Abstract
Objective:
To determine whether peripheral immune responses as measured by serum antigen arrays are linked to cerebral MRI measures of disease severity in multiple sclerosis (MS).
Methods:
In this cross-sectional study, serum samples were obtained from patients with relapsing-remitting MS (n = 21) and assayed using antigen arrays that contained 420 antigens including CNS-related autoantigens, lipids, and heat shock proteins. Normalized compartment-specific global brain volumes were obtained from 3-tesla MRI as surrogates of atrophy, including gray matter fraction (GMF), white matter fraction (WMF), and total brain parenchymal fraction (BPF). Total brain T2 hyperintense lesion volume (T2LV) was quantified from fluid-attenuated inversion recovery images.
Results:
We found serum antibody patterns uniquely correlated with BPF, GMF, WMF, and T2LV. Furthermore, we identified immune signatures linked to MRI markers of neurodegeneration (BPF, GMF, WMF) that differentiated those linked to T2LV. Each MRI measure was correlated with a specific set of antibodies. Strikingly, immunoglobulin G (IgG) antibodies to lipids were linked to brain MRI measures. Based on the association between IgG antibody reactivity and each unique MRI measure, we developed a lipid index. This comprised the reactivity directed against all of the lipids associated with each specific MRI measure. We validated these findings in an additional independent set of patients with MS (n = 14) and detected a similar trend for the correlations between BPF, GMF, and T2LV vs their respective lipid indexes.
Conclusions:
We propose serum antibody repertoires that are associated with MRI measures of cerebral MS involvement. Such antibodies may serve as biomarkers for monitoring disease pathology and progression.
Multiple sclerosis (MS) is characterized by immune dysfunction and inflammation, leading to focal lesions, brain and spinal cord atrophy, and progressive neurologic dysfunction. The known heterogeneity likely reflects myriad and complex underlying pathogenic mechanisms that make specific and unique contributions to MS.1
MRI-defined T2 hyperintense brain lesions are key to diagnosis and therapeutic monitoring. However, such lesions are nonspecific for the underlying pathology and have limited clinical predictive value.2,3 Measurement of brain atrophy provides the potential to detect destructive disease effects and show better associations with clinical status than can be obtained with lesion measures.2 Atrophy begins early in MS and can be monitored by MRI segmentation.2–4 Gray matter (GM) atrophy is more closely linked to clinical status than white matter (WM) atrophy, whole brain atrophy, or conventional lesion assessments.5,6 This likely reflects the functional importance of GM and the contention that pseudoatrophy confounds the use of whole brain or WM atrophy to monitor progressive neurodegeneration.7
Immune processes have a central role in both the pathogenesis and treatment of MS.8–12 The ability to link such changes to MRI presents the opportunity to provide new biomarkers and better understanding of disease pathophysiology.13–19
Antigen microarrays are newly developed tools for the high-throughput characterization of the immune response20,21 that have been used to identify biomarkers and mechanisms of disease pathogenesis in several autoimmune disorders including MS.22–31 In the present study, we investigated the relationship between antigen arrays and both GM and WM cerebral MRI involvement in MS.
METHODS
Patients.
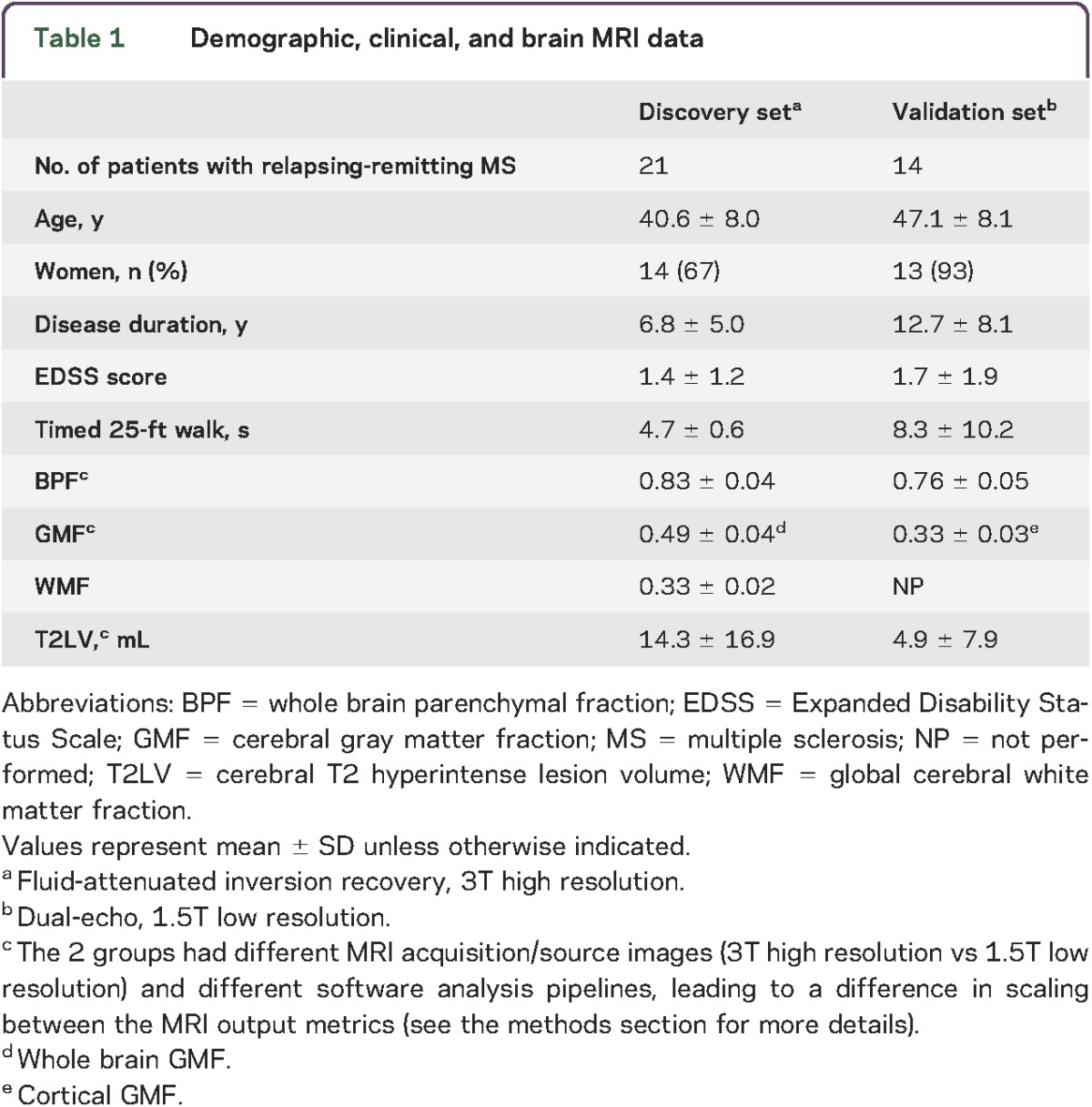
Table 1 summarizes the patients' demographic and clinical characteristics of the discovery and validation sets. All serum samples were collected from the ongoing cohort of patients being followed in the CLIMB study (Comprehensive Longitudinal Investigation of MS at Brigham and Women's Hospital32) in which participants are followed with comprehensive clinical and imaging assessments to monitor disease progression and response to therapy on a yearly basis. Samples were collected within (mean ± SD) 5.0 ± 3.2 months of MRI acquisition. Patients were free of relapses or changes in disease-modifying therapy during the interval between blood collection and MRI. This was a consecutive sample meeting the following criteria: (1) age 18 to 55 years; (2) diagnosis of relapsing-remitting MS33; (3) absence of other major medical, neurologic, or neuropsychiatric disorders; (4) lack of any relapse or corticosteroid use in the 4 weeks before MRI or start of disease-modifying therapy 6 months before MRI (to reduce confounding effects on MRI); and (5) no history of smoking or substance abuse. The majority of patients were receiving disease-modifying treatment at the time of MRI. Within 3 months of MRI, each patient received an examination by an MS specialist-neurologist, including evaluation of neurologic disability on the Expanded Disability Status Scale and a timed 25-foot walk.
Table 1.
Demographic, clinical, and brain MRI data
Standard protocol approvals, registrations, and patient consents.
Our study received approval from the ethical standards committee on human experimentation at our institution (The Partners Health Care Institutional Review Board). All participants gave written informed consent for their participation in the study.
MRI acquisition and analysis.
All participants in the discovery set underwent MRI on the same scanner (3T Signa; General Electric Healthcare, Milwaukee, WI) using a receive-only phase array head coil with the same MRI protocol. The scan acquisition protocol has been detailed previously.34 Contiguous slices covering the whole brain were acquired in high-resolution protocols using 3-dimensional modified driven equilibrium Fourier transform and T2-weighted fast fluid-attenuated inversion recovery sequences. Patients in the validation set underwent brain MRI on a 1.5T scanner (GE Signa) including a 2-dimensional axial conventional spin-echo dual-echo T2-weighted series (voxel sizes 0.94 × 0.94 × 3 mm). Analysis of these scans was performed by operators who were unaware of clinical and biomarker information. In the discovery set, we obtained normalized compartment-specific global brain volumes as surrogates of atrophy, including GM fraction (GMF), WM fraction (WMF), and total brain parenchymal fraction (BPF), using statistical parametric mapping version 8 (Wellcome Department of Cognitive Neurology, London, UK, http://www.fil.ion.ucl.ac.uk/spm), after manual correction of (1) misclassifications of tissue compartments due to MS lesion, and (2) ineffective contouring of the deep GM structures.34 In the validation set, BPF and GMF were obtained in statistical parametric mapping version 8 from the dual-echo images. Because the source images did not show effective contrast for segmentation of the deep gray structures, we performed manual masking to derive only the cerebral cortical GMF. Quantification of total brain T2 hyperintense lesion volume (T2LV) was performed using Jim (Xinapse Systems Ltd., West Bergholt, UK, http://www.xinapse.com) by the consensus of 2 experienced observers from the fluid-attenuated inversion recovery (discovery set) or dual-echo (validation set) images using a semiautomated technique. For the measurement of these atrophy and lesion surrogates from MRI scans, our methods are well established regarding their operational procedures, validity, and reliability.34–39
Antigens.
Peptides were synthesized at the Biopolymers Facility of the Department of Biological Chemistry and Molecular Pharmacology of Harvard Medical School. Recombinant proteins and lipids were purchased from Sigma (St. Louis, MO), Abnova (Taipei City, Taiwan), Matreya LLC (Pleasant Gap, PA), Avanti Polar Lipids (Alabaster, AL), Calbiochem (San Diego, CA), Chemicon (Temecula, CA), GeneTex (San Antonio, TX), Novus Biologicals (Littleton, CO), Assay Designs (Ann Arbor, MI), ProSci Inc. (Poway, CA), EMD Biosciences (San Diego, CA), Cayman Chemical (Ann Arbor, MI), HyTest (Turku, Finland), Meridian Life Science (Memphis, TN), and Biodesign International (Saco, ME). The antigens used in the construction of antigen microarrays are listed in table e-1 at Neurology.org/nn.
Antigen microarray production, development, and data analysis.
The antigens listed in table e-1 were spotted in replicates of 6 on SuperEpoxy 2 slides (TeleChem, Sunnyvale, CA) using an Arrayit NanoPrint 2 LM210 microarray printer (Arrayit Corporation, Sunnyvale, CA) and optimized spotting conditions as described.28,30,31,40 The microarrays were hybridized using an HS 4800Pro Hybridization Station (Tecan, Männedorf, Switzerland), in which they were blocked with 1% bovine serum albumin for 1 hour and incubated for 2 hours at 37°C with the test serum at a 1:100 dilution in blocking buffer. The arrays were then washed and incubated for 45 minutes with a 1:500 dilution of goat anti-human immunoglobulin G (IgG) Cy3-conjugated and goat anti-human IgM Cy5-conjugated detection antibodies (Jackson ImmunoResearch Labs, West Grove, PA). The arrays were scanned with a Tecan PowerScanner. Repeated measurements indicate that our antigen microarray technique is reproducible, exhibiting a coefficient of variation of 13.3 ± 1.2.31
Background signal was subtracted and raw data were normalized and analyzed using the GeneSpring software (Silicon Genetics, Redwood City, CA). Antigen reactivity was defined by the mean intensity of binding to the replicates of that antigen on the microarray and expressed as relative fluorescence units. Scatter plots were generated using linear regression models in the R statistical package. Pearson product-moment correlation coefficients were calculated between the MRI measures (BPF, GMF, WMF, T2LV) and the weighted average of a group of statistically significant lipids. The weighted average of significant lipids was calculated using the formula (ΣWiAi/ΣWi) (ΣWi = 1, i = 1, 2, 3 …), where Wi is the proportion of intensity of antigen, Ai, Ai is the observed intensity of an antigen, and ΣWi is the sum of the weights.
RESULTS
Serum IgG antibodies correlate with brain MRI measures of disease severity.
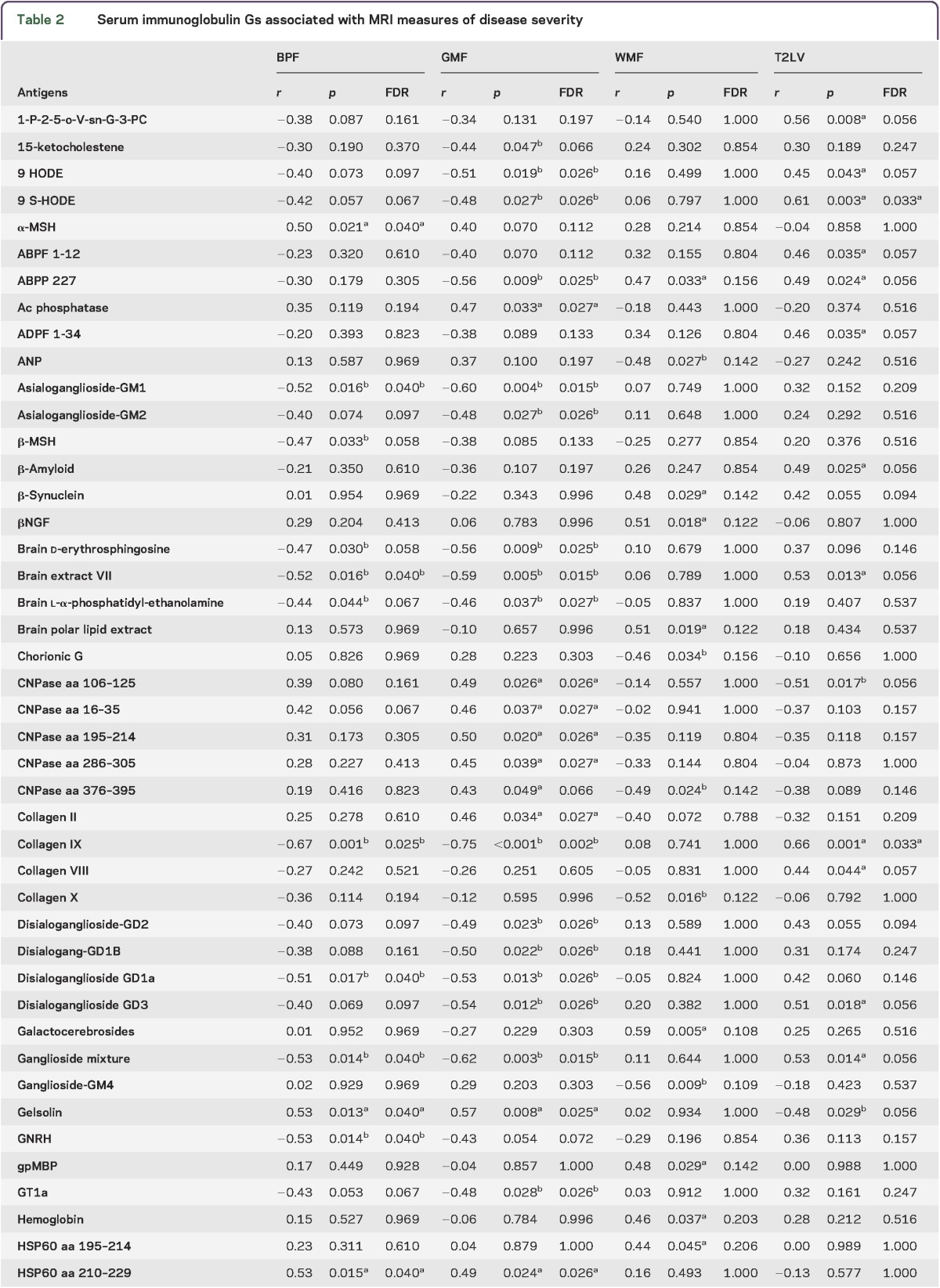
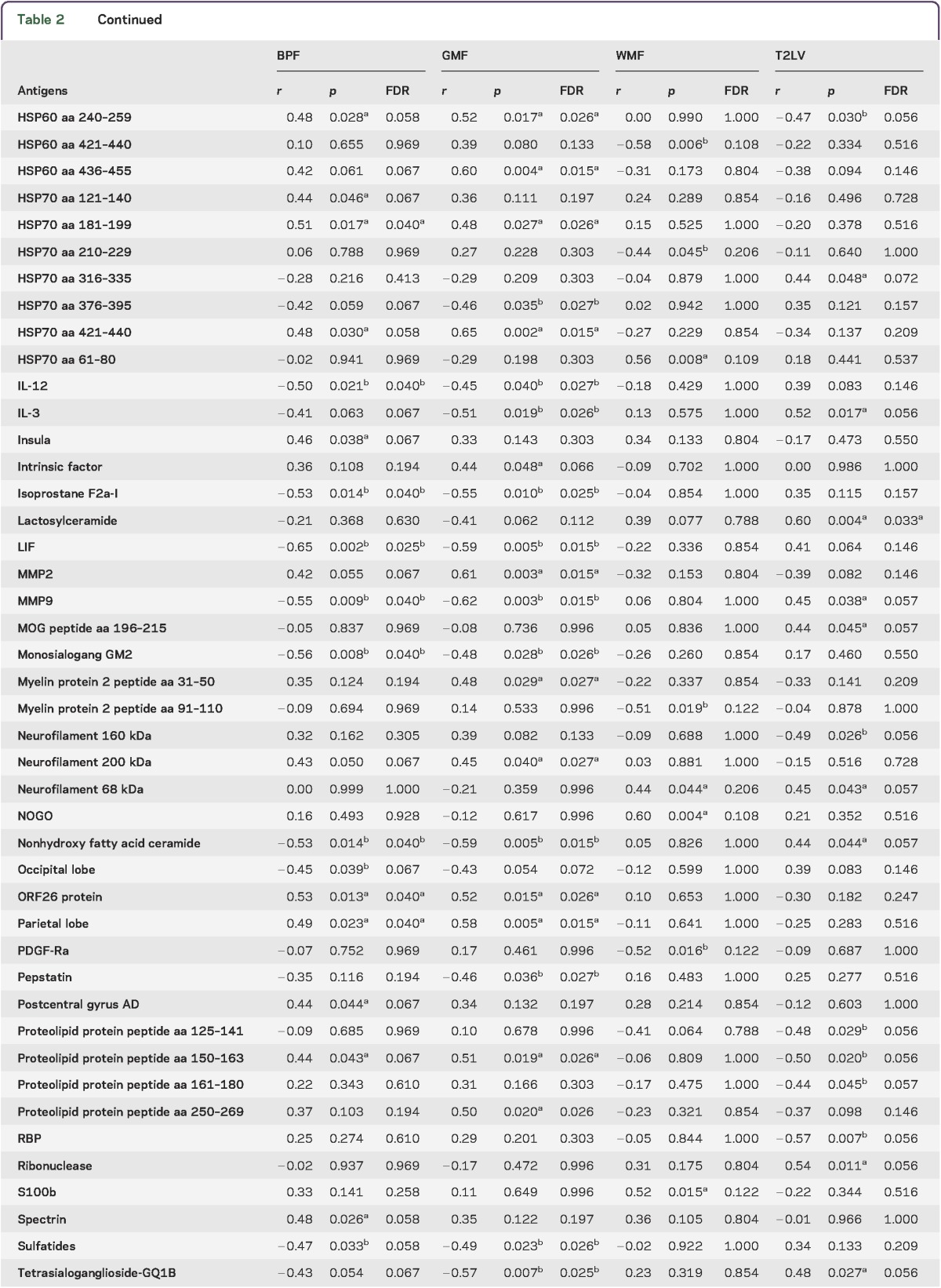
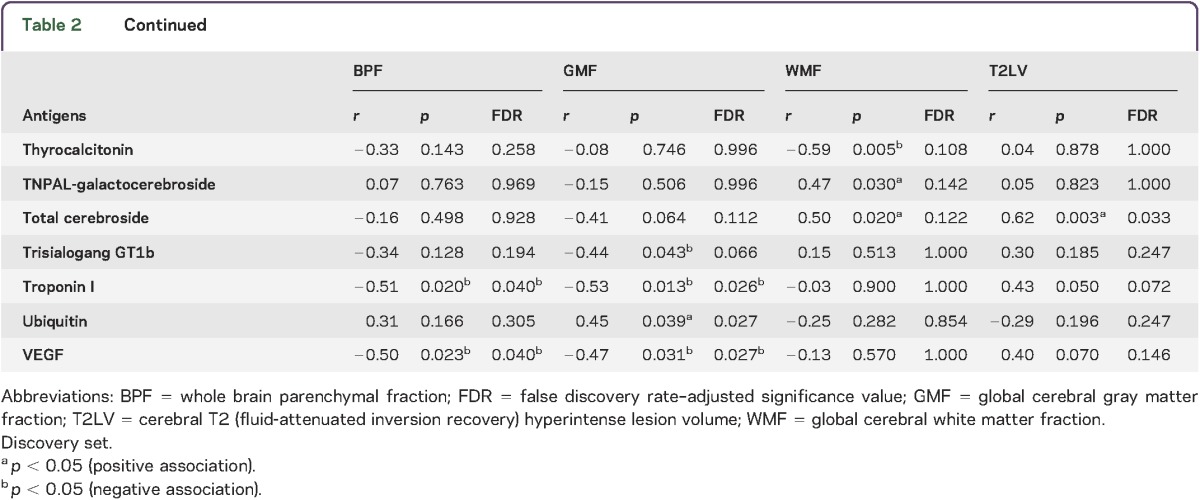
To study the relationship between the peripheral immune response and MRI measures of disease severity, we analyzed serum antibody reactivity in those MS samples. We analyzed IgG serum antibodies using a panel of antigens including CNS antigens, heat shock proteins, and lipids. The association between the antibody reactivity against each antigen and 4 MRI measures of disease (T2LV, BPF, WMF, and GMF) was investigated using Spearman correlation tests.
We found significant associations between each MRI measure of disease and different sets of IgG antibody reactivity, which are shown in table 2. Similar patterns of antibody reactivity were linked to BPF and GMF, consistent with the known dominant contribution of GM atrophy to whole brain atrophy measures.37,41 Strikingly, there was little overlap between the antibody reactivity associated with GMF and WMF, suggesting that different immunopathologic processes contribute to tissue degeneration in these areas of the brain. Similarly, these profiles of antibody reactivity were also different from those associated with T2LV.
Table 2.
Serum immunoglobulin Gs associated with MRI measures of disease severity
Serum lipid-reactive IgGs are associated with increased disease pathogenesis determined by MRI.
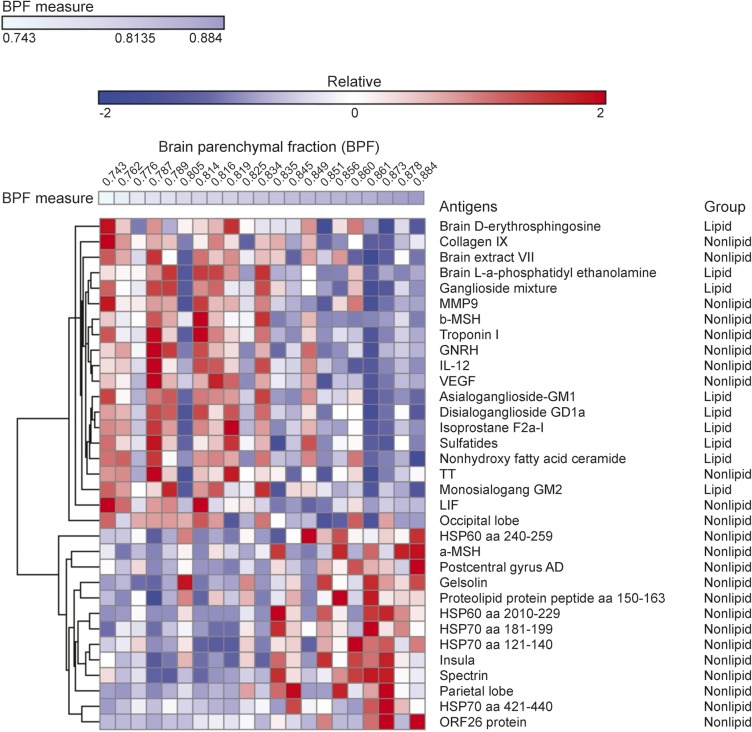
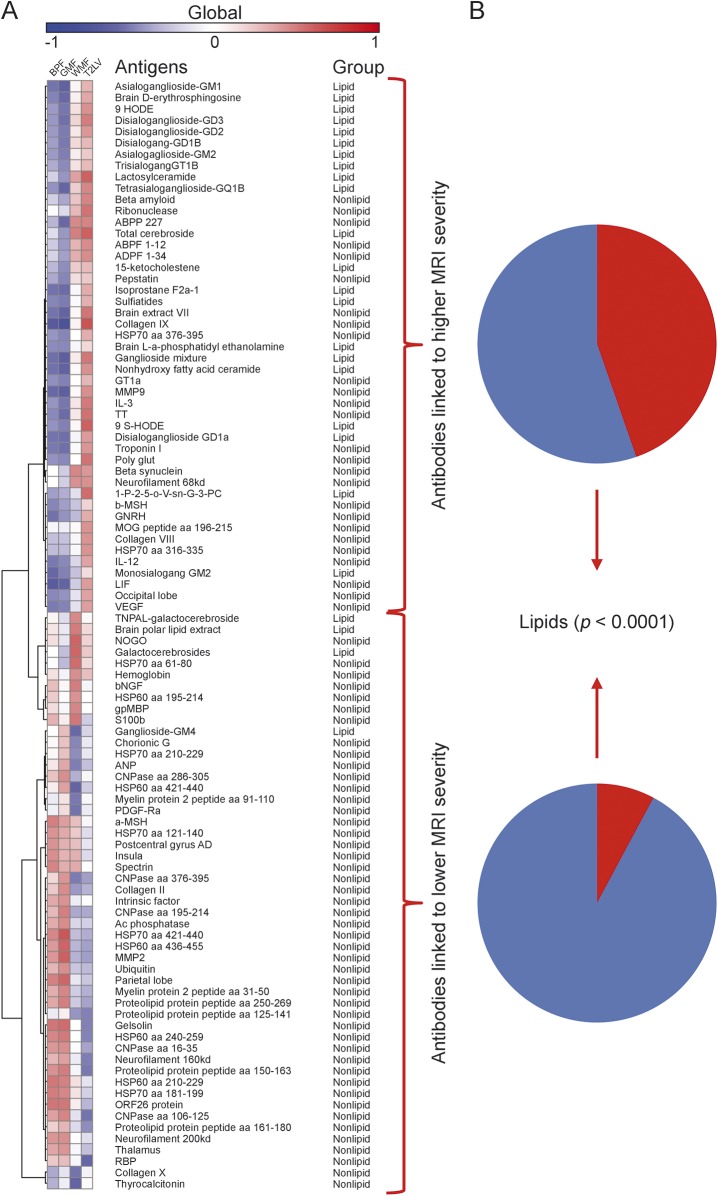
In evaluation of MRI measures and their link to disease, BPF, GMF, and WMF decrease with disease progression while T2LV increases with disease progression. Accordingly, we analyzed the linkage between each MRI measure and the significant antibody reactivities shown in table 2. We identified IgG antibody reactivities associated with increased tissue destruction, as evidenced by decreased BPF values (figure 1). Strikingly, we found that antibodies linked to disease pathology as measured by BPF were enriched for reactivity against lipids. Furthermore, a selective increase in lipid-reactive antibodies linked to disease severity was also observed when we analyzed all available MRI measures (figure 2). Of note, we did not detect an enrichment in lipid-reactive IgM in the group of antibodies associated with increased disease pathogenesis (not shown).
Figure 1. Antibody reactivity in serum is associated with decreased brain volume.
Heatmap in which each column represents the mean immunoglobulin G antibody reactivity in a serum sample from a patient with multiple sclerosis, sorted according to the whole brain parenchymal fraction (BPF) (indicated at the top—a lower BPF indicates more brain atrophy), and each row represents the antibody reactivity to an antigen according to the colorimetric scale shown. The antibody reactivities included in this heatmap are listed in table 2.
Figure 2. Association of serum IgG reactivity with MRI measures of disease severity.
(A) Heatmap in which each column represents an MRI measure of either atrophy (BPF, GMF, WMF) or lesions (T2LV), and each row represents the correlation to IgG serum antibody reactivity according to a colorimetric scale. (B) Frequency of lipid-reactive antibodies linked to higher or lower MRI disease severity. BPF = whole brain parenchymal fraction; GMF = global cerebral gray matter fraction; IgG = immunoglobulin G; T2LV = cerebral T2 (fluid-attenuated inversion recovery) hyperintense lesion volume; WMF = global cerebral white matter fraction.
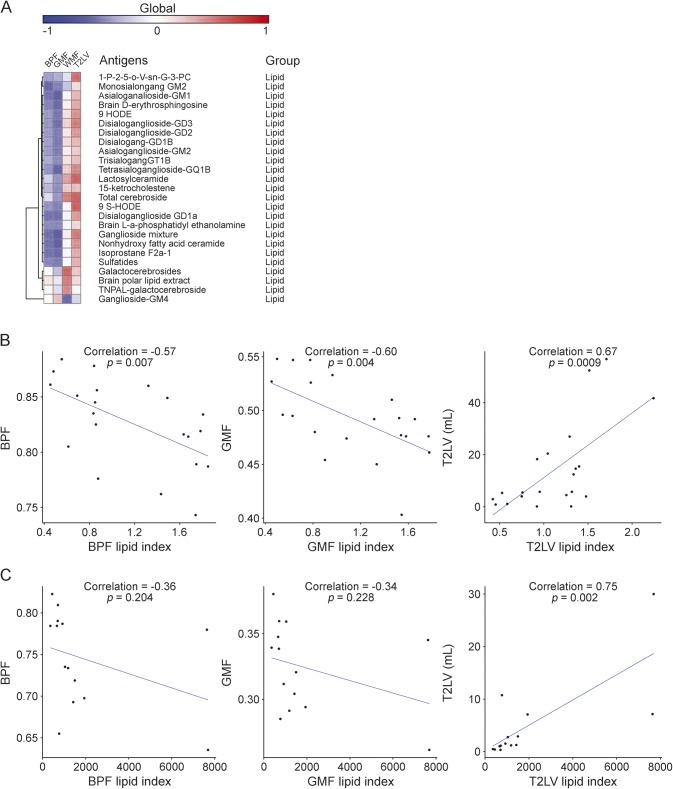
An index of serum IgG reactivity to lipids is associated with MRI measures of disease severity.
To further investigate the association between serum IgG reactivity to lipids and MRI measures of disease severity, we calculated an IgG anti-lipid index for each patient corresponding to the information on each lipid-specific antibody listed in table 2, normalized by the strength of its correlation with that MRI measure under investigation. The antibody reactivities to lipids used to calculate the lipid index are shown in figure 3A. With this approach, we calculated one index for each of the MRI measures analyzed in this study. We found a significant correlation between each IgG lipid antibody index and BPF, GMF, and T2LV (figure 3B); no significant correlation was found with WMF. Moreover, no significant correlations were found between BPF, WMF, GMF, and T2LV measures and anti-lipid antibody indexes based on IgM reactivity (not shown).
Figure 3. Correlation of lipid indexes to MRI measures of disease severity in multiple sclerosis.
(A) Heatmap in which each column represents an MRI measure and each row represents the correlation to immunoglobulin G serum antibody reactivity to lipids according to a colorimetric scale. (B) Scatter plots depicting the correlation between each lipid index and MRI measures (BPF, GMF, and T2LV) in the discovery set. (C) Scatter plots depicting the correlation between each lipid index and MRI measures (BPF, GMF, and T2LV) in the validation set. BPF = whole brain parenchymal fraction; GMF = cerebral gray matter fraction (see methods section for details); T2LV = cerebral T2 hyperintense lesion volume; WMF = global cerebral white matter fraction.
Finally, we evaluated the performance of the lipid antibody indexes linked to BPF, GMF, and T2LV on an additional set of independent MS samples (validation set, table 1). We detected a similar trend to the one detected in the discovery set with regard to the correlation between BPF and GMF and their respective lipid indexes, and we validated the correlation between T2LV and its lipid index (figure 3C).
DISCUSSION
Previous studies have investigated the association of MRI with immune activity in MS.13–19 These studies have included cellular immune measures as well as oligoclonal bands or neurofilament-specific antibodies in CSF.42–44 In the present study, we analyzed the association between serum antibody profiles detected with antigen microarrays and MRI measures of disease, including cerebral atrophy and T2 hyperintense lesions. We found that specific antibody patterns are associated with different aspects of MRI-defined disease pathology. Our major finding was that increased reactivity to lipids is associated with different aspects of brain MRI measures of disease severity. Of note, the specific set of lipids associated with atrophy differed from those associated with lesions. Taken together, these data suggest that anti-lipid antibodies in serum are related to MRI measures of disease. Our findings highlight the important role of lipid and lipid-specific immunity in the pathogenesis of MS.
Although a potential limitation of these studies is the relatively small number of samples analyzed, we have validated our findings in an independent validation cohort, strengthening their significance. In addition, further studies in large cohorts from patients affected by non-MS neurodegenerative diseases and healthy controls may indicate whether the antibody reactivities and lipid indexes identified in these studies are exclusive to MS, or are associated with additional biological processes. Indeed, some of these antibody reactivities contributing to the immune signatures described in this work have been found in newborns.45
Different sets of antibody reactivities were associated with GMF, WMF, BPF, and T2LV. The different antibody link between lesions and atrophy is in keeping with the long-held view that brain atrophy in patients with MS is multifactorial and is only weakly related to lesions.41 This has been underscored in recent studies showing the complementary information obtained by combining lesion and atrophy measures.46,47
Among the cerebral atrophy measures, there was a striking overlap between those antibodies associated with GMF and BPF, which separated them from WMF results. These findings are in keeping with previous observations that whole brain atrophy is dominated by GM loss.37,41 In the early stages of MS, this GM atrophy selectively involves the deep GM nuclei.34 Thus, the similarities observed in the autoantibodies linked to GMF and BPF might reflect the dominant contribution and colinearity of GM atrophy to total brain atrophy. The set of antibodies linked to GMF may be the most relevant given that MRI studies have shown that GM volume is more closely linked to physical disability37 and cognitive impairment38 than are WM volume or lesion measures.
Using antigen microarrays, antibody reactivity to lipids has been detected in the CSF and serum of patients with MS at different stages of the disease and such antibodies in the CSF have been linked to disease progression and MRI involvement.44 Although several mechanisms are thought to drive brain inflammation and atrophy in MS,4,6 the connection between these mechanisms and the antibodies detected with our antigen microarrays is yet unknown. One possibility is that the serum autoantibodies identified in our studies are not pathogenic and reflect the result of immunization against self-antigens released from the CNS during the course of the disease. Indeed, neurofilament is released from damaged axons during the course of MS, and both neurofilament light chains48 and antibodies against it42 have been linked to MRI measures of disease. Moreover, heat shock proteins are upregulated in different cell types during inflammation and have been shown to have an important role as immunomodulators when released to the extracellular medium.49,50 Thus, it is possible that heat shock protein–reactive antibodies reflect changes in the production and release of these immunomodulators during the course of disease pathogenesis. Similarly, bioactive lipids and their products are released from damaged myelin, and lipid-specific antibodies have been detected in patients with MS.27,29,30,51 Lipids have an important role in the immune response, both as bioactive molecules with immunomodulatory properties and also as targets of the adaptive immune response.52 Moreover, lipids have significant effects on the murine model of MS experimental autoimmune encephalomyelitis.27,29,30,51,53 Indeed, we recently found that the glycolipid lactosylceramide activates a broad set of biological processes in astrocytes, promoting neurodegeneration and inflammation.53 Thus, the lipid-reactive antibodies detected in this work may reflect the release of myelin lipids in the context of MS pathogenesis, and/or may directly contribute to immune-mediated damage in CNS tissue. Of note, lipid-reactive antibodies are highly cross-reactive.52 In addition, the lipid-reactive antibodies detected in this work as associated with MRI measures of disease activity were of the IgG class. These are important points to consider when evaluating a potential role of lipid-reactive antibodies in MS pathogenesis. Further studies are warranted to investigate whether bioactive lipids offer new therapeutic targets in MS.
We have found that unique patterns of immune reactivity determined with antibody arrays are associated with specific MRI measures of disease severity. These patterns agree with the interpretation that different pathogenic mechanisms drive diverse disease processes reflected by these MRI measures. These patterns also suggest a predominant role for lipids and lipid-specific immunity in MS pathology. Further studies are warranted to determine whether the early appearance of anti-lipid antibodies predicts the subsequent development of clinical and MRI-defined disease worsening.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Mohit Neema, Dr. Antonella Ceccarelli, and Dr. Ashish Arora for valuable assistance at the early stages of this project.
GLOSSARY
- BPF
brain parenchymal fraction
- GM
gray matter
- GMF
gray matter fraction
- IgG
immunoglobulin G
- MS
multiple sclerosis
- T2LV
T2 hyperintense lesion volume
- WM
white matter
- WMF
white matter fraction
Footnotes
Supplemental data at Neurology.org/nn
AUTHOR CONTRIBUTIONS
Rohit Bakshi: drafting/revising the manuscript, study concept or design, analysis or interpretation of data, acquisition of data, study supervision, obtaining funding. Ada Yeste: study concept or design, acquisition of data. Bonny Patel: analysis or interpretation of data, statistical analysis. Shahamat Tauhid: analysis or interpretation of data. Subhash Tummala: analysis or interpretation of data. Roya Rahbari: analysis or interpretation of data, statistical analysis. Renxin Chu: analysis or interpretation of data. Keren Regev: analysis or interpretation of data, acquisition of data. Pia Kivisäkk: drafting/revising the manuscript, study concept or design, contribution of vital reagents/tools/patients, acquisition of data, study supervision. Howard L. Weiner: study concept or design, obtaining funding. Francisco J. Quintana: drafting/revising the manuscript, study concept or design, contribution of vital reagents/tools/patients, study supervision, obtaining funding.
STUDY FUNDING
This work was supported in part by grants to F.J.Q. and H.L.W. from EMD Serono, RG4111A1 and JF2161-A-5 from the National Multiple Sclerosis Society, PA0069 from the International Progressive MS Alliance, and by grants from the NIH (5R01NS055083-04) and NMSS (RG3798A2) to R.B.
DISCLOSURE
F.J. Quintana serves on the editorial board for Systems Biomedicine, Inmunologia, American Journal of Clinical and Experimental Immunology, is an associate editor for Immunology (UK), is an advisory board member for Seminars in Immunopathology; received research support from Harvard Medical School, BADERC, NMSS. A. Yeste and B. Patel report no disclosures. S. Tauhid is managing editor for Journal of Neuroimaging. S. Tummala, R. Rahbari, R. Chu, and K. Regev report no disclosures. P. Kivisäkk received research support from EMD Serono. R. Bakshi is editor-in-chief for Journal of Neuroimaging; received consulting fees from AbbVie, Alkermes, Biogen, Novartis, Questcor; received research support from Biogen, EMD-Serono, Novartis, Sanofi-Genzyme, Teva; his spouse holds stock in Biogen, Inc. H. Weiner served on the advisory board for The Guthy-Jackson Charitable Foundation, Teva Pharmaceuticals Industries Ltd., Biogen Idec, Novartis, Sanofi-Aventis; has consulted for Therapix, Bioven, Novartis, Serono, Teva, Sanofi; received research support from National Multiple Sclerosis Society. Go to Neurology.org/nn for full disclosure forms.
REFERENCES
- 1.Weiner HL. The challenge of multiple sclerosis: how do we cure a chronic heterogeneous disease? Ann Neurol 2009;65:239–248. [DOI] [PubMed] [Google Scholar]
- 2.Bakshi R, Thompson AJ, Rocca MA, et al. MRI in multiple sclerosis: current status and future prospects. Lancet Neurol 2008;7:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccarelli A, Bakshi R, Neema M. MRI in multiple sclerosis: a review of the current literature. Curr Opin Neurol 2012;25:402–409. [DOI] [PubMed] [Google Scholar]
- 4.Filippi M, Rocca MA, Barkhof F, et al. Association between pathological and MRI findings in multiple sclerosis. Lancet Neurol 2012;11:349–360. [DOI] [PubMed] [Google Scholar]
- 5.Geurts JJ, Calabrese M, Fisher E, Rudick RA. Measurement and clinical effect of grey matter pathology in multiple sclerosis. Lancet Neurol 2012;11:1082–1092. [DOI] [PubMed] [Google Scholar]
- 6.Klaver R, De Vries HE, Schenk GJ, Geurts JJ. Grey matter damage in multiple sclerosis: a pathology perspective. Prion 2013;7:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoury S, Bakshi R. Cerebral pseudoatrophy or real atrophy after therapy in multiple sclerosis. Ann Neurol 2010;68:778–779. [DOI] [PubMed] [Google Scholar]
- 8.Hauser SL, Chan JR, Oksenberg JR. Multiple sclerosis: prospects and promise. Ann Neurol 2013;74:317–327. [DOI] [PubMed] [Google Scholar]
- 9.McFarland HF, Martin R. Multiple sclerosis: a complicated picture of autoimmunity. Nat Immunol 2007;8:913–919. [DOI] [PubMed] [Google Scholar]
- 10.Nylander A, Hafler DA. Multiple sclerosis. J Clin Invest 2012;122:1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sospedra M, Martin R. Immunology of multiple sclerosis. Ann Rev Immunol 2005;23:683–747. [DOI] [PubMed] [Google Scholar]
- 12.Steinman L. Immunology of relapse and remission in multiple sclerosis. Ann Rev Immunol 2014;32:257–281. [DOI] [PubMed] [Google Scholar]
- 13.Khoury SJ, Guttmann CR, Orav EJ, Kikinis R, Jolesz FA, Weiner HL. Changes in activated T cells in the blood correlate with disease activity in multiple sclerosis. Arch Neurol 2000;57:1183–1189. [DOI] [PubMed] [Google Scholar]
- 14.Khoury SJ, Orav EJ, Guttmann CR, Kikinis R, Jolesz FA, Weiner HL. Changes in serum levels of ICAM and TNF-R correlate with disease activity in multiple sclerosis. Neurology 1999;53:758–764. [DOI] [PubMed] [Google Scholar]
- 15.Laplaud DA, Berthelot L, Miqueu P, et al. Serial blood T cell repertoire alterations in multiple sclerosis patients: correlation with clinical and MRI parameters. J Neuroimmunol 2006;177:151–160. [DOI] [PubMed] [Google Scholar]
- 16.Makhlouf K, Weiner HL, Khoury SJ. Increased percentage of IL-12+ monocytes in the blood correlates with the presence of active MRI lesions in MS. J Neuroimmunol 2001;119:145–149. [DOI] [PubMed] [Google Scholar]
- 17.Prat A, Biernacki K, Saroli T, et al. Kinin B1 receptor expression on multiple sclerosis mononuclear cells: correlation with magnetic resonance imaging T2-weighted lesion volume and clinical disability. Arch Neurol 2005;62:795–800. [DOI] [PubMed] [Google Scholar]
- 18.Rinaldi L, Gallo P, Calabrese M, et al. Longitudinal analysis of immune cell phenotypes in early stage multiple sclerosis: distinctive patterns characterize MRI-active patients. Brain 2006;129:1993–2007. [DOI] [PubMed] [Google Scholar]
- 19.Tortorella P, Lagana MM, Saresella M, et al. Determinants of disability in multiple sclerosis: an immunological and MRI study. Biomed Res Int 2014;2014:875768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson WH, DiGennaro C, Hueber W, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med 2002;8:295–301. [DOI] [PubMed] [Google Scholar]
- 21.Yeste A, Quintana FJ. Antigen microarrays for the study of autoimmune diseases. Clin Chem 2013;59:1036–1044. [DOI] [PubMed] [Google Scholar]
- 22.Li QZ, Xie C, Wu T, et al. Identification of autoantibody clusters that best predict lupus disease activity using glomerular proteome arrays. J Clin Invest 2005;115:3428–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueber W, Kidd BA, Tomooka BH, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum 2005;52:2645–2655. [DOI] [PubMed] [Google Scholar]
- 24.Goldschmidt Y, Sharon E, Quintana FJ, Cohen IR, Brandt A. Adaptive methods for classification of biological microarray data from multiple experiments. Report no MCS03-07 Rehovot, Israel: The Arthur and Rochelle Belfer Institute of Mathematics and Computer Science; 2003. [Google Scholar]
- 25.Quintana FJ, Getz G, Hed G, Domany E, Cohen IR. Cluster analysis of human autoantibody reactivities in health and in type 1 diabetes mellitus: a bio-informatic approach to immune complexity. J Autoimmun 2003;21:65–75. [DOI] [PubMed] [Google Scholar]
- 26.Lalive PH, Menge T, Barman I, Cree BA, Genain CP. Identification of new serum autoantibodies in neuromyelitis optica using protein microarrays. Neurology 2006;67:176–177. [DOI] [PubMed] [Google Scholar]
- 27.Kanter JL, Narayana S, Ho PP, et al. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nat Med 2006;12:138–143. [DOI] [PubMed] [Google Scholar]
- 28.Quintana FJ, Farez MF, Izquierdo G, Lucas M, Cohen IR, Weiner HL. Antigen microarrays identify CNS-produced autoantibodies in RRMS. Neurology 2012;78:532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farez MF, Quintana FJ, Gandhi R, Izquierdo G, Lucas M, Weiner HL. Toll-like receptor 2 and poly(ADP-ribose) polymerase 1 promote central nervous system neuroinflammation in progressive EAE. Nat Immunol 2009;10:958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana FJ, Farez MF, Viglietta V, et al. Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc Natl Acad Sci USA 2008;105:18889–18894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintana FJ, Patel B, Yeste A, et al. Epitope spreading as an early pathogenic event in pediatric multiple sclerosis. Neurology 2014;83:2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauthier SA, Glanz BI, Mandel M, Weiner HL. A model for the comprehensive investigation of a chronic autoimmune disease: the multiple sclerosis CLIMB study. Autoimmun Rev 2006;5:532–536. [DOI] [PubMed] [Google Scholar]
- 33.Polman CH, Reingold SC, Edan G, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria.” Ann Neurol 2005;58:840–846. [DOI] [PubMed] [Google Scholar]
- 34.Dell'Oglio E, Ceccarelli A, Glanz BI, et al. Quantification of global cerebral atrophy in multiple sclerosis from 3T MRI using SPM: the role of misclassification errors. J Neuroimaging 2015;25:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceccarelli A, Jackson JS, Tauhid S, et al. The impact of lesion in-painting and registration methods on voxel-based morphometry in detecting regional cerebral gray matter atrophy in multiple sclerosis. AJNR Am J Neuroradiol 2012;33:1579–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen AB, Neema M, Arora A, et al. The relationships among MRI-defined spinal cord involvement, brain involvement, and disability in multiple sclerosis. J Neuroimaging 2012;22:122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanfilipo MP, Benedict RH, Sharma J, Weinstock-Guttman B, Bakshi R. The relationship between whole brain volume and disability in multiple sclerosis: a comparison of normalized gray vs. white matter with misclassification correction. Neuroimage 2005;26:1068–1077. [DOI] [PubMed] [Google Scholar]
- 38.Sanfilipo MP, Benedict RH, Weinstock-Guttman B, Bakshi R. Gray and white matter brain atrophy and neuropsychological impairment in multiple sclerosis. Neurology 2006;66:685–692. [DOI] [PubMed] [Google Scholar]
- 39.Stankiewicz JM, Glanz BI, Healy BC, et al. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging 2011;21:e50–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quintana FJ, Hagedorn PH, Elizur G, Merbl Y, Domany E, Cohen IR. Functional immunomics: microarray analysis of IgG autoantibody repertoires predicts the future response of mice to induced diabetes. Proc Natl Acad Sci USA 2004;101(suppl 2):14615–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bermel RA, Bakshi R. The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol 2006;5:158–170. [DOI] [PubMed] [Google Scholar]
- 42.Eikelenboom MJ, Petzold A, Lazeron RH, et al. Multiple sclerosis: neurofilament light chain antibodies are correlated to cerebral atrophy. Neurology 2003;60:219–223. [DOI] [PubMed] [Google Scholar]
- 43.Magraner MJ, Bosca I, Simo-Castello M, et al. Brain atrophy and lesion load are related to CSF lipid-specific IgM oligoclonal bands in clinically isolated syndromes. Neuroradiol 2012;54:5–12. [DOI] [PubMed] [Google Scholar]
- 44.Villar LM, Sadaba MC, Roldan E, et al. Intrathecal synthesis of oligoclonal IgM against myelin lipids predicts an aggressive disease course in MS. J Clin Invest 2005;115:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merbl Y, Zucker-Toledano M, Quintana FJ, Cohen IR. Newborn humans manifest autoantibodies to defined self molecules detected by antigen microarray informatics. J Clin Invest 2007;117:712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sormani MP, Arnold DL, De Stefano N. Treatment effect on brain atrophy correlates with treatment effect on disability in multiple sclerosis. Ann Neurol 2014;75:43–49. [DOI] [PubMed] [Google Scholar]
- 47.Tauhid S, Neema M, Healy BC, Weiner HL, Bakshi R. MRI phenotypes based on cerebral lesions and atrophy in patients with multiple sclerosis. J Neurol Sci 2014;346:250–254. [DOI] [PubMed] [Google Scholar]
- 48.Gnanapavan S, Grant D, Morant S, et al. Biomarker report from the phase II lamotrigine trial in secondary progressive MS: neurofilament as a surrogate of disease progression. PLoS One 2013;8:e70019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintana FJ, Cohen IR. Heat shock proteins as endogenous adjuvants in sterile and septic inflammation. J Immunol 2005;175:2777–2782. [DOI] [PubMed] [Google Scholar]
- 50.Quintana FJ, Cohen IR. The HSP60 immune system network. Trends Immunol 2011;32:89–95. [DOI] [PubMed] [Google Scholar]
- 51.Ho PP, Kanter JL, Johnson AM, et al. Identification of naturally occurring fatty acids of the myelin sheath that resolve neuroinflammation. Sci Transl Med 2012;4:137ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quintana FJ, Yeste A, Weiner HL, Covacu R. Lipids and lipid-reactive antibodies as biomarkers for multiple sclerosis. J Neuroimmunol 2012;248:53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayo L, Trauger SA, Blain M, et al. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat Med 2014;20:1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.