Abstract
Large genome-wide association studies of glycemic traits have identified genetics variants that are associated with insulin resistance (IR) in the general population. It is unknown whether people with genetic enrichment for these IR variants respond differently to interventions that aim to improve insulin sensitivity. We built a genetic risk score (GRS) based on 17 established IR variants and effect sizes (weighted IR-GRS) in 2,713 participants of the Diabetes Prevention Program (DPP) with genetic consent. We tested associations between the weighted IR-GRS and insulin sensitivity index (ISI) at baseline in all participants, and with change in ISI over 1 year of follow-up in the DPP intervention (metformin and lifestyle) and control (placebo) arms. All models were adjusted for age, sex, ethnicity, and waist circumference at baseline (plus baseline ISI for 1-year ISI change models). A higher IR-GRS was associated with lower baseline ISI (β = −0.754 [SE = 0.229] log-ISI per unit, P = 0.001 in fully adjusted models). There was no differential effect of treatment for the association between the IR-GRS on the change in ISI; higher IR-GRS was associated with an attenuation in ISI improvement over 1 year (β = −0.520 [SE = 0.233], P = 0.03 in fully adjusted models; all treatment arms). Lifestyle intervention and metformin treatment improved the ISI, regardless of the genetic burden of IR variants.
Introduction
Genome-wide association studies (GWAS) have yielded the identities of almost 100 common genetic variants associated with type 2 diabetes (T2D) and glycemic traits (1). Though most of these variants are associated with β-cell dysfunction (2–4), a concerted search for genetic associations with measures of insulin resistance (IR) has identified 19 single nucleotide polymorphisms (SNPs) that have reached genome-wide levels of significance for association with fasting insulin (FI) levels, as a proxy for IR, in large population-based studies (5–7). It is unknown whether these SNPs predict changes over time in IR in the context of interventions designed to ameliorate IR.
The Diabetes Prevention Program (DPP), a randomized controlled trial of metformin and lifestyle versus placebo/control, showed large beneficial effects on IR. We constructed a genetic risk score (GRS) that was composed of known IR-associated variants (5–7), and tested whether it was associated with IR at baseline in DPP participants and with change in IR over 1 year, accounting for potential interactions between the IR-GRS and treatment.
Research Design and Methods
Description of Participants
The DPP study design and characteristics of the participants at baseline have been described in detail previously (8,9). In brief, the DPP was a U.S. multicenter trial (27 centers) that tested intensive lifestyle modification and pharmacologic intervention to prevent progression to diabetes in glucose-intolerant individuals. Enrolled participants had fasting plasma glucose levels between 95 and 125 mg/dL (between 5.3 and 6.9 mmol/L), and 2-h plasma glucose levels between 140 and 199 mg/dL (between 7.8 and 11.0 mmol/L) during a standard 75-g oral glucose tolerance test (OGTT). A total of 3,234 participants were randomized to intensive lifestyle modification (goal >7% weight loss and >150 min/week of physical activity), metformin treatment (850 mg twice daily), or placebo treatment. The primary end point of the DPP was diabetes incidence. The diagnosis of diabetes was defined according to American Diabetes Association guidelines (10) as a fasting glucose level of ≥126 mg/dL or a 2-h glucose level of ≥200 mg/dL during the OGTT, which were confirmed on a second test within 6 weeks.
Institutional review board approval was obtained at each clinical center and the coordinating center. The 2,713 participants included in this report provided written informed consent for the main study and for subsequent genetic investigations.
Measurements at Baseline and 1 Year
Demographics were collected at baseline; 95% of participants completed the 1-year follow-up. We derived glycemic regulation indices from validated equations based on glucose and insulin levels during the OGTT at baseline and the 1-year follow-up. Participants did not take metformin/placebo on the morning of the OGTT. Methods for glucose and insulin assays are described elsewhere (9). For our primary insulin sensitivity outcome, we calculated the insulin sensitivity index (ISI) as the reciprocal of HOMA-IR, determined as 22.5/[(FI × fasting glucose)/18.01] (11). We estimated the insulin response by the insulinogenic index using the formula [(insulin at 30 min) − (insulin at 0 min)]/[(glucose at 30 min) − (glucose at 0 min)] (12). The oral disposition index was calculated using the formula [insulinogenic index/FI] (13). We also calculated the change in insulin sensitivity over time (ISI at 1 year − ISI at baseline). We chose 1 year because weight loss in the intervention arms was the most pronounced at that time point, and to evaluate ISI changes with the largest sample size for longitudinal analyses.
Genotyping
We extracted DNA from peripheral blood leukocytes. Genotyping was performed on the customized Metabochip (Illumina, San Diego, CA), containing ∼200,000 SNPs chosen based on previous GWAS meta-analyses of 23 metabolic traits related to T2D, obesity, and/or cardiovascular diseases. We excluded study participants with sex inconsistency or familial relatedness. We excluded SNPs with a call rate <95% or if they failed Hardy-Weinberg equilibrium testing (P < 1.0 × 10−7) within each ethnic group. The overall genotyping success rate was >99.85%.
Selection of the Variants Associated With IR
We identified 19 variants that had been associated with FI at the accepted level of genome-wide significance (P < 5 × 10−8) in GWAS previously published by MAGIC investigators (5–7). We did not include TCF7L2, because the association of the T2D risk allele with lower FI levels is considered to be an artifact of ascertainment driven by the determination of this association in nondiabetic persons (i.e., carriers of the T2D risk allele have an associated reduction in β-cell function that must be compensated for by greater insulin sensitivity in order to remain diabetes free, as observed at baseline in DPP participants and in other studies including a nondiabetic population) (2). We built the GRS with and without FTO because its effect on diabetes-related traits occurs mainly via its effect on adiposity; the results were essentially the same, so we decided to present our main analyses using an IR-GRS, including 17 SNPs primarily discovered as representing IR based on MAGIC reports (not including FTO) (Table 1).
Table 1.
Frequencies of genetic variants associated with FI in previous MAGIC reports (European descent) and in DPP participants, overall and by ethnic groups
| Putative genes | SNP | Risk allele based on MAGIC | Effect size based on FI(log) in MAGIC* (β) | Risk allele frequency MAGIC | Frequency DPP overall | Frequency DPP white | Frequency DPP African American | Frequency DPP Hispanic | Frequency DPP Asian/Pacific Islander | Frequency DPP American Indian |
|---|---|---|---|---|---|---|---|---|---|---|
| COBLL1/GRB14 | rs7607980 | T | 0.0270 | 88 | 88 | 88 | 85 | 89 | 98 | 97 |
| IRS1 | rs2943634 | C | 0.0210 | 66 | 67 | 68 | 45 | 77 | 92 | 92 |
| PPP1R3B | rs4841132 | A | 0.0240 | 10 | 12 | 8 | 12 | 22 | 6 | 24 |
| UHRF1BP1 | rs4646949 | T | 0.0180 | 75 | 58 | 71 | 20 | 65 | 40 | 71 |
| PDGFC | rs4691380 | C | 0.0170 | 67 | 59 | 67 | 29 | 63 | 65 | 71 |
| LYPLAL1 | rs2785980 | T | 0.0150 | 67 | 70 | 69 | 86 | 56 | 74 | 44 |
| GCKR | rs780094 | C | 0.0190 | 60 | 66 | 59 | 82 | 63 | 65 | 90 |
| IGF1 | rs35767 | G | 0.0220 | 85 | 77 | 85 | 56 | 78 | 66 | 76 |
| PPARG | rs18001282 | C | 0.0220 | 87 | 91 | 90 | 98 | 91 | 90 | 80 |
| TET2 | rs9884482 | C | 0.0120 | 39 | 35 | 38 | 11 | 45 | 57 | 40 |
| HIP1 | rs1167800 | A | 0.0130 | 54 | 65 | 57 | 84 | 65 | 68 | 75 |
| RSPO3 | rs2745353 | T | 0.0110 | 51 | 55 | 50 | 64 | 56 | 59 | 66 |
| PEPD | rs731839 | G | 0.0140 | 34 | 37 | 34 | 38 | 40 | 58 | 47 |
| ARL15 | rs4865796 | A | 0.0110 | 67 | 74 | 70 | 75 | 82 | 81 | 94 |
| YSK4 | rs1530559 | A | 0.0074 | 52 | 40 | 49 | 25 | 32 | 13 | 47 |
| ANKRD55-MAP3K1 | rs459193 | G | 0.0130 | 73 | 70 | 75 | 58 | 73 | 53 | 74 |
| FAM13A | rs3822072 | A | 0.0130 | 48 | 49 | 49 | 53 | 43 | 54 | 40 |
| Total number of DPP participants | 2,713 | 1,503 | 554 | 458 | 120 | 78 | ||||
| IR-GRS mean (SD) | 20.2 (2.8) | 20.5 (2.7) | 18.4 (2.5) | 20.8 (2.6) | 20.8 (2.4) | 22.6 (2.6) |
Data are reported as %, unless otherwise indicated.
*Effect size per risk allele based on Manning et al. (6) and publicly available on MAGIC website; modes are adjusted for sex, age, and BMI.
Building the IR-GRS Score
We computed the GRS based on the assumption of an additive genetic effect and using published effect size on log-FI per risk allele (adjusted for age, sex, and BMI) based on MAGIC publicly available data (http://www.magicinvestigators.org/downloads/). Each subject was assigned an aggregate GRS based on the number of risk alleles × effect size for the respective 17 SNPs under investigation. We excluded 281 individuals with more than three missing SNPs. For participants with one, two, or three missing SNPs (total 120 individuals), we calculated their GRS by multiplying the GRS from the available SNPs by 34 and dividing by twice the number of successfully genotyped SNPs.
Statistical Analyses
We present qualitative characteristics as frequency (percentage), and continuous variables as mean ± SD, if normally distributed, or as median with interquartile range otherwise. We log transformed the ISI to achieve normal distribution. We used linear regression models to estimate the association of IR-GRS with baseline ISI and the 1-year change in ISI, after adjusting for age, sex, ethnicity, and waist circumference (we included waist circumference because it is the anthropometric measure most strongly associated with outcomes in DPP [14] and is based on our experience from previously conducted genetic analyses in DPP [15]). We further adjusted the 1-year change in ISI model for the baseline ISI, treatment group, and change in waist circumference. We used proportional hazards regression to estimate the effect of IR-GRS on the risk of the development of diabetes, after adjusting for baseline covariates. We also checked for interaction effects between treatment and GRS. Furthermore, for easier interpretation and illustration, we computed tertiles of IR-GRS and conducted the same analyses as when using GRS as a continuous variable; we presented participant baseline characteristics in each tertile, and we assessed differences between tertile groups using ANOVA for continuous variables with symmetric distributions, the nonparametric Kruskal-Wallis test for continuous variables with skewed distributions, and χ2 tests for categorical variables. We conducted sensitivity analyses in white participants only. All tests performed are two sided, and an α-level of 0.05 was used to determine statistical significance. The Statistical Analysis Software (SAS) version 9.3 was used for all analyses (SAS Institute, Inc., Cary, NC).
Results
The 2,713 DPP participants analyzed in this study had a mean (±SD) age of 50.7 ± 10.7 years; 67% were women, and 45% were nonwhite. At baseline, the mean (±SD) BMI was 34.1 ± 6.7 kg/m2 and waist circumference was 105.4 ± 14.6 cm. Based on the selected 17 known IR genetic variants and their published effect size on FI (Table 1), the mean weighted IR-GRS was 0.34 ± 0.05 in the DPP population.
The baseline characteristics of DPP participants in each tertile of the IR-GRS are shown in Table 2.
Table 2.
Characteristics of DPP participants at baseline by tertile of the IR-GRS, with each risk allele weighted effect size based on original MAGIC publication
| Tertile 1 (n = 900) | Tertile 2 (n = 908) | Tertile 3 (n = 905) | P value | |
|---|---|---|---|---|
| IR-GRS (weighted) | 0.29 ± 0.03 | 0.34 ± 0.01 | 0.39 ± 0.02 | <0.001 |
| Demographic | ||||
| Age (years) | 51.2 ± 10.7 | 50.8 ± 10.4 | 50.2 ± 11.0 | 0.16 |
| Race | <0.001 | |||
| White | 427 (47.4) | 523 (57.6) | 553 (61.1) | |
| African American | 337 (37.4) | 156 (17.2) | 61 (6.7) | |
| Hispanic | 109 (12.1) | 160 (17.6) | 189 (20.9) | |
| Asian | 23 (2.6) | 48 (5.3) | 49 (5.4) | |
| American Indian | 4 (0.4) | 21 (2.3) | 53 (5.9) | |
| Sex | 0.431 | |||
| Male | 290 (32.2) | 286 (31.5) | 310 (34.3) | |
| Female | 610 (67.8) | 622 (68.5) | 595 (65.7) | |
| Anthropometry | ||||
| Weight (kg) | 97.4 ± 20.2 | 94.0 ± 20.2 | 92.8 ± 19.7 | <0.001 |
| BMI (kg/m2) | 35.0 ± 6.9 | 34.0 ± 6.6 | 33.4 ± 6.4 | <0.001 |
| Waist (cm) | 106.9 ± 14.8 | 104.7 ± 14.5 | 104.6 ± 14.3 | <0.001 |
| Glucose | ||||
| Fasting glucose (mg/dL) | 106.9 ± 8.0 | 106.8 ± 8.2 | 106.7 ± 8.3 | 0.80 |
| log-ISI | −1.81 ± 0.54 | −1.81 ± 0.58 | −1.83 ± 0.57 | 0.58 |
| HOMA-IR | 7.03 ± 4.13 | 7.13 ± 4.23 | 7.28 ± 4.25 | 0.45 |
| Insulinogenic index | 104.48 [68.29, 158.49] | 104.49 [66.15, 154.44] | 104.92 [70.24, 161.70] | 0.76 |
| Proinsulin/insulin ratio | 0.17 [0.13, 0.24] | 0.17 [0.12, 0.23] | 0.18 [0.13, 0.24] | 0.02 |
| Oral disposition index | 4.47 [3.02, 6.53] | 4.43 [2.98, 6.52] | 4.39 [2.94, 6.55] | 0.67 |
Data are reported as the mean ± SD or median [interquartile range] for continuous variables and frequency (%) for categorical variables.
Individuals in the lowest tertile of the IR-GRS were less likely to be white and more likely to be African American than those in the highest tertile; they also had a higher BMI and waist circumference. A higher IR-GRS was associated with lower baseline insulin sensitivity (β = −0.754 log-ISI per GRS unit increase [SE = 0.229], P = 0.001) after adjustment for age, sex, ethnicity, and baseline waist circumference.
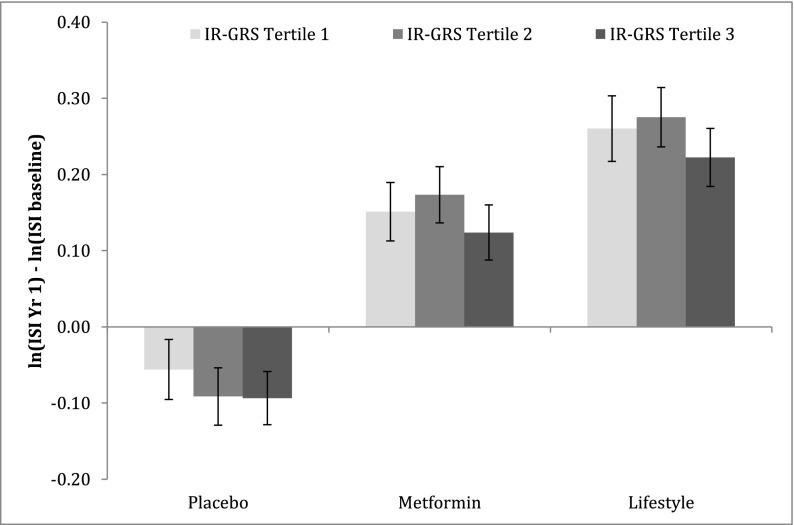
We evaluated the association between the IR-GRS and change in insulin sensitivity over the first year of the trial (Fig. 1). The interaction among treatment arms (placebo/metformin/lifestyle) and the effect of IR-GRS on the 1-year change in ISI was not significant (P = 0.98), so we analyzed all participants together, including treatment assignment as a covariate in the models. A higher IR-GRS was associated with attenuation or lack of improvement in insulin sensitivity over 1 year after adjustment for treatment arm, age, sex, ethnicity, and baseline ISI and waist (β = −0.520 change in log-ISI per GRS unit increase [SE = 0.233], P = 0.026); this association remained significant after we further adjusted for change in waist circumference over the first year (β = −0.456 change in log-ISI per GRS unit increase [SE = 0.220], P = 0.038). In subsidiary analyses of an IR-GRS, which also included the FTO risk variant at rs9939609, we found essentially the same results.
Figure 1.
Change in ISI over 1 year for DPP participants in each arm according to tertile of the weighted IR-GRS. All values are adjusted for baseline ISI, age, sex, ethnicity, and waist circumference at baseline. The y-axis represents the change in ISI (ln-transformed with SE) over the first year of the DPP. P value for interaction treatment × IR-GRS per tertile = 0.54. Yr, year.
We also evaluated the association between IR-GRS and diabetes incidence over the course of the main trial (mean follow-up time 3.2 years). We found no significant association after adjusting for treatment arms, age, sex, ethnicity, and baseline waist circumference (hazard ratio 3.52 per GRS unit increase [95% CI 0.51, 24.52], P = 0.204).
In our sensitivity analyses in white DPP participants only, the mean weighted IR-GRS was 0.35 ± 0.04. We found consistent associations with ISI in multivariable adjusted models: higher IR-GRS was associated with lower baseline insulin sensitivity (β = −1.065 log-ISI per GRS unit increase [SE = 0.294], P = 0.0003) and with attenuation or lack of improvement in insulin sensitivity over 1 year (β = −0.800 change in log-ISI per GRS unit increase [SE = 0.299], P = 0.008). We found no association of the IR-GRS with diabetes incidence.
Discussion
Lifestyle and metformin treatments during the DPP produced great improvement in insulin sensitivity, especially over the first year. This improvement in insulin sensitivity was associated with reduction of the risk of the development of diabetes overall and in each treatment arm (16). This is concordant with other diabetes prevention randomized controlled trials showing that improvement in insulin sensitivity induced by lifestyle intervention is a strong predictor of risk reduction in individuals with diabetes (17). In the current report, we showed that DPP participants carrying a higher genetic burden for IR were indeed less insulin sensitive at baseline, and less likely to improve indices of insulin sensitivity at 1 year after taking into account adiposity and demographic characteristics. More importantly, we showed that lifestyle and metformin treatment improved insulin sensitivity independent of the genetic burden of the participants. Taken together, this means that, among high-risk populations, a GRS can predict who is likely to become more insulin resistant over time, but that treatment by either metformin or lifestyle modification can significantly improve their insulin sensitivity independent of their IR genetic burden.
A higher IR-GRS was associated with less improvement in ISI independent of waist circumference change over 1 year in our study; change in weight was correlated with change in IR, and change in both weight and change in ISI were independently correlated with diabetes incidence in each treatment arm (16). We have previously shown that a GRS derived from established T2D variants predict diabetes incidence in DPP participants (15); in contrast, our IR-GRS was not associated with diabetes incidence in the current analyses. This may be because the contributions of these genetic variants to diabetes risk in the DPP are below the level that can be detected in this population; further, the majority of risk alleles at loci associated directly with FI are not associated with T2D in large population-based studies (5–7). Our results are in keeping with the critical role of the β-cell in the pathogenesis of T2D (18) and the greater predictive power of loci identified to be associated with β-cell responses (19). Indeed, in the Framingham Heart Study and the CARDIA population-based studies, a GRS based on T2D risk alleles representing IR pathways was not associated with diabetes incidence, whereas a similar score based on genes potentially affecting β-cell function was significantly associated with diabetes over >25 years of follow-up (19).
We observed a counterintuitive association of our IR-GRS with lower adiposity at baseline that is likely driven by the constraint on ascertainment induced by enrolling DPP participants within a narrow range of glycemia, as those participants with a greater degree of genetically influenced IR must be protected by other features lest diabetes develops and they are not eligible for enrollment. We also noted a difference in the ethnic composition of each tertile of IR-GRS; putative ethnic differences in the genetic architecture of insulin secretion and sensitivity merit further exploration. Our sensitivity analyses in white participants gave similar results, but smaller sample sizes in other ethnic groups limited our ability to conduct analyses in each specific ethnicity represented in the DPP.
Our study is strengthened by its standardized measurements of anthropometry and of insulin sensitivity indices at baseline and over time, and that we assessed the genetics of IR in the context of interventions shown to improve insulin sensitivity that are clinically recommended. We acknowledge that the gold standard for IR measurement is the euglycemic-hyperinsulinemic clamp and that our results might have been different if we had access to such measures. Power was limited by our sample size, especially in each treatment arm and for interaction testing (20).
Conclusion
We demonstrated that a GRS informed by prior knowledge of established genetic determinants of IR in population-based studies was associated with IR at baseline and the change in IR in DPP participants. Of high clinical importance, we showed that metformin treatment and lifestyle improve insulin sensitivity independent of the IR genetic burden estimated based on current knowledge. Other genetic markers might predict the intervention response, but these are challenging to detect with current methods and statistical approaches; thus, novel approaches are necessary to reveal genetic predictors of response to diabetes-preventive interventions to overcome the issue of limited power related to the relatively small sample size included in intervention trials.
Supplementary Material
Article Information
Acknowledgments. The Diabetes Prevention Program Research Group acknowledges the commitment and dedication of the participants of the Diabetes Prevention Program and the Diabetes Prevention Program Outcomes Study.
Funding. During the DPPOS, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding (grant DK-48489) to the clinical centers and the Coordinating Center for the design and conduct of the study, and the collection, management, analysis, and interpretation of the data. The Southwestern American Indian Centers were supported directly by the NIDDK, including its Intramural Research Program and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Funding was also provided by the National Institute of Child Health and Human Development; the National Institute on Aging; the National Eye Institute; the National Heart, Lung, and Blood Institute; the Office of Research on Women’s Health; the National Institute on Minority Health and Health Disparities; the Centers for Disease Control and Prevention; and the American Diabetes Association. Lipha (Merck-Sante) provided medication, and LifeScan, Inc. donated materials during the DPP and DPPOS.
Duality of Interest. Bristol-Myers Squibb and Parke-Davis provided additional funding and material support during the Diabetes Prevention Program (DPP). No other potential conflicts of interest relevant to this article were reported.
Author Contributions. M.-F.H. wrote the article, conceived the analytic plans, led the interpretation of results, and edited and reviewed the article before submission. C.A.C. and K.A.J. conducted statistical analyses, contributed to the interpretation of the results, and edited and reviewed the article before submission. P.W.F., D.A.E., S.E.K., E.S.H., T.I.P., K.J.M., L.P., and E.B.-C. contributed to the interpretation of the results, and edited and reviewed the article before submission. W.C.K. and J.C.F. conceived the study design, contributed to the interpretation of the results, and edited and reviewed the article before submission. J.C.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-0950/-/DC1.
Clinical trial reg. no. NCT00004992, clinicaltrials.gov.
A complete list of centers, investigators, and staff of the Diabetes Prevention Program Research Group can be found in the Supplementary Data online.
The opinions expressed in this article are those of the investigators and do not necessarily reflect the views of the funding agencies.
References
- 1.Hivert MF, Vassy JL, Meigs JB. Susceptibility to type 2 diabetes mellitus—from genes to prevention. Nat Rev Endocrinol 2014;10:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia 2008;51:1100–1110 [DOI] [PubMed] [Google Scholar]
- 3.Dimas AS, Lagou V, Barker A, et al.; MAGIC Investigators . Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes 2014;63:2158–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingelsson E, Langenberg C, Hivert MF, et al.; MAGIC investigators . Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes 2010;59:1266–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning AK, Hivert MF, Scott RA, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium . A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 2012;44:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Diabetes Prevention Program . The Diabetes Prevention Program. Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care 1999;22:623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Diabetes Prevention Program Research Group . The Diabetes Prevention Program: baseline characteristics of the randomized cohort. Diabetes Care 2000;23:1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowler WC, Barrett-Connor E, Fowler SE, et al.; Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 12.Byrne CD, Wareham NJ, Brown DC, et al. Hypertriglyceridaemia in subjects with normal and abnormal glucose tolerance: relative contributions of insulin secretion, insulin resistance and suppression of plasma non-esterified fatty acids. Diabetologia 1994;37:889–896 [DOI] [PubMed] [Google Scholar]
- 13.Utzschneider KM, Prigeon RL, Faulenbach MV, et al. Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care 2009;32:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diabetes Prevention Program Research Group . Relationship of body size and shape to the development of diabetes in the diabetes prevention program. Obesity (Silver Spring) 2006;14:2107–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hivert MF, Jablonski KA, Perreault L, et al.; DIAGRAM Consortium; Diabetes Prevention Program Research Group . Updated genetic score based on 34 confirmed type 2 diabetes Loci is associated with diabetes incidence and regression to normoglycemia in the diabetes prevention program. Diabetes 2011;60:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitabchi AE, Temprosa M, Knowler WC, et al.; Diabetes Prevention Program Research Group . Role of insulin secretion and sensitivity in the evolution of type 2 diabetes in the diabetes prevention program: effects of lifestyle intervention and metformin. Diabetes 2005;54:2404–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uusitupa M, Lindi V, Louheranta A, Salopuro T, Lindström J, Tuomilehto J; Finnish Diabetes Prevention Study Group . Long-term improvement in insulin sensitivity by changing lifestyles of people with impaired glucose tolerance: 4-year results from the Finnish Diabetes Prevention Study. Diabetes 2003;52:2532–2538 [DOI] [PubMed] [Google Scholar]
- 18.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia 2003;46:3–19 [DOI] [PubMed] [Google Scholar]
- 19.Vassy JL, Hivert MF, Porneala B, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes 2014;63:2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jablonski KA, McAteer JB, de Bakker PI, et al.; Diabetes Prevention Program Research Group . Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the Diabetes Prevention Program. Diabetes 2010;59:2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.