are defined by specific gene expression programs, which are governed by core transcription factors. These factors interact with other transcription factors co-occupying specific regulatory elements of target genes to exhibit transcriptional cooperativity. They also recruit other transcriptional co-regulators with chromatin remodeling activities (e.g., epigenetic proteins, such as histone and DNA readers, writers, and erasers) to regulate chromatin accessibility at specific DNA sequences, as well as transcriptional cofactors to activate or repress the activity of transcriptional machinery. These factors collaboratively modulate the frequency, specificity, and strength of gene expression to determine a particular cell fate.
To reprogram and stably establish a cell to a new fate, the balance of the original transcriptional network must be broken. Conventionally, disrupting this balance occurs through genetic approaches, such as overexpressing or knocking down/out core transcription factors. The generation of induced pluripotent stem (iPS) cells by ectopic expression of four transcription factors (iPSC-TFs) exemplifies such approach in this field [1]. Recently, small molecules have proven useful in regulating cell fate and function, especially cellular reprogramming.
Compared to conventional genetic approaches, small molecules provide several distinct advantages to reprogramming. For example, small molecules modulate specific protein targets rapidly and often reversibly, and thus can regulate cell functions with higher precision in a temporal manner. Additionally, small molecules can be applied at various concentrations and combinations so that their effects are highly tunable. These features can improve their specificity and efficacy, alleviate safety concerns, and potentially overcome hurdles in clinical applications that genetic methods cannot.
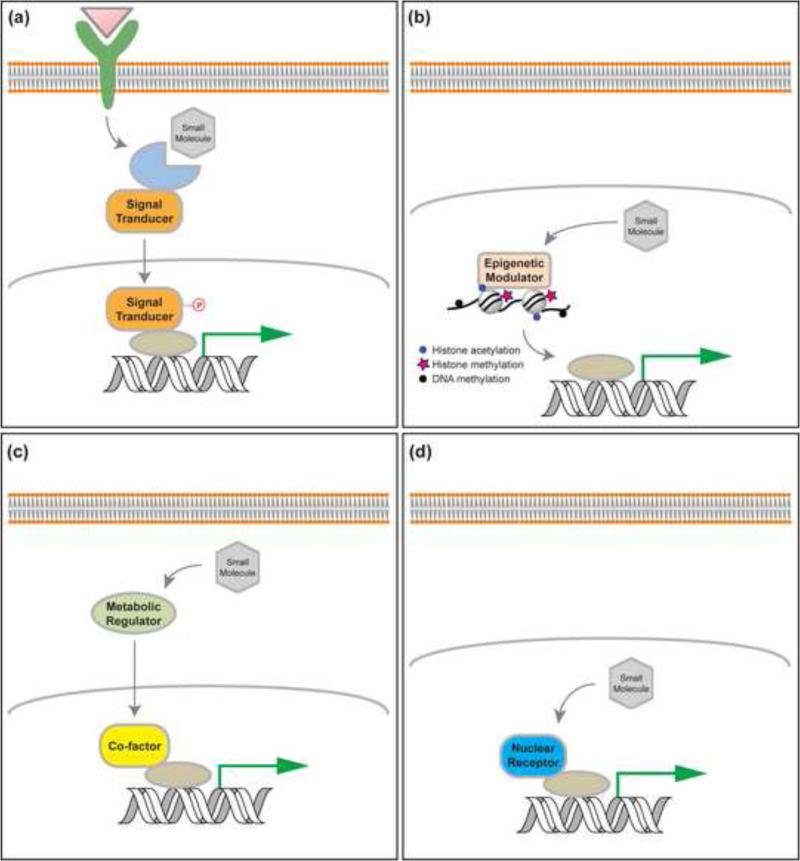
Small molecules can regulate gene transcription typically through four classes of mechanisms: signaling pathway modulators, which activate or repress components of signal transduction to regulate downstream transcription activity; modulators of epigenetic proteins, which regulate the activity of epigenetic complexes; metabolic regulators, which adjust cell state and shift balance of metabolites that serve as ligands for proteins (e.g., GPCRs and nuclear receptors) and cofactors for epigenetic proteins; and nuclear receptor agonists and antagonists, which directly modulate transcription by regulating the activity of nuclear receptors (Figure 1). Here, we will review each of these categories for applying small molecules to reprogramming. We will also discuss the transdifferentiation paradigm and its possible mechanisms of action.
Figure 1.
Small molecules regulate gene transcription mainly through four classes of mechanisms: signaling pathway modulators, which activate or repress components of signal transduction to regulate downstream transcription activity (a); modulators of epigenetic proteins, which regulate the activity of epigenetic complexes to modify epigenetic marks of certain chromatin region and its gene transcription (b); metabolic regulators, which adjust cell state and shift balance of metabolites that serve as ligands for proteins (e.g., GPCRs and nuclear receptors) and cofactors for epigenetic proteins (c); and nuclear receptor agonists and antagonists, which directly modulate transcription by regulating the activity of nuclear receptors (d).
Signaling pathway modulators
Signaling pathway modulators represent a major group of small molecules regulating reprogramming. Some signaling pathways directly target the pluripotency transcriptional network to positively affect iPS cell generation. For example, a glycogen synthase kinase (GSK) 3 inhibitor, CHIR99021, was shown to promote maintenance of pluripotency and enhance reprogramming [2-4]. This is consistent with the mechanism that under Wnt stimulation, T-cell factor (TCF), a downstream component of the Wnt pathway could act in an activating complex to bind many pluripotency genes in ES cells, including Oct4 Sox2 and Nanog [5]. The LIF-Stat3 pathway, well characterized to sustain self-renewal of mouse embryonic stem (ES) cells [6], was shown to enhance late stage reprogramming using a system that excludes interference from two other LIF-dependent pathways, PI3K/Akt and MAPK/Erk [7].
Some signaling pathway modulators regulate essential events during reprogramming. For example, during iPS cell reprogramming fibroblasts lose mesenchymal characteristics and acquire epithelial features. Such changes indicate that reprogramming requires the mesenchymal-to-epithelial transition (MET), a key early barrier in this process [8,9]. Thus, small molecule inhibitors of the transforming growth factor (TGF)-β pathway, which induces EMT, would enhance reprogramming. Indeed, small molecule inhibitors of TGF-β pathway, such as SB431542, A-83-01, and E616452, greatly enhance iPS cell reprogramming or helped to functionally replace iPSC-TFs in various contexts [10-12]. These effects could be through the inhibition of TGF-β downstream transcriptional repressor Snail, and the following enhanced transcription of epithelial genes such as E-cad [8].
Signaling pathway modulators also help identify novel reprogramming mechanisms. For example, many cytoplasmic macromolecules and organelles are drastically turned over during reprogramming. This transformation may involve autophagy, a catabolic process that recycles cell components by degrading proteins and organelles [13]. Chen et al. showed that several autophagy activators, such as rapamycin and PP242 (mTOR inhibitors), enhanced reprogramming [14]. Additionally, Wang et al. demonstrated that ATG5-dependent autophagy is essential to iPS cell reprogramming via Sox2-mediated repression of mammalian target of rapamycin (mTOR) in the early stage, which stimulates a transient onset of autophagy [15]. The mechanism for this effect relies on Sox2 to recruit the nucleosome remodeling deacetylase (NuRD) complex to a repressive region in mTOR's promoter.
Epigenetic Enzyme Inhibitors
In reprogramming, a key question is how iPSC-TFs remodel somatic chromatin and activate transcription of silenced pluripotency genes. This process involves recruiting chromatin modifiers that modulate the 3D structure of chromatin and accessibility of loci in silenced genes [16]. Studies continue to identify chromatin modulators that function in this process. For example, Mbd3, a member of repressive NuRD complex, was found to modulate reprogramming in a stage-dependent manner [17,18]. Additionally, inhibition of HDACs (histone deacetylases), another component of this complex, by small molecules such as Valproic acid and sodium butyrate, promoted reprogramming [19-21]. This class of enzymes regulates lysine acetylation of histones and usually condenses chromatin and represses transcription.
Other histone-modifying enzymes switch the active or repressive marks of both pluripotency and somatic genes to establish a pluripotency program during reprogramming. Utx, a H3K27 demethylase, binds Oct4 and co-occupies many regions of target genomes to help them keep their active marks in ES cells [22]. Interestingly, another H3K27 demethylase, Jmjd3, inhibits reprogramming, in part by activation of the Ink4α/Arf locus [23]. PHF20, which participates reprogramming and reactivation of endogenous Oct4 is also inhibited by Jmjd3. Additionally, Parnate, an inhibitor of the H3K4/9 histone demethylase LSD1, as well as several histone methyltransferase inhibitors, such as Dot1L inhibitor EPZ004777, G9a inhibitor Bix01294, and Ezh2 inhibitor DZNep, were shown to promote iPS cell generation [10,24,25].
Another critical epigenetic mechanism that controls gene expression is DNA methylation. Gene promoter methylation can stably inactivate gene expression by blocking binding of transcription factors. Inhibitors of DNA methyltransferases (that mediate transfer of methyl groups to DNA), such as RG108 and 5-zaz, were shown to increase reprogramming [25,26]. More recently, studies uncovered enzymes that mediate DNA demethylation in reprogramming. For example, ten eleven translocation (Tet) enzymes convert 5-mC to 5-hmC in DNA, the initial step in active DNA demethylation [27,28]. Gao et al. showed that Tet1 not only promoted Oct4 demethylation and reactivation, but also functionally replaced exogenous Oct4 [29]. Hu et al. demonstrated that knocking out Tet1, 2, and 3 completely blocked MET and prevented reprogramming [30]. These and other studies found that TET enzymes regulate reprogramming by activating pluripotency genes and regulating intermediate events [31,32]. Interestingly, a recent study demonstrated that Vitamin C increased Tet1 activity, and induced a rapid and global increase of 5hmC in mouse ES cells [33]. This may in part explain how Vitamin C enhances reprogramming [34].
Metabolic Regulators
Compared with somatic cells, many stem cells and highly proliferative cells rely more heavily on aerobic glycolysis to support their proliferation. For example, studies showed a correlation between the self-renewal ability of ES cells, reduced oxidative phosphorylation, and increased glycolysis [35,36], suggesting that the transition from oxidative phosphorylation to glycolysis be a barrier in reprogramming. In fact, the finding that hypoxic condition improved reprogramming efficiency and kinetics supports this hypothesis [37]. PS48, an activator of 3’ phosphoinositide-dependent kinase 1, which facilitated metabolic conversion to glycolysis, improved human iPS cell generation via ectopic expression of a single transcription factor (OCT4) [38]. Consistently, many small molecules that promote glycolytic metabolism and act more directly on metabolic pathways, such as fructose 2, 6-bisphosphate (an activator of phosphofructokinase 1) and Quercetin (which promotes HIF-1 activity) also promote reprogramming. Conversely, a glycolysis inhibitor, 2-Deoxy-D-glucose, inhibits reprogramming [39].
Nuclear Receptor Agonists
Nuclear receptors can directly bind DNA and regulate gene expression. The ligand-dependent nuclear receptors can be modulated by small molecule agonists and antagonists. An orphan nuclear receptor Nr5a2 was shown to functionally replace Oct4 in iPS cell reprogramming in the presence of Sox2, Myc, and Klf4 [40]. Another orphan nuclear receptor, Esrrb, a direct target of Nanog and a key component of pluripotency transcriptional program [41], worked with Oct4 and Sox2 to activate pluripotency genes [42]. Together with Nr5a2, another type of nuclear receptors, RARa/g greatly enhanced reprogramming efficiency and kinetics [40]. Consistent with this, some RAR agonists, such as AM580, CD437, and TTNPB, were shown to enhance reprogramming in various contexts [10,43].
Transdifferentiation
Besides using cell type specific transcription factors and miRNAs to induce lineage-specific reprogramming (i.e., transdifferentiation), an alternative approach using the paradigm of cell-activation and signaling-directed (CASD) strategy has been developed. This strategy employs temporal and transient overexpression of iPSC-TFs or treatment with reprograming inducing small molecules (cell activation, CA) in conjunction with tissue patterning cues (signal-directed, SD) to reprogram somatic cells into diverse lineage-specific cell types without entering the pluripotent state [44,45]. This strategy in part mimics epimorphic regeneration in some humbler organisms (e.g., newts), in that cells at the injury site undergo a deprogramming process to generate lineage-specific precursor cells that can re-differentiate to replace lost cells. Using this strategy, cardiac, neural, endothelial, pancreatic and hepatic cells have been generated from fibroblasts via a corresponding tissue-specific multipotent precursor stage [46-50]. Recent studies have also identified cocktails of small molecules that enhance and/or functionally replace the reprogramming factors in the CASD transdifferentiation paradigm. For example, a chemical cocktail containing SB435142, CHIR99021, Parnate and Forskolin was identified to enable converting mouse fibroblasts into cardiac cells through a cardiac precursor stage, but not a pluripotent state, in conjunction with a single transcription factor Oct4 during the cell activation/deprogramming step [51].
Compared to conventional transdifferentiation [52], CASD-based transdifferentiation might be advantageous because a single set of TFs may be used for all cell types . Additionally, transient gene expression might be more easily replaced with safer and more convenient methods without risking genetic modifications. Furthermore, CASD-based transdifferentiation generate lineage-specific progenitor cells that can be expanded, therefore, more advantageous for various applications to regenerative medicine.
Although the detailed mechanism of the CASD paradigm remains to be elucidated, it was hypothesized that the initial cell activation occurs by a universal deprogramming mechanism induced by iPSC-TFs and small molecules. The transient expression of reprogramming factors, especially those functioning as pioneer factors, initiates a wave of epigenetic remodeling by binding and recruiting epigenetic modifiers to regulatory elements, and mainly deprograms starting cell's transcriptional network. Interestingly, increasing numbers of small molecules have been shown to contribute to this universal mechanism of cell activation. Some of these small molecules include epigenetic modifiers (e.g. parnate and NaB) that induce open chromatin state and initiate epigenetic remodeling, as well as some that effectively signal to destabilize a fibroblast phenotype (e.g. CHIR99021 and SB431542) [51]. It is conceivable that the effects induced by these small molecules will have broad applications in CASD-based transdifferentiation, as well as various other processes of cellular reprogramming.
After initial cell activation, a destabilized cellular state can respond to patterning signals to complete lineage-specific transdifferentiation. This specification mechanism has not been well understood. Considering that iPSC-TFs participate in the initial differentiation programs of pluripotent stem cells, it might be that under the influence of the soluble differentiation signals, the transient expression of exogenous iPSC-TFs assume a similar specifying role during lineage specification [53-55]. In deprogrammed cells, transcription factors downstream of patterning signals may orchestrate with iPSC-TFs and other endogenous TFs to induce lineage specific transcription programs.
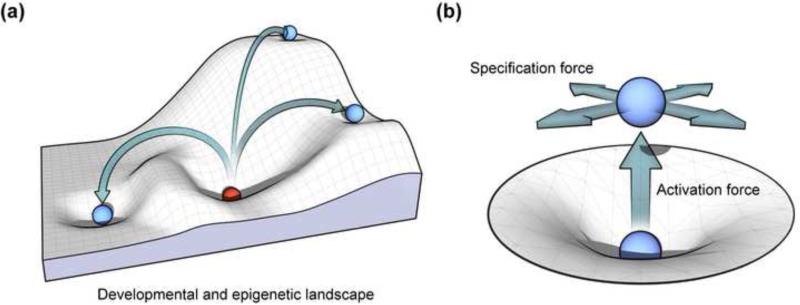
In analogy, the CASD mechanism may be conceptualized to a simple model of law of motion. In this model, pluripotent cells can roll down (differentiate) a slope of the differentiation landscape and settle into an energy minimum (that is stabilized by epigenetic barriers), where such differentiated cell type becomes relative static (Figure 2a). A static cell type is relatively motionless and does not move until an external force is applied to it. One such external force is activities exerted by reprogramming factors and small molecules, which destabilize the cellular state in an upright direction to push cell out of the minimum, while the force exerted by soluble specifying signals guides the cell toward another minimum in the landscape with a distinct epigenetic characteristics (Figure 2b). The synergy of these two forces directs lineage-specific transdifferentiation from one minimum to another on the landscape without the cell entering the pluripotent state.
Figure 2.
Conceptual model of cellular reprogramming. (a) In the developmental and epigenetic landscape, pluripotent cells can roll down (differentiate) a slope and settle into an energy minimum (that is stabilized by epigenetic barriers), where such differentiated cell type becomes relatively static. A static cell type can be reprogrammed to other cell types under external forces. (b) These reprogramming processes are induced by a composition of an upright activation force pushing the cell out of the energy minimum (destabilizing the cellular state) and a specification force guiding the cell toward another minimum in the landscape.
Conclusion
Major developments in cellular reprogramming have continued to advance stem cell biology toward translations. Future directions for cellular reprogramming would envision more efficient and precise control toward therapeutic applications. Small molecules will continue to play essential roles in controlling cell fate and improving our mechanistic understanding of cellular reprogramming. They also naturally promise to be developed as new generation of regenerative medicine aimed at stimulating tissue repair and regeneration through modulating cell fate and function in vivo.
Acknowledgements
Sheng Ding is supported by funding from NICHD, NHLBI, NEI, NIMH/NIH, California Institute for Regenerative Medicine, and the Gladstone Institute. We apologize to all scientists whose work could not be properly discussed and cited here due to limited space.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Schöler HR, Hayek A, Ding S. Generation of Human-Induced Pluripotent Stem Cells in the Absence of Exogenous Sox2. STEM CELLS. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JCR, Smith A. Stat3 Activation Is Limiting for Reprogramming to Ground State Pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 10••.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [This study showed the feasibility of reprogramming fibroblasts into iPSCs by only small molecule combinations.] [DOI] [PubMed] [Google Scholar]
- 11.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A Small-Molecule Inhibitor of Tgf-β Signaling Replaces Sox2 in Reprogramming by Inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menendez JA, Vellon L, Oliveras-Ferraros C, Cufi S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–3677. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- 14.Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, Long Y, Zhao J, Pei G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–911. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Xia P, Ye B, Huang G, Liu J, Fan Z. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13:617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 18•.dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15:102–110. doi: 10.1016/j.stem.2014.04.019. [These two papers showed that Mbd3, a NuRD component, is a key factor to determine reprogramming efficiency and kinetics.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, et al. NuRD Blocks Reprogramming of Mouse Somatic Cells into Pluripotent Stem Cells. STEM CELLS. 2013;31:1278–1286. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 20•.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [This paper is the first study to show that small molecules can functionally replace reprogramming factors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, Chou B-K, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate Greatly Enhances Derivation of Human Induced Pluripotent Stem Cells by Promoting Epigenetic Remodeling and the Expression of Pluripotency-Associated Genes. STEM CELLS. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, Chen Y, Wang Helen Y, Wang R-F. Jmjd3 Inhibits Reprogramming by Upregulating Expression of INK4a/Arf and Targeting PHF20 for Ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of Pluripotent Stem Cells from Mouse Embryonic Fibroblasts by Oct4 and Klf4 with Small-Molecule Compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Bagci H, Fisher AG. DNA demethylation in pluripotency and reprogramming: the role of tet proteins and cell division. Cell Stem Cell. 2013;13:265–269. doi: 10.1016/j.stem.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. Vitamin C Enhances the Generation of Mouse and Human Induced Pluripotent Stem Cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Xia Y, Son MY, Jin G, Seol B, Kim MJ, Son MJ, Do M, Lee M, Kim D, et al. A novel small molecule facilitates the reprogramming of human somatic cells into a pluripotent state and supports the maintenance of an undifferentiated state of human pluripotent stem cells. Angew Chem Int Ed Engl. 2012;51:12509–12513. doi: 10.1002/anie.201206691. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folmes Clifford DL, Nelson Timothy J, Martinez-Fernandez A, Arrell DK, Lindor Jelena Z, Dzeja Petras P, Ikeda Y, Perez-Terzic C, Terzic A. Somatic Oxidative Bioenergetics Transitions into Pluripotency-Dependent Glycolysis to Facilitate Nuclear Reprogramming. Cell Metabolism. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heng J-CD, Feng B, Han J, Jiang J, Kraus P, Ng J-H, Orlov YL, Huss M, Yang L, Lufkin T, et al. The Nuclear Receptor Nr5a2 Can Replace Oct4 in the Reprogramming of Murine Somatic Cells to Pluripotent Cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson Simon R, Chambers I. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Li K, Wei W, Ding S. Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C, Yu C, Ding S. Toward directed reprogramming through exogenous factors. Curr Opin Genet Dev. 2013;23:519–525. doi: 10.1016/j.gde.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 47.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, Cooke JP, Ding S. Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol. 2013;33:1366–1375. doi: 10.1161/ATVBAHA.112.301167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, Willenbring H, Ding S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [This study developed a stepwise protocol for the generation of hepatocytes without going through pluripotent state with higher engraftment efficiency than hepatocytes derived from iPSC or ESC.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Li K, Zhu S, Russ Holger A, Xu S, Xu T, Zhang Y, Ma T, Hebrok M, Ding S. Small Molecules Facilitate the Reprogramming of Mouse Fibroblasts into Pancreatic Lineages. Cell Stem Cell. 2014;14:228–236. doi: 10.1016/j.stem.2014.01.006. [This paper Used a combination of soluble molecules and a step wise transdifferentiation protocol to obtain large numbers of islets cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D, Ding S. Small Molecules Enable Cardiac Reprogramming of Mouse Fibroblasts with a Single Factor, Oct4. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ladewig J, Koch P, Brustle O. Leveling Waddington: the emergence of direct programming and the loss of cell fate hierarchies. Nat Rev Mol Cell Biol. 2013;14:225–236. doi: 10.1038/nrm3543. [DOI] [PubMed] [Google Scholar]
- 53.Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 54.Radzisheuskaya A, Silva JC. Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24:275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Radzisheuskaya A, Chia Gle B, dos Santos RL, Theunissen TW, Castro LF, Nichols J, Silva JC. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [This study showed that the defined Oct4 expression level functions in commitment to all embryonic lineages.] [DOI] [PMC free article] [PubMed] [Google Scholar]