Abstract
Scale insects (Hemiptera: Coccoidea) are small herbivorous insects found on all continents except Antarctica. They are extremely invasive, and many species are serious agricultural pests. They are also emerging models for studies of the evolution of genetic systems, endosymbiosis and plant-insect interactions. ScaleNet was launched in 1995 to provide insect identifiers, pest managers, insect systematists, evolutionary biologists and ecologists efficient access to information about scale insect biological diversity. It provides comprehensive information on scale insects taken directly from the primary literature. Currently, it draws from 23 477 articles and describes the systematics and biology of 8194 valid species. For 20 years, ScaleNet ran on the same software platform. That platform is no longer viable. Here, we present a new, open-source implementation of ScaleNet. We have normalized the data model, begun the process of correcting invalid data, upgraded the user interface, and added online administrative tools. These improvements make ScaleNet easier to use and maintain and make the ScaleNet data more accurate and extendable.
Database URL: http://scalenet.info
Introduction
Scale insects (Hemiptera: Coccoidea)
Scale insects are sap-sucking plant parasites that can be found almost anywhere that plants grow. They get their name from the protective waxy exudates produced by most species. Currently, there are at least 8194 described species, classified among 50 families. Scale insects play key roles in ecosystems. They, along with most other plant-feeding members of the order Hemiptera, are the only insects that feed exclusively on phloem sap (although armored scale insects feed primarily on parenchyma cells) (1). Phloem is rich in sugars but poor in amino acids, and phloem-feeding is an inefficient process. The waste is copious amounts of honeydew, i.e. sugar-rich excrement that is an important food source for birds, mammals and especially other insects (1). The availability of honeydew can affect insect communities in ways that alter ecosystem processes such as herbivore assemblage, soil structure, and predation (2, 3). Many scale insect species are agricultural pests, damaging plants through sap loss, encouraging the growth of sooty molds and vectoring plant diseases. Scale insects can be difficult to detect, and are extremely invasive. For example, scale insects account for only 1% of the total insect fauna of the United States, but for 13% of the introduced insect fauna, and on average one new invasive species is established as a pest in the USA per year (4). The host plant associations of scale insects have been exceptionally well documented, and the breadth of these associations is unusually variable. As is the case for other plant-feeding insects, most scale insect species are host-plant specialists. However, some species are among the most polyphagous insect species known. For example, the brown soft scale, Coccus hesperidum, can successfully develop on host plants in at least 121 plant families, and 325 plant genera. Scale insects are also noteworthy for the unparalleled diversity of their genetic systems, and for the diversity and complexity of their relationships with endosymbionts (5). In addition to being a taxing problem for applied biologists, they are emerging as models for research addressing questions about the evolution of reproductive modes, genetic conflict and collaboration, and niche breadth evolution. There is high demand for synoptic information about the biological diversity of scale insects. That demand is met by ScaleNet.
ScaleNet
ScaleNet is a manually-curated, web-accessible database that models the biological diversity of scale insects through ∼300 years of published research. ScaleNet manages information about the systematics, ecological associations (host plants, natural enemies and mutualists), geographic distributions, life histories, economic importance and morphology of each scale insect species. As a model of the scale insect literature, the core of ScaleNet is an exhaustive bibliography. The rest of the information in the database can be thought of as annotations of that literature. ScaleNet began as a collaboration between Yair Ben-Dov (Agricultural Research Organization, Israel Department of Entomology), Douglass R. Miller (US Department of Agriculture) and Gary A.P. Gibson (Agriculture and Agri-Food Canada), with funding from the USA–Israel Binational Agricultural Research and Development Fund. It was developed as a Microsoft FoxPro application, using the BASIS (Biological and Systematic Information System) database schema engineered by Gary Gibson and Jennifer Read (Agriculture and Agri-Food Canada) to manage taxonomic bibliographies. It first went online in 1995 (6). For 20 years, the ScaleNet data grew and evolved, but the ScaleNet application did not. By 2015, ScaleNet was running on an unsupported, insecure, closed-source software platform and was no longer tenable. Here, we describe a new version of ScaleNet.
Methods
Redeveloping ScaleNet
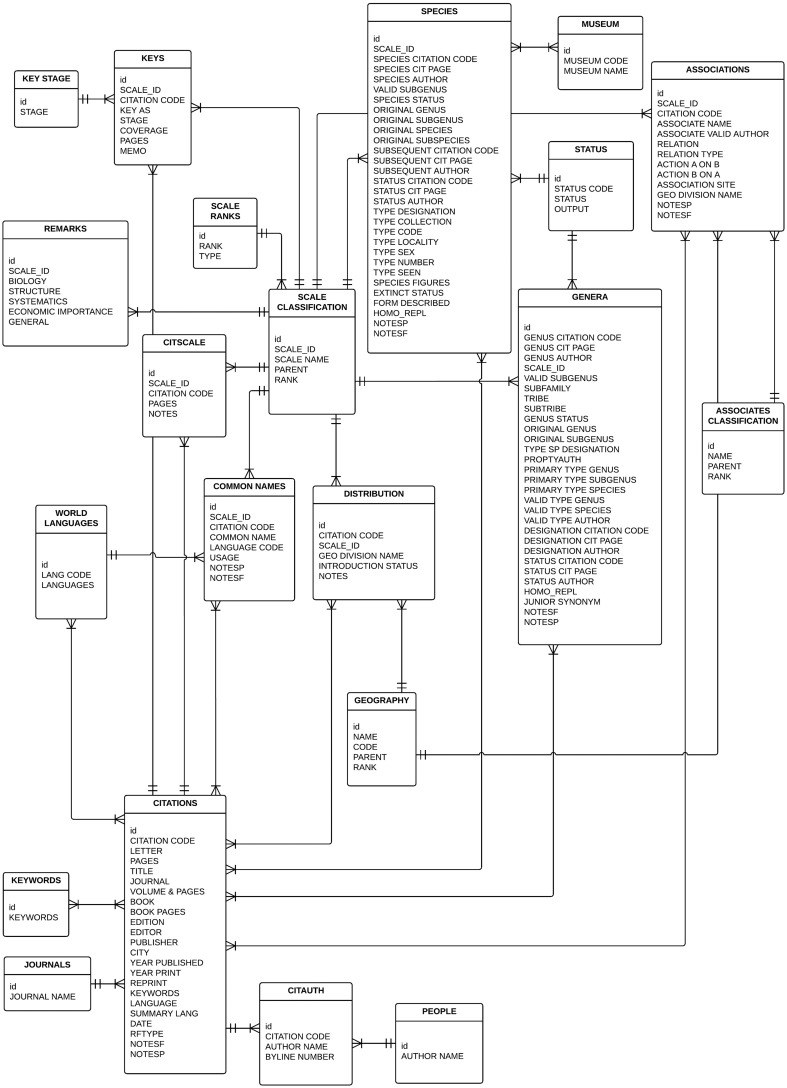
Our overarching goals for the redevelopment of ScaleNet were to (i) keep it online, (ii) make the software and data store easier to maintain, (iii) improve quality control and (iv) make it easier to extend and articulate with other biodiversity resources. Our new version of ScaleNet is a Django application (a Python web framework: https://www.djangoproject.com/) with an SQLite database engine (https://www.sqlite.org/), that currently runs on Linux, behind an Apache web server (http://httpd.apache.org/), but which can be configured to run in other environments. Django follows a Model-View-Controller architecture, i.e. the controller (logic) receives user requests and fetches information from the model (data store) to be displayed in a view (HTML). We normalized the data model (Figure 1) and performed the data migrations with a set of custom Python scripts. As part of the migration we performed a number of data cleaning and standardization routines. We standardized the valid scientific names and classifications of all ecological associates following the schema of the Catologue of Life (CoL: http://www.catalogueoflife.org/) 2015 annual checklist. To amend spelling errors in the names of ecological associates, we used the fuzzy matching feature of the Global Names Resolver API (http://resolver.globalnames.biodinfo.org/). In addition to adding the CoL classification of ecological associates to the ScaleNet schema, we added a class for the classification of scale insect taxa (absent from the original schema) and another for nested relationships of the geopolitical and zoogeographical units that are used to describe the geographic distributions of scale insects.
Figure 1.
Diagram of the ScaleNet data model. The central tables to the database are the Citations, Scale Classification, Species and Genera tables. Together these tables validate the currently accepted valid names of scale insects, which are then used throughout the database to track ecological associations, distributions, taxonomic keys, etc. The figure depicts relationships between the tables using Crow’s Foot Notation. The symbol ‖ represents a one-and-only-one relationship. The crow’s foot symbol represents a one-or-many relationship. Relationships can be asymmetrical, and the nature of the relationship of object A to object B is specified at the connection with B. For example, the relationship between Keys and Keys Stages would be read as ‘One key can have one and only one key stage; a key stage can be in one or many keys.’
As ScaleNet is a model of the scale insect primary literature, all ScaleNet data need to be associated with a publication. However, early in its initial development, ScaleNet was seeded with information from databases compiled by Y.B-D. to summarize the biological diversity of the scale insect families Coccidae and Pseudococcidae (7, 8). At that time validation sources for host and distribution records were not being recorded. These data are invalid in the new ScaleNet, and were not migrated. Instead, they were flagged and given to the ScaleNet curators to be manually restructured and added to the new database.
Results and discussion
Database overview
Currently, ScaleNet contains 23 477 bibliographic records, pertaining to 9509 currently valid scale insect names (8194 of which are species combinations). Complete nomenclatural histories are available for each genus and species name, and ScaleNet associates 1955 common names with 1161 valid scientific names. Because of the agricultural importance of scale insects, the ScaleNet information about ecological associations and geographic ranges are particularly rich. There are 47 341 records of ecological associations between scale insects and their hosts, natural enemies and mutualists. The geographic ranges of scale insect species are described by 32 641 records of occurrence in specific geopolitical or zoogeographic regions.
User interface
The public user interface exposes five major queries: (i) In the catalog query, users submit an available genus or species name to retrieve all of the information in the database associated with the valid form of that name. According to the rules of zoological nomenclature, a valid name is defined as the oldest available name for a genus or species, i.e. the one that has priority. An available species name is defined as any published binomial that is linked to a type specimen, and an available genus name is any published name that is linked to a type species. Users entering any of the available names associated with a species or genus will retrieve the data for the current valid name. The returned data view presents a nomenclatural history, lists of ecological associates and geopolitical units in which the taxon occurs, remarks on economic importance, biology, systematics and morphology and a complete bibliography. (ii) The places query allows users to retrieve a checklist of all of the scale insect species known to occur in a specified geopolitical or zoogeographic region. It is possible to constrain these searches to particular scale insect subgroups, e.g. specific genera. (iii) The ecological associates query returns a list of scale insect species associated with a specified host plant, natural enemy or mutualist. As in the places query, the results can be constrained to a scale insect subgroup. (iv) The references query gives users the ability to search the scale insect literature by author, year and keywords. (v) The common names query helps users make the connection between common and scientific names of scale insect species. Users can also peruse the taxonomic diversity of scale insects and access catalogs by drilling down (and up) through a searchable scale insect classification.
Administrative interface
Previously, ScaleNet data were managed through FoxPro desktop clients. In the new version of ScaleNet, data management is through online administrative interfaces that Django automatically generates from the model metadata. The new ScaleNet affords curators considerably more flexibility in terms of where they work on ScaleNet. It is also more flexible in terms of who can manage the data. For most of its history, the ScaleNet curators were Y. Ben-Dov and D.R. Miller. Currently, ScaleNet is curated primarily by B.D. Denno. For a period of time following the retirement of Y. Ben-Dov and D.R. Miller, no one maintained the ScaleNet data. By the time B.D. Denno started her tenure as curator, ScaleNet was several years out of date, and many known data errors had gone uncorrected. Should there be a period in the future in which no one is able to assume a major responsibility for the curation of ScaleNet data, it may be possible to open the administrative interface up to the community of scale insect workers at large.
Data curation
Scale insect papers are added to ScaleNet after they have been identified through weekly Internet searches, or have been sent directly to ScaleNet curators by authors. Updates to the database will be performed on a monthly basis. We aim for ScaleNet to include all published papers that deal with scale insects, but data entry is prioritized by subject, with the top priority going to papers that deal with the taxonomy and systematics of scale insects. Once a paper has been added to ScaleNet, curators extract information from that paper about the biological diversity of scale insects, and use that paper as a validation source for new records in various ScaleNet data classes (e.g. species names and geographic distributions). ScaleNet is meant to be a faithful representation of the literature; as a result, the data in ScaleNet is only as good as the data in the published literature. For the most part, ScaleNet curators do not judge the quality of the published information. If published information is erroneous, it needs to be corrected in a subsequent publication before that error will be corrected in ScaleNet. Nevertheless, ScaleNet curators may exercise their discretion on issues of nomenclature and classification. Nomenclature changes in ScaleNet must comply with the International Code of Zoological Nomenclature, and if a taxonomic paper fails to do so, the proposed changes will not be committed to ScaleNet. Furthermore, ScaleNet is a comprehensive resource for a global fauna. It may be impossible for ScaleNet curators to commit published changes to scale insect systematics that apply to non-monophyletic groups, e.g. a family-level reclassification of only the Palaearctic species of a global radiation.
The future of ScaleNet
One impetus for the normalization of the ScaleNet data model was to increase the quality of the data through structural validations. However, because these validations were lacking in the original application, a considerable amount of the data was invalid, and failed to be successfully migrated to the new platform. At the time of writing, manual restructuring and addition of these data is underway. Another impetus was to make ScaleNet more easily extendable, that is, increase the kinds of information accessible through ScaleNet. Some of what ScaleNet models, e.g. geographic ranges, can be more accurately modeled from specimen data, i.e. the metadata associated with physical insect specimens within natural history collections. Increasingly, these specimen data are available through web resources, such as the Global Biodiversity Information Facility’s data portal (http://www.gbif.org/) and Discover Life (http://www.discoverlife.org/). In the past few years, data from hundreds of thousands of hemipteran specimens held in non-federal insect collections in the USA have been digitized by the Tri-Trophic Database project, an NSF-funded effort in the Advancing Digitization of Biological Collections program. In the future, we aim to include specimen-level data in ScaleNet’s characterizations of scale insect biology.
ScaleNet is used heavily by insect identifiers as a diagnostic tool. The extreme invasiveness of scale insect species stems in part from high propagule pressure, i.e. the sheer number of individuals which are brought along with plant materials to ports of entry. Scale insect species identifications are among the highest volume and most difficult jobs performed by inspection services. In the future we plan to make ScaleNet more useful as a diagnostic aid, by adding habitus images, taxonomic illustrations and complete taxonomic descriptions to catalog entries. ScaleNet is used increasingly by ecologists and evolutionary biologists. For example, recent studies have used ScaleNet data to address questions about the evolution of parthenogenesis (9) and diet breadth (10, 11). To facilitate the compilation of comparative datasets from ScaleNet data, we plan to develop a ScaleNet web service API, i.e. more machine-friendly mechanisms for getting information from ScaleNet.
Acknowledgements
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA; USDA is an equal opportunity provider and employer.
Funding
This work was supported by a Specific Cooperative Agreement between N.B.H. at Auburn University and G.L.M. at the United States Department of Agriculture, Systematic Entomology Lab.
Conflict of interests. None declared.
References
- 1.Douglas A.E. (2006) Phloem-sap feeding by animals: problems and solutions. J. Exp. Bot., 57, 747–754. [DOI] [PubMed] [Google Scholar]
- 2.Styrsky J.D., Eubanks M.D. (2007) Ecological consequences of interactions between ants and honeydew-producing insects. Proc. R. Soc. B, 274, 151–164. doi: 10.1098/rspb.2006.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadler B., Michalzik B., Muller T. (1998) Linking aphid ecology with nutrient fluxes in a coniferous forest. Ecology, 79, 1514–1525. [Google Scholar]
- 4.Miller D.R., Miller G.L., Hodges G.S. et al. (2005) Introduced scale insects (Hemiptera: Coccoidea) of the United States and their impact on U.S. agriculture. Proc. Entomol. Soc. Wash., 107, 123–158. [Google Scholar]
- 5.Normark B.B. (2003) The evolution of alternative genetic systems in insects. Annu. Rev. Entomol., 48, 397–423. [DOI] [PubMed] [Google Scholar]
- 6.Miller D.R., Ben-Dov Y., Gibson G.A.P. (1999) ScaleNet: A Searchable Information System on Scale Insects. Entomologica, 33, 37–46. [Google Scholar]
- 7.Ben-Dov Y. (1994) A Systematic Catalogue of the Mealybugs of the World (Insecta: Homoptera: Coccoidea: Pseudococcidae: Putoidae). Intercept Ltd., Andover, UK, p. 686. [Google Scholar]
- 8.Ben-Dov Y. (1993) A systematic catalogue of the soft scales of the world (Insecta: Homoptera: Coccoidea: Coccidae) Flora and Fauna Handbook No. 9, Sandhill Crane Press, Gainesville, Florida, USA, p. 536. [Google Scholar]
- 9.Ross L., Shuker D.M., Normark B.B. et al. (2012) The role of endosymbionts in the evolution of haploid-male genetic systems in scale insects (Coccoidea). Ecol. Evol., 2, 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y.P., Cook D.H., Gullan P.J. et al. (2015) Does host-plant diversity explain species richness in insects? A test using Coccidae (Hemiptera). Ecol. Entomol., 40, 299–306. doi: 10.1111/een.12191 [Google Scholar]
- 11.Peterson D.A., Hardy N.B., Morse G.E. et al. (2015) Phylogenetic analysis reveals positive correlations between adaptations to diverse hosts in a group of pathogen-like herbivores. Evolution, 69, doi: 10.1111/evo.12772 [DOI] [PubMed] [Google Scholar]