Abstract
We performed a meta-analysis of five genome-wide association studies to identify common variants influencing colorectal cancer (CRC) risk comprising 8,682 cases and 9,649 controls. Replication analysis was performed in case-control sets totalling 21,096 cases and 19,555 controls. We identified three novel CRC risk loci at 6p21 (rs1321311, near CDKN1A; P=1.14×10−10), 11q13.4 (rs3824999, intronic to POLD3; P=3.65×10−10) and Xp22.2 (rs5934683, near SHROOM2; P=7.30×10−10) This brings to 20 the number of independent loci associated with CRC risk, and provides further insight into the genetic architecture of inherited susceptibility to CRC.
Many colorectal cancers (CRCs) develop in genetically susceptible individuals, most of whom are not carriers of germ-line mismatch repair or APC mutations1-3. Genome-wide association studies (GWASs) have validated the hypothesis that part of the heritable risk of CRC is attributable to common, low-risk variants identifying CRC susceptibility loci at 17 loci4-10. The statistical power of individual GWASs is limited by the modest effect sizes of genetic variants and financial constraints on the numbers of variants that can be followed up. Meta-analysis of existing GWAS data offers the opportunity to discover additional disease loci given current projections for the number of independent regions harbouring common variants associated with CRC risk11. In this study, we conducted a meta-analysis of GWAS data, followed by validation in multiple independent case-control series, identifying three novel susceptibility loci for CRC.
The discovery phase comprised five GWAS datasets from the UK population, totalling 8,682 cases and 9,649 controls (Supplementary Table 1). The Scotland1 GWAS consisted of genotyping 1,012 early-onset Scottish CRC cases and 1,012 controls using the Illumina HumanHap300 and HumanHap240S arrays (COGS Study). The London phase 1 (UK1) was based on genotyping 940 cases with familial colorectal neoplasia and 965 controls ascertained through the Colorectal Tumour Gene Identification (CoRGI) consortium using Illumina HumanHap550 arrays. Scotland2 was based on an additional 2,057 cases and 2,111 controls (SOCCS Study) and UK2 samples comprised an additional 2,873 CRC cases and 2,871 controls ascertained through the National Study of Colorectal Cancer Genetics (NSCCG). Scotland2 and UK2 samples were genotyped using Illumina Infinium-iSelect and GoldenGate arrays for a common set of 43,140 SNPs: the 14,982 most strongly associated SNPs from UK1; the 14,972 most strongly associated SNPs from Scotland1 and 13,186 SNPs showing the strongest association from a joint analysis of all CRC cases and controls from both phase 1 datasets. The VQ58 GWAS comprised 1,800 CRC cases from the UK-based VICTOR and QUASAR2 adjuvant chemotherapy clinical trials. The VQ58 cases were genotyped using the Illumina Hap300 and Hap370 arrays. The 2,690 controls, typed on the Illumina Human-1.2M-Duo Custom_v1 array, were from the UK population-based 1958 Birth Cohort.
Prior to undertaking the meta-analysis of all GWAS datasets, we searched for potential biases in each case-control series (Supplementary Figure 1). Comparison of the observed and expected distributions showed little evidence for an inflation of the test statistics (Supplementary Figure 2), thereby excluding the possibility of significant hidden population substructure, cryptic relatedness among subjects or differential genotype calling. Principal component analysis showed that the cases and controls were genetically well matched (Supplementary Figure 3; Supplementary Note). Any outliers or related individuals were excluded (Supplementary Methods; Supplementary Figure 1).
We also made use of data on 260 SNPs from 2,183 cases and 2,501 controls which had been genotyped as part of the COINNBS series. These SNPs had been selected as showing some evidence of association with CRC in a previous meta-analysis of the 5 GWAS datasets in which a smaller set of VQ cases had been genotyped8 (Supplementary Table 1).
Using data from the above six studies, we derived for each SNP joint odds ratios (ORs) and confidence intervals (CIs) under a fixed-effects model, and the associated P-values. We identified two SNPs, rs1321311 and rs3824999, showing good evidence of association (P<5.0×10−5) and mapping to distinct loci not previously associated with CRC risk. This threshold did not exclude the possibility that other SNPs represented genuine association signals, but was simply a pragmatic strategy for prioritizing replication.
To validate our findings, we conducted a replication study of rs1321311 and rs3824999, genotyping samples from nine additional case-control series: Colon Cancer Family Registry (CCFR1), UK NSCCG (UK3), UK CORGI (UK4), Edinburgh (Scotland3), Cambridge (Cambridge), Croatian (Croatia), Finnish Colorectal Cancer Predisposition Study (Helsinki), and Swedish (Sweden), together with a Japanese study (Japan) (Supplementary Table 1). In the combined analysis, both rs1321311 (P=1.14×10−10; Phet=0.55, I2=0%) and rs3824999 (P=3.65×10−10; Phet=0.05, I2=41%) showed evidence for an association with CRC at genome-wide significance (i.e., P<5.0×10−8) (Table 1, Supplementary Table 2).
Table 1.
Summary results for the SNPS: rs1321311 (6p21), rs3824999 (11q13.4) and rs5934683 (Xp22.2) associated with CRC risk.
| SNP | STUDY | ORa | 95% CIb | P-value |
|---|---|---|---|---|
| rs1321311 | Discovery | 1.09 | 1.05-1.14 | 4.79×10−5 |
| Replication | 1.09 | 1.05-1.14 | 5.74×10−6 | |
| Japan | 1.18 | 1.03-1.36 | 1.71×10−2 | |
| Combined | 1.10 | 1.07-1.13 | 1.14×10−10 (Phet = 0.55, I2 =0%) | |
| rs3824999 | Discovery | 1.08 | 1.05-1.13 | 1.77×10−5 |
| Replication | 1.07 | 1.04-1.11 | 2.06×10−5 | |
| Japan | 1.09 | 0.99-1.19 | 8.46×10−2 | |
| Combined | 1.08 | 1.05-1.10 | 3.65×10−10 (Phet = 0.05, I2 = 41%) | |
| rs5934683 | Discovery | 1.08 | 1.04-1.12 | 8.19×10−5 |
| Replication | 1.07 | 1.04-1.10 | 2.16×10−6 | |
| Japan | 1.04 | 0.93-1.16 | 5.38×10−1 | |
| Combined | 1.07 | 1.04-1.10 | 7.30×10−10 (Phet = 0.31, I2 = 13%) |
Odds ratio.
95% Confidence Interval.
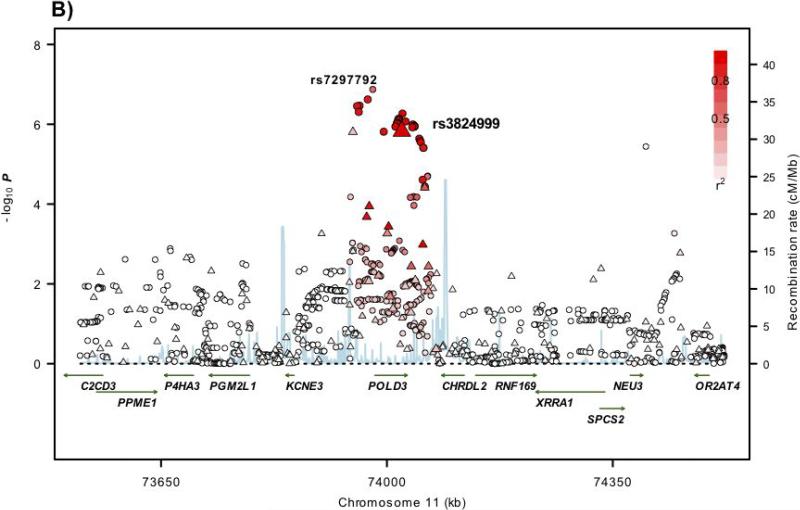
rs3824999 maps to 11q13.4 at 74,023,198bps, within intron 9 of the POLD3 gene (polymerase DNA-directed delta 3; MIM 611415; Figure 1). POLD3 is a component of the DNA polymerase-δ complex which comprises proliferating cell nuclear antigen (PCNA), the multisubunit replication factor C and the 4-subunit polymerase complex. As well as being involved in suppression of homologous recombination, the DNA polymerase-δ complex participates in DNA mismatch and base excision repair, key processes shown to be defective in Mendelian CRC susceptibility disorders12.
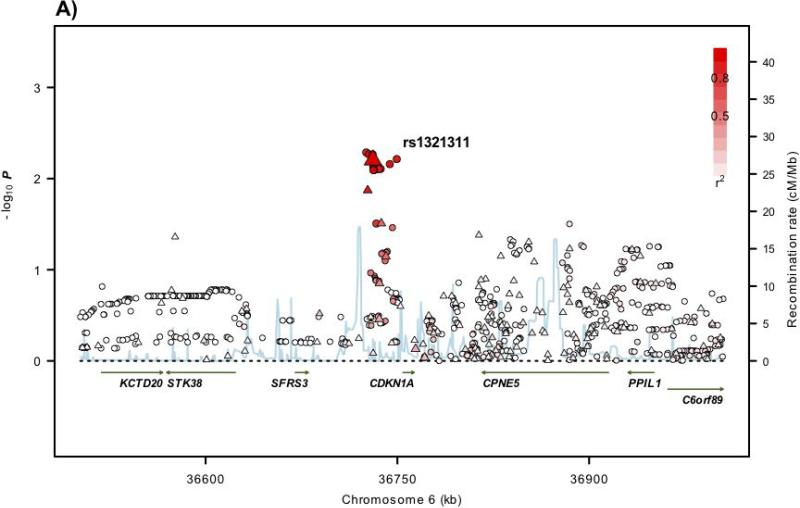
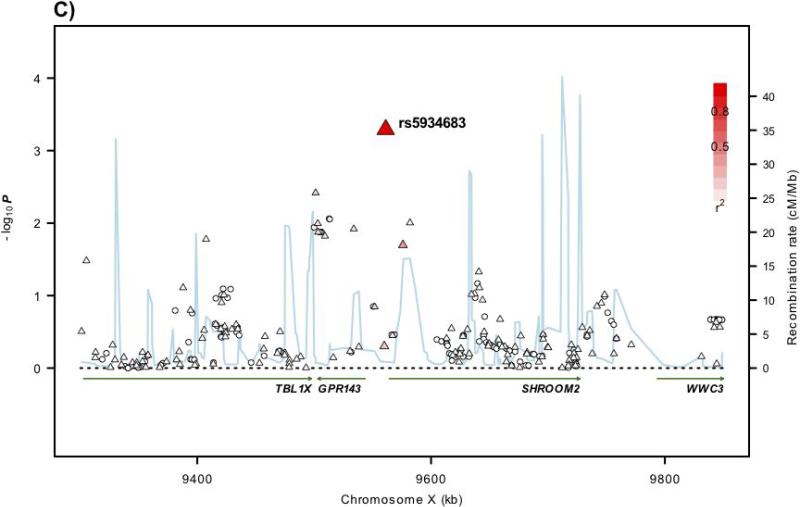
Figure 1. Regional plots of association results and recombination rates for the 6p21, 11q13.4, Xp22.2 susceptibility loci.
(a-d) Association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates within the loci: (a) 6p21, (b), 11q13.4, (c) Xp22.2. For each plot, −log10 P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The top genotyped SNP in each combined analysis is a large triangle and is labelled by its rsID. The colour intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2=0) through to dark red (r2=1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on NCBI build 36 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale.
rs1321311 maps to 6p21 at 36,730,878bps within a region of linkage disequilibrium (LD) that encompasses the CDKN1A gene (cyclin-dependent kinase inhibitor 1A; MIM 116899; Figure 1). Intriguingly, rs1321311 has been shown to be associated with electrocardiographic QRS duration13. CDKN1A encodes p21WAF1/Cip1 which mediates p53-dependent G1 growth arrest14. Moreover, p21 acts as a master effector of multiple tumour suppressor pathways which are independent of classical p53 tumour suppression. Also, by binding to PCNA, p21 interferes with PCNA-dependent DNA polymerase activity, thereby inhibiting DNA replication and modulating PCNA-dependent DNA repair14. Through binding to PCNA, p21 also competes for PCNA binding with DNA polymerase-δ and several other proteins involved in DNA synthesis, thus directly inhibiting DNA synthesis14. Similarly, p21 represses MYC-dependent transcription and in turn, MYC disrupts the PCNA–p21 interaction, thus alleviating p21-dependent inhibition of PCNA and DNA synthesis14. Decreased p21 expression has been reported to be a feature of dysplastic aberrant crypt foci in colonic mucosa and adenomas. The finding that p21 down-regulation inversely correlates with MSI status in CRC, irrespective of p53 status, again invokes a relationship with defective DNA repair and genomic instability14.
Including the two newly-discovered SNPs, a total of 19 independent risk SNPs for CRC have been identified, all mapping to autosomal regions of the genome. The risk of sporadic CRC is higher for males in both economically developed and less-developed countries. Furthermore, males are at greater overall CRC risk and have earlier age at onset in Lynch Syndrome15-17. It is therefore possible that some of these differences in risk are attributable to sex chromosome genetic variation. To explore this hypothesis, we studied the relationship between SNPs mapping to the sex-specific region of the X-chromosome and CRC risk. Genotypes were analysed using an extension to the standard Cochran-Armitage test for trend18 (Supplementary Methods).
rs5934683 was the only SNP showing strong evidence of association in the meta-analysis of UK1, UK2, Scotland1, Scotland2, and VQ58. We then genotyped rs5934683 in UK3, Scotland3, UK4, CCFR1, Cambridge, Croatia, Helsinki, Sweden, and Japan (Supplementary Table 1). In the combined analysis, rs5934683 showed evidence for an association with CRC at genome-wide significance (P=7.30×10−10, Phet=0.31, I2=13%; Table 1; Supplementary Table 2).
rs5934683 maps to Xp22.2 within a 43Kb region of LD (9,711,474bps, Figure 1). Two genes map to this region, GPR143 (G protein-coupled receptor 143; MIM300808) which is expressed by melanocytes and retinal pigment epithelium and SHROOM2 (shroom family member 2; MIM 300103) a human homolog of the Xenopus laevis APX gene. rs5934683 is situated between GPR143 and SHROOM2 and appears to be within the distal promoter region of SHROOM2. There is also evidence of longer, less abundant GPR143 transcripts extending into the SHROOM2 promoter. SHROOM2 is known to have broad roles in cell morphogenesis, during endothelial and epithelial tissue development19. Missense mutations in SHROOM2 have been detected in large-scale screens for recurring mutations in cancer cell lines20. Like GPR143, SHROOM2 regulates melanosome biogenesis and localisation in the retinal pigment epithelium21. Intriguingly, abnormal retinal pigmentation, similar to the congenital hypertrophy of retinal pigment epithelium (CHRPE) lesions that are a component of the familial adenomatous polyposis syndrome, has been previously been shown to be an extracolonic feature of non-FAP CRC22,23. To our knowledge, the relationship between Xp22.2 and CRC risk represents the first evidence for the role of X-chromosome variation in predisposition to a non-sex-specific cancer.
Next we assessed associations between clinico-pathological variables (sex, age at diagnosis, family history of CRC, tumour site, stage or microsatellite instability) and genotype at rs1321311, rs3824999 and rs5934683 through case-only logistic regression (Supplementary Table 3). After adjusting for multiple testing, we did not find any significant association.
To analyse comprehensively the associations at 6p21, 11q13.4 and Xp22.2, we imputed genotypes in GWAS cases and controls using HapMap3 and 1000genomes data for the autosomal regions and HapMap release21 for Xp22.2 (Supplementary Methods; Figure 1). We did not find substantive evidence of stronger associations at the 6p21.2 and Xp22.2 risk loci. However, at the 11q13.4 locus, rs72977282, mapping 3,188bps 5′ to POLD3, was more strongly associated with CRC than rs3824999 (Figure 1; Supplementary Table 4). No non-synonymous SNPs showing strong LD (i.e. r2>0.4/D′>0.8) with rs1321311, rs3824999 or rs5934683 at 6p21, 11q13.4 and Xp22.2 loci were identified. These data make it likely that the associations between 6p21, 11q13.4 and Xp22.2 and CRC risk are mediated through changes that influence gene expression rather than impacting on protein sequence.
To examine if any directly typed or imputed SNPs lie within or very close to a putative transcription factor binding/enhancer element, we conducted a bioinformatic search using Transfac24, ENCODE CHIP-Seq and ENCODE UW DNAaseI Hypersensitivity data. These analyses did not provide evidence that rs1321311, rs3824999 and rs5934683or any closely correlated SNP maps to a known or predicted region of transcriptional regulation (Supplementary Table 4).
To explore whether the rs1321311, rs3824999 and rs5934683 associations (or SNP proxies) reflect cis-acting regulatory effects on POLD3, CDKN1A, GPR143 or SHROOM2, we conducted expression studies using Illumina HT-12 arrays using RNA extracted from 42 samples of normal colonic epithelium (Supplementary Table 5). We also analyzed publicly-available mRNA expression data from fibroblasts, lymphoblastoid cell lines (LCL), T-cells, adipose tissue and CRC25,26 (Supplementary Table 5). In silico analysis revealed a statistically significant relationship between rs1321311 genotype and expression of CDKN1A. However, this was observed only in the LCLs and T-cell data, with no evidence of an effect in colon (Supplementary Table 5). We also found that the risk allele at rs5934683 was associated with a striking reduction in SHROOM2 expression in both normal colonic-epithelium and CRC tissue (Supplementary Figure 4). The relationship between SHROOM2 expression in normal colonic epithelium and rs5934683 genotype was very strong (P=1.3×10−7) and was significant after accounting for all genes tested on the HT-12 array (P=9.0×10−4). Indeed, rs5934683 genotype accounted for 55% of the variation in SHROOM2 expression. Exploring the relationship between SHROOM2 expression, rs5934683 risk genotype and CRC causation will be of considerable interest, not least because of the observations of an association between excess pigmented lesions in the retinal pigment epithelium and CRC22,23. There was no significant difference in the observed MAF of rs5934683 between female and male cases raising the possibility that skewed X-inactivation might underscore the associated CRC risk. Favored X-inactivation producing a normal phenotype has been documented in X-linked dominant disease27 and skewed X-inactivation has been implicated as a risk factor for breast cancer28. The expression data were consistent with full dosage compensation but, due to sample and effect sizes, we are currently unable to confirm or refute a dosage effect on risk. There was no detectable relationship between rs3824999 and POLD3 expression from any of the expression studies. It should be noted that these exploratory analyses could only detect >5% difference in RNA expression by genotype with 80% power at a single time point and hence we could not exclude any subtle effects of genotype on target tissues relevant to CRC.
By pooling GWAS data and conducting extensive replication analyses, we have identified three new loci influencing CRC susceptibility. The loci are of modest effect size, which is unsurprising given that common alleles with a larger impact on CRC were likely to have been discovered in previous studies. While additional analyses are required to determine the functional consequences that lead to CRC, our findings highlight the importance of variation in genes encoding components of the p21WAF1/Cip1 signalling pathway in CRC. This pathway, elucidated through the extended interaction network of CDKN1A, incorporates not only POLD3 discovered as a CRC locus here, but also MYC and other genes (including SMADs and other TGF-β pathway genes) that we have previously identified as risk factors for CRC.
URLs
The R suite can be found at http://www.r-project.org
Detailed information on the tag SNP panel can be found at http://www.illumina.com
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP
HapMap: http://www.hapmap.org
1000Genomes: http://www.1000genomes.org
SNAP http://www.broadinstitute.org/mpg/snap
IMPUTE: https://mathgen.stats.ox.ac.uk/impute/impute.html
SNPTEST: http://www.stats.ox.ac.uk/~marchini/software/gwas/snptest.html
Transfac Matrix Database: http://www.biobase-international.com/pages/index.php?id=transfac
Wellcome Trust Case Control Consortium: www.wtccc.org.uk
Mendelian Inheritance In Man: http://www.ncbi.nlm.nih.gov/omim
SIFT: http://sift.jcvi.org/
PolyPhen: http://genetics.bwh.harvard.edu/pph
Globocan: http://globocan.iarc.fr
Cancer Genome Atlas project: http://cancergenome.nih.gov
The ENCODE Project: ENCyclopedia Of DNA Elements: http://www.genome.gov
Genevar (GENe Expression VARiation): http://www.sanger.ac.uk/resources
Catalogue Of Somatic Mutations In Cancer: http://www.sanger.ac.uk/genetics/CGP/cosmic
METHODS
Ethics statement
Collection of blood samples and clinico-pathological information from subjects was undertaken with informed consent and ethical review board approval at all sites in accordance with the tenets of the Declaration of Helsinki.
Datasets, sample preparation and genotyping
Full details of each dataset are provided in the Supplementary Note.
DNA was extracted from samples using conventional methods and quantified using PicoGreen (Invitrogen). The VQ, UK1, and Scotland1 GWA cohorts were genotyped using Illumina Hap300, Hap240S, Hap370, or Hap550 arrays. 1958BC and NBS genotyping was performed as part of the WTCCC2 study on Hap1.2M-Duo Custom arrays. The CCFR1 samples were genotyped using Illumina Hap1M or Hap1M-Duo arrays. In UK2 and Scotland2, genotyping was conducted using custom Illumina Infinium arrays according to the manufacturer's protocols. Some COIN SNPs were typed on custom Illumina Goldengate arrays. To ensure quality of genotyping, a series of duplicate samples was genotyped, resulting in 99.9% concordant calls in all cases. Other genotyping was conducted using competitive allele-specific PCR KASPar chemistry (KBiosciences Ltd, Hertfordshire, UK), Taqman (Life Sciences, Carlsbad, California) or MassARRAY (Sequenom Inc., San Diego, USA). All primers, probes and conditions used are available on request. Genotyping quality control was tested using duplicate DNA samples within studies and SNP assays, together with direct sequencing of subsets of samples to confirm genotyping accuracy. For all SNPs, >99% concordant results were obtained.
Quality control and sample exclusion
We excluded SNPs from analysis if they failed one or more of the following thresholds: GenCall scores <0.25; overall call rates <95%; MAF<0.01; departure from Hardy-Weinberg equilibrium (HWE) in controls at P<10−4 or in cases at P<10−6; outlying in terms of signal intensity or X:Y ratio; discordance between duplicate samples; and, for SNPs with evidence of association, poor clustering on inspection of X:Y plots. We excluded individuals from analysis if they failed one or more of the following thresholds: duplication or cryptic relatedness to estimated identity by descent (IBD) >6.25%; overall successfully genotyped SNPs<95%; mismatch between predicted and reported gender; outliers in a plot of heterozygosity versus missingness; and evidence of non-white European ancestry by PCA-based analysis in comparison with HapMap samples (http://hapmap.ncbi.nlm.nih.gov). Details of all sample exclusions are provided in Supplementary Figure 1.
To identify individuals who might have non-northern European ancestry, we merged our case and control data from all sample sets with the 60 European (CEU), 60 Nigerian (YRI), and 90 Japanese (JPT) and 90 Han Chinese (CHB) individuals from the International HapMap Project. For each pair of individuals, we calculated genome-wide identity-by-state distances based on markers shared between HapMap2 and our SNP panel, and used these as dissimilarity measures upon which to perform principal components analysis. Principal components analysis was performed in R using CEU, YRI and HCB HapMap samples as reference. The first two principal components for each individual were plotted and any individual not present in the main CEU cluster (that is, >5% of the PC distance from HapMap CEU cluster centroid) was excluded from subsequent analyses (Supplementary Figure 3).
We had previously shown the adequacy of the case-control matching and possibility of differential genotyping of cases and controls using Q-Q plots of test statistics. The inflation factor λGC was calculated by dividing the mean of the lower 90% of the test statistics by the mean of the lower 90% of the expected values from a χ2 distribution with 1 d.f. Deviation of the genotype frequencies in the controls from those expected under HWE was assessed by χ2 test (1 d.f.), or Fisher's exact test where an expected cell count was <5.
Statistical and bioinformatic analysis
Main analyses were undertaken using R (v2.6), Stata v.11 (College Station, Texas, US) and PLINK (v1.06) software29. The association between each SNP and risk of CRC was assessed by the Cochran-Armitage trend test. Odds ratios (ORs) and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression. Meta-analysis was conducted using standard methods30. Cochran's Q statistic to test for heterogeneity30 and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated31. I2 values ≥75% are considered characteristic of large heterogeneity31,32. Associations by sex, age and clinic-pathological phenotypes were examined by logistic regression in case-only analyses.
For SNPs on the non-pseudoautosomal region of X chromosome, males carry only one copy and in females most loci are subject to X inactivation33. To test for X chromosome associations we used an extension to the standard, 1df Cochran-Armitage test for trend, proposed by Clayton (2008)18 whereby males can be regarded as homozygous females. This 1df trend test adjusts for the different variances for males and females.
Prediction of the untyped SNPs was carried out using IMPUTEv2, based on HapMap Phase III haplotypes release 2 (HapMap Data Release 27/phase III Feb 2009 on NCBI B36 assembly, dbSNP26) and 1000genomes. Imputation of the X chromosome loci was only possible using IMPUTEv1 with HapMap Data Release 21 on NCBI Build 35. Imputed data were analysed using SNPTEST v2 to account for uncertainties in SNP prediction. An imputation info score of 0.95 was used to remove SNPs with poor imputation quality. LD metrics between HapMap SNPs were based on Data Release 27/phase III (Feb 2009) on NCBI B36 assembly, dbSNP26, viewed using Haploview software (v4.2) and plotted using SNAP. LD blocks were defined on the basis of HapMap recombination rate (cM/Mb) as defined using the Oxford recombination hotspots34 and on the basis of distribution of confidence intervals defined by Gabriel et al 35. To annotate potential regulatory sequences within disease loci we implemented in silico searches using Transfac Matrix Database v7.29 24, and PReMod1036 36software. We used the in silico algorithms SIFT and PolyPhen to predict the impact of amino acid substitutions.
Relationship between SNP genotype and mRNA expression
Expression studies in colonic epithelium
To examine for a relationship between SNP genotype and mRNA expression in colonic epithelium, 42 samples were collected fresh immediately after surgical resection of specimens for colorectal cancer (n=34), solitary adenoma (n=5) or benign conditions (not inflammatory bowel disease) (n=3). For 2 of the 42 subjects, 3 samples of mucosa were harvested from different locations of the fresh resected bowel. Normal epithelium was dissected from muscularis propria, and samples snap frozen and placed in RNAlater (Applied Biosystems) and kept at 4°C overnight before storage at −80°C. Tissue was disrupted and homogenised using TissueLyser LT (Qiagen) and RNA extracted using Ribopure kit (Applied Biosystems). RNA integrity and concentration were assessed on an Agilent Bioanalyzer, RNA purity (A260/A280 and A260/A230) on Nanodrop. RT-PCR products were analysed on HumanHT-12 Expression BeadChip which were scanned using the Illumina HiScan. Array data processing and analysis was performed using Illumina GenomeStudio software (version 2011.1). Microarray data were exported from Illumina Beadstudio software, processed and normalized using the R, Bioconductor beadarray and limma packages37,38. Prior to normalization probes that were not detected (detection P-value>0.01) on the microarrays were removed. Microarrays were Quantile normalized to remove technical variation. Three mucosa samples were available for 2 of the 42 subjects and in which we used the average signal of the replicates in the analysis. The limma package was used to find differential expressed genes, using the functions lmFit, eBayes and topTable. To test all associations between SNPs and expression, a linear model was fitted to the expression level of each probe, using this genotype value as effect. For SNPs associations with gene expression on the X chromosome, gender was added to the model. Significant associations were considered as < 0.05 using P-values adjusted for multiple testing using the Benjamini, Hochberg method from R's p.adjust function
In silico analysis of publicly available expression data
We analysed expression data generated from: (1) Fibroblast, lymphoblastoid cell lines (LCL) and T-cells derived from the umbilical cords of 75 Geneva GenCord individuals25; (2) 166 adipose, 156 LCL and 160 skin samples derived from a subset of healthy female twins of the MuTHER resource26 using Sentrix Human-6 Expression BeadChips (Illumina, San Diego, USA)39,40 (3) AgilentG4502A_07_3 custom gene expression data on 154 CRCs as part of the Cancer Genome Atlas project: http://cancergenome.nih.gov. Power of assays to establish a relationship between genotype and expression we made using STATA software.
Assignment of microsatellite instability (MSI) in colorectal cancers
Tumour MSI status in CRCs was determined using the mononucleotide microsatellite loci BAT25 and BAT26, which are highly sensitive MSI markers. Briefly, 10 mm sections were cut from formalin-fixed paraffin-embedded CRC tumours, lightly stained with toluidine blue and regions containing at least 60% tumour microdissected. Tumour DNA was extracted using the QIAamp DNA Mini kit (Qiagen, Crawley, UK) according to the manufacturer's instructions and genotyped for the BAT25 and BAT26 loci using either 32P–labelled or fluorescently-labelled oligonucleotide primers (UK2/3 and COINNBS studies respectively). Samples showing more than or equal to five novel alleles, when compared with normal DNA, at either or both markers were assigned as MSI-H (corresponding to MSI-high)41.
Supplementary Material
ACKNOWLEDGEMENTS
Cancer Research UK provided principal funding for this study individually to R.S.H. (C1298/A8362 - Bobby Moore Fund for Cancer Research UK), I.P.M.T., and M.G.D. At the Institute of Cancer Research additional funding was provided a Centre grant from CORE as part of the Digestive Cancer Campaign, the National Cancer Research Network and the NHS via the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. S,L., was in receipt of a PhD studentship from Cancer Research UK, I.C., a Clinical Research Training Fellowship from St. George's Hospital Medical School and N.W., is a PhD Studentship from the Institute of Cancer Research. M.H. was in receipt of a Post-Doctoral Training post from Leukaemia Lymphoma Research Fund.
In Oxford additional funding was provided by the Oxford Comprehensive Biomedical Research Centre (E. Domingo, C.P. and I.P.M.T.) and the EU FP7 CHIBCHA grant (A.J. L.G.C.-C. and I.P.M.T.). Core infrastructure support to the Wellcome Trust Centre for Human Genetics, Oxford was provided by grant (090532/Z/09/Z).
We are grateful to many colleagues within UK Clinical Genetics Departments (for CORGI) and to many collaborators who participated in the VICTOR and QUASAR2 trials. We also thank colleagues from the UK National Cancer Research Network (for NSCCG).
In Edinburgh funding was provided by a Cancer Research UK Programme Grant (C348/A12076) and a Centre Grant from the CORE Charity. E.T. was funded by a Cancer Research UK Fellowship (C31250/A10107). LYO is supported by a Cancer Research UK Research Training Fellowship (C10195/A12996). Claire S is supported by an MRC Research Studentship to the MRC HGU. We gratefully acknowledge the work of Marion Walker and Stuart Reid for technical support; Ruth Wilson (SOCCS3 and COGS study coordinator), Gisela Barr for data entry in SOCCS studies, and the research nurse recruitment teams; the Wellcome Trust Clinical Research Facility for sample preparation; and to all surgeons, oncologists and pathologists throughout Scotland at contributing centres. Lothian Birth Cohort Illumina genotyping was supported by the UK's Biotechnology and Biological Sciences Research Council (BBSRC). Phenotype collection in the Lothian Birth Cohort 1921 was supported by the BBSRC, The Royal Society and The Chief Scientist Office of the Scottish Government. Phenotype collection in the Lothian Birth Cohort 1936 was supported by Research into Ageing (continues as part of Age UK's The Disconnected Mind project). The work on the Lothian Birth Cohorts was undertaken in the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Funding from the BBSRC, EPSRC, ESRC and MRC is gratefully acknowledged.
For the Cambridge study, we thank the SEARCH study team and all the participants in the study. SEARCH is funded by a grant from Cancer Research UK (C490/A10124).
In Cardiff, the work was supported by the Kidani Trust, Tenovus, Cancer Research Wales, The Bobby Moore Fund from CRUK (ref C10314/A4886), the Wales Assembly Government NISCHR Cancer Genetics BRU and the Wales Gene Park (all to J Cheadle). We acknowledge the use of DNA from the blood samples collected from COIN and COIN-B funded by Cancer Research UK and the MRC. We also acknowledge the use of DNA from the NBS (UKBS) collection, funded by the Wellcome Trust grant 076113/C/04/Z, by the Juvenile Diabetes Research Foundation grant WT061858, and by the National Institute of Health Research of England. The COIN-B Collaborative Group includes H Wasan, T Maughan, R Adams, R Wilson, A Madi, E Hodgkinson, M Pope, P Rogers, J Cassidy.
The Swedish sample and data resource was funded by the Swedish Cancer Society, the Swedish Scientific Research Council and the Stockholm Cancer Foundation. We acknowledge the contribution to recruitment and data collection of the Swedish Low-Risk Colorectal Cancer Study Group (list of contributing surgeons below).
For the Helsinki study, the work was supported by grants from Academy of Finland (Finnish Centre of Excellence Program 2006-2011), the Finnish Cancer Society and the Sigrid Juselius Foundation.
This work of the Colon Cancer Family Registry CFR was supported by the National Cancer Institute, National Institutes of Health under RFA #CA-95-011 and through cooperative agreements with members of the Colon CFR and Principal Investigators. Collaborating centers include the Australasian Colorectal Cancer Family Registry (U01 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01 CA074799), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783) and the Seattle Colorectal Cancer Family Registry (U01 CA074794). The Colon CFR GWAS was supported by funding from the National Cancer Institute, National Institutes of Health (U01CA122839 to GC).
The Japanese study was conducted as a part of the BioBank Japan Project that was supported by the Ministry of Education, Culture, Sports, Science and Technology of the Japanese government.
This study made use of genotyping data from the 1958 Birth Cohort and NBS samples, kindly made available by the Wellcome Trust Case Control Consortium 2. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk/. Finally, we would like to thank all individuals who participated in the study.
Footnotes
Full lists of members of The CORGI Consortium, The COIN Collaborative Group, The Swedish Low-Risk Colorectal Cancer Study Group are provided in the Supplementary Material.
AUTHOR CONTRIBUTIONS
The study was designed and financial support was obtained by R.S.H., I.P.M.T., and M.G.D. The manuscript was drafted by R.S.H., I.P.M.T., and M.G.D. Statistical and bioinformatic analyses were conducted by S.E.D., N.W, with contributions from Y.M, M.H., M.F., C.S., G.G., R.S.H, P.B, S.S and I.P.M.T.
Institute of Cancer Research and local collaborators: subject recruitment and sample acquisition to NSCCG were undertaken by S.P. The coordination of sample preparation and genotyping was performed by P.B. Sample preparation and genotyping were performed by A.L. B.O. and N.W. Tumour pathology analyses were performed by I.C. and S.L.
Oxford and local collaborators: subject recruitment and sample acquisition were done by E.B., M.G., L.M., and members of the CORGI Consortium, and by R. Midgley and D.J.K. Genotyping was performed and coordinated by L.G.C.-C., A.W. and E. Domingo.
Colon Cancer Genetics Group, Edinburgh and local collaborators: subject recruitment and sample acquisition were performed by S.F. S.H., J.M.S., I.D., H.C., and M.G.D., as well as members of SOCCS and COGS recruitment teams. Sample preparation was coordinated by S.F. Genotyping was performed and coordinated by S.M.F., and M.G.D. Data curation and analysis in Edinburgh was conducted by A.T., S.M., E.T., L.Z., J.P., and S.B. Recruitment sample preps, wet lab expression analysis and genotyping was performed by L.Y.O., and C.S., G.G., and C.S., performed the bioinformatic analyses.
The following authors from collaborating groups conceived the local or national study, undertook assembly of case-control series in their respective regions, collected data and samples and variously undertook genotyping and analysis: C.G.S., H.W., S.I., T.M. and J. Cheadle in Cardiff; I.N., S.T. and L.A.A. in Finland; and P.P. in Cambridge. All other authors undertook sample collection and phenotype data collection and collation in the respective centres.
Note: Supplementary information is available on the Nature Genetics website
Consortium members
The CORGI Consortium comprises: Huw Thomas23, Eamonn Maher24, Gareth Evans25, Lisa Walker26, Dorothy Halliday26, Anneke Lucassen27, Joan Paterson28, Shirley Hodgson29, Tessa Homfray29, Lucy Side30, Louise Izatt31, Alan Donaldson32, Susan Tomkins32, Patrick Morrison33, Carole Brewer34, Alex Henderson35, Rosemarie Davidson36 and Victoria Murday36, Jaqueline Cook37, Neva Haites38, Timothy Bishop39, Eamonn Sheridan39, Andrew Green40, Christopher Marks41, Sue Carpenter41, Mary Broughton41, Lynn Greenhalge42, Mohnish Suri43.
23. Family Cancer Clinic, St.Mark's Hospital, Harrow and Imperial College, UK
24. Dept. of Clinical Genetics, University of Birmingham, UK
25. Dept. of Clinical Genetics, University of Manchester, UK
26. Oxford Regional Genetics Service, Churchill Hospital, Oxford, UK
27. Wessex Regional Genetics Service, Princess Anne Hospital, Southampton, UK
28. Anglia Regional Genetics Service, Addenbrooke's Hospital, Cambridge, UK
29. South-West Thames Regional Genetics Service, St George's Hospital, Tooting, London, UK
30. North-East Thames Regional Genetics Service, Great Ormond Street Hospital, London, UK
31. South-East Thames Regional Genetics Service, Guy's Hospital, London, UK
32. South-West Regional Genetics Service, Bristol, UK
33. Northern Ireland Regional Genetics Service, City Hospital, Belfast, UK
34. South-West Regional Genetics Service, Royal Devon and Exeter Hospital, Exeter, UK
35. Northern Regional Genetics Service, International Centre for Life, Newcastle, UK
36. West of Scotland Regional Genetics Service, Yorkhill Hospital, Glasgow, UK
37. Sheffield Regional Genetics Service, Children's Hospital, Sheffield, UK
38. North of Scotland Regional Genetics Service, Foresterhill Hospital, Aberdeen, UK
39. Yorkshire Regional Genetics Service, St James's Hospital, Leeds, UK
40. Republic of Ireland Genetics Service, Our Lady's Hospital for Sick Children, Dublin, Republic of Ireland
41. The Royal Surrey County Hospital, Egerton Road, Guildford, Surrey
42. Department of Clinical Genetics, Royal Liverpool Children's Hospital, Eaton Road, Alderhay, Liverpool
43. Department of Clinical Genetics, City Hospital, Hucknall Road, Nottingham
The Swedish Low-Risk Colorectal Cancer Study Group comprises; David Edler44, Claes Lenander45, Johan Dalén46, Fredrik Hjern47, Nils Lundqvist48, Ulrik Lindforss49, Lars Påhlman50, Kennet Smedh51, Anders Törnqvist52, Jörn Holm53, Martin Janson54, Magnus Andersson55, Susanne Ekelund56, Louise Olsson57.
44. Karolinska Universitetssjukhuset/Solna, 171 76 Stockholm
45. Mag-tarm-centrum, Ersta sjukhus, Box 4622, 116 91 Stockholm
46. S:t Görans sjukhus, 112 81 Stockholm
47. Danderyds sjukhus, 182 88 Danderyd
48. Norrtälje sjukhus, Box 905, 761 29 Norrtälje
49. Södertälje sjukhus, 152 86 Södertälje
50. Akademiska sjukhuset, 751 85 Uppsala
51. Centrallasarettet, 721 89 Västerås
52. Centralsjukhuset, 651 85 Karlstad
53. Länssjukhuset Gävle-Sandviken, 801 87 Gävle
54. Karolinska Universitetssjukhuset/Huddinge, 141 86 Huddinge
55. Universitetssjukhuset, 703 62 Örebro
56. Södersjukhuset, 118 83 Stockholm
57. Mälarsjukhuset, 631 88 Eskilstuna
The COIN Collaborative Group comprises:
Investigators and Centres: G MacDonald58, L M Samuel58, A Ahmad59, P Corrie59, D Joell59, C Palmer59, C Wilson59, J O'Hagan60, D Smith60, R McDermott61, J Walsh61, (J Cassidy62, A McDonald62, N Mohammed62, J White62, H Yosef62, (O Breathnach63, L Grogan63, R Thomas64, M Eatock65, P Henry65, R Houston65, P Johnston65, R Wilson65, I Geh66, F Danwata67, A Hindley67, S Susnerwala67, C Bradley68, A Conn68, A Raine68, C Twelves68, S Falk69, K Hopkins69, S Tahir70, A Dhadda71, A Maraveyas71, J Sgouros71, M Teo71, R Ahmad72, S Cleator72, A Creak72, C Lowdell72, P Riddle72, K Benstead73, D Farrugia73, N Reed73, S Shepherd73, E Levine74, S Mullamitha74, M Saunders74, J Valle74, G Wilson74, A Jones75, A Weaver75, P I Clark76, B Haylock76, MI Iqbal76, S Myint76, D Smith76, S Beesley77, T Sevitt77, J Nicoll78, F Daniel79, V Ford79, T Talbot79, M Butt80, A Hamid80, P Mack80, R Roy80, R Osborne81, F McKinna82, H Alsab83, D Basu83, P Murray83, B Sizer83, FA Azam84, S Gollins84, R Neupane84, A Waterston85, J Glaholm86, C Blesing87, S Lowndes87, A Medisetti87, A Gaya88, M Leslie88, N Maisey88, P Ross88, G Dunn89, H Yosef89, O Al-Salihi90, HS Wasan90, C Palmer91, LT Tan91, J Dent92, U Hofmann92, JK Joffe92, E Sherwin93, R Soomal93, A Chakraborti94, S Joseph94, J Van der Voet94, NJ Wadd94, D Wilson94, S Anjarwalia95, J Hall95, R Hughes96, A Polychronis96, JH Scarffe96, M Hill97, RD James97, R Shah97, J Summers97, A Hartley98, D Carney99, J Mc Caffrey99, D Carney100, J Mc Caffrey100, B Bystricky101, S O'Reilly101, R Gupta102, T Al-Mishlab103, F Gidden103, R O'Hara103, K Patel103, J Stewart103, D Ashford104, R Glynne-Jones104, M Harrison104, S Mawdsley104, L Melcher104, H Barlow105, M Tighe105, J Walther105, J Neal106, C Rees106, J Bridgewater107, S Karp107, U McGovern107, PJ Atherton108, H Eldeeb109, C Macmillan109, K Patel109, EM Bessell110, PD Dickinson110, V Potter110, C Jephcott111, K McAdam111, J Wrigley111, T Hickish112, R Osborne112, SR Muthuramalingam113, A O'Callaghan113, J Bridgewater114, L Melcher114, J Hall115, C Bracon116, I Geh116, D Palmer116, P Narayana116, N Steven116, A Gaya117, N Maisey117, S Rudman117, P Chakraborti118, K Kelly119, C Macgregor119, D Whillis119, A Freebairn120, J Gildersleve120, S Sharif120, G Astras121, T Hickish121, D Beech122, R Ellis122, R Kulkarni123, K Shankland123, R Begent124, A Mayer124, T Meyer124, S Strauss124, V Hall125, S Raj125, I Chau126, D Cunningham126, I Chau127, D Cunningham127, A Birtle128, A Biswas128, S Susnerwala128, M Wise128, S Cummins129, S Essapen129, G Middleton129, C Topham129, R Langley130, F McKinna130, A Webb130, M Wilkins130, TJ Iveson131, A Dhadda132, A Hamid133, C Askill134, J Wagstaff134, A Azzabi135, (A Bateman136, T Iveson136, J Prejbisz137, D Tsang137, N Ali138, A Jones138, S Myint138, P O'Neill138, C Cottrill139, D Propper139, F J Lofts140, J Kennedy141, A Anthoney142, R Cooper142, A Crellin142, A Melcher142, M Seymour142, C Baughan143, A Alexander144, S Cleator144, J Crown145, D Fennelly145, F Adab146, S Giridharan146, A Azzabi147, I Pedley147, K Wright147, P Bliss148, G Cogill148, N Lo148, L Toy148, J Bridgewater149, D Hochhauser149, M Keane149, J Ledermann149, F Adab150, A Brewster151, T Maughan151, D Mort151, S Mukherjee151, W Dobrowsky152, P Calvert153, G Leonard153, R Ahmad154, P Riddle154, H Ford155, M Moody155, S Goriah156, M Wilkins156, S Clive157, L Dawson157, C McLean157, H A Phillips157, K Gopi158, M Tomlinson158, S Clenton159, D Furniss159, J Hornbuckle159, S Pledge159, J Wadsley15, Abbas160, E Marshall160, J Hall161, C Harper-Wynne161, A Barnes162, S Kumar162, V Vigneswaran162, D Farrugia163, A Webb164, S Gollins164, S Falk165, M Genton165, G Sparrow165, C Bale166, C Fuller166, A Mullard166, N Stuart166, R Williams166.
Trial Management Group (T Maughan151, M Seymour142, R Wilson65, H Wasan90, R Adams151, A Madi151, J Cassidy62, J Kennedy141, E Hodgkinson167, P Rogers168, M Pope151)
Medical Research Council Clinical Trials Unit (R Kaplan169, A Meade169, M Parmar169, S Kenny169, D Fisher169, L Harper169, J Mitchell169, L Nichols169, B Sydes169, L Clement169, E Kay169, C Courtney169, M Gallagher169, C Murphy169, L Thompson169, S Beall169, S Hassan169, R Gracie169, G Griffiths169)
Trial Steering Committee (M Mason151, C Parker170, R Rudd171, P Johnson172, J Whelan173)
Independent Data Monitoring Committee (J Northover174, J Brown175, M Aapro176, R Stout177)
58. Aberdeen Royal Infirmary
59. Addenbrooke's Hospital
60. Aintree, University Hospital
61. AMNCH, Tallaght Hospital, Dublin
62. Beatson Oncology Centre, Glasgow
63. Beaumont Hospital, Dublin
64. Bedford Hospital NHS Trust
65. Belfast City Hospital
66. Birmingham Heartlands Hospital
67. Blackpool Victoria Hospital
68. Bradford Royal Infirmary
69. Bristol Oncology Centre
70. Broomfield Hospital, Essex
71. Castle Hill Hospital, Hull
72. Charing Cross Hospital
73. Cheltenham General Hospital
74. Christie Hospital
75. Churchill Hospital, Oxford
76. Clatterbridge Centre for Oncology
77. Conquest Hospital
78. Cumberland Infirmary
79. Derriford Hospital, Plymouth
80. Diana, Princess of Wales Hospital, Grimsby
81. Dorset County Hospital
82. Eastbourne District General
83. Essex County Hospital
84. Glan Clwyd Hospital
85. Glasgow Royal Infirmary
86. Good Hope Hospital
87. Great Western Hospital, Swindon
88. Guys & St Thomas’ Hospitals
89. Hairmyres
90. Hammersmith Hospital
91. Hinchingbrooke Hospital
92. Huddersfield Royal Infirmary
93. Ipswich Hospital NHS Trust
94. James Cook University Hospital
95. Kent and Canterbury Hospital
96. Lister Hospital
97. Maidstone Hospital
98. Manor Hospital, Walsall
99. Mater Private Hospital, Dublin
100. Mater Public Hospital, Dublin
101. Mercy Hospital, Cork
102. Midwestern Regional Hospital, Limerick
103. Milton Keynes General Hospital
104. Mount Vernon Hospital
105. Musgrove Park Hospital, formerly Taunton and Somerset
106. North Hampshire Hospital, Basingstoke
107. North Middlesex Hospital
108. North Tyneside General Hospital
109. Northampton General
110. Nottingham City Hospital
111. Peterborough District
112. Poole Hospital, Dorset
113. Portsmouth Oncology Centre, Queen Alexandra Hospital
114. Princess Alexandra Hospital
115. Queen Elizabeth, The Queen Mother Hospital
116. Queen Elizabeth, University Hospital, Birmingham
117. Queen Elizabeth, Woolwich
118. Queens, Burton Upon Trent
119. Raigmore Hospital
120. Royal Berkshire / Berkshire Cancer Centre
121. Royal Bournemouth Hospital
122. Royal Cornwall
123. Royal Derby Hospital
124. Royal Free Hospital
125. Royal Hampshire County Hospital, Winchester
126. Royal Marsden, Fulham Road
127. Royal Marsden, Sutton
128. Royal Preston Hospital
129. Royal Surrey County Hospital, St Lukes
130. Royal Sussex County Hospital, Brighton
131. Salisbury District
132. Scarborough Hospital
133. Scunthorpe General
134. Singleton Hospital
135. South Tyneside
136. Southampton Hospital
137. Southend Hospital
138. Southport & Formby DGH
139. St. Bartholomew's Hospital
140. St. George's Hospital, London
141. St. James' Hospital, Dublin
142. St. James' University Hospital, Leeds (Cookridge)
143. St. Mary's Hospital, Isle Of Wight
144. St. Mary's, London
145. St. Vincent's Hospital, Dublin
146. Staffordshire General Hospital
147. Sunderland Royal Hospital
148. Torbay Hospital
149. University College Hospital London
150. University Hospital of North Staffs NHS Trust
151. Velindre Hospital, Cardiff
152. Wansbeck General Hospital
153. Waterford Regional Hospital, Waterford
154. West Middlesex and Charing Cross
155. West Suffolk Hospital
156. West Wales General
157. Western General, Edinburgh
158. Weston General Hospital, Weston Super Mare
159. Weston Park Hospital, Sheffield
160. Whiston Hospital
161. William Harvey Hospital
162. Withybush General Hospital
163. Worcestershire Royal Hospital
164. Worthing Hospital
165. Yeovil District Hospital
166. Ysbyty Gwynedd
167. Pharmacy Department, Weston Park Hospital, Sheffield
168. Radiotherapy Department, Charing Cross Hospital, London
169. Medical Research Council Clinical Trials Unit, London, UK
170. Institute of Cancer Research, Sutton
171. Barts Mesothelioma Research, 54 New Cavendish Street, London
172. Queen's University, Belfast
173. UCL Hospitals NHS Foundation Trust, London
174. The London Clinic, Consulting Rooms, 116 Harley Street, London
175. Section of clinical trials research, university of leeds
176. Institut Multidisciplinaire d'Oncologie, Clinique de Genolier
177. Christie Hospital, Manchester
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
REFERENCES
- 1.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R. Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin Cancer Res. 2007;13:356–61. doi: 10.1158/1078-0432.CCR-06-1256. [DOI] [PubMed] [Google Scholar]
- 3.Lubbe SJ, Webb EL, Chandler IP, Houlston RS. Implications of familial colorectal cancer risk profiles and microsatellite instability status. J Clin Oncol. 2009;27:2238–44. doi: 10.1200/JCO.2008.20.3364. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houlston RS, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger E, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–8. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 11.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353–8. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 12.Miquel C, et al. Frequent alteration of DNA damage signalling and repair pathways in human colorectal cancers with microsatellite instability. Oncogene. 2007;26:5919–26. doi: 10.1038/sj.onc.1210419. [DOI] [PubMed] [Google Scholar]
- 13.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–22. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 14.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–14. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlop MG, et al. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet. 1997;6:105–10. doi: 10.1093/hmg/6.1.105. [DOI] [PubMed] [Google Scholar]
- 16.Quehenberger F, Vasen HF, van Houwelingen HC. Risk of colorectal and endometrial cancer for carriers of mutations of the hMLH1 and hMSH2 gene: correction for ascertainment. J Med Genet. 2005;42:491–6. doi: 10.1136/jmg.2004.024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baglietto L, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102:193–201. doi: 10.1093/jnci/djp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clayton DG. Testing for association on the X chromosome. Biostatistics. 2008:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farber MJ, Rizaldy R, Hildebrand JD. Shroom2 regulates contractility to control endothelial morphogenesis. Mol Biol Cell. 22:795–805. doi: 10.1091/mbc.E10-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2010;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fairbank PD, et al. Shroom2 (APXL) regulates melanosome biogenesis and localization in the retinal pigment epithelium. Development. 2006;133:4109–18. doi: 10.1242/dev.02563. [DOI] [PubMed] [Google Scholar]
- 22.Houlston RS, et al. Congenital hypertrophy of retinal pigment epithelium in patients with colonic polyps associated with cancer family syndrome. Clin Genet. 1992;42:16–8. doi: 10.1111/j.1399-0004.1992.tb03128.x. [DOI] [PubMed] [Google Scholar]
- 23.Dunlop MG, et al. Extracolonic features of familial adenomatous polyposis in patients with sporadic colorectal cancer. Br J Cancer. 1996;74:1789–95. doi: 10.1038/bjc.1996.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin JH, Kaler SG. Non-random maternal X-chromosome inactivation associated with PHACES. Clin Genet. 2007;72:345–50. doi: 10.1111/j.1399-0004.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- 28.Kristiansen M, et al. High incidence of skewed X chromosome inactivation in young patients with familial non-BRCA1/BRCA2 breast cancer. J Med Genet. 2005;42:877–80. doi: 10.1136/jmg.2005.032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettiti D. Meta-analysis decision analysis and cost-effectiveness analysis. . Oxford University Press. 1994 [Google Scholar]
- 31.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–8. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 33.Chow JC, Yen Z, Ziesche SM, Brown CJ. Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet. 2005;6:69–92. doi: 10.1146/annurev.genom.6.080604.162350. [DOI] [PubMed] [Google Scholar]
- 34.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 35.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 36.Ferretti V, et al. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–6. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–4. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 38.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. ‘Bioinformatics and Computational Biology Solutions using R and Bioconductor’. Springer; New York: 2005. pp. 397–-420. [Google Scholar]
- 39.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boland CR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.