Abstract
Flavin-dependent, lysine-specific protein demethylases (KDM1s) are a subfamily of amine oxidases that catalyze the selective posttranslational oxidative demethylation of methyllysine side chains within protein and peptide substrates. KDM1s participate in the widespread epigenetic regulation of both normal and disease state transcriptional programs. Their activities are central to various cellular functions, such as hematopoietic and neuronal differentiation, cancer proliferation and metastasis, and viral lytic replication and establishment of latency. Interestingly, KDM1s function as catalytic subunits within complexes with coregulatory molecules that modulate enzymatic activity of the demethylases and coordinate their access to specific substrates at distinct sites within the cell and chromatin. Although several classes of KDM1 -selective small molecule inhibitors have been recently developed, these pan-active site inhibition strategies lack the ability to selectively discriminate between KDM1 activity in specific, and occasionally opposing, functional contexts within these complexes. Here we review the discovery of this class of demethylases, their structures, chemical mechanisms, and specificity. Additionally, we review inhibition of this class of enzymes as well as emerging interactions with coregulatory molecules that regulate demethylase activity in highly specific functional contexts of biological and potential therapeutic importance.
Keywords: KDM1A, KDM1B, Epigenetic, LSD1, LSD2
INTRODUCTION
The functional capacity of genetically encoded proteins can be powerfully expanded by reversible posttranslational modifications (PTMs). Protein side chain covalent modifications such as methylation, phosphorylation, acetylation, glycosylation, ubiquitination, sumoylation, ADP-ribosylation, nitrosylation, carboxylation, and sulfonation, among others, can potentiate local and global protein architecture and generate novel reactive functional groups for binding and catalysis. Additionally, these modifications can generate signals for cellular compartmentalization and confer alterations to physiochemical properties such as stability, solubility, and resistance to proteolytic degradation. Such modifications are intimately integrated into all aspects of modulating cellular proliferation and biological recognition.
Allfrey first linked histone acetylation and methylation to the control of RNA synthesis in 1964.1,2 Since that time, it has become clear that protein and histone methylation is a pervasive cellular regulatory mechanism. Specifically, protein lysine methylation status plays a critical role in biology and pathobiology by influencing transcriptional activation and repression, chromatin remodeling, normal and oncogenic signaling, viral pathogenesis, and protein and transcription factor recruitment, among other functions. S-adenosylmethionine-dependent methyltransferases (SAM-dependent MTases) catalyze the installation of lysine methyl marks, forming mono-, di-, and tri-methylated side chain PTMs. These marks are removed by two mechanistically-distinct classes of lysine-specific demethylases (KDMs) that utilize either a flavin adenine dinucleotide (FAD) cofactor (KDM subfamily 1) or iron(II) and α-ketoglutarate cofactors (KDM subfamily 2–6) to catalyze the oxidative demethylation of protein side chains.3–5
For decades, methylation marks on proteins like histones were presumed to be immutable, reversed only through protein turnover and degradation. The first hints of a candidate histone demethylase enzyme emerged in 1998 and again in 2001, when several histone deacetylase (HDAC)-containing complexes were discovered to associate with KIAA0601 (aka KDM1A/LSD1/ BHC110/AOF2), a protein with homology to human FAD-dependent oxidoreductases (MAO-A/B) and maize polyamine oxidase (mPAO) (Figure 1a), enzymes that catalyze the oxidative cleavage of C-N bonds.6–8 In 2003, Shiekhattar reported that BHC110/KIAA0601 formed a stable complex with several proteins, including HDACs 1 and 2, CoREST, BRCA2-associated factor, BCH80, TFII-1, KIAA0182/GSE-1, ZNF217, ZNF198/FIM and ZNF261/X-FIM.9
FIGURE 1.
Structural overview of KDM1 demethylases. (a) Sequence alignment of a portion of the amine oxidase domains of KDM1A, KDM1B, MAO-A, MAO-B, and maize polyamine oxidase (mPAO). Sequence conserved active site Lys residue is starred. (b) Domain maps of KDM1A and KDM1B. SWI3p, Rsc8p, and Moira (SWIRM) domains shown in green, amine oxidase domains shown in blue, tower domain shown in lavender, linker domain shown in teal, C4H2C2 domain shown in purple, and Zf-CW domain shown in orange. (c) Ribbon representation of the structure of KDM1A. Domains follow same color scheme as outlined above (PDB 2IW5). (d) Ribbon representation of the structure of KDM1B. Domains follow same color scheme as outlined above (PDB 4HSU). (e) Venn diagram of domain conservation between KDM1A and KDM1B.
In 2003 Amasino and coworkers independently discovered that the gene FLOWERING LOCUS D (FLD) encoded a plant homolog of KIAA0601.10 FLD is one of six genes in the Arabidopsis thaliana autonomous floral-promotion pathway that initiates the transition from a vegetative to reproductive state by repression of the MADS-box transcription factor FLOWERING LOCUS C (FLC). FLD not only contains a KIAA0601-like amine oxidase domain, but also possesses an N-terminal SWI3p, Rsc8p, and Moira (SWIRM) domain similar to that associated with a range of proteins involved in chromatin remodeling, including KIAA0601.11,12 Deletion of FLD in A. thaliana results in hyperacetylation of histones in the FLC locus, upregulation of FLC expression, and extremely delayed flowering, suggesting that the autonomous pathway involves regulation of histone deacetylase activity by FLD.10 Deletion of FLD also resulted in increased histone methylation levels (R. Amasino, personal communication), implicating FLD (and by analogy KIAA0601) as the elusive human histone demethylase enzyme.
In early 2004, Shi and coworkers provided the first direct evidence that KIAA0601 (from here on referred to as KDM1A) functions as a histone demethylase and transcriptional corepressor.13 The authors reveal that KDM1A specifically demethylates mono- and di-methylated histone H3 lysine 4 (H3K4me1/ 2), a histone mark linked to active transcription, and that lysine demethylation occurrs via an oxidative reaction that generates formaldehyde. In addition, loss of KDM1A through siRNA knockdown results in increased H3K4 methylation and concomitant derepression of several neuronal-associated target genes. Shi’s discovery of KDM1A activity implied that lysine methylation might be dynamically controlled.
The second human flavin-dependent histone demethylase, KDM1B (aka LSD2/AOF1) was identified by Shi and coworkers in 2004 through a domain homology search of genomic databases.13 Mattevi and coworkers first isolated and confirmed the flavin-dependent demethylation activity of KDM1B, noting specificity for H3K4me1/2, despite the relatively low sequence identity (<25%) with KDM1A.14 Unlike KDM1A, KDM1B does not form a biochemically-stable complex with the C-terminal domain of the corepressor CoREST, but does possess both CW-type and C4H2C2-type zinc finger motifs. This suggests that KDM1B may interact with different targets or coregulatory molecules and may be involved in transcriptional programs distinct from those of KDM1A.
Herein we review the biological function, biochemical characterization, and inhibition of this enzyme class to date. First, we briefly discuss the biological importance and therapeutic potential of KDM1s. This is followed by the structural organization, chemical mechanism, and substrate specificity of the KDM1 enzymes. We then outline the various inhibitor classes that have been developed for these demethylases, specifically highlighting the utility of peptide-based inhibitors. Finally, we describe the known interactions between the KDM1s and regulatory biomolecules, which direct their activity toward specific cellular pathways. Given the wide-ranging and ubiquitous functions of this enzyme class, probes with the ability to target KDM1 activity in a manner that is pathway-specific would be of heuristic and therapeutic value. We see these coregulatory molecules as a valuable starting point for the development of such probes, as underscored by the recent development of peptide inhibitors of this enzyme class.
KDM1A AND KDM1B BIOLOGYAND THERAPEUTIC POTENTIAL
KDM1A is involved in a wide variety of cellular processes and pathologies, including signal transduction, chromatin remodeling, transcriptional regulation, development, differentiation, viral pathogenesis, and cancer proliferation and metastasis.3,15–23 As such, KDM1 demethylases have emerged as potential therapeutic targets. Although their clinical value is only beginning to be explored, at the time of this review’s publication, five clinical trials involving KDM1A were either planned or currently recruiting subjects. Three trials will investigate KDM1A inhibition as a therapeutic strategy in acute myeloid leukemia (AML), one will evaluate the same strategy in small cell lung cancer, and one seeks to correlate the status of KDM1A with cardiovascular responses to changes in sodium intake (see http://clinicaltrials.gov). We expect the scope of clinical trials to expand with our growing knowledge of KDM1A function.
Cellular Differentiation
Epigenetic modifications are critical for the maintenance of pluripotent stem cells and their subsequent differentiation into specialized tissues.24–26 Specifically, KDM1A is an integral part of the cellular machinery that governs differentiation processes in various cell types, including embryonic,27,28 hematopoietic,23,29–33 neuronal,34–38 pituitary,39 and osteogenic.40 While KDM1A activity is critical for proper development, aberrant regulation can contribute to tissue-specific disease states. For example, KDM1A activity has been linked to neuronal cell proliferation and survival41,42 and long-term memory formation,43 and is therefore an emerging therapeutic target for some neurological and cognitive disorders. Similarly, recent work has also identified KDM1A as a key regulator of leukemia stem cells,44 sparking interest in its potential as a therapeutic target in AML (see below), among other hematopoietic-related diseases.45
Cancer Development and Progression
KDM1A has also been widely implicated in the development and progression of cancer. In addition to the malignancies described in detail below, KDM1A is emerging as a potential therapeutic target in various other cancer types.46–56
Neuroblastoma
Overexpression of KDM1A in neuroblastic tumors correlates with poor prognosis57 and may be related to the aberrant downregulation of KDM1A-silencing micro-RNAs.58,59 Schulte and coworkers first noted in 2009 that KDM1A inhibition in neuroblastoma cells leads to increased H3K4 methylation, decreased proliferation in cell culture, and reduced tumor growth in a xenograft model.57 Similarly, KDM1A inhibitors synergize with retinoic acid, a currently approved treatment for neuroblastoma.60 A later report demonstrated similar results in the closely-related medulloblastoma tumor type, where KDM1A inhibition decreases proliferation and initiates apoptosis.61
Colorectal Cancer
KDM1A is also an emerging target in colorectal cancers, where it is markedly overexpressed in tumor samples compared to matched normal tissue62 and exhibits a positive correlation with metastatic potential.63,64 Although genetic deletion of KDM1A does not result in a global increase of H3K4me2, it does alter gene expression programs and reduces proliferation in cell culture and in vivo.62,65 KDM1A may also promote colorectal cancer progression by derepressing the Wnt/β-catenin signaling pathway, a known regulator of tumorigenesis.62
Leukemia
KDM1A is also a promising target for pharmacological intervention in leukemia. For example, KDM1A inhibition alone provokes cytotoxic effects in AML cell lines,66 and when used in conjunction with HDAC inhibitors, reduces tumor engraftment and improves survival in mice.67 KDM1A inhibition also unlocks the anti-leukemic effects of all-transretinoic acid (ATRA), a differentiation-inducing agent that otherwise is ineffective for the treatment of AML.68 KDM1A has been implicated in other leukemia subtypes as well. In a model of MLL-AF9 leukemia cells, KDM1A cooperates with the MLL MTase fusion protein to maintain an oncogenic transcription program, and KDM1A inhibitors preferentially reduce the repopulation potential of MLL-AF9 cells over normal hematopoietic cells.44 Interestingly, Spruüssel and coworkers evaluated the potential side effects of KDM1A inhibition in leukemia and found that while a conditional KDM1A knockdown significantly alters existing pools of normal hematopoietic tissues, these effects are largely reversible following reinstatement of the demethylase.31 For a more detailed discussion of KDM1A’s function and therapeutic potential in leukemia, we refer readers to a recent review by Mould and colleagues.69
Breast Cancer
KDM1A has also been implicated as a regulator of breast cancer development and progression. The majority of the breast tumors are estrogen-dependent, making estrogen antagonists one of the most commonly employed therapies. However, circumvention of estrogen signaling, such as through loss of estrogen-receptor alpha (ERα), can lead to hormone-resistant tumors, reduced treatment options, and worse prognosis. KDM1A is generally associated with a more aggressive breast cancer phenotype and may be overexpressed in both ERα-positive and -negative cells,70–73 possibly due to aberrant stabilization of the demethylase.74 Correspondingly, several groups have reported that KDM1A inhibition decreases proliferation of breast cancer cells.70,72,75 Notably, at least two groups have reported contradicting results that suggest KDM1A may be downregulated in breast cancer tissue and that inhibition affects metastasis with no observable effects on proliferation.76,77 KDM1A inhibition may also have therapeutic effects when used in conjunction with anti-estrogen treatment78 and HDAC inhibitors.79,80
Prostate Cancer
A similarly vague relationship exists between KDM1A and prostate cancer. One study reported a correlation between KDM1A expression and risk of recurrence,81 while at least two other studies have noted little to no overexpression of KDM1A in prostate cancer cell lines and tumor samples.82,83 However, Metzger and coworkers have linked KDM1A activity to transcriptional regulation with the androgen receptor (AR), a nuclear hormone receptor closely linked with prostate cancer.84 Likewise, KDM1A inhibition appears to impede AR-mediated transcription and prostate cancer proliferation.84–87 A review detailing further the roles of demethylases in prostate cancer has recently been published.88
The Epithelial-to-Mesenchymal Transition
In addition to these roles in specific cancer types, KDM1A is also intimately linked with the epithelial-to-mesenchymal transition (EMT), the process by which a polarized, adhesive epithelial cell transitions into a motile, invasive mesenchymal phenotype. The importance of epigenetic mechanisms during the EMT is only beginning to be appreciated.89 KDM1A is an established functional partner of the EMT master regulator SNAI1 and cooperates to silence epithelial genes.90 KDM1A also regulates upstream inducers of the EMT,91 including TGFβ,76,92,93 Wnt,62,94 NF-κB,80 and Notch.95,96 Through these and potentially other mechanisms, KDM1A participates in large-scale chromatin remodeling events that accompany the EMT.92
The EMT is a hallmark of aggressive cancers with a high risk of metastasis, potentially implicating KDM1A as a therapeutic target.89 Accordingly, KDM1A inhibition in AML models produces morphological features of differentiation in primary and cultured cells,67 derepresses differentiation-associated genes, 66,68 and reduces engraftment of primary cells in a mouse model.68 Similar effects are observed in MLL-AF9 leukemia and neuroblastoma models.44,57 Furthermore, KDM1A activity is associated with increased metastatic potential and its inhibition or knockdown decreases migration and invasion in several cancer types,48,49,63,64 with again conflicting reports concerning breast cancer.76,97,98
Viral Pathogenesis
Lastly, KDM1A is associated with herpes simplex virus (HSV) infection and replication, as HSV hijacks KDM1A-containing machinery required for human neuronal gene repression during host infection. Cooption of KDM1A demethylase activity helps promote early stage (α) viral gene activation, regulates late stage (β/γ) viral gene repression, and has been linked to gene repression during the development of HSV latency, the process by which viruses enter and emerge from dormancy in infected host cells.99 Kristie and coworkers recently demonstrated that interruption of KDM1A activity potently represses HSV immediate-early (IE) gene expression and genome replication, resulting in suppression of primary infections and subsequent reactivation from latency in murine infection models.100–102 In addition, KDM1A activity may play a similar role in the infection processes of several other viruses.101,103,104 Collectively, these studies underscore the emerging importance of KDM1A as an antiviral target.
Emerging Biological Roles for KDM1B
The biological roles of KDM1B are beginning to emerge; however, our current understanding of this enzyme lags far behind that of KDM1A. In general, KDM1B is believed to function as an enhancer of gene transcription that functions in highly transcribed coding regions of chromatin.105 Although mechanistic data is not yet available, KDM1B has been linked to genomic imprinting,106 liver development,107, somatic cell reprogramming,108,109 and NF-κB signaling.110 Future research will likely reveal a broader set of KDM1B-dependent biological processes.
DOMAIN ORGANIZATION AND STRUCTURAL FEATURES OF KDM1 DEMETHYLASES
KDM1A Demethylase
KDM1A is an 852 amino acid (aa) polypeptide composed of three domains: an amine oxidase catalytic domain (AOD) (residues 272–415 and 515–852), an ~100 aa insert known as the “tower” domain (residues 415–515), and a SWIRM domain (residues 172–272) (Figure 1b,c).11,111,112 The N-terminal region of the enzyme (~170 aa) has no predicted conserved structural elements, but does contain a nuclear localization signal (RRKRAK; residues 112–117) for nuclear import via importin-α translocases.113,114 The three-dimensional structure of KDM1A has been solved by X-ray crystallographic methods in the unliganded form and in complex with varying ligands, including peptides and inhibitors.111,112,115–126 The AOD of the enzyme contains a single non-covalently bound FAD molecule and bears similarity to human MAO-A/B and mPAO (17.6%, 17.6%, and 22.4% sequence identity, respectively; Figure 1a).127 Consistent with other flavin-dependent amine oxidases,128,129 the AOD can be further subdivided into two separate lobes, where residues 357–415, 515–558, and 658–769 are involved in substrate binding and residues 272–356, 559–657, and 770–852 form an expanded Rossmann fold used to bind FAD.112,115
The tower domain of KDM1A is an antiparallel coiled-coil composed of two α-helices (termed TαA and TαB) bridged by a tight turn formed by a KPPRD sequence.112 This domain projects almost 100 Å from the AOD and functionally is the site of association with corepressor CoREST (aka RCOR1/KIAA0071), among other proteins.112,115,130 The SWIRM domain of KDM1A is predominantly a bundle of α-helices, with a long central α-helix that separates two smaller helixturn-helix motifs (Figure 1c).111 This domain also engages the C-terminal tail of the AOD with a highly hydrophobic interface that buries a surface area of ~1,700 Å2.111 Mutations of conserved residues along this interface have been shown to reduce the catalytic activity of KDMA1 and its stability.111
Although SWIRM domains are highly conserved amongst chromatin associating proteins and have been shown to bind DNA, ,12,131 this is not the case in KDM1A. The residues that compose the typical DNA-binding interface (α6; residues 247–259) are not conserved and are partially blocked by their interaction with the AOD.111,115 Accordingly, neither KDM1A nor the isolated SWIRM domain binds DNA.115,132
In addition, Luka and coworkers showed that KDM1A binds to the small molecule tetrahydrofolate with a Kd of 2.8 µM, and recently reported the crystal structure of the KDM1A-CoREST-tetrahydrofolate complex.126,133 In this complex, the folate binds in the active site in close proximity to the FAD cofactor, and overlaps with the peptide-binding site observed by Forneris.117 Luka and coworkers also detected the production of 5,10-methylene-tetrahydrofolate by mass spectrometry following incubation of KDM1A with a H3K4-derived methylated peptide substrate and tetrahydrofolate. From this data, it was postulated that tetrahydrofolate binding might protect the cell from potentially toxic formaldehyde or regulate access of the substrate to the FAD within the active site. It is not known at this time if KDM1B exhibits a similar ability to bind tetrahydrofolate.
KDM1B Demethylase
The closely related KDM1B is an 822 aa flavoenzyme amine oxidase that contains a predicted unordered N-terminus and a nuclear localization signal (~50 aa) responsible for its residence within the nucleus of human cells.106 KDM1B is organized into three domains: an N-terminal dual zinc finger domain (residues 50–264), a SWIRM domain (residues 264– 372), and a C-terminal catalytic AOD (residues 372–822; Figure 1b,d). 134–136 In the substrate-free structure of KDM1B, the zinc fingers and catalytic AOD are packed closely against a central SWIRM domain (Figure 1d), collectively resembling a boot.134 Comparison of the AOD between KDM1A and KDM1B (33% sequence identity) shows that many secondary structural elements are conserved, including strong conservation of the catalytic architecture around the FAD-binding site. 134 However, when comparing the AODs through structural alignment, there is a 2.0 Å RMSD difference in the overall polypeptide backbone conformation. These structural differences are mostly attributed to the length and conformation of solvent exposed loops within the domain.134 Despite these differences, the similarity of the AODs of KDM1A and KDM1B argues for a common chemical mechanism for catalysis.
Structural Differences Between KDM1A and KDM1B
Despite catalyzing identical chemical reactions and sharing significant AOD structural similarity, KDM1A and KDM1B have several structurally important differences. For example, the SWIRM domains of both enzymes only share 24% sequence identity. Although helices within this domain are qualitatively positioned, significant differences have been observed at both solvent exposed interfaces along the helical termini. Since the entrance to the AOD active site lies at the interface of the AOD and SWIRM domains, structural and sequence differences at this interface are suspected to impact substrate specificity of each isoform.134
Indeed, at this interface the SWIRM-AOD subdomain interactions differ in several aspects when comparing KDM1A to KDM1B. Notably, the SWIRM domain of KDM1B closely packs against the N-terminal zinc finger domain, burying a surface area of 1,872 Å2. 134 As compared to KDM1A, the SWIRM domain of KDM1B lacks a C-terminal helix (KDM1A; residues 171–181), but is replaced by an extended, coiled loop (KDM1B; residues 271–281).136 This substitution in KDM1B leads to the formation of two additional hydrogen bonds (E452 with N266, and Y268 with D571) and one electrostatic interaction (E323 with K323) that may influence the degree of association among these domains.136 Not only does this loop provide additional intramolecular interactions, but also forms a second binding site for the N-terminal tail of histone H3.135
The most striking structural difference between KDM1A and KDM1B is the inclusion of distinct domains that mediate interactions with other biomolecules (Figures 1b,e). KDM1A contains an α-helical, antiparallel coiled-coil tower domain that is absent in KDM1B (Figure 1c – e). DALI137 structural similarity analyses indicate that the tower domain fold appears infrequently among known eukaryotic protein structures, with the PI3Kα-p85α subunit complex (PDB 3HHM)138 and the DNA double-strand break repair ATPase RAD50 (PDB 1L8D)139 bearing the closest structural similarity. In contrast, the tower fold was heavily represented among a subset of prokaryotic proteins known to utilize such motifs as part of inter-molecular protein recruitment, membrane docking, and membrane translocation functions.140
Although KDM1B does not contain a tower domain, it does contain two individual zinc fingers and two linker domains that are not reflected in the structure of KDM1A (Figure 1b,e). At the N-terminus of KDM1B, a C4H2C2-type zinc finger is joined to a second CW-type zinc finger (Zf-CW) by a linker formed from two α-helices (Figure 1b,d). Immediately following the Zf-CW domain, a second linker formed by four α-helices joined through several large loops bridges these domains to the AOD.134–136 The surface of the C4H2C2-type zinc finger shows a marked concentration of basic residues, and thus may impact demethylase substrate specificity or positioning within nucleosomal DNA. Additionally, these residues may facilitate interactions with coregulatory molecules or serve to recruit transcriptional machinery, such as phosphorylated RNA polymerase II.105,134 Interestingly, it appears that the zinc finger domain of KDM1B is required for histone demethylation activity. Wong and coworkers suggest that mutations which disrupt the zinc finger domains or relays of interactions among the zinc finger-SWIRM-AOD likely lead to subtle conformational alterations in the AOD that in turn impair the incorporation of FAD, and consequently its enzymatic activity.134
Furthermore, although zinc fingers are common folding motifs, the Cys2A-Cys2B-His2A-Cys2B sequence within KDM1B folds into a cross-brace topology that has not been previously observed, raising the possibility that this domain may participate in biomolecular interactions that are exclusive to KDM1B.134 On the other hand, the Zf-CW of KDM1B bears significant sequence similarity to CW-type zinc finger domains, a class linked to histone H3 binding. Despite this, as an isolated recombinant domain, the KDM1B Zf-CW fails to bind histone H3-derived peptides (residues 1–21 containing H3K4me1/2 or unmethylated H3K4), which is atypical to behavior exhibited by other isolated Zf-CW domains.134,136 However, since ligand binding within Zf-CW domains is mediated by interactions within a well-conserved hydrophobic cleft, which happens to be absent in the KDM1B Zf-CW domain structure, this may partially explain its inability to bind histone H3 peptides.
Taken together, the exclusive presence of the tower domain in KDM1A and the zinc finger motifs in KDM1B strongly suggest that these two demethylases share a related chemical mechanism, but likely participate in associations with non-identical sets of interacting partner biomolecules. These partners may influence individual catalytic activity, substrate specificity, or localization of these enzymes within genetic loci.
CHEMICAL MECHANISM OF KDM1-FAMILY FLAVIN-DEPENDENT DEMETHYLASES
KDM1A and KDM1B are Mechanistically Distinct from Iron(II)-Dependent Lysine-Demethylases
KDM1 family human demethylases utilize a FAD cofactor to oxidize C-N bonds with subsequent production of a demethylated lysine residue and formaldehyde by product.141 By contrast, KDM subfamilies 2–6 (Jumonji-C domain-containing demethylases) are mechanistically distinct from the KDM1s since they utilize non-heme iron(II) and α-ketoglutarate (α-KG) cofactors to demethylate lysine residues with the concomitant production of succinate and formaldehyde.4 KDM families 2–6 are also distinguished from KDM1A and KDM1B by their unique ability to demethylate trimethylated lysine residues.142 However, KDM1 flavoenzymes have a mechanistic requirement of a protonated iminium intermediate, achievable only with mono- and dimethylated lysines.
Involvement of Flavin Adenine Dinucleotide as a Cofactor
Experimental and theoretical mechanistic studies have been conducted on flavin-utilizing KDMs, focusing almost exclusively on KDM1A. Mattevi and coworkers first confirmed the participation of the non-covalent FAD in the demethylation reaction using two truncated forms of KDM1A lacking the first N-terminal 157 or 184 aa.143 They visualized production of the two-electron fully reduced flavin, noting the absence of intermediate one-electron reduced forms of the cofactor. This suggested that KDM1A was likely related by mechanism to the broader family of flavoenzyme oxidases that catalyze two-election oxidation of substrates with concomitant cofactor reduction.143 Once the substrate is oxidized, hydrolysis by bulk solvent of the iminium intermediate produces an unstable hemiaminal that spontaneously decomposes into formaldehyde and the mono-demethylated product. Additionally, reoxidiation of the FAD cofactor by molecular oxygen produces a molar equivalent of H2O2 and the fully oxidized quinone (Scheme 1).13,143
SCHEME 1.
Proposed chemical mechanism of FAD-dependent demethylases KDM1A and KDM1B. Oxidation of the C-N bond of the methylated lysine sidechain to an iminium ion, with concurrent reduction of the flavin enables hydrolysis via bulk water. Collapse of the hemiaminal (not shown) yields formaldehyde and the demethylated amine sidechain. The reduced flavin is reoxidized by molecular oxygen, generating a molar equivalent of hydrogen peroxide.
Evidence for a Direct Hydride Transfer Mechanism
McCafferty, Fitzpatrick, and coworkers conducted a detailed investigation of the chemical mechanism of KDM1A.144,145 The mechanism was examined using the effects of pH and isotopic substitution on steady-state and pre-equilibrium kinetic parameters. Using a 21-mer derived from histone H3 (H3K4me2, residues 1–21) at pH 7.5, the rate constant for flavin reduction in the transient phase, kred, was shown to equal kcat, establishing the reductive half-reaction as rate-limiting at physiological pH. Deuteration of the lysyl Nε-methyl groups (H3K4me2-d6) produced identical kinetic isotope effects of 3.2 ± 0.1 on the kred, kcat, and kcat/Km values for the peptide, thus establishing C2H bond cleavage as rate-limiting with this substrate. The D(kcat/Km) value for the peptide was pH-independent, suggesting that the observed value is the intrinsic deuterium kinetic isotope effect for oxidation of this substrate.146 No intermediates between oxidized and reduced flavin forms were detected by stopped-flow spectroscopy, consistent with the expectation for a direct hydride transfer catalytic mechanism for amine oxidation. Additionally, the kcat/Km value for the peptide was bell-shaped, consistent with a requirement that the nitrogen at the site of oxidation be uncharged and that at least one of the other lysyl residues be charged for catalysis.144,145,147
A subsequent theoretical investigation by Karasulu and coworkers has confirmed that the H3K4me2 residue of the substrate has to be deprotonated in order for catalysis to occur, with K661 of KDM1A acting as the proton acceptor in the active site.148 Additional support for a direct hydride transfer mechanism was obtained through inhibition studies with tranylcypromine (2-PCPA, Parnate, 1).118,127 Parnate, which covalently inactivates KDM1A and KDM1B, is known to inactivate FAD-dependent enzymes that function through either the SET149 or direct hydride transfer mechanisms.150,151
As a further point of clarification, in KDM1A, reoxidation of the FAD cofactor by O2 is a relatively fast process compared to the turnover number measured under steady-state conditions.152 This entails that reoxidation is not the rate-limiting step of the chemical mechanism. Additionally, the FAD reoxidation rate is not perturbed by the presence of the product peptide152 and suggests that KDM1A can remain bound to the demethylated product and possibly functions through a ternary complex kinetic mechanism.153 Using computational approaches, Baron and coworkers suggest that molecular O2 diffuses though the enzyme for the purpose of reoxidaton while the demethylated product remains bound.123 Despite this prediction, there is no chemical requirement for the demethylated product to remain bound to the enzyme (i.e. the apparent rate of reoxidation is not faster or slower in the presence of the peptide product). Therefore, KDM1A might also function through a ‘ping-pong’ or double displacement kinetic mechanism, in which the release of the demethylated product occurs before reoxidation. Interestingly, in cell culture KDM1A removes a single methyl mark from p53-K370me2 to produce K370me1; however, in vitro it completely demethylates this residue.154,155 Thus, one may argue that the release of the demethylated product may be influenced by coregulatory molecules or other contributing factors capable of tuning its catalytic proficiency or substrate specificity.
Using classical molecular dynamics (MD) and quantum mechanics/molecular mechanics (QM/MM) approaches, Karasulu and coworkers provided theoretical support for a hydride transfer mechanistic pathway for methyl lysine oxidation over SET, carbanion, or polar-nucleophilic mechanisms.148 Proper alignment of the substrate, transitionstate stabilization (due to the protein environment and favorable orbital interactions), and product stabilization via adduct formation were found to be crucial for facilitating the oxidative C–H bond cleavage.148 Similarly, Truhlar and coworkers provided theoretical support for a hydride transfer mechanism, but also noted that because of the magnitude of the calculated free energy, a mechanism involving concerted transfer of a hydrogen atom and an electron could not be ruled out.156 Lastly, Kong and coworkers used theoretical calculations to provide support for a hydride transfer mechanism, but also used MD to implicate a conserved Y761 and K661-water-flavin motif in orienting FAD with respect to the substrate.157 Collectively, these theoretical analyses are in good agreement with the experimentally determined hydride transfer mechanism for KDM1A.148,156,157
KDM1B Chemical Mechanism
Tentatively, we suspect that KDM1B also adopts a similar hydride transfer chemical mechanism given the overall structural homology it maintains with the AOD of KDM1A, the conservation of active site residues, and subsequent, nearly identical, substrate specificity towards peptide substrates in vitro.14 Additional support for this hypothesis is reflected by the similar pH-rate profile behavior of the kinetic parameters as compared to KDM1A, non-covalent nature of the FAD cofactor, and highly similar catalytic efficiency parameters, with KDM1B being slightly less efficient in vitro.14,134–136
Chemical Mechanism Implications
When comparing KDM1A and KDM1B to other amine oxidases, it is important to note that the rate constant of substrate amine oxidation is 2–5 orders of magnitude slower than that of the MAOs and PAOs.158–164 This suggests that KDM1s likely evolved for substrate specificity rather than catalytic proficiency. Given the physiological role of KDM1 demethylases in the regulation of gene expression, high catalytic efficiency, which is necessary for metabolic enzymes to maintain balance among metabolic pathways, would be much less critical than very high specificity.
SUBSTRATE SPECIFICITY
Specificity for Histone Substrates
Shi and coworkers first reported that KDM1A was capable of demethylating methyllysine peptides derived from the highly conserved N-terminus of histone H3, as well as full-length H3 in vitro.13 No cross-reactivity for polyamine substrates was observed, despite the similarity of KDM1A to the PAO superfamily.13 They also reported that KDM1A is specific for histone H3K4me1/2, as no other methyllysine sites were processed.13 Forneris and coworkers demonstrated that histone H3 tail peptides greater than 16 aa in length are necessary to achieve demethylase efficiency in vitro. Additionally, they revealed that KDM1A demethylated H3K4me1 and H3K4me2 with similar kinetic parameters in vitro, illustrating a lack of a strong kinetic preference for either substrate.147
Mattevi and coworkers have also examined the contribution of residues within the H3 N-terminal tail to the efficiency of KDM1A demethylase activity, as well as the effects of epigenetic PTMs within this sequence.144,147,152 For example, Mattevi showed that methylation of H3K9 does not affect enzyme catalysis, yet acetylation at H3K9 increases the Km 6-fold for H3 peptide substrates methylated on K4. Similarly, phosphorylation of H3S9 abolishes demethylase activity, suggesting that KDM1A ‘reads’ PTMs along the histone tail.147,152 It was also proposed that electrostatic interactions likely contribute significantly to substrate binding, since demethylation activity was shown to be sensitive to ionic strength as well as hyperacetylation in vitro.147,152 Additionally, Mattevi showed that unmethylated H3-derived product peptides exhibit inhibition, which may serve to regulate KDM1A activity or localization.147 Parallel experiments conducted with KDM1B revealed a similar substrate specificity profile,14 which was anticipated given the significant active site structural similarity shared between the two enzymes in the AOD.112,134,136
While the ability of both KDM1A and KDM1B to demethylate H3K4me1/2 is quite striking, both enzymes do not demethylate H3K9me1/2 peptides in vitro.13,14,147 Despite lacking this activity towards peptide substrates, it has been suggested that KDM1A association with AR shifts its substrate specificity to H3K9me1/2 demethylation.84 However, the mechanisms behind these observations are not yet clear. A simple explanation is that AR association with KDM1A physically occludes its access to H3K4 or triggers recruitment of another demethylase, such as the H3K9me2/3-specific KDM4C (JMJD2C) demethylase.165 Alternatively, modifications on surrounding histone residues,166 a conformational change induced by a protein-protein interaction, or a PTM to KDM1A could potentiate this switch in specificity.167
As a member of the flavin-dependent oxidases, the KDM1 demethylases exhibit a strict requirement for proper substrate positioning in relation to the FAD cofactor to promote catalysis.129 However, KDM1s differ from other amine oxidases because their active sites are significantly expanded in order to accommodate the N-terminal residues of the histone H3 peptide (i.e. 1,245 Å3 in KDM1A).112 Within KDM1A, for example, there are four major invaginations lined with distinct groupings of residues to form complimentary interactions with H3 substrate side chains as well as for recognition of PTMs.111 No structural features within the active site of either enzyme have been found that might suggest that the methylation state of H3K4 can be distinguished sterically or electronically, and discrimination against H3K4me3 substrates is due to the inherent chemical mechanism of the enzymes.111,134 Although more than 16 N-terminal residues of H3 are required for efficient catalysis, a significant portion of the C-terminus of substrates likely extends from the active site and potentially interact along a cleft formed at the interface of the AOD and SWIRM domain.111,112,115,141 Mutations of residues in this cleft in KDM1A abolish or abrogate demethylation activity, suggesting that interactions with substrates within this region may provide an additional specificity determinant.111 Additionally, surface plasmon resonance interaction studies have demonstrated that the SWIRM domain of KDM1A interacts with H3-derived peptides.132 In KDM1B, the surface landscape of this region is quite different and binds the coregulatory protein Nuclear Protein 60kDa/Glyoxylate Reductase 1 Homolog (NPAC/GLYR1).135,136
In order to dissect the molecular basis of KDM1A and KDM1B substrate specificity, several three-dimensional structures have been determined of enzyme-substrate like complexes where short peptides occupy the active site in two general conformations (Figure 2).116,117,123,134–136 The structure of KDM1A bound to a substrate-like peptide inhibitor containing a N-methylpropargyl derivatized lysine residue at position four revealed density for residues 1–7 of the peptide and adopted an unusually compacted backbone with three consecutive γ-turns (Figure 2a).116 Subsequently, a H3 peptide inhibitor containing a K4M mutation (H3K4M, pK4M) was co-crystallized with KDM1A and CoREST,117 which revealed density for the first 16 residues of the peptide with the first five adopting an α-helical conformation (Figure 2b). It is important to note that the C-termini of each of these peptides points in opposing directions upon exiting the active site. Additionally, the N-terminal region of SNAI1 and related peptide derivatives have been co-crystallized with KDM1A and display a nearly identical conformation to that of the H3K4M peptide (Figure 2c), thus suggesting some form of molecular mimicry in KDM1A inhibition (Figure 2e).123,124 Finally, when KDM1B was co-crystallized in the presence of the same H3K4M peptide, it exhibited a similar binding conformation to that found in KDM1A (Figure 2d).134–136 Interestingly, elucidation of the structure of KDM1B bound to the cofactor NPAC/GLYR1 revealed a secondary binding site occupied by the C-terminal region of the H3 peptide tail.135
FIGURE 2.
Overview of structural conformations of bound peptide ligands. (a) Conformation of covalently bound N-methypropargyl-Lys4 H3 peptide in KDM1A (PDB 2UXN). (b) Conformation of non-covalently bound H3K4M (pK4M) in KDM1A (PDB 2V1D). (c) Conformation of non-covalently bound SNAI1 in KDM1A (PDB 2Y48). (d) Conformation of non-covalently bound pK4M in KDM1B (PDB 4HSU). (e) Structural alignment of pK4M and SNAI1 (b and c, respectively) bound to KDM1A, illustrating the overall fold conservation of the peptides and relative position of the non-covalent FAD and active-site Lys661.
Due to the above conformational differences and identification of the folate binding site within the KDM1A active site, there is some controversy as to which binding mode positions the K4 residue in the proper geometry that allows catalysis to proceed.4,117,126,167 Despite this discrepancy, both conformations reveal that the N-terminus of the peptides bind in an anionic pocket with a maximum of three residues N-terminal to the methyllysine residue.4,141 Additionally, both conformations make extensive contacts with a large set of conserved, negatively charged residues in the active site cavity.116,117 Overall, these structures exemplify the apparent plasticity of the active sites of KDM1A and KDM1B and only hint at the richness of substrate specificity mechanisms available to this enzyme class. Based on these data, it is speculated that KDM1A and KDM1B can recognize and demethylate different substrates in vastly different binding modes.4
Specificity for Non-Histone Substrates
In addition to histone H3, non-histone proteins have been shown to be substrates for KDM1 family enzymes. KDM1A has been shown to demethylate p53, DNMT1, E2F1, MYPT1, STAT3, HIV Tat, HSP90, MTA1, and potentially ERα (Table I).103,154,155,168–174 To our knowledge, no non-histone substrates have yet been identified for KDM1B; however, we suspect that future work will also define multiple, albeit different substrates and roles for this enzyme. The existence of multiple non-histone substrates for KDM1A has strong implications in apoptosis, cell cycle progression, and the transcriptional activation or repression of associated genes.
Table I.
KDM1 Demethylase Family
| Name | Synonyms | Histone Substrates | Non-Histone Substrates |
|---|---|---|---|
| KDM1A | LSD1, AOF2, BHC110, KIAA0601 | H3K4me1/2, H3K9me1/2 | p53 (K370), DNMT1 (K1096),a E2F1 (K185), ERα (K266),b HIV Tat (K51), HSP90 (K615h/K614m),c,d MTA1 (K532), MYPT1 (K442), STAT3 (K140) |
| KDM1B | LSD2, AOF1, C6orf193 | H3K4me1/2 |
Not verified to be major site of demethylation.
Inferred site of demethylation, not confirmed.
Confirmed in vitro.
Numbering system conventions between H. sapiens (K615h) and M. musculus (K614m).
The first non-histone substrate of KDM1A identified was the tumor-suppressor p53.154,155 Like histone proteins, p53 is subject to regulation by various PTMs. In particular, dimethylation of p53 at K370 (K370me2) promotes its association with 53BP1 (p53-binding protein 1), a protein that coactivates p53 transcriptional programs.175–177 Although in vitro KDM1A completely removes dimethylation at K370, in cell culture it preferentially removes a single methylation mark, generating the monomethylated species. This mark abrogates transcription through modulating p53-DNA interactions that subsequently repress the pro-apoptotic functions of p53. Further complicating this interaction, Barton and coworkers observed that KDM1A localizes only to genes where p53 represses transcription.154 This suggests some type of inherent regulation of the KDM1A-p53 interaction and may ensure repression of desired target genes.178
In contrast, in p53-deficient tumors, Set7/9 and KDM1A regulate DNA damage-induced cell death in a manner opposite to that observed in p53+/+ cells, via modulation of the stability of the transcription factor E2F1.169 Kontaki and Talianidis illustrated that Set7/9 methylates E2F1 at K185 and prevents E2F1 accumulation during DNA damage and subsequent activation of its pro-apototic target gene, p73. This PTM is removed by KDM1A, and is required for E2F1 stabilization and downstream pro-apoptotic function. Interestingly, the molecular mechanism of these events involves crosstalk between lysine methylation and other PTMs that affect E2F1 stability and turnover.
Recently, Sakane and coworkers have identified KDM1A as a HIV Tat K51me1-specific demethylase that is required for the activation of HIV transcription in latently infected T cells.103 The RNA-binding domain of Tat is a known region containing PTMs179 in which K51 is methylated by Set7/9.180 This PTM corresponds to an early event in the Tat transactivation cycle and strengthens the interaction of Tat with TAR RNA.180 On the other hand, acetylation of K50 is mediated by p300/ KAT3B, which dissociates the complex formed by Tat, TAR RNA, and the cyclin T1 subunit of the positive transcription elongation factor b (P-TEFb).181,182 Subsequent to K50 acetylation, the histone acetyltransferase, PCAF/KAT2B, is recruited to the elongating RNA polymerase II complex.183–185 Interestingly, the association of the KDM1A/CoREST complex with the HIV promoter subsequently activated Tat transcriptional activity in a K51-dependent manner. In accordance with Tat transcriptional activity, shRNAs directed at KDM1A or its inhibition by phenelzine suppressed the activation of HIV transcription in latently infected cells. The study again illustrates the dual nature of KDM1A and its ability to function as both a transcriptional suppressor and activator depending on the context of the substrate and interacting partners.
In addition to p53 and E2F1, Chen and coworkers have identified DNA methyltransferase 1 (DNMT1) as a novel substrate of KDM1A.168 Targeted deletion of the gene encoding KDM1A (aof2) in mouse embryonic stem cells induces the progressive loss of DNA methylation in addition to an increase in the methylation of DNMT1 and a decrease in the DNMT1 protein level. Chen and coworkers illustrated that Set7/9 methylation of K1096 of DNMT1 can be reversed by KDM1A in vitro. Despite this evidence, it remains to be determined if K1096 is the major targeted site of KDM1A, since DNMT1 is methylated on multiple sites in vivo. By acting on and demethylating DNMT1, KDM1A may play a role in the coordination of DNA and histone methylation.
In another example, like E2F1, the transcription factor STAT3 is subject to PTMs that modulate downstream events in response to different cytokines and growth factors.186 Following phosphorylation, STAT3 is methylated on K140 by SET7/9 and subsequently demethylated by KDM1A when it is bound to a subset of promoters that it activates.171 Timing for this process occurs in an ordered sequence on the SOC3 promoter, illustrating the underlying temporal and spatial control of the methylation and demethylation events. The authors conclude that lysine methylation of promoter-bound STAT3 leads to biologically important downregulation of the dependent responses. This work illustrates the ability of methyltransferases and demethylases to control the methylation status of a transcription factor in a promoter-specific manner and provide a way to modulate the time of residence and promoter occupancy of the protein. Additionally, the transcription factors or the proteins recruited during these events may also provide a docking site for other proteins to carry out important functions, a role suggested for KDM1A by Mattevi and coworkers.152
Myosin phosphatase target subunit 1 (MYPT1) is an example of a KDM1A non-histone substrate that has a downstream affect on cell cycle progression.170 MYPT1, a known regulator of phosphorylation of the transcription factor retinoblastoma protein 1 (RB1), is methylated in vitro and in cell culture by SET7/9 and subsequently demethylated by KDM1A, in which K442 is the target residue. The methylation status of MYPT1 influences the affinity the protein maintains for the ubiquitin-proteasome pathway and subsequent protein turnover. Interestingly, overexpression of KDM1A, which is prevalent in many cancer subtypes (see above), could activate RB1 phosphorylation by inducing a destabilization of the MYPT1 protein. Subsequently, the release of E2F could activate transcription of genes required for S phase to enhance cell cycle progression.
An extremely unique target of KDM1A is metastatic tumor antigen 1 (MTA1), a core subunit of the NURD complex (discussed in detail later). Interestingly, it is the only known dual function coregulator with an expected corepressor activity, but unusual ability to also stimulate transcription.187,188 MTA1 is mono-methylated at residue K532 by G9a (EHMT2) and demethylated by KDM1A.173 This demethylation event is required for the stable interaction of the two proteins (i.e. KDM1A and MTA1), but also represents a molecular switch, as the methylation state of MTA1 potentiates the nucleation of either the NuRD or NURF complexes. Reconstitution of either complex occurs in a cyclical manner that alternates between opposing biological functions. This activity further compounds the role of KDM1A as both a transcriptional activator and repressor via the demethylation of a non-histone substrates and modulation of subsequent downstream targets.
The least well characterized KDM1A non-histone substrates are HSP90 and ERα.172,174 Using proteomics and biochemical approaches, Abu-Farha and colleagues determined that HSP90 is methylated at K209 and K615 by SMYD2,172 the latter of which is present in the dimerization domain (DD) of HSP90. Coincidentally HOP, a co-chaperone known to interact with the DD,189 blocks methylation at K615 in vitro. Although demethylation at this residue was performed in vitro by KDM1A, it has not yet been demonstrated in vivo. Furthermore, the methylation status of K615 has been implicated in the ability of HSP90 to associate with the sarcomeric protein titin, thereby affecting the maintenance and function of skeletal muscle.190,191 The role of HSP90 methylation status may also have potential, yet undiscovered, significance in carcinoma maintenance.192,193 On the other hand, the direct demethylation of ERα at K266 by KDM1A has not been explicitly demonstrated. However, KDM1A seems to negatively regulate the methylation status at this residue.174 It is interesting to suspect that the presence of KDM1A on estrogen responsive elements (EREs) has a dual role in the demethylation of H3K9me2 and ERα,194 or possibly uses this interaction to recruit an H3K9-selective demethylase enzyme to perform this function.
Interestingly, either SMYD2 (KMT3C) or SET7/9 (KMT7) methylates all but one of the current non-histone substrates that have been identified for KDM1A. In the future it will be informative to investigate more non-histone substrates of KDM1A and KDM1B to determine if they too maintain this same relationship with specific MTases. Such analyses may provide insight into the interplay between ‘writers’ and ‘erasers’ of PTMs in vivo.
INHIBITION OF FLAVIN-DEPENDENT KDM1 DEMETHYLASES
Discovery and Optimization of Mechanism-Based Inhibitors
The discovery and development of KDM1 family inhibitors has been the subject of several recent comprehensive reviews; therefore, an abbreviated overview of KDM1 inhibitors is presented, which includes many recent developments.15,21,69,142,195–199 Motivated by the relationship between the AOD of KDM1A to that of MAO-A/B and PAO, the first KDM1A inhibitors were discovered by McCafferty and coworkers.200 Through screening a focused group of irreversible and reversible amine oxidase inhibitors, tranylcypromine (trans– 2-phenylcyclopropylamine, 2-PCPA, Parnate™, 1) was found to exhibit the highest KDM1A inhibitory activity towards methylated bulk histones as well as methylated nucleosomal substrates in vitro. Treatment of P19 embryonal carcinoma cells with 1 resulted in the global increase of H3K4 methylation as well as transcriptional derepression of two KDM1A target genes, Egr1 and the pluripotent stem cell marker Oct4. This was the first example of small molecule inhibition of histone demethylation and interruption of transcriptional programs regulated by KDM1A.200
In a subsequent study, our group determined that tranylcypromine was a mechanism-based inactivator of KDM1A, forming a covalent adduct with the FAD cofactor and demonstrating inactivation kinetic parameters of KI = 242 µM and a kinact = 0.0106 s−1.127 Soon after, the laboratories of Cole,118 Yokohama,119,122 and Mattevi121 provided additional support for this inhibitory mechanism, including defining the structure of the tranylcypromine-KDM1A adduct using X-ray methods, which revealed the N5 of the FAD isoalloxazine ring as the site of covalent attachment of the inhibitor.
In addition to tranylcypromine (1), our study revealed less potent KDM1A inhibition from the hydrazine-containing phenelzine (2) and the propargylamines, represented by pargyline (3) and clorgyline (4) (Figure 3).200 KDM1A inhibitors 1 and 2 achieved complete inhibition in vitro at significantly lower concentrations than are physiologically relevant.127,201 However, propargylamines were not inhibitory at 5 mM, and only partial inhibition of KDM1A was observed at 30 mM.147,200,201 Exposure of cells to such non-physiologically relevant concentrations of 3 causes inhibition of proliferation and alteration of transcriptional programs via mechanisms that do not necessarily involve KDM1A activity.72 As such, one should approach the use of members of this inhibitor class with caution when attempting to attribute phenotypic effects exclusively to KDM1A inhibition.
FIGURE 3.
Chemical structures of representative mechanism-based, irreversible inhibitors.
Analysis of the crystal structures of KDM1A, MAOs, and PAOs, lead our group to conclude that KDM1A may be distinguished from related amine oxidases since MAO-A/B and PAO have significantly restricted active site volume when compared to KDM1A. As such, we designed, synthesized, and characterized analogues of 1 for the purpose of increasing potency and specificity towards KDM1A through aromatic substitutions and heteroaromatic exchanges.202 Within this initial group of inhibitors, improved selectivity and potency was observed, and these analogues were subsequently utilized to probe the role of KDM1A activity in estrogen receptor signaling.72 Since that time, numerous research groups, in addition to our own, have further optimized the potency and selectivity of arylcyclopropylamines for KDM1A inhibition.121,122,196,203–205
In fact, several members of the arylcyclopropylamine inhibitor class have achieved pre-clinical and clinical development status industrially. For example, Oryzon researchers reported the discovery of KDM1A inhibitor 5 that exhibited a 100-fold improved potency over the parent inhibitor 1. In a recent review, it was reported that Oryzon optimized 5 to produce ORY-1001 (structure not yet released), which possesses 1000-fold greater potency than 1 and is highly selective for KDM1A over KDM1B, MAO-A/B, IL4I1, and SMOX (SMO/PAO/PAO-1/PAOH1).198 Additionally, GlaxoSmithKline recently reported the discovery, development, and optimization of arylcycloproylamine-based KDM1A inhibitors (lead compound, 6) that achieve exclusive selectivity for KDM1A and also exhibit good oral bioavailability.206
Peptide-Based Inhibitors
Protein-protein and protein-ligand interactions provide auspicious starting points for the development of high affinity peptide and peptidomimetic inhibitors. Although this class of molecules has traditionally suffered from poor stability and cell permeability, modifications to the native peptide structure can greatly increase their pharmacokinetic properties and therapeutic value. This inhibition strategy is especially appealing in regard to KDM1A, as it is involved in multiprotein complexes, several of which are beginning to be biophysically characterized in detail (see below). Here we outline the existing peptide-based active site inhibitors of the KDM1 class demethylases with the hope that they will catalyze the development of peptide-based inhibitors that target a wider range of KDM1 interfaces.
In addition to the small molecules discussed above, unmethylated product peptides (7) of the KDM1A reaction as well as a related substrate-like inhibitor containing a methionine point mutant (8; H3K4M; pK4M) have been shown to be competitive inhibitors of KDM1A with enhanced binding over methylated substrates.117,147 Recently, Kumarasignhe and Woster have also developed a series of cyclic peptides based on the substrate-like inhibitor pK4M that were more hydrolytically stable than the acyclic analogues. Additionally, moderate antitumor effects were observed in MCF7 and Calu-6 cancer cell lines.207 On the other hand, Mattevi and coworkers have developed a series of short peptide reversible inhibitors based on the N-terminal region of SNAI1, some of which exhibited anti-proliferative activity.123,124 Both classes of peptides show promise and may serve to lay the foundation for the development of peptidomimetic small molecule inhibitors.
In addition to reversible peptide inhibitors, several groups have also adopted an inhibitor design strategy whereby flavin-reactive moieties are grafted as ‘warheads’ onto peptides derived from histone H3, including reactive N-propargyl, -cyclopropyl, -aziridine, -phenelzine, -vinylchloride, or -tranylcypromine substituents (compounds 9–17; Figure 4).201,208,209 Within this series, phenelzine-containing H3K4-derived peptide (15) exhibited the most potent inhibitory activity in vitro;201 however, none of these compounds exhibited significant KDM1A inhibitor activity in cellular models, presumably due to poor cellular uptake or susceptibility to proteolytic degradation.210 Additionally, although alone pargyline is a very poor inhibitor of KDM1A,147,200,201 presentation of the propargylamine group as a pendant substituent within a peptide derived from histone H3 (9, 10) significantly enhanced its activity as a mechanism-based inactivator (Figure 4).116,201,208
FIGURE 4.
Structures of substrate-based peptide inhibitors.
Emerging KDM1A Inhibitor Classes
Recently, Jung and coworkers have reported the development of lysine mimetics that possess a propargyl warhead for covalent modification of KDM1A (18,19; Figure 5).211 Although these compounds were inhibitory at low micromolar concentrations, they did not exhibit selectivity over the MAOs and PAO in this first generation series. This class of molecules may likely gain selectivity through future optimization efforts.
FIGURE 5.
Structures of nonpeptidic, warhead small molecule inhibitors that are substrate derived mimics.
Similarly, Suzuki and coworkers have reported a series of KDM1A inactivators in which tranylcypromine (1) is coupled to a lysine carrier moiety at the nitrogen atom (20; Figure 5).212,213 The nonpeptidic small molecules selectively and efficiently delivered 1 to the KDM1A active site creating the inactivated, FAD-adduct in a time-dependent manner. Additionally, the molecules exhibited potent cell growth inhibitory activities against cancer cell lines. Together, the high potency and selectivity towards KDM1A allows this class of molecules to be considered good candidates for further optimization and implementation as chemical biology probes.
The considerable homology KDM1A shares with PAOs, including spermine oxidase (SMOX/SMO/PAO/PAO-1/ PAOH1) and others, spurred Woster and Casero to develop a class of reversible KDM1A antagonists capable of derepressing tumor suppressor genes, as well as inducing other beneficial anticancer activities. The first class of polyamine analogues contained bisguanide and bisguanidine functional groups (21, 22) that inhibited KDM1A activity in cell and animal models of cancer.214–216 Since that time, they have expanded the KDM1A inhibitor repertoire to also include (bis)urea and (bis)thiourea analogues (23–25) that have exhibited excellent potency, cell permeability, and selectivity for KDM1A inhibition.217 In a recent example,66 the polyamine (1,15-bis(N5-[3,3-(diphenyl)propyl]-N1-biguanido)–4,12-diazapentadecane) (22; Figure 6) induced cytotoxicity and inhibited KDM1A activity in human AML cell lines. Exposure to this agent increased global levels of H3K4me1/2 and exhibited reexpression of the tumor suppressor E-cadherin.
FIGURE 6.
Structures of bisguanide and bisguanidine polyamine inhibitors.
Polyamine analogues have enormous potential as therapeutics for many classes of human cancers, including breast, prostate, and AML. Also, there is potential for these molecules as probes for understanding the contribution of KDM1A to specific epigenetic demethylation events that contribute to cancer pathobiology. As such, the discovery and development of polyamine analogues as reversible KDM1A inhibitors has been the subject of several recent seminal reviews.19,197,218–222
Merging key pharmacophoric features of reported KDM1A inhibitors, Dulla and coworkers recently reported the development of 3-amino/guanidine substituted phenyl oxazole-containing inhibitors.223 Following treatment of cultured cells with nanomolar inhibitor concentrations, viability decreased as compared to the low micromolar range observed for inhibition of KDM1A in vitro. Although the basis of the enhanced reactivity in cell culture and the degree of KDM1A selectivity remains to be fully explored, one inhibitor (26; Figure 7) avoided zebra-fish embryo apoptosis and toxicity and may constitute a lead for further optimization. Additionally, it remains to be determined if this class is selective for KDM1A since these compounds were designed based on existing broad spectrum amine oxidase inhibitors.
FIGURE 7.
Structures of bifunctional small molecule inhibitors that incorporate multiple pharmacaphores.
Other examples of bifunctional KDM1A inhibitors have also recently been described. First, Zheng and coworkers reported a novel triazole-dithiocarbamate scaffold that inhibits KDM1A more potently than 1.54 The 1,2,3-triazole moiety within these inhibitors is readily accessible through the Cu(I)-catalyzed Huisgen 1,3-dipolar cycloaddition of azides with alkynes (i.e. Sharpless “click” chemistry).224,225 Additionally, this pharmacophore has been shown to possess numerous biological activities,226,227 including inhibitory activity towards MAO-A.228,229 On the other hand, the dithiocarbamate pharmacophore was chosen for its intrinsic inhibitory activity against fungi, bacteria, and malignant cancer.230–232 One compound synthesized in the series (27; Figure 7) effectively reduced tumor growth of human gastric cancer cells in vivo.
In another example, Rotili and coworkers synthesized hybrid KDM1A/JmjC bifunctional inhibitors. This was achieved by coupling the skeleton of the KDM1A inhibitor tranylcypromine (1), with 4-carboxy-4’-carbomethoxy-2,2’-bipyridine or 5-carboxy-8-hydroxyquinoline chelating groups that were developed as competitive inhibitors for the iron(II)/ α-KG-dependent JmjC demethylases.86 Two compounds in this series (28,29; Figure 7) increase H3K4 and H3K9 methylation levels in cells and cause growth arrest and substantial apoptosis in LNCaP prostate and HCT116 colon cancer cells.86
Fortuitously, Yang and coworkers decided to test natural polyphenols as potential inhibitors of KDM1A.233 Dietary polyphenols are known to have beneficial effects against diabetes, cancer, and cardiovascular disease, and exposure has been linked to both antioxidant activity and modulation of signaling pathways.234–236 Yang and coworkers demonstrated that resveratrol (30), curcumin (31), and quercetin (32) displayed inhibitory activity against KDM1A in vitro, which was independent of their antioxidant proproperties (Figure 8). However, since quercetin forms colloid-like aggregates in aqueous solution it may inhibit KDM1A in vitro non-specifically due to aggregate formation.237 Based on this precedence, further insight into its mode of action as well as structurally-related compounds is needed.
FIGURE 8.
Natural polyphenol inhibitors.
Screening has also been a means for identifying novel scaffolds against KDM1A and KDM1B and has yielded several anti-KDM1A inhibitor candidates. By combining protein structure similarity clustering and in vitro compound screening, Beuttner and coworkers discovered the KDM1A inhibitory activity of the γ-pyrone, Namoline, in vivo and in vitro (33, Figure 9).85 The IC50 of this compound was reported to be 51 µM against KDM1A and was fully reversible. Namoline administration to prostate cancer cells led to silencing of AR-regulated gene expression and impairment of androgen-dependent proliferation in vitro and in vivo. Namoline may be a promising starting point (i.e. core structure) for the development of therapeutics to treat androgen-dependent prostate cancer.
FIGURE 9.
Structures of screen identified and validated inhibitors.
On the other hand, Sharma and coworkers recently reported a structure-based virtual screen of a compound library containing ~2 million small molecules against KDM1A.238 Computational docking and scoring followed by biochemical screening led to the identification of a novel N’-(1-phenylethylidene)-benzohydrazide series of inhibitors. Hit-to-lead optimization and structure-activity relationship studies aided the discovery of a specific, reversible inhibitor (34) of KDM1A with a reported Ki of 31 nM. Compound 34 inhibits proliferation and survival in breast and colorectal cancer cell lines.
Zha and coworkers have also garnered recent success with a pharmacophore-based virtual screening approach to identify novel inhibitors of KDM1A.239 Using a moderate database of a commercial compound library and molecular docking tools, the group was able to identify nine candidate KDM1A inhibitors that lacked structural similarity to previously identified inhibitor classes. These compounds were subsequently tested in vitro against KDM1A and showed IC50 values in the low micromolar range. One compound, XZ09 (35), which showed the highest potency against KDM1A, was also tested versus MAO-A/B and showed moderate selectivity. The XZ09 core structure represents a lead compound that, with further modification, could serve as another tool in probing KDM1A function.
Finally, Cole and coworkers recently reported the design, synthesis, and characterization of the KDM1A inhibitory properties of phenelzine analogues.240 Bizine (36; Figure 10), a novel phenylzine analogue containing a phenyl-butyrylamide appendage, exhibited good in vitro activity against KDM1A (KI = 0.06 µM; kinact = 0.15 min−1; kinact/KI = 2.5 µM−1min−1) and was selective versus MAO-A/B (26- and 60-fold, respectively) and KDM1B (>100-fold). Bizine exhibited anti-proliferative effects in some cancer cells and was found to be effective at modulating bulk histone methylation. Additionally, these compounds synergistically inhibited cellular proliferation in combination with several histone deacetylase inhibitors. Interestingly, neurons exposed to oxidative stress were protected by the presence of bizine.
FIGURE 10.
Structures of mechanism-based phenelzine analogs.
FUNCTIONALLY IMPORTANT INTERACTIONS OF KDM1A AND KDM1B DEMETHYLASES WITH BIOMOLECULES
KDM1 family demethylases typically operate as catalytic subunits within specific stable complexes formed with additional biomolecules and enzymes that perform coregulatory or scaffolding functions. Such interactions link the catalytic activity of KDM1s to distinctive biological occupations, and have been shown to influence the degree of catalytic activity, substrate specificity, and/or localization of these enzymes within chromatin.
Interaction of KDM1A with CoREST and CoREST-like Proteins in Heteromeric Complexes
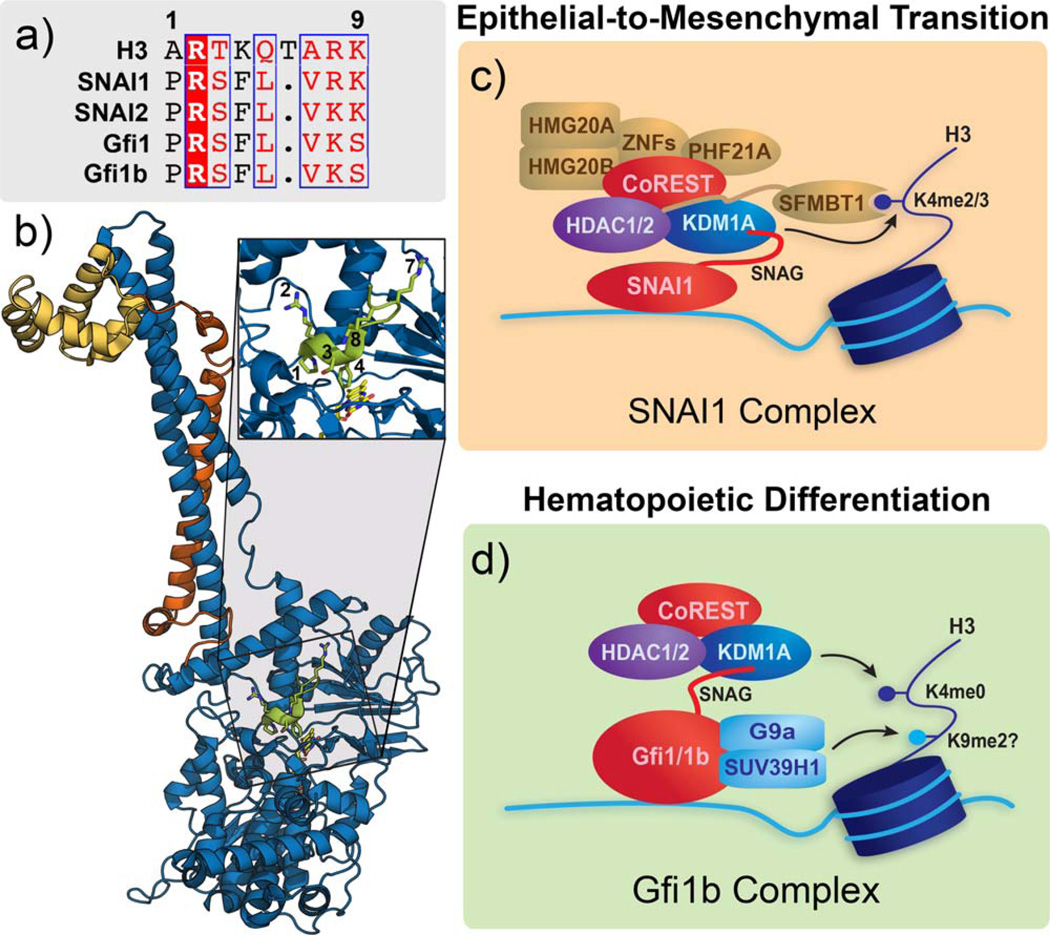
CoREST is one of the earliest identified interaction partners of KDM1A and this protein complex assembly (i.e. KDM1A/ CoREST complex) is frequently found within several distinct, larger multi-protein complexes.8,9,34,241–243 Most likely due to this early association, the structural and functional characterization of this particular interaction far outstrips that available for any other KDM1A interaction. CoREST consists of an N-terminal ELM2 domain followed by dual SANT domains (SANT1 and SANT2) with an intervening region colloquially known as the linker domain (Figure 11f). The N-terminal region spanning the ELM2 domain and first SANT domain have been mapped as a binding region for HDAC1,8 and a crystal structure of HDAC1 bound to a similar ELM2/SANT region in the CoREST-related protein, MTA1, has recently been reported.244 Furthermore, Shi and colleagues first mapped the interaction of KDM1A to the CoREST linker domain,34 indicating that CoREST likely acts as a scaffolding protein that joins deactylase and demethylase activity into a single catalytic sub-complex. The CoREST SANT2 domain, and possibly the SANT1 domain, has been shown to facilitate demethylation of nucleosomes, presumably by acting as a bridge between KDM1A and its substrate.34,115,242 A similar mechanism is observed in the SMRT protein, where the C-terminal SANT2 domain stabilizes the protein complex on chromatin by interacting with histone tails.245 As a point of further clarification, Yang and coworkers highlighted the similarity between the SANT2 domains of CoREST and the viral DNA-binding protein v-Myb, and demonstrated that an isolated CoREST SANT2 domain binds directly to DNA with modest affinity (Kd = 84 µM).115
FIGURE 11.
KDM1A complexes that utilize CoREST and CoREST-like interactions. (a) Ribbon representation of KDM1A (blue) in complex with the linker (orange) and SANT2 (yellow) domains of CoREST (PDB 2IW5). Insert highlights the regions of contact between CoREST and the KDM1A tower domain. (b) CoREST recruits a KDM1A complex to suppress neuronal genes through its interaction with REST, which also recruits the Sin3 complex. (c) The CoREST core complex is recruited by a direct interaction between TLX and KDM1A. This complex forms a negative feedback loop with the microRNA miR-137. (d) The transcription factor TAL1 recruits the CoREST complex, and potentially other coregulators, to repress gene transcription as an effector of hematopoietic differentiation. (e) The CtBP proteins associate with DNA-binding proteins for genomic localization and recruit chromatin remodelers, including KDM1A, to regulate the EMT. (f) Domain maps of CoREST and MTA1 are representative of their respective isoforms and show a similar ELM2/SANT domain organization. g) KDM1A is recruited by its tower domain to the MTA subunit of the NuRD complex, and in this context suppresses the EMT in breast cancer.
Seeking more detailed molecular characterization, Yang and colleagues reported the first crystal structure of KDM1A in complex with the linker and SANT2 domains of CoREST in 2006 (Figure 11a).115 In this model, the CoREST linker domain forms an L-shaped helical conformation, with the short helix contacting the base of the KDM1A tower domain, the longer helix extending up the TaB helix, and the SANT2 domain contacting the top turn of the tower (Figure 11a, insert). These interactions are mainly hydrophobic in nature, although a few electrostatic interactions may facilitate proper alignment of the CoREST linker with the KDM1A tower domain.115,130 A second crystal structure of KDM1A and CoREST with a histone substrate-like peptide bound (pK4M) showed little overall conformational change as compared to the non-peptide-bound structure.117 However, the model does illustrate that the L-shaped short arm formed by the CoREST linker may have an indirect effect on substrate binding by stabilizing the helix formed from KDM1A residues 372–395.
As noted above, although the CoREST SANT2 domain contacts the KDM1A tower in reported crystal structures,115,117 McCafferty and coworkers have demonstrated that the Co-REST linker domain is chiefly responsible for high affinity binding with KDM1A.130 Specifically, a CoREST fragment consisting of the linker domain (residues 293–380) exhibits low nanomolar binding affinity towards KDM1A (Kd = 7.78 nM), while inclusion of the SANT2 domain does not substantially alter binding affinity and in isolation lacks significant affinity towards KDM1A.130 Our group also used mutagenesis and isothermal titration calorimetry to reveal that the energy of binding along the CoREST/KDM1A helix-helix interface is not concentrated into “hotspots,” but is instead distributed almost uniformly along the interaction interface.246 As CoREST is required for nucleosomal demethylation,34,242 this argues that inhibitors of the CoREST linker/KDM1A tower interaction may inhibit KDM1A functionality in the cell.
In addition, MD studies by Baron and colleagues have suggested that the KDM1A/CoREST interaction may be dynamic. Specifically, their simulations indicate that the KDM1A SWIRM and CoREST SANT2 domains oscillate in distance and rotational angle in relation to each other, and that substrate binding allosterically rigidifies this fluctuation, favoring an “open” state.247–249 They further suggest that blocking these ‘hinge sites’ may prevent KDM1A/CoREST association with chromatin or other protein partners and therefore provide another potential strategy for modulating KDM1A function.250
In addition to the regulatory mechanisms enforced on KDM1A by CoREST, two CoREST paralogs, CoREST2 (RCOR2, 523 aa) and CoREST3 (RCOR3, four isoforms of 436, 449, 495, and 553 aa), have recently been revealed as further governors of KDM1A activity.251,252 These paralogs share a similar domain organization, including conserved ELM2 and dual SANT domains. Not surprisingly, CoREST2 and Co-REST3 both interact with KDM1A and are capable of incorporation into larger protein complexes.251,252 Similarly, the crystal structure of KDM1A bound to the linker-SANT2 domain of CoREST3 (residues 273–405; PDB 4CZZ) exhibited a nearly identical conformation as the KDM1A/CoREST (residues 308–440) complex (PDB 2V1D).251 However, CoREST2 displays a reduced ability to facilitate KDM1A nucleosomal demethylation252 and transcriptional repression as compared to CoREST.251 This weakening of repressive activity is attributed to residue L165 in the CoREST2 SANT1 domain, which diminishes its affinity for HDACs 1 and 2.251 However, CoR-EST3 plays an even more antagonistic role, as it competitively inhibits KDM1A nucleosomal demethylation. A CoREST3 chimera with the SANT2 domain replaced by the corresponding domain in CoREST reverses this phenotype and reinstates KDM1A nucleosomal demethylation.252 Collectively, these studies clearly indicate that CoREST and its paralogs play important roles in a subset of KDM1A-dependent transcriptional regulatory events by acting as a scaffold for the assembly and recruitment of deacetylase/demethylase-containing complexes, and by serving as a bridge to link epigenetic enzymes to nucleosomal substrates.
REST Complex
RE1-silencing transcription factor (REST) is a supervisor of cell fate decisions during neuronal differentiation, and functions as a transcription repressor that localizes to RE1 motifs in the promoters of neuron-specific genes (Figure 11b).253 In multipotent progenitor cells, these genes are held in a ‘primed-repressed’ state, in which REST recruits corepressor complexes mSin3254 and CoREST255–257 to its N- and C-terminal repressor domains, respectively, while simultaneously maintaining an activating chromatin environment and basal-level gene transcription.258 Upon differentiation into a neuron cell, REST is largely degraded to activate a neuronal expression program, although the CoREST complex sustains a role in tempering the expression of certain genes.258 Alternately, upon differentiation into a non-neuronal cell type, the REST/mSin3/ CoREST repressor machinery persists on RE1-containing promoters and adjusts chromatin in the vicinity of neuron-specific genes to a repressive state.258 Specifically, H3K4 demethylation is executed by KDM1A,13 and H3K9 methylation is likely mediated by the histone methyltransferases (HMTs) G9a and EHMT1, which are bridged to REST by the corepressor protein CDYL.259 The HMTs SUV39H1 and SETDB1 have also been linked to the CoREST complex and may play a role in H3K9 methylation.257,260 Increased CpG methylation (mCpG) in RE1 motifs has also been observed in differentiated cells compared to embryonic stem cells,258 although the DNA methyltransferase responsible has not yet been identified.
The H3K4 and H3K9 methylation status within chromatin encompassing repressed neuron-specific genes further recruits additional corepressors. For example, demethylated H3K4 is bound by PHF21A (BHC80), which stabilizes KDM1A on chromatin and enhances repression.261,262 However, PHF21A exists as multiple isoforms,263 is expressed tissue-specifically,241 and has been reported to paradoxically inhibit KDM1A activity34 and REST-mediated repression,264 indicating that its activity may be context-specific. Similarly, REST-associated G9a recruits CBX3 (HP1) to methylated H3K9me2 residues,265 where it is known to bind through its chromodomain and maintain a heterochromatic state.266 In another example, the mCpG-binding protein MeCP2 is also recruited to the larger corepressor complex for stable gene silencing,257 although it also appears to play a role in transient gene repression between neuron depolarization events.258
Other accessory proteins that may not directly interact with chromatin are also recruited to regulate gene repression.34,241 For example, REST repression is reportedly dependent upon sumoylation of HMG20B (BRAF35)267 and its recruitment to the CoREST complex.34,241 Previous studies have suggested that sumoylated regulators may bind the CoREST/KDM1A interaction surface,268 although it is unclear if this is the case for HMG20B. In contrast, the repressive activity of the REST complex is antagonized by HMG20A (iBRAF), which forms a heterodimer with HMG20B to prevent its sumoylation and incorporation into the CoREST complex.267 In fact, HMG20A is a critical regulator of neuronal differentiation that promotes neuron-specific gene expression by recruitment of the H3K4 HMT, MLL.269
TLX Orphan Nuclear Receptor Complex
KDM1A also regulates neuronal processes through its interaction with the orphan nuclear hormone receptor TLX (NR2E1). In line with KDM1A’s role in neuronal differentiation, TLX maintains the proliferative and self-renewal properties of adult neural stem cells and regulates neurogenesis.270 Yokoyama and coworkers first reported that TLX interacts directly with the AOD and SWIRM domains of KDM1A and further demonstrated that TLX recruits the core CoREST/KDM1A/HDAC1 complex (Figure 11c).271 Additionally, recruitment of the complex occurs in a KDM1A-dependent manner for H3K4 demethylation, H3 deacetylation, and downstream target gene repression. A subsequent report revealed that HDAC5 also associates with this complex.41 Interestingly, immunoprecipitation of TLX also coprecipitates several other proteins, including ZMYM3 and two that are also found in the SNAI1 complex (see below),93 namely GSE1 and ZMYM2.271 Despite these results, it is currently unclear whether any crosstalk exists between these complexes.
Within these complexes, Sun and coworkers also reported that the TLX/KDM1A ensemble regulates neural stem cell proliferation.41 Specifically, the microRNA, miR-137, antagonizes this proliferative activity and encourages differentiation by downregulating KDM1A. On the other hand, while in complex with KDM1A, TLX represses miR-137 expression. This feedback regulation likely contributes to timely neuronal differentiation events in which KDM1A is intimately involved.272
TAL1 Complex
TAL1 is a transcription factor that is involved in normal hematopoieisis and leukemogenesis and can act as both a transcriptional activator and repressor, depending upon the genomic context and stage of cellular differentiation.273 Specifically, TAL1 promotes transcription of erythroid-specific genes through association with p300 and PCAF,274,275 but can also associate with the mSin3A and ETO2 complexes to induce gene repression.276–280 KDM1A is recruited by TAL1 along with the core CoREST/HDAC complex to repress erythroid-specific genes in progenitor cells prior to differentiation (Figure 11d), but is dismissed from TAL1 promoters during the early stages of differentiation.281 The PTM of TAL1 may regulate this gene activation, as phosphorylation of S172 by protein kinase A (PKA) abolishes TAL1 interaction with KDM1A and results in recruitment of the H3K4 HMT, hSET1, activation of target genes, and initiation of erythroid differentiation.282 It is currently unclear if the KDM1A/CoREST core complex cooperates with any of the other known TAL1-interacting corepressor complexes to stimulate gene repression.
CtBP Complex
C-terminal binding proteins (CtBPs) are implicated widely as repressors of mammalian gene expression, and likely function in these varied contexts by recruiting different subsets of corepressors.283,284 KDM1A was first coprecipitated with the CtBP complex prior to discovery of its demethylase activity243 and has since been associated with several CtBP functions, including pituitary gland development,285 suppression of the tumor suppressor BRCA1,286 and activation of tissue-specific genes in gastrointestinal endocrine cells.287 However, the best-established role of KDM1A in the CtBP complex is its control of the EMT through suppression of the epithelial gene E-cadherin.243,288,289
On the molecular level, CtBPs interact with coregulators (Figure 11e) mainly through two interaction domains: a hydrophobic pocket that canonically binds proteins containing a Pro-X-Asp-Leu-Ser (PXDLS) sequence motif and a structurally distinct surface crevice that binds an Arg-Arg-Thr (RRT) motif.290 Additionally, CtBP proteins form extensive scaffolds by dimerization in a manner that positions the PXDLS binding pockets at opposite ends of the dimer, and in such a manner that the PXDLS pocket of one CtBP protein is in relatively close proximity to the RRT binding crevice of the second CtBP molecule.290 Initial studies indicated that the PXDLS-binding region mediates CtBP genomic localization by associating with DNA-binding transcription factors containing PXDLS motifs.291 However, later studies revealed that this site also recruits accessory proteins with histone-modification activity.292 For example, deacetylase activity is recruited by PXDLS motifs in class II HDACs293 and possibly by class I HDACs bridged through other PXDLS-containing corepressor proteins, such as NRIP140 and LCoR.294,295 Similarly, PXDLS-like motifs in Wiz recruit the HMTs G9a and EHMT1,296 and possibly CDYL and HDAC1/2 to the CtBP complex.292,297 Several other proteins co-precipitate with CtBP, although not all have well-defined functions.243
Interestingly, direct binding of CtBP1 to CoREST and HDAC2, respectively, has been reported despite the lack of PXDLS motifs in these proteins, and seem to bridge KDM1A to the complex.292 However, KDM1A recruitment to the CtBP complex also appears to be mediated by ZNF217,288,292 a large DNA-binding zinc finger protein that spans the CtBP dimer interface to interact with both PXDLS and RRT domains simultaneously.290 ZNF217 localizes the CtBP complex for transcriptional repression of several disease-related genes, such as E-cadherin and BRCA1.286,288 Other DNA-binding proteins have also been reported to bridge KDM1A to the PXDLS-binding pocket and appear to dramatically alter the demethylase’s biological function. For example, recruitment by ZEB1 can extend gene repression programs associated with pituitary differentiation,285 while recruitment by RREB1 can actually promote CtBP/KDM1A-mediate gene activation.287 Clearly, KDM1A activity within the CtBP complex may have farreaching biological implications that are most likely determined by the presence of specific coregulatory subunits.
NuRD Complex
The nucleosome remodeling and histone deacetylase (NuRD) complex is a large, multiprotein complex that has been implicated as a regulator of a wide variety of cellular processes, from human development to the progression of several cancer types.298 Since its discovery, the majority of its core protein constituents have been well defined: the ATP-dependent chromatin remodelers Mi-2 α/β (CHD3/4), histone deacetylases HDAC1/2, methyl-CpG-binding proteins MDB2/ 3, retinoblastoma-binding proteins RBBP4/7, corepressors GATAD2A/B (p66α/β), and metastasis-associated proteins MTA1/2/3 (Figure 11g).299
Interestingly, the MTA proteins share striking similarities with the CoREST proteins. Specifically, the MTA isoforms possess, from N- to C-termini, BAH, ELM2, SANT and GATA-like zinc finger domains (Figure 11f).299 As previously mentioned, the ELM2 and SANT domains serve to recruit HDACs 1 and 2,8,244 presumably in similar fashion to CoREST. Wang and coworkers first established that the MTA2 protein, like Co-REST, is sufficient to stimulate KDM1A demethylation of nucleosome substrates and mapped an interaction interface involving the KDM1A tower domain and MTA1/2/3 SANT domains.76 The authors further demonstrated that this interaction recruits H3K4 demethylase activity to the NuRD complex for enhanced gene repression, and that KDM1A association with each MTA isoform corresponds to a unique transcriptional outcome. Functionally, the authors linked KDM1A in the NuRD complex to silencing of the TGFβ signaling pathway and repression of the EMT and metastatic potential in a breast cancer cell line. Since this initial study, KDM1A has been associated as a corepressor of the NuRD complex in a subset of its biological functions, such as lipid homeostasis,300 aberrant gene repression in Ewing sarcoma,301 and enhancer decommissioning during embryonic stem cell differentiation.27 It is, however, noteworthy that a recent mass spectrometry-proteomics coupled study did not identify KDM1A as a stable component of the NuRD complex in HeLa cell extracts, indicating that it likely interacts in a context-specific manner.302
While the MTA proteins resemble CoREST in their ability to recruit HDAC/KDM1A, several domains unique to MTA suggest a unique functionality, possibly through altered interactions with chromatin or coregulatory proteins. In fact, the KDM1A/NuRD complex has been shown to also associate with the α-KG dependent histone demethylase KDM5B.303 In this context, KDM5B and KDM1A cooperate to sequentially demethylate H3K4 and repress the metastasis and angiogenesis-associated CCL14 chemokine pathway. However, it remains unclear if KDM5B recruitment is dependent upon a specific MTA isoform. A recent report from Nair and colleagues suggests a regulatory role of KDM1A in NuRD activity.173 The authors show that G9a methylation of MTA K532 is required for its incorporation into the repressive NuRD complex, while KDM1A-mediated demethylation of the same residue triggers its dissociation from the NuRD complex and recruitment to the coactivator complex, NURF. Although these studies provide much needed insight into the function of KDM1A within the NuRD complex, further investigation into the demethylase activity of KDM1A within the heteromeric complex is needed.
KDM1A Cooption During Herpes Simplex Virus Infection
Cooption of host KDM1A activity plays a role in infection and latency of herpes simplex virus (HSV) and has recently been reviewed.18,99 Briefly, HSV establishes an active infection at the portal of entry (typically a mucosal membrane), where the host cell initially silences the inserted viral genome. Through a series of coordinated derepression events, viral immediate-early (IE) stage (α) genes are first transcribed and eventually give rise to transcription of latestage (β and γ) genes. Although the primary infection produces massive viral titers that are destructive toward the host tissue, the virus also establishes a latent infection in neuronal cells, in which viral genes are present but silenced. This state is non-destructive to the host cell and allows for periodic reemergence of the virus through poorly understood mechanisms.
The HSV genome is parsimonious, lacking much of the cellular machinery required for transcription and regulation of viral genes. The virus instead appropriates host cell transcriptional regulators, including KDM1A, to establish active and latent infections. Gu and coworkers first associated the REST/ CoREST/HDAC repressor complex,304 and later KDM1A, with HSV infection (Figure 12).305 Consistently, Pinnoji and colleagues identified REST-binding RE1 motifs in the viral genome, suggesting a potential mechanism for cooption of the complex.306 Indeed, REST appears to aid in repression of viral genes during latent infection, as genomic insertion of a REST mutant incapable of binding corepressor proteins produces a more virulent strain compared to wild-type REST.307 Similarly, HDAC inhibitors reactivate silenced viral gene expression, while REST antagonizes reactivation.308
FIGURE 12.
KDM1A involvement in HSV infection and latency. KDM1A, along with other host cell epigenetic machinery, is coopted by HSV for viral gene activation. KDM1A and the CoREST/ REST/HDAC complex may also participate in viral gene repression, where KDM1A may demethylate H3K4 residues.
KDM1A also plays a role in viral gene activation. In order to induce transcription, the viral protein VP16 recruits host proteins HCF1 and OCT1 to target gene promoters.309 Liang and coworkers report that HCF1 recruits KDM1A and HMTs, SET1 and MLL1, for H3K9 demethylation and H3K4 methylation, respectively (Figure 12).100 Accordingly, the authors also report that inhibition of KDM1A prevents activation of viral genes and viral reemergence from latency.100–102 Interestingly, the JMJD2 family H3K9 HDMs work synergistically with KDM1A to activate α viral genes and inhibition similarly prevents viral activation and emergence from latency.310,311 Additionally, the histone acetyl transferase, CLOCK, also appears to promote viral gene expression and may interact with HCF1 through the transcription factor BMAL1.312 The dihydropyrido-indole natural product Harmaline reportedly represses α gene activation, possibly by disrupting this viral-host hybrid protein complex.313 Gu and coworkers have also suggested that the viral α protein ICP0 interacts with CoREST and may compete HDAC1 away from the REST/CoREST complex to initiate the switch to β/γ genes.304,314 However, the idea that CoREST plays a role in viral gene repression has been thrown into question by a study that showed no change in virulence when Co-REST is knocked-down in an ICP0-deficient HSV strain.315 Given that KDM1A is a promising target for preventing primary HSV infections and suppressing emergence from latency, it is likely that future work will shed light on such discrepancies and further detail KDM1A’s mechanistic role.
KDM1A Involvement in Nuclear Hormone Signaling
AR Complexes
The AR is a nuclear hormone receptor that is intimately linked to prostate function, regulating processes ranging from normal tissue development to the genesis and progression of prostate cancers, including castration-resistant tumors.316 Metzger and colleagues first reported KDM1A association with AR, suggesting that a large portion of the two proteins are involved in their mutual interaction and that the demethylase facilitates hormone-dependent AR-mediated transcription by demethylating H3K9 residues.84 Subsequent studies demonstrated that the Jumonji-domain demethylase, KDM4C, also interacts with KDM1A/AR and works in concert with KDM1A to demethylate H3K9me3 residues.165 Concurrently, hybrid inhibitors that simultaneously target both KDM1A and KDM4C exhibit anti-proliferative effects in prostate cancer cells.86 Although KDM1A canonically demethylates H3K4 residues, Metzger and colleagues proposed a mechanism for the observed switch in substrate specificity by which protein kinase Cβ1 (PKCβ1) associates with AR/KDM1A at target genes following hormone treatment and phosphorylates H3T6, a modification that prevents KDM1A demethylation of H3K4 on peptide substrates (Figure 13a).166
FIGURE 13.
KDM1A as an effector of gene activation and repression with nuclear hormone receptors. (a) KDM1A promotes AR gene activation following hormone stimulation at androgen-responsive gene promoters. At high hormone levels, KDM1A, and potentially other corepressors, repress AR transcription by demethylating H3K4 residues in the corresponding enhancer region. (b) KDM1A and PELP1 are recruited to ERα-occupied promoters. PELP1 reads H3K4me2 marks, and may position KDM1A for H3K9 demethylation. Association of CAC1 with this ERα/KDM1A complex causes dissociation from the promoter, accumulation of H3K9me2 marks, and gene repression.
KDM1A may also play a role in repression of AR target genes. Unlike AR, KDM1A resides on AR target gene promoters even in the absence of androgen treatment, when these gene are presumably repressed.84,165 Furthermore, the KDM1A/AR complex forms a negative feedback loop at high androgen levels, in which AR recruits KDM1A to an enhancer of the AR gene and mediates gene repression by demethylation of H3K4 (Figure 13a).317 Although the role of KDM1A as a coregulator of AR signaling is fairly well established, further work will hopefully better delineate its opposing roles as both an activator and repressor.
ERα Complexes
Paralleling the function of the AR, the ERα is the main modulator of estrogen (E2) signaling in estrogen-responsive tissues. As such, its dysregulation results in the development and progression of several cancer types.318 Recently, much attention has focused on the epigenetic mechanisms that accompany ERα signaling.319 As observed for KDM1A participation in AR signaling, the demethylase seems to function as both an activator and repressor of ERα signaling. Perillo and coworkers reported that in a ‘resting state’ with no hormone stimulation, KDM1A is constitutively bound to ERα target gene promoters and demethylates H3K4.194 However, E2 addition causes the recruitment of ERα to these promoters and KDM1A-dependent gene activation, most likely by catalyzing the demethylation of inhibitory H3K9 methyl marks.194,320 HMTs such as SET9 and SUV39H1 may also participate in E2-stimulated gene activation by methylating H3K4 (Figure 13b).194
The molecular mechanisms governing the activating and repressive roles of KDM1A are beginning to be described. For example, PELP1, an ERα coregulator with no known enzymatic activity, can interact directly with KDM1A, localizes on known KDM1A/ERα-bound genes, and aids in gene activation.321 When associated with KDM1A/ERα, the histone binding preference of PELP1 switches from H3K9me2 to H3K4me2, presumably to allow the subsequent demethylation of H3K9me2 by KDM1A (Figure 13b).321 Interestingly, a recent report suggests that targeting this KDM1A/PELP1 activity may be of therapeutic benefit in breast cancer.78 KDM1A may also demethylate ERα to promote transcriptional activation, although further validation of this activity is required.174 More indirectly, KDM1A demethylation events and the accompanying production hydrogen peroxide by products also reportedly induces controlled DNA damage that attracts chromatin remodelers to enhance chromatin plasticity and promote target gene transcription.194 In opposition to PELP1-mediated activation, KDM1A also interacts with the ERα corepressor CAC1, which binds KDM1A/ERα, reduces occupancy of the complex on target genes, allows H3K9me2 accumulation, and represses gene transcription (Figure 13b).322 Additionally, retinoic acid may interfere with the KDM1A-mediated E2 response, as it appears to prevent activation of protein kinase A (PKA) and prohibits H3K9 demethylation, although the mechanism is not yet clear.323
In addition to these roles in ERα signaling, KDM1A may also perform ERα-independent functions in breast cancer. For example, inhibition of the demethylase resensitizes hormone-refractory breast cancer cells to anti-estrogen treatment.78 In ERα-negative breast cancer, an interplay between KDM1A and HDACs is apparently necessary for tumor progression, as both affect the activity of the other towards histone substrates and simultaneous administration of KDM1A and HDAC inhibitors synergistically inhibits proliferation.79,80 Also, overexpression of the ERα corepressor, CAC1, induces sensitivity to paclitaxel in ERα-positive, paclitaxel-resistant breast cancer cells.322 These studies collectively imply that KDM1A functions in a wide range of ERα activities, and is additionally involved in various resistance mechanisms.
SNAG Family Proteins Interact with KDM1A Using Product Mimicry
The Snail/Gfi1 (SNAG) family of proteins encompasses over 70 different members characterized by a conserved SNAG domain and between four to six C2H2 zinc finger domains.324 Zinc finger domains are generally located near the C-terminal regions of SNAG proteins and function as DNA-recognition domains that frequently localize to E-box sequences (CANNTG), although recruitment of individual proteins is likely tailored by minor variations in zinc fingers domains and DNA sequences.324 In all but one known example, SNAG domains begin at the most N-terminal residue and are composed of a conserved “PRSFLV” sequence, where only the fourth residue is variable (Figure 14a).324 Proteins in the SNAG family of transcription factors have been reported to regulate KDM1A activity through product-like interactions, in which the SNAG domain mimics the histone H3 N-terminal tail sequence and binds competitively to the KDM1A active site.
FIGURE 14.
SNAG family proteins recruit KDM1A using product mimicry. (a) Alignment of the H3 tail with the first 8 residues of the SNAI1/2 and Gfi1/1b SNAG domains shows striking similarity, with conserved residues highlighted in red and chemically similar residues boxed in blue. (b) Ribbon representation of the ternary complex formed by KDM1A (blue), CoREST (orange/yellow), and SNAG (green; PDB 2Y48). The insert shows an enlarged view of the SNAG peptide in the KDM1A active site with numbered residues starting with the N-terminal proline. (c) SNAI1 recruits the CoREST core complex and other proteins through its SNAG domain to regulate the EMT. SFMBT1 may also help localize the complex by binding H3K4me2/3. (d) Gfi1 and Gfi1b also recruits the CoREST core complex and cooperates with HMTs to regulate hematopoietic processes.
Lin and colleagues first identified an interaction between the SNAG domain of SNAI1 and the AOD of KDM1A and demonstrated that it could be disrupted with small molecule inhibitors.97 Soon after, they showed that SNAI1 recruits KDM1A to target promoters where demethylation of H3K4 is required for gene repression.90 Following this discovery, Baron and colleagues determined the crystal structure of KDM1A and CoREST in complex with the 20 N-terminal residues of SNAI1 encompassing the SNAG domain, revealing density for the first nine residues (Figure 14b).123 In this report, the authors indicate that the SNAG domain mimics histone H3 through several conserved, positively charged residues and binds in the active site of KDM1A, resulting in competitive inhibition. Similar to the H3 tail, the first three SNAI1 residues form hydrogen-bonds deep in the active site, while Phe4 (F4), corresponding to Lys4 on H3 (H3K4), forms edge-to-face interactions with the flavin cofactor and KDM1A residue Y761. On the outer rim of the active site, Arg7 and Lys8 (R7 and K8) of SNAI1 seem to occupy the same positions as Arg8 and Lys9 of H3 (H3R8 and H3K9), respectively. Although direct evidence has not yet been provided, KDM1A presumably interacts with other SNAG proteins, including SNAI2, Gfi1, and Gfi1b, in a similar manner. In fact, treatment with either the KDM1A inhibitor tranylcypromine (1) or a cell-permeable SNAG peptide phenocopies down-regulation of either KDM1A or SNAI2 in decreasing the EMT.325
However, these studies raise an interesting question: if the SNAG domain is inhibitory toward KDM1A, how does it recruit the enzyme for demethylation of its target genes? One possible scenario is that the SNAG domain recruits KDM1A, but then relinquishes its hold in order to transfer binding to the H3 tail. In a recent study by Tortorici and colleagues, the first six residues of the SNAI1 SNAG domain were found to be sufficient for KDM1A binding, while residues 10–20 increased affinity only marginally and decreased ligand efficiency.124 On the other hand, the corresponding residues of H3 form multiple interactions with KDM1A,117 and hence may allow H3 to displace the SNAG domain.
SNAI1/2 Complexes
SNAIl (Snail, Snail1) and SNAI2 (Slug) are master regulators of the EMT and bind to E-box motifs in the promoters of epithelial genes, such as E-cadherin, to repress transcription and provoke an invasive, mesenchymal phenotype.91,326 KDM1A recruitment to target genes and demethylation of H3K4me2 is required for SNAI1-mediated gene repression.90 As discussed above, the SNAI1/2 SNAG domains recruit KDM1A via its active site.97,123 As such, small molecule inhibitors or isolated SNAG domain peptides can displace SNAI1, derepress target genes, and decrease invasiveness and motility in colon and neuroblastoma cancer cell lines without affecting proliferation.97,325 Interestingly, the chemotherapeutic drug doxorubicin has the opposite effect, enhancing the interaction between SNAI1 and KDM1A by inducing poly(ADP-ribosyl)ation of SNAI1.327 The authors further suggest that this increased interaction causes repression of the tumorsupressor gene, PTEN, and may facilitate drug-resistance in cancer.
SNAI1 also coprecipitates the core KDM1A/CoREST/ HDAC complex, as well as several auxiliary proteins such as HMG20A/B and PHF21A, which are associated with the REST complex, and CtBP, GSE1, and several zinc finger proteins, which are associated with the CtBP complex.93,97 It is currently unclear if SNAI1 recruits these larger KDM1A-containing complexes to further regulate gene expression. Recent studies have also identified the chromatin ‘reader’ SFMBT1, previously identified with the CoREST complex and other repressive complexes,328,329 as a member of the SNAI1/KDM1A complex. SFMBT1 binds directly to the CoR-EST linker region and mediates recruitment of KDM1A to SNAI1, gene repression, and induction of the EMT.93 Additionally, it recognizes H3K4me2/3 marks,93 and therefore may help to target the complex to active genes for subsequent repression (Figure 14c).
Gfi1/1b Complexes
Gfi1 and Gfi1b are SNAG-containing transcription factors that are critical for hematopoiesis. While Gfi1 regulates the differentiation programs of hematopoietic stem cells, small lymphocytes (B and T cells), dendritic cells, granulocytes and macrophages, Gfi1b is involved in the development of megakaryocytes and erythrocytes.330 As might be expected, both proteins are also involved in hematopoietic-derived cancers, although their roles as promoters or repressors of cancer progression seem to vary significantly depending upon tissue origin.331–336
As with the SNAI1 and 2 proteins, Gfi1 and Gfi1b recruit the core KDM1A/CoREST/HDAC complex via their SNAG domains,29 presumably by binding the KDM1A active site. Furthermore, Gfi1b recruits KDM1A in this manner to the majority of its target genes for demethylation of H3K4me2 residues and transcriptional repression.29 However, paralleling the tissue-dependent roles of Gfi1/1b, KDM1A differentially affects proliferation depending on the hematopoietic cell type and differentiation status.29 For example, splice variant Gfi1b p32, is required for erythroid differentiation and recruits the CoREST/ KDM1A complex more efficiently than the major Gfi1b isoform when dimethylated on Lys8 (K8) in its SNAG domain.337 Similarly, the three CoREST isoforms all interact with Gfi1b but have varying effects on its activity (see above).252 Gfi1b has also been associated with the H3K9 HMTs SUV29H1 and G9a,338 although these proteins have not been explicitly shown to co-localize with Gfi1b/CoREST (Figure 14d).
KDM1A Association with Long Non-Coding RNAs
A new class of interactions has recently been identified between KDM1A and long noncoding RNAs (lncRNAs).339–341 LncRNAs, broadly defined as non-protein coding RNA transcripts greater than 200 nucleotides, are differentially expressed in a number of human cancers and can regulate transcription by acting as scaffolds for chromatin modifying complexes.342–346 The HOTAIR transcript, which is dramatically overexpressed in several cancers347–350 and can be induced by estradiol,351 recruits the PRC2 and KDM1A/CoREST/REST complexes for downregulation of tumor suppressor genes at the HOXD locus.352,353 Binding interactions for EZH2 and KDM1A were observed at the 5’- and 3’-terminal domains of HOTAIR, respectively, and HOTAIR expression was required for co-immunoprecipitation and genomic co-localization of PRC2 protein complex and KDM1A (Figure 15a).339 These interactions and the corresponding changes in histone methylation states were also observed in vivo.353 The HOTAIR/PRC2/ KDM1A complex has also been implicated as a positive regulator of NFAT5, a transcription factor associated with enhanced metastasis and angiogenesis in breast cancer.354 In contrast, glioblastoma cell cycle progression appears to be dependent upon HOTAIR and PRC2 but not KDM1A, implying these corepressors may play specific, non-interdependent roles in conjunction with HOTAIR.355 More extensive reviews of HOTAIR and its role in cancer have recently been published.356,357
FIGURE 15.
KDM1A forms multi-component complexes with lncRNAs. (a) The lncRNA HOTAIR colocalizes the RE1-silencing transcription factor (REST)/CoREST/KDM1A and PRC2 complexes within the genome for subsequent gene repression. (b) While TRF2 protects telomere ends from degradation, loss of TRF2 allows recruitment of KDM1A and the telomeric repeat-containing lncRNA TERRA to the nuclease MRE11 and promotes telomere end processing.
In 2013, a second breast cancer-associated lncRNA, SRA (steroid receptor RNA activator), was reported to associate with KDM1A-containing repressive complexes, though in this case the interaction was mediated by progesterone receptor (PR).340,358 Most recently, Porro and colleagues have demonstrated an interaction between KDM1A and the telomeric lncRNA TERRA.341 The authors report that TERRA, which consists of UUAGGG repeats and has been implicated in the progressive shortening of telomeres, recruits KDM1A to telomeric DNA in the absence of the protective telomere repeat-binding factor 2 (TRF2). KDM1A does not seem to affect histone methylation levels in this context but instead promotes the removal of telomere 3′ guanine overhangs by enhancing the nuclease activity of MRE11, an enzyme involved in telomere 3′ end processing. Both TERRA and MRE11 interact with the SWIRM and N-terminal AOD region of KDM1A, but do not interact directly themselves. The authors therefore speculate that TERRA enhances the affinity of KDM1A for MRE11 and hence promotes telomeric shortening, although a direct substrate of KDM1A was not identified (Figure 15b). Given that the biological significance of lncRNAs is just beginning to be appreciated, future reports of KDM1A interactions with lncRNAs are anticipated.
Interaction of KDM1B with Proteins and Multi-Protein Complexes
Compared to the identified roles of KDM1A described above, much less is known about the interactions and biological significance of KDM1B. In contrast to KDM1A, KDM1B does not possess a coiled-coil tower domain, and hence does not interact with CoREST14 nor cooperate with HDACs in breast cancer.79 However, it has been reported as an effector of genomic imprinting,106 and its activity is required for somatic cell reprogramming108,109 and liver development.107 KDM1B has more recently been implicated as a regulator of breast cancer proliferation, where inhibition works synergistically with DNMT and KDM1B inhibitors to derepress aberrantly silenced genes and decrease cell growth.359
KDM1B has been reported to function as a positive regulator of gene transcription by binding to chromatin in the highly transcribed, H3K36me3-enriched coding regions down-stream of gene promoters.105 Here it helps to maintain H3K4 and H3K9 methylation by associating with a larger complex that includes Pol II and other elongation factors, as well as the SET-family HMTs NSD3 and G9a, which methylate histone H3K36 and H3K9 sites, respectively (Figure 16a).105 Later studies indicated that the H3K36me3 reader NPAC/GLYR1 is likely part of this complex as well, and enhances KDM1B demethylation of H3K4me1/2 by binding at its AOD/SWIRM interface (Figure 16b).136 Finally, KDM1B is recruited to the promoters of inflammatory genes by the NF-κB subunit c-Rel and is required for subsequent H3K9 demethylation, recruitment of NF-κB, and gene expression.110 Given the intricate mechanisms involved in regulating and modulating KDM1A activity, it is likely that future work will uncover an analogous but unique set of protein partners that govern KDM1B activity.
FIGURE 16.
KDM1B forms a multiprotein complex that epigenetically regulates gene expression by altering histone modifications in the coding regions of genes. (a) KDM1B associates with the chromatin reader Nuclear Protein 60kDa/Glyoxylate Reductase 1 Homolog (NPAC/GLYR1), RNA polymerase II, and other elongation factors within active genes. (b) A ribbon representation of KDM1B (colored as described in Figure 1) in complex with a NPAC/GLYR1 fragment peptide (pink). The insert shows an enlarge view of NPAC binding in the cleft between the amine oxidase catalytic domain and SWIRM domains of KDM1B.
CONCLUSIONS
KDM1s function as oxidative demethylases, removing methyl marks on protein substrates that provide chemical cues for the regulation of transcription. As such, KDM1A and KDM1B are central to numerous transcriptional programs in normal and disease-associated biology. These include regulating differentiation processes of both hematopoietic and neuronal tissues, cancer proliferation and metastasis, and viral replication and emergence from latency. Within these enzymes exist interaction motifs unique to each KDM1 isoform that facilitate interactions with biomolecules that regulate catalysis, promote KDM1 enzyme recruitment, and direct access to specific substrates at explicit sites within the cell. These coregulatory molecules include proteins, product-like inhibitors, and lncRNAs. Recently, transcriptional outcomes have been correlated to specific interactions of KDM1A or KDM1B with coregulatory biomolecules. In fact, interactions with differing subsets of coregulators can lead to KDM1 involvement in opposing transcriptional programs. Although several classes of KDM1-selective small molecule inhibitors have been recently developed, pan-inhibition strategies lack the selectivity capable of discriminating between KDM1 activities in these specific functional contexts. The development of next-generation chemical biology probes and therapeutics that modulate these interactions may further deconvolute the complex biology of epigenetic regulation. We suspect that due to the shear size of these multiprotein complexes that these probes will either be peptides, peptoids, or peptide-like in nature.
Acknowledgments
We wish to thank the members of the McCafferty and Hargrove laboratories for their thoughtful insights during the preparation of this manuscript.
Contract grant sponsors: U.S. Department of Defense Office of Congressionally Directed Medical Research Programs for Breast Cancer Research Grant W81XWH-13-1-0400 (to D.G.M.), by the U.S. National Institutes of Health (NIH) for Research Grants R01-GM65539 (to D.G.M), by the NIH Predoctoral Science Training Grant T32-GM 7105-39 in Pharmacological Sciences (to J.E.L.), and by the NIH Predoctoral Training Grant T32-GM 8487-19 in Structural Biology and Biophysics (to J.M.B. and B.S.M.), and by Duke University. This manuscript is dedicated to Professor William F. DeGrado on the occasion of his 60th birthday.
Footnotes
This manuscript is dedicated to Professor William F. DeGrado on the occasion of his 60th birthday.
REFERENCES
- 1.Allfrey VG, Mirsky AE. Science. 1964;144:559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- 2.Allfrey VG, Faulkner R, Mirsky AE. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kooistra SM, Helin K. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 4.Hou H, Yu H. Curr Opin Struct Biol. 2010;20:739–748. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian C, Zhou MM. Cell Mol Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 7.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 8.You A, Tong JK, Grozinger CM, Schreiber SL. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. J Biol Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Michaels SD, Amasino RM. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Iyer LM. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0039. RESEARCH0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Proc Natl Acad Sci USA. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. J Biol Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spannhoff A, Hauser AT, Heinke R, Sippl W, Jung M. ChemMedChem. 2009;4:1568–1582. doi: 10.1002/cmdc.200900301. [DOI] [PubMed] [Google Scholar]
- 16.Tian X, Fang J. Acta Biochim Biophys Sin (Shanghai) 2007;39:81–88. doi: 10.1111/j.1745-7270.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- 17.Nottke A, Colaiacovo MP, Shi Y. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roizman B, Zhou G, Du T. J Neurovirol. 2011;17:512–517. doi: 10.1007/s13365-011-0058-x. [DOI] [PubMed] [Google Scholar]
- 19.Rotili D, Mai A. Genes Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Jie W, Yan W, Zhou K, Xiao Y. Crit Rev Eukaryot Gene Expr. 2012;22:53–59. doi: 10.1615/critreveukargeneexpr.v22.i1.40. [DOI] [PubMed] [Google Scholar]
- 21.Lynch JT, Harris WJ, Somervaille TC. Expert Opin Ther Targets. 2012;16:1239–1249. doi: 10.1517/14728222.2012.722206. [DOI] [PubMed] [Google Scholar]
- 22.Amente S, Lania L, Majello B. Biochim Biophys Acta. 2013;1829:981–986. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Dent SY, Chandra J. Elife. 2013;2:e00963. doi: 10.7554/eLife.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meissner A. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 25.Voigt P, Tee WW, Reinberg D. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puri D, Gala H, Mishra R, Dhawan J. FEBS J. 2014 doi: 10.1111/febs.13165. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- 29.Saleque S, Kim J, Rooke HM, Orkin SH. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 30.Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O’Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, Pajtler KW, Schramm A, Konig K, Diehl L, Mestdagh P, Vandesompele J, Speleman F, Jastrow H, Heukamp LC, Schule R, Duhrsen U, Buettner R, Eggert A, Gothert JR. Leukemia. 2012;26:2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 32.Butler JS, Dent SY. Blood. 2013;121:3076–3084. doi: 10.1182/blood-2012-10-451237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang RF, Zhao GW, Liang ST, Chen HZ, Liu DP. Chin Med Sci J. 2013;28:82–87. doi: 10.1016/s1001-9294(13)60027-9. [DOI] [PubMed] [Google Scholar]
- 34.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Kim HJ, Rosenfeld MG. Arch Pharm Res. 2010;33:1467–1473. doi: 10.1007/s12272-010-1001-z. [DOI] [PubMed] [Google Scholar]
- 36.Fuentes P, Canovas J, Berndt FA, Noctor SC, Kukuljan M. Cereb Cortex. 2012;22:1431–1441. doi: 10.1093/cercor/bhr218. [DOI] [PubMed] [Google Scholar]
- 37.He Y, Yu H, Sun S, Wang Y, Liu I, Chen Z, Li H. Int J Dev Biol. 2013;57:365–373. doi: 10.1387/ijdb.120227hl. [DOI] [PubMed] [Google Scholar]
- 38.Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X, Wang J, Ju BG, Rosenfeld MG. Curr Opin Cell Biol. 2007;19:605–611. doi: 10.1016/j.ceb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge W, Liu Y, Chen T, Zhang X, Lv L, Jin C, Jiang Y, Shi L, Zhou Y. Biomaterials. 2014;35:6015–6025. doi: 10.1016/j.biomaterials.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 41.Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Mol Cell Biol. 2010;30:1997–2005. doi: 10.1128/MCB.01116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jie Z, Li T, Jia-Yun H, Qiu J, Ping-Yao Z, Houyan S. Brain Res Bull. 2009;80:79–84. doi: 10.1016/j.brainresbull.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Neelamegam R, Ricq EL, Malvaez M, Patnaik D, Norton S, Carlin SM, Hill IT, Wood MA, Haggarty SJ, Hooker JM. ACS Chem Neurosci. 2012;3:120–128. doi: 10.1021/cn200104y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, Miller CJ, Ogilvie DJ, Somervaille TC. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Niebel D, Kirfel J, Janzen V, Holler T, Majores M, Gutgemann I. Blood. 2014;124:151–152. doi: 10.1182/blood-2014-04-569525. [DOI] [PubMed] [Google Scholar]
- 46.Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, Hamamoto R. Int J Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 47.Schildhaus HU, Riegel R, Hartmann W, Steiner S, Wardelmann E, Merkelbach-Bruse S, Tanaka S, Sonobe H, Schule R, Buettner R, Kirfel J. Hum Pathol. 2011;42:1667–1675. doi: 10.1016/j.humpath.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 48.Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. PLoS One. 2012;7:e35065. doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y, Wang B, Zhang K, Lei Z, Guo Y, Xiao H, Wang J, Fan L, Lan C, Wei Y, Ma Q, Lin L, Mao C, Yang X, Chen X, Li Y, Bai Y, Chen D. Biochem Biophys Res Commun. 2013;437:192–198. doi: 10.1016/j.bbrc.2013.05.123. [DOI] [PubMed] [Google Scholar]
- 50.Konovalov S, Garcia-Bassets I. J Ovarian Res. 2013;6:75. doi: 10.1186/1757-2215-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding D, Liu X, Guo SW. Fertil Steril. 2014;101:740–749. doi: 10.1016/j.fertnstert.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 52.Singh MM, Manton CA, Bhat KP, Tsai WW, Aldape K, Barton MC, Chandra J. Neuro Oncol. 2011;13:894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sareddy GR, Nair BC, Krishnan SK, Gonugunta VK, Zhang QG, Suzuki T, Miyata N, Brenner AJ, Brann DW, Vadlamudi RK. Oncotarget. 2013;4:18–28. doi: 10.18632/oncotarget.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng YC, Duan YC, Ma JL, Xu RM, Zi X, Lv WL, Wang MM, Ye XW, Zhu S, Mobley D, Zhu YY, Wang JW, Li JF, Wang ZR, Zhao W, Liu HM. J Med Chem. 2013;56:8543–8560. doi: 10.1021/jm401002r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, Beckerle MC, Sharma S, Lessnick SL. Clin Cancer Res. 2014;20:4584–4597. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Theisen ER, Gajiwala S, Bearss J, Sorna V, Sharma S, Janat-Amsbury M. BMC Cancer. 2014;14:752. doi: 10.1186/1471-2407-14-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, Metzger E, Schule R, Eggert A, Buettner R, Kirfel J. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 58.Althoff K, Beckers A, Odersky A, Mestdagh P, Koster J, Bray IM, Bryan K, Vandesompele J, Speleman F, Stallings RL, Schramm A, Eggert A, Sprussel A, Schulte JH. Int J Cancer. 2013;133:1064–1073. doi: 10.1002/ijc.28091. [DOI] [PubMed] [Google Scholar]
- 59.Yang H, Li Q, Zhao W, Yuan D, Zhao H, Zhou Y. FEBS Lett. 2014;588:192–197. doi: 10.1016/j.febslet.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 60.Xu G, Xiao Y, Hu J, Xing L, Zhao O, Wu Y. Cell Physiol Biochem. 2013;31:854–862. doi: 10.1159/000350103. [DOI] [PubMed] [Google Scholar]
- 61.Pajtler KW, Weingarten C, Thor T, Kunkele A, Heukamp LC, Buttner R, Suzuki T, Miyata N, Grotzer M, Rieb A, Sprussel A, Eggert A, Schramm A, Schulte JH. Acta Neuropathol Commun. 2013;1:19. doi: 10.1186/2051-5960-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Z, Li S, Song W, Li X, Li Q, Zhang Z, Han Y, Zhang X, Miao S, Du R, Wang L. PLoS One. 2013;8:e70077. doi: 10.1371/journal.pone.0070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jie D, Zhongmin Z, Guoqing L, Sheng L, Yi Z, Jing W, Liang Z. Dig Dis Sci. 2013;58:1581–1589. doi: 10.1007/s10620-012-2552-2. [DOI] [PubMed] [Google Scholar]
- 64.Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, Zhang Y, Yan ZS. Br J Cancer. 2013;109:994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin L, Hanigan CL, Wu Y, Wang W, Park BH, Woster PM, Casero RA. Biochem J. 2013;449:459–468. doi: 10.1042/BJ20121360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murray-Stewart T, Woster PM, Casero RA., Jr Amino Acids. 2014;46:585–594. doi: 10.1007/s00726-013-1485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fiskus W, Sharma S, Shah B, Portier BP, Devaraj SG, Liu K, Iyer SP, Bearss D, Bhalla KN. Leukemia. 2014;28:2155–2164. doi: 10.1038/leu.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, Casero RA, Jr, Marton L, Woster P, Minden MD, Dugas M, Wang JC, Dick JE, Muller-Tidow C, Petrie K, Zelent A. Nat Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mould DP, McGonagle AE, Wiseman DH, Williams EL, Jordan AM. Med Res Rev. 2014 doi: 10.1002/med.21334. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 70.Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 71.Serce N, Gnatzy A, Steiner S, Lorenzen H, Kirfel J, Buettner R. BMC Clin Pathol. 2012;12:13. doi: 10.1186/1472-6890-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pollock JA, Larrea MD, Jasper JS, McDonnell DP, McCafferty DG. ACS Chem Biol. 2012;7:1221–1231. doi: 10.1021/cb300108c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Derr RS, van Hoesel AQ, Benard A, Goossens-Beumer IJ, Sajet A, Dekker-Ensink NG, de Kruijf EM, Bastiaannet E, Smit VT, van de Velde CJ, Kuppen PJ. BMC Cancer. 2014;14:604. doi: 10.1186/1471-2407-14-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, Evers BM, Zhou BP. Cell Rep. 2013;5:224–236. doi: 10.1016/j.celrep.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA., Jr Amino Acids. 2012;42:887–898. doi: 10.1007/s00726-011-1004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 77.Patani N, Jiang WG, Newbold RF, Mokbel K. Anticancer Res. 2011;31:4115–4125. [PubMed] [Google Scholar]
- 78.Cortez V, Mann M, Tekmal S, Suzuki T, Miyata N, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Vadlamudi RK. Breast Cancer Res. 2012;14:404. doi: 10.1186/bcr3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Breast Cancer Res Treat. 2012;131:777–789. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y. Carcinogenesis. 2013;34:1196–1207. doi: 10.1093/carcin/bgt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, Schule R, Buettner R. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 82.Urbanucci A, Waltering KK, Suikki HE, Helenius MA, Visakorpi T. BMC Cancer. 2008;8:219. doi: 10.1186/1471-2407-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suikki HE, Kujala PM, Tammela TL, van Weerden WM, Vessella RL, Visakorpi T. Prostate. 2010;70:889–898. doi: 10.1002/pros.21123. [DOI] [PubMed] [Google Scholar]
- 84.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 85.Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, Kirfel J, Waldmann H, Schule R, Buettner R. Int J Cancer. 2012;131:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- 86.Rotili D, Tomassi S, Conte M, Benedetti R, Tortorici M, Ciossani G, Valente S, Marrocco B, Labella D, Novellino E, Mattevi A, Altucci L, Tumber A, Yapp C, King ON, Hopkinson RJ, Kawamura A, Schofield CJ, Mai A. J Med Chem. 2014;57:42–55. doi: 10.1021/jm4012802. [DOI] [PubMed] [Google Scholar]
- 87.Kashyap V, Ahmad S, Nilsson EM, Helczynski L, Kenna S, Persson JL, Gudas LJ, Mongan NP. Mol Oncol. 2013;7:555–566. doi: 10.1016/j.molonc.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crea F, Sun L, Mai A, Chiang YT, Farrar WL, Danesi R, Helgason CD. Mol Cancer. 2012;11:52. doi: 10.1186/1476-4598-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Craene B, Berx G. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 90.Lin T, Ponn A, Hu X, Law BK, Lu J. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peinado H, Olmeda D, Cano A. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 92.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Nat Struct Mol Biol. 2011;18:867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang M, Shen H, Jin Y, Lin T, Cai Q, Pinard MA, Biswas S, Tran Q, Li G, Shenoy AK, Tongdee E, Lin S, Gu Y, Law BK, Zhou L, McKenna R, Wu L, Lu J. J Biol Chem. 2013;288:27680–27691. doi: 10.1074/jbc.M113.482349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Proc Natl Acad Sci USA. 2012;109:16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, Mostoslavsky R, Gygi SP, Gill G, Dyson NJ, Naar AM. Mol Cell. 2011;42:689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. Mol Cell. 2012;48:445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, Skibinski A, Shirley SH, Perou CM, Gill G, Gupta PB, Kuperwasser C. Stem Cell Reports. 2014;2:633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou G, Du T, Roizman B. Viruses. 2013;5:1208–1218. doi: 10.3390/v5051208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Nat Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. MBio. 2013;4:e00558–e00512. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. Sci Transl Med. 2014;6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sakane N, Kwon HS, Pagans S, Kaehlcke K, Mizusawa Y, Kamada M, Lassen KG, Chan J, Greene WC, Schnoelzer M, Ott M. PLoS Pathog. 2011;7:e1002184. doi: 10.1371/journal.ppat.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Le Douce V, Colin L, Redel L, Cherrier T, Herbein G, Aunis D, Rohr O, Van Lint C, Schwartz C. Nucleic Acids Res. 2012;40:1904–1915. doi: 10.1093/nar/gkr857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 107.Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD. Epigenetics. 2012;7:914–929. doi: 10.4161/epi.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Nucleic Acids Res. 2011;39:1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bourguignon LY, Wong G, Earle C, Chen L. J Biol Chem. 2012;287:32800–32824. doi: 10.1074/jbc.M111.308528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Essen D, Zhu Y, Saccani S. Mol Cell. 2010;39:750–760. doi: 10.1016/j.molcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 111.Stavropoulos P, Blobel G, Hoelz A. Nat Struct Mol Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 112.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Proc Natl Acad Sci USA. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kubicek S, Jenuwein T. Cell. 2004;119:903–906. doi: 10.1016/j.cell.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 114.Jin Y, Kim TY, Kim MS, Kim MA, Park SH, Jang YK. J Biochem. 2014;156:305–313. doi: 10.1093/jb/mvu042. [DOI] [PubMed] [Google Scholar]
- 115.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Mol Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 116.Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H. Nat Struct Mol Biol. 2007;14:535–539. doi: 10.1038/nsmb1255. [DOI] [PubMed] [Google Scholar]
- 117.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. J Biol Chem. 2007;282:20070–20074. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 118.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 119.Mimasu S, Sengoku T, Fukuzawa S, Umehara T, Yokoyama S. Biochem Biophys Res Commun. 2008;366:15–22. doi: 10.1016/j.bbrc.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 120.Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. J Neurosci. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, Minucci S, Mattevi A, Mai A. J Am Chem Soc. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 122.Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Biochemistry. 2010;49:6494–6503. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- 123.Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A. Structure. 2011;19:212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tortorici M, Borrello MT, Tardugno M, Chiarelli LR, Pilotto S, Ciossani G, Vellore NA, Bailey SG, Cowan J, O’Connell M, Crabb SJ, Packham G, Mai A, Baron R, Ganesan A, Mattevi A. ACS Chem Biol. 2013;8:1677–1682. doi: 10.1021/cb4001926. [DOI] [PubMed] [Google Scholar]
- 125.Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, Mattevi A, Battaglioli E. J Neurochem. 2014;128:603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- 126.Luka Z, Pakhomova S, Loukachevitch LV, Calcutt MW, Newcomer ME, Wagner C. Protein Sci. 2014;23:993–998. doi: 10.1002/pro.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schmidt DM, McCafferty DG. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 128.De Colibus L, Mattevi A. Curr Opin Struct Biol. 2006;16:722–728. doi: 10.1016/j.sbi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 129.Fraaije MW, Mattevi A. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 130.Hwang S, Schmitt AA, Luteran AE, Toone EJ, McCafferty DG. Biochemistry. 2011;50:546–557. doi: 10.1021/bi101776t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Nat Struct Mol Biol. 2005;12:1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- 132.Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, Ikari M, Sato M, Terada T, Nagase T, Ohara O, Shirouzu M, Tanaka A, Kigawa T, Yokoyama S. Structure. 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 133.Luka Z, Moss F, Loukachevitch LV, Bornhop DJ, Wagner C. Biochemistry. 2011;50:4750–4756. doi: 10.1021/bi200247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Q, Qi S, Xu M, Yu L, Tao Y, Deng Z, Wu W, Li J, Chen Z, Wong J. Cell Res. 2013;23:225–241. doi: 10.1038/cr.2012.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen F, Yang H, Dong Z, Fang J, Wang P, Zhu T, Gong W, Fang R, Shi YG, Li Z, Xu Y. Cell Res. 2013;23:306–309. doi: 10.1038/cr.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fang R, Chen F, Dong Z, Hu D, Barbera AJ, Clark EA, Fang J, Yang Y, Mei P, Rutenberg M, Li Z, Zhang Y, Xu Y, Yang H, Wang P, Simon MD, Zhou Q, Li J, Marynick MP, Li X, Lu H, Kaiser UB, Kingston RE, Xu Y, Shi YG. Mol Cell. 2013;49:558–570. doi: 10.1016/j.molcel.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Holm L, Rosenstrom P. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mandelker D, Gabelli SB, Schmidt-Kittler O, Zhu J, Cheong I, Huang CH, Kinzler KW, Vogelstein B, Amzel LM. Proc Natl Acad Sci USA. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 140.Barta ML, Dickenson NE, Patil M, Keightley A, Wyckoff GJ, Picking WD, Picking WL, Geisbrecht BV. J Mol Biol. 2012;417:395–405. doi: 10.1016/j.jmb.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Culhane JC, Cole PA. Curr Opin Chem Biol. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hojfeldt JW, Agger K, Helin K. Nat Rev Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 143.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 144.Gaweska HM. University of Pennsylvania; 2009. [Google Scholar]
- 145.Gaweska H, Henderson Pozzi M, Schmidt DM, McCafferty DG, Fitzpatrick PF. Biochemistry. 2009;48:5440–5445. doi: 10.1021/bi900499w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cook PF, Cleland WW. Biochemistry. 1981;20:1797–1805. doi: 10.1021/bi00510a014. [DOI] [PubMed] [Google Scholar]
- 147.Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. J Biol Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- 148.Karasulu B, Patil M, Thiel W. J Am Chem Soc. 2013;135:13400–13413. doi: 10.1021/ja403582u. [DOI] [PubMed] [Google Scholar]
- 149.Silverman RB. Accounts of Chemical Research. 1995;28:335–342. [Google Scholar]
- 150.McCann AE, Sampson NS. Journal of the American Chemical Society. 2000;122:35–39. [Google Scholar]
- 151.Chen ZW, Zhao G, Martinovic S, Jorns MS, Mathews FS. Biochemistry. 2005;44:15444–15450. doi: 10.1021/bi0515422. [DOI] [PubMed] [Google Scholar]
- 152.Forneris F, Binda C, Dall’Aglio A, Fraaije MW, Battaglioli E, Mattevi A. J Biol Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 153.Mattevi A. Trends Biochem Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 154.Tsai WW, Nguyen TT, Shi Y, Barton MC. Mol Cell Biol. 2008;28:5139–5146. doi: 10.1128/MCB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 156.Yu T, Higashi M, Cembran A, Gao J, Truhlar DG. J Phys Chem B. 2013;117:8422–8429. doi: 10.1021/jp404292t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kong X, Ouyang S, Liang Z, Lu J, Chen L, Shen B, Li D, Zheng M, Li KK, Luo C, Jiang H. PLoS One. 2011;6:e25444. doi: 10.1371/journal.pone.0025444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Henderson Pozzi M, Gawandi V, Fitzpatrick PF. Biochemistry. 2009;48:1508–1516. doi: 10.1021/bi802227m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Polticelli F, Basran J, Faso C, Cona A, Minervini G, Angelini R, Federico R, Scrutton NS, Tavladoraki P. Biochemistry. 2005;44:16108–16120. doi: 10.1021/bi050983i. [DOI] [PubMed] [Google Scholar]
- 160.Massey V, Curti B. J Biol Chem. 1967;242:1259–1264. [PubMed] [Google Scholar]
- 161.Massey V, Gibson QH. Fed Proc. 1964;23:18–29. [PubMed] [Google Scholar]
- 162.Walker MC, Edmondson DE. Biochemistry. 1994;33:7088–7098. doi: 10.1021/bi00189a011. [DOI] [PubMed] [Google Scholar]
- 163.Emanuele JJ, Fitzpatrick PF. Biochemistry. 1995;34:3710–3715. doi: 10.1021/bi00011a028. [DOI] [PubMed] [Google Scholar]
- 164.Basran J, Bhanji N, Basran A, Nietlispach D, Mistry S, Meskys R, Scrutton NS. Biochemistry. 2002;41:4733–4743. doi: 10.1021/bi025519h. [DOI] [PubMed] [Google Scholar]
- 165.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 166.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R. Nature. 2010;464:792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 167.Forneris F, Binda C, Battaglioli E, Mattevi A. Trends Biochem Sci. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 168.Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 169.Kontaki H, Talianidis I. Mol Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 170.Cho HS, Suzuki T, Dohmae N, Hayami S, Unoki M, Yoshimatsu M, Toyokawa G, Takawa M, Chen T, Kurash JK, Field HI, Ponder BA, Nakamura Y, Hamamoto R. Cancer Res. 2011;71:655–660. doi: 10.1158/0008-5472.CAN-10-2446. [DOI] [PubMed] [Google Scholar]
- 171.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, An L, Wang Q, Lu T, Zhang X, Wang Z, Stark GR. Proc Natl Acad Sci USA. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Abu-Farha M, Lanouette S, Elisma F, Tremblay V, Butson J, Figeys D, Couture JF. J Mol Cell Biol. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- 173.Nair SS, Li DQ, Kumar R. Mol Cell. 2013;49:704–718. doi: 10.1016/j.molcel.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang X, Tanaka K, Yan J, Li J, Peng D, Jiang Y, Yang Z, Barton MC, Wen H, Shi X. Proc Natl Acad Sci USA. 2013;110:17284–17289. doi: 10.1073/pnas.1307959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Iwabuchi K, Li B, Massa HF, Trask BJ, Date T, Fields S. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 176.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 178.Nicholson TB, Chen T. Epigenetics. 2009;4:129–132. doi: 10.4161/epi.4.3.8443. [DOI] [PubMed] [Google Scholar]
- 179.Hauber J, Malim MH, Cullen BR. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pagans S, Kauder SE, Kaehlcke K, Sakane N, Schroeder S, Dormeyer W, Trievel RC, Verdin E, Schnolzer M, Ott M. Cell Host Microbe. 2010;7:234–244. doi: 10.1016/j.chom.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E. Curr Biol. 1999;9:1489–1492. doi: 10.1016/s0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- 182.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dorr A, Kiermer V, Pedal A, Rackwitz HR, Henklein P, Schubert U, Zhou MM, Verdin E, Ott M. EMBO J. 2002;21:2715–2723. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM. Mol Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- 185.Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M. Mol Cell. 2003;12:167–176. doi: 10.1016/s1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- 186.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 187.Manavathi B, Kumar R. J Biol Chem. 2007;282:1529–1533. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- 188.Li DQ, Pakala SB, Reddy SD, Ohshiro K, Peng SH, Lian Y, Fu SW, Kumar R. J Biol Chem. 2010;285:10044–10052. doi: 10.1074/jbc.M109.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pratt WB, Toft DO. Exp Biol Med (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 190.Donlin LT, Andresen C, Just S, Rudensky E, Pappas CT, Kruger M, Jacobs EY, Unger A, Zieseniss A, Dobenecker MW, Voelkel T, Chait BT, Gregorio CC, Rottbauer W, Tarakhovsky A, Linke WA. Genes Dev. 2012;26:114–119. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Voelkel T, Andresen C, Unger A, Just S, Rottbauer W, Linke WA. Biochim Biophys Acta. 2013;1833:812–822. doi: 10.1016/j.bbamcr.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 192.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Trepel J, Mollapour M, Giaccone G, Neckers L. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 195.Itoh Y, Suzuki T, Miyata N. Mol Biosyst. 2013;9:873–896. doi: 10.1039/c3mb25410k. [DOI] [PubMed] [Google Scholar]
- 196.Khan MN, Suzuki T, Miyata N. Med Res Rev. 2013;33:873–910. doi: 10.1002/med.21269. [DOI] [PubMed] [Google Scholar]
- 197.Lohse B, Kristensen JL, Kristensen LH, Agger K, Helin K, Gajhede M, Clausen RP. Bioorg Med Chem. 2011;19:3625–3636. doi: 10.1016/j.bmc.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 198.Helin K, Dhanak D. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 199.Stavropoulos P, Hoelz A. Expert Opin Ther Targets. 2007;11:809–820. doi: 10.1517/14728222.11.6.809. [DOI] [PubMed] [Google Scholar]
- 200.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 201.Culhane JC, Wang D, Yen PM, Cole PA. J Am Chem Soc. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gooden DM, Schmidt DM, Pollock JA, Kabadi AM, McCafferty DG. Bioorg Med Chem Lett. 2008;18:3047–3051. doi: 10.1016/j.bmcl.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. J Am Chem Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 204.Vianello P, Botrugno OA, Cappa A, Ciossani G, Dessanti P, Mai A, Mattevi A, Meroni G, Minucci S, Thaler F, Tortorici M, Trifiro P, Valente S, Villa M, Varasi M, Mercurio C. Eur J Med Chem. 2014;86:352–363. doi: 10.1016/j.ejmech.2014.08.068. [DOI] [PubMed] [Google Scholar]
- 205.Benelkebir H, Hodgkinson C, Duriez PJ, Hayden AL, Bulleid RA, Crabb SJ, Packham G, Ganesan A. Bioorg Med Chem. 2011;19:3709–3716. doi: 10.1016/j.bmc.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 206.Johnson NW, Kasparec J, Rouse MB, Tian X, Suarez DP, McNulty KC, Blackledge CW, Van Aller GS, Schneck J, Carson JD, Kruger RG, Mohammad HP, Crouthamel MH, Smitheman KN, Liu Y, Gorman S, McHugh CF, Bonnette W, Concha NO, Patel M, Tummino PJ, Miller WH. 246th ACS National Meeting and Exposition: Indiana Convention Center; 2013. [Google Scholar]
- 207.Kumarasinghe IR, Woster PM. ACS Med Chem Lett. 2014;5:29–33. doi: 10.1021/ml4002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. J Am Chem Soc. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- 209.Szewczuk LM, Culhane JC, Yang M, Majumdar A, Yu H, Cole PA. Biochemistry. 2007;46:6892–6902. doi: 10.1021/bi700414b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gongora-Benitez M, Tulla-Puche J, Albericio F. Chem Rev. 2014;114:901–926. doi: 10.1021/cr400031z. [DOI] [PubMed] [Google Scholar]
- 211.Schmitt ML, Hauser AT, Carlino L, Pippel M, Schulz-Fincke J, Metzger E, Willmann D, Yiu T, Barton M, Schule R, Sippl W, Jung M. J Med Chem. 2013;56:7334–7342. doi: 10.1021/jm400792m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ogasawara D, Itoh Y, Tsumoto H, Kakizawa T, Mino K, Fukuhara K, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N, Suzuki T. Angew Chem Int Ed Engl. 2013;52:8620–8624. doi: 10.1002/anie.201303999. [DOI] [PubMed] [Google Scholar]
- 213.Itoh Y, Ogasawara D, Ota Y, Mizukami T, Suzuki T. Comput Struct Biotechnol J. 2014;9:e201402002. doi: 10.5936/csbj.201402002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Clin Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Proc Natl Acad Sci USA. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bi X, Lopez C, Bacchi CJ, Rattendi D, Woster PM. Bioorg Med Chem Lett. 2006;16:3229–3232. doi: 10.1016/j.bmcl.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 217.Sharma SK, Wu Y, Steinbergs N, Crowley ML, Hanson AS, Casero RA, Woster PM. J Med Chem. 2010;53:5197–5212. doi: 10.1021/jm100217a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sharma SK, Hazeldine S, Crowley ML, Hanson A, Beattie R, Varghese S, Senanayake TM, Hirata A, Hirata F, Huang Y, Wu Y, Steinbergs N, Murray-Stewart T, Bytheway I, Casero RA, Jr, Woster PM. Medchemcomm. 2012;3:14–21. doi: 10.1039/C1MD00220A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pachaiyappan B, Woster PM. Bioorg Med Chem Lett. 2014;24:21–32. doi: 10.1016/j.bmcl.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Huang Y, Marton LJ, Woster PM, Casero RA. Essays Biochem. 2009;46:95–110. doi: 10.1042/bse0460007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang Z, Patel DJ. Q Rev Biophys. 2013;46:349–373. doi: 10.1017/S0033583513000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nowotarski SL, Woster PM, Casero RA., Jr Expert Rev Mol Med. 2013;15:e3. doi: 10.1017/erm.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dulla B, Kirla KT, Rathore V, Deora GS, Kavela S, Maddika S, Chatti K, Reiser O, Iqbal J, Pal M. Org Biomol Chem. 2013;11:3103–3107. doi: 10.1039/c3ob40217g. [DOI] [PubMed] [Google Scholar]
- 224.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 225.Kolb HC, Sharpless KB. Drug Discov Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 226.Zificsak CA, Hlasta DJ. Tetrahedron. 2004;60:8991–9016. [Google Scholar]
- 227.Davies JR, Kane PD, Moody CJ. Tetrahedron. 2004;60:3967–3977. [Google Scholar]
- 228.Jia Z, Zhu Q. Bioorg Med Chem Lett. 2010;20:6222–6225. doi: 10.1016/j.bmcl.2010.08.104. [DOI] [PubMed] [Google Scholar]
- 229.Reck F, Zhou F, Girardot M, Kern G, Eyermann CJ, Hales NJ, Ramsay RR, Gravestock MB. J Med Chem. 2005;48:499–506. doi: 10.1021/jm0400810. [DOI] [PubMed] [Google Scholar]
- 230.Len C, BoulogneMerlot AS, Postel D, Ronco G, Villa P, Goubert C, Jeufrault E, Mathon B, Simon H. Journal of Agricultural and Food Chemistry. 1996;44:2856–2858. [Google Scholar]
- 231.Imamura H, Ohtake N, Jona H, Shimizu A, Moriya M, Sato H, Sugimoto Y, Ikeura C, Kiyonaga H, Nakano M, Nagano R, Abe S, Yamada K, Hashizume T, Morishima H. Bioorg Med Chem. 2001;9:1571–1578. doi: 10.1016/s0968-0896(01)00044-x. [DOI] [PubMed] [Google Scholar]
- 232.Ronconi L, Marzano C, Zanello P, Corsini M, Miolo G, Macca C, Trevisan A, Fregona D. J Med Chem. 2006;49:1648–1657. doi: 10.1021/jm0509288. [DOI] [PubMed] [Google Scholar]
- 233.Abdulla A, Zhao X, Yang F. J Biochem Pharmacol Res. 2013;1:56–63. [PMC free article] [PubMed] [Google Scholar]
- 234.Yu W, Fu YC, Wang W. J Cell Biochem. 2012;113:752–759. doi: 10.1002/jcb.23431. [DOI] [PubMed] [Google Scholar]
- 235.Zhou H, Beevers CS, Huang S. Curr Drug Targets. 2011;12:332–347. doi: 10.2174/138945011794815356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Boots AW, Haenen GR, Bast A. Eur J Pharmacol. 2008;585:325–337. doi: 10.1016/j.ejphar.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 237.Seidler J, McGovern SL, Doman TN, Shoichet BK. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 238.Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, Sharma S. J Med Chem. 2013;56:9496–9508. doi: 10.1021/jm400870h. [DOI] [PubMed] [Google Scholar]
- 239.Zhou C, Kang D, Xu Y, Zhang L, Zha X. Chem Biol Drug Des. 2014 doi: 10.1111/cbdd.12461. [DOI] [PubMed] [Google Scholar]
- 240.Prusevich P, Kalin JH, Ming SA, Basso M, Givens J, Li X, Hu J, Taylor MS, Cieniewicz AM, Hsiao PY, Huang R, Roberson H, Adejola N, Avery LB, Casero RA, Jr, Taverna SD, Qian J, Tackett AJ, Ratan RR, McDonald OG, Feinberg AP, Cole PA. ACS Chem Biol. 2014;9:1284–1293. doi: 10.1021/cb500018s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. Proc Natl Acad Sci USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lee MG, Wynder C, Cooch N, Shiekhattar R. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 243.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 244.Millard CJ, Watson PJ, Celardo I, Gordiyenko Y, Cowley SM, Robinson CV, Fairall L, Schwabe JW. Mol Cell. 2013;51:57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA. EMBO J. 2003;22:3403–3410. doi: 10.1093/emboj/cdg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hwang S. Duke University; 2012. [Google Scholar]
- 247.Baron R, Vellore NA. Proc Natl Acad Sci USA. 2012;109:12509–12514. doi: 10.1073/pnas.1207892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Baron R, Vellore NA. Biochemistry. 2012;51:3151–3153. doi: 10.1021/bi300068r. [DOI] [PubMed] [Google Scholar]
- 249.Vellore NA, Baron R. BMC Biophys. 2013;6:15. doi: 10.1186/2046-1682-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Robertson JC, Hurley NC, Tortorici M, Ciossani G, Borrello MT, Vellore NA, Ganesan A, Mattevi A, Baron R. PLoS Comput Biol. 2013;9:e1003158. doi: 10.1371/journal.pcbi.1003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Barrios AP, Gomez AV, Saez JE, Ciossani G, Toffolo E, Battaglioli E, Mattevi A, Andres ME. Mol Cell Biol. 2014;34:2760–2770. doi: 10.1128/MCB.00083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Upadhyay G, Chowdhury AH, Vaidyanathan B, Kim D, Saleque S. Proc Natl Acad Sci USA. 2014;111:8071–8076. doi: 10.1073/pnas.1404292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ballas N, Mandel G. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 254.Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T. Journal of Biological Chemistry. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 255.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 257.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 258.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 259.Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, Shi YJ, Barretina J, Liu J, Howley PM, Elledge SJ, Shi Y. Mol Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kim BJ, Kang KM, Jung SY, Choi HK, Seo JH, Chae JH, Cho EJ, Youn HD, Qin J, Kim ST. Biochem Biophys Res Commun. 2008;372:298–304. doi: 10.1016/j.bbrc.2008.05.056. [DOI] [PubMed] [Google Scholar]
- 261.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kim HG, Kim HT, Leach NT, Lan F, Ullmann R, Silahtaroglu A, Kurth I, Nowka A, Seong IS, Shen Y, Talkowski ME, Ruderfer D, Lee JH, Glotzbach C, Ha K, Kjaergaard S, Levin AV, Romeike BF, Kleefstra T, Bartsch O, Elsea SH, Jabs EW, MacDonald ME, Harris DJ, Quade BJ, Ropers HH, Shaffer LG, Kutsche K, Layman LC, Tommerup N, Kalscheuer VM, Shi Y, Morton CC, Kim CH, Gusella JF. Am J Hum Genet. 2012;91:56–72. doi: 10.1016/j.ajhg.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Iwase S, Januma A, Miyamoto K, Shono N, Honda A, Yanagisawa J, Baba T. Biochem Biophys Res Commun. 2004;322:601–608. doi: 10.1016/j.bbrc.2004.07.163. [DOI] [PubMed] [Google Scholar]
- 264.Klajn A, Ferrai C, Stucchi L, Prada I, Podini P, Baba T, Rocchi M, Meldolesi J, D’Alessandro R. J Neurosci. 2009;29:6296–6307. doi: 10.1523/JNEUROSCI.5943-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 266.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 267.Ceballos-Chavez M, Rivero S, Garcia-Gutierrez P, Rodriguez-Paredes M, Garcia-Dominguez M, Bhattacharya S, Reyes JC. Proc Natl Acad Sci USA. 2012;109:8085–8090. doi: 10.1073/pnas.1121522109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, Shi Y, Shi Y, Gill G. Proc Natl Acad Sci USA. 2006;103:5308–5313. doi: 10.1073/pnas.0601066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wynder C, Hakimi MA, Epstein JA, Shilatifard A, Shiekhattar R. Nat Cell Biol. 2005;7:1113–1117. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- 270.Islam MM, Zhang CL. Biochim Biophys Acta. 2015;1849:210–216. doi: 10.1016/j.bbagrm.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yokoyama A, Takezawa S, Schule R, Kitagawa H, Kato S. Mol Cell Biol. 2008;28:3995–4003. doi: 10.1128/MCB.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, Ma X, Shi Y. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Hu X, Ybarra R, Qiu Y, Bungert J, Huang S. Epigenetics. 2009;4:357–361. doi: 10.4161/epi.4.6.9711. [DOI] [PubMed] [Google Scholar]
- 274.Huang SM, Qiu Y, Stein RW, Brandt SJ. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- 275.Huang S, Qiu Y, Shi Y, Xu Z, Brandt SJ. EMBO J. 2000;19:6792–6803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Huang S, Brandt SJ. Mol Cell Biol. 2000;20:2248–2259. doi: 10.1128/mcb.20.6.2248-2259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 278.Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ. Biochem J. 2006;399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Schuh AH, Tipping AJ, Clark AJ, Hamlett I, Guyot B, Iborra FJ, Rodriguez P, Strouboulis J, Enver T, Vyas P, Porcher C. Mol Cell Biol. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Cai Y, Xu Z, Xie J, Ham AJ, Koury MJ, Hiebert SW, Brandt SJ. Biochem Biophys Res Commun. 2009;390:295–301. doi: 10.1016/j.bbrc.2009.09.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hu X, Li X, Valverde K, Fu X, Noguchi C, Qiu Y, Huang S. Proc Natl Acad Sci USA. 2009;106:10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Li Y, Deng C, Hu X, Patel B, Fu X, Qiu Y, Brand M, Zhao K, Huang S. Oncogene. 2012;31:5007–5018. doi: 10.1038/onc.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chinnadurai G. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 284.Chinnadurai G. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 286.Banck MS, Li S, Nishio H, Wang C, Beutler AS, Walsh MJ. Epigenetics. 2009;4:100–106. doi: 10.4161/epi.4.2.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Ray SK, Li HJ, Metzger E, Schule R, Leiter AB. Mol Cell Biol. 2014;34:2308–2317. doi: 10.1128/MCB.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Cowger JJ, Zhao Q, Isovic M, Torchia J. Oncogene. 2007;26:3378–3386. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 289.Gocke CB, Yu H. PLoS One. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, Bolognesi M, Crossley M. Mol Cell Biol. 2006;26:8159–8172. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Quinlan KG, Verger A, Kwok A, Lee SH, Perdomo J, Nardini M, Bolognesi M, Crossley M. Mol Cell Biol. 2006;26:8202–8213. doi: 10.1128/MCB.00445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, Ryerse J, Chinnadurai G. Mol Cell Biol. 2008;28:269–281. doi: 10.1128/MCB.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang CL, McKinsey TA, Lu JR, Olson EN. J Biol Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 294.Castet A, Boulahtouf A, Versini G, Bonnet S, Augereau P, Vignon F, Khochbin S, Jalaguier S, Cavailles V. Nucleic Acids Res. 2004;32:1957–1966. doi: 10.1093/nar/gkh524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 296.Ueda J, Tachibana M, Ikura T, Shinkai Y. J Biol Chem. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 297.Caron C, Pivot-Pajot C, van Grunsven LA, Col E, Lestrat C, Rousseaux S, Khochbin S. EMBO Rep. 2003;4:877–882. doi: 10.1038/sj.embor.embor917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Basta J, Rauchman M. Transl Res. 2015;165:36–47. doi: 10.1016/j.trsl.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Allen HF, Wade PA, Kutateladze TG. Cell Mol Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Ouyang H, Qin Y, Liu Y, Xie Y, Liu J. PLoS One. 2013;8:e62192. doi: 10.1371/journal.pone.0062192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, Lessnick SL. Oncogene. 2013;32:5089–5100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D, Han X, Yao Z, Shang Y. Cancer Res. 2011;71:6899–6908. doi: 10.1158/0008-5472.CAN-11-1523. [DOI] [PubMed] [Google Scholar]
- 304.Gu H, Liang Y, Mandel G, Roizman B. Proc Natl Acad Sci USA. 2005;102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Gu H, Roizman B. J Virol. 2009;83:4376–4385. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Pinnoji RC, Bedadala GR, George B, Holland TC, Hill JM, Hsia SCV. Virology Journal. 2007;4:56–66. doi: 10.1186/1743-422X-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Du T, Zhou G, Khan S, Gu H, Roizman B. Proc Natl Acad Sci USA. 2010;107:15904–15909. doi: 10.1073/pnas.1010741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Zhou G, Du T, Roizman B. Proc Natl Acad Sci USA. 2013;110:E498–E506. doi: 10.1073/pnas.1222497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Wysocka J, Herr W. Trends Biochem Sci. 2003;28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 310.Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. Sci Transl Med. 2013;5:167ra165. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rai G, Kawamura A, Tumber A, Liang Y, Vogel JL, Arbuckle JH, Rose NR, Dexheimer TS, Foley TL, King ON, Quinn A, Mott BT, Schofield CJ, Oppermann U, Jadhav A, Simeonov A, Kristie TM, Maloney DJ. Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD: National Center for Biotechnology Information; 2012. [PubMed] [Google Scholar]
- 312.Kalamvoki M, Roizman B. Proc Natl Acad Sci USA. 2010;107:17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bag P, Ojha D, Mukherjee H, Halder UC, Mondal S, Biswas A, Sharon A, Van Kaer L, Chakrabarty S, Das G, Mitra D, Chattopadhyay D. Antiviral Res. 2014;105:126–134. doi: 10.1016/j.antiviral.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 314.Gu H, Roizman B. Proc Natl Acad Sci USA. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Everett RD. J Virol. 2010;84:3695–3698. doi: 10.1128/JVI.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Zhou Y, Bolton EC, Jones JO. J Mol Endocrinol. 2015;54:R15–R29. doi: 10.1530/JME-14-0203. [DOI] [PubMed] [Google Scholar]
- 317.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Chen S, Nelson PS, Liu XS, Brown M, Balk SP. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Thomas C, Gustafsson JA. Nat Rev Cancer. 2011;11:597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 319.Hervouet E, Cartron PF, Jouvenot M, Delage-Mourroux R. Epigenetics. 2013;8:237–245. doi: 10.4161/epi.23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, Brann DW, Tekmal RR, Vadlamudi RK. EMBO Rep. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kim J, Park UH, Moon M, Um SJ, Kim EJ. FEBS Lett. 2013;587:17–22. doi: 10.1016/j.febslet.2012.10.054. [DOI] [PubMed] [Google Scholar]
- 323.Ombra MN, Di Santi A, Abbondanza C, Migliaccio A, Avvedimento EV, Perillo B. Biochim Biophys Acta. 2013;1829:480–486. doi: 10.1016/j.bbagrm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 324.Chiang C, Ayyanathan K. Cytokine Growth Factor Rev. 2013;24:123–131. doi: 10.1016/j.cytogfr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S, Manzotti G, Grisendi G, Dominici M, Pieraccioli M, Raschella G, Chiodoni C, Colombo MP, Calabretta B. Cancer Res. 2013;73:235–245. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Lin Y, Dong C, Zhou BP. Curr Pharm Des. 2014;20:1698–1705. doi: 10.2174/13816128113199990512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Lin Y, Kang T, Zhou BP. Cell Cycle. 2014;13:1708–1716. doi: 10.4161/cc.28619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lin S, Shen H, Li JL, Tang S, Gu Y, Chen Z, Hu C, Rice JC, Lu J, Wu L. J Biol Chem. 2013;288:6238–6247. doi: 10.1074/jbc.M112.429605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zhang J, Bonasio R, Strino F, Kluger Y, Holloway JK, Modzelewski AJ, Cohen PE, Reinberg D. Genes Dev. 2013;27:749–766. doi: 10.1101/gad.210963.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.van der Meer LT, Jansen JH, van der Reijden BA. Leukemia. 2010;24:1834–1843. doi: 10.1038/leu.2010.195. [DOI] [PubMed] [Google Scholar]
- 331.Kazanjian A, Gross EA, Grimes HL. Crit Rev Oncol Hematol. 2006;59:85–97. doi: 10.1016/j.critrevonc.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Huang M, Hu Z, Chang W, Ou D, Zhou J, Zhang Y. Acta Haematol. 2010;123:1–5. doi: 10.1159/000253856. [DOI] [PubMed] [Google Scholar]
- 333.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Lowenberg B, Delwel R. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 334.Vassen L, Khandanpour C, Ebeling P, van der Reijden BA, Jansen JH, Mahlmann S, Duhrsen U, Moroy T. Int J Hematol. 2009;89:422–430. doi: 10.1007/s12185-009-0286-5. [DOI] [PubMed] [Google Scholar]
- 335.Xu W, Kee BL. Blood. 2007;109:4406–4414. doi: 10.1182/blood-2006-08-043331. [DOI] [PubMed] [Google Scholar]
- 336.Chowdhury AH, Ramroop JR, Upadhyay G, Sengupta A, Andrzejczyk A, Saleque S. PLoS One. 2013;8:e53666. doi: 10.1371/journal.pone.0053666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Laurent B, Randrianarison-Huetz V, Frisan E, Andrieu-Soler C, Soler E, Fontenay M, Dusanter-Fourt I, Dumenil D. J Cell Sci. 2012;125:993–1002. doi: 10.1242/jcs.095877. [DOI] [PubMed] [Google Scholar]
- 338.Vassen L, Fiolka K, Moroy T. EMBO J. 2006;25:2409–2419. doi: 10.1038/sj.emboj.7601124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, Reyes D, Beato M. Genes Dev. 2013;27:1179–1197. doi: 10.1101/gad.215293.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Porro A, Feuerhahn S, Lingner J. Cell Rep. 2014;6:765–776. doi: 10.1016/j.celrep.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 342.Lee JT. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 343.Moran VA, Perera RJ, Khalil AM. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Prensner JR, Chinnaiyan AM. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tsai MC, Spitale RC, Chang HY. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Spitale RC, Tsai MC, Chang HY. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Li X, Wu Z, Mei Q, Li X, Guo M, Fu X, Han W. Br J Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 351.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Li JT, Wang LF, Zhao YL, Yang T, Li W, Zhao J, Yu F, Wang L, Meng YL, Liu NN, Zhu XS, Gao CF, Jia LT, Yang AG. Breast Cancer Res. 2014;16:454. doi: 10.1186/s13058-014-0454-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 355.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, Zhang A, Ye M, Wang Q, Liu C, Wei J, Ren Y, Yang J, Zhang J, Pu P, Li M, Kang C. Oncotarget. 2015;6:537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Cai B, Song XQ, Cai JP, Zhang S. Neoplasma. 2014;61:379–391. doi: 10.4149/neo_2014_075. [DOI] [PubMed] [Google Scholar]
- 357.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 358.Beato M, Vicent GP. Transcription. 2013;4:167–171. doi: 10.4161/trns.25777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y. Breast Cancer Res Treat. 2014;146:99–108. doi: 10.1007/s10549-014-3012-9. [DOI] [PubMed] [Google Scholar]