Abstract
Background. Staphylococcus aureus can cause life-threatening infections. Human susceptibility to S. aureus infection may be influenced by host genetic variation.
Methods. A genome-wide association study (GWAS) in a large health plan–based cohort included biologic specimens from 4701 culture-confirmed S. aureus cases and 45 344 matched controls; 584 535 single-nucleotide polymorphisms (SNPs) were genotyped on an array specific to individuals of European ancestry. Coverage was increased by imputation of >25 million common SNPs, using the 1000 Genomes Reference panel. In addition, human leukocyte antigen (HLA) serotypes were also imputed.
Results. Logistic regression analysis, performed under the assumption of an additive genetic model, revealed several imputed SNPs (eg, rs115231074: odds ratio [OR], 1.22 [P = 1.3 × 10−10]; rs35079132: OR, 1.24 [P = 3.8 × 10−8]) achieving genome-wide significance on chromosome 6 in the HLA class II region. One adjacent genotyped SNP was nearly genome-wide significant (rs4321864: OR, 1.13; P = 8.8 × 10−8). These polymorphisms are located near the genes encoding HLA-DRA and HLA-DRB1. Results of further logistic regression analysis, in which the most significant GWAS SNPs were conditioned on HLA-DRB1*04 serotype, showed additional support for the strength of association between HLA class II genetic variants and S. aureus infection.
Conclusions. Our study results are the first reported evidence of human genetic susceptibility to S. aureus infection.
Keywords: Staphylococcus aureus, host genetics, HLA
Staphylococcus aureus is both a harmless colonizer and a leading cause of life-threatening infections. A number of observations suggest a genetic basis for human susceptibility to S. aureus, including variable genetic susceptibility to S. aureus infection in inbred mice [1, 2], cattle [3], and sheep [4]; familial clusters of S. aureus infection [5]; and genetic conditions conferring susceptibility to S. aureus (eg, Job syndrome and Chediak-Higashi syndrome) [6, 7]. Two previous investigations have used genome-wide association studies (GWAS) to evaluate human genetic susceptibility to S. aureus infection [8, 9]. Both were likely underpowered to detect effects at genome-wide significance. As a result, the impact of host genetic variation on the susceptibility to S. aureus infection is largely unknown.
Our goal for the current study was to determine which host genetic polymorphisms were associated with (1) all S. aureus infections and (2) a subset of community-acquired skin and soft tissue S. aureus infections (SSTIs), using genomic data from >50 000 white individuals. We hypothesized that genetic variants that encode proteins with immune functions would be associated with an increased (or decreased) risk of S. aureus infection. A second objective was to determine whether including imputed genetic variants [10] would reveal additional gene–S. aureus infection associations. A third objective was to evaluate the strength of potential significant associations from the HLA regions of chromosome 6 by conditioning on imputed HLA serotypes [11].
MATERIALS AND METHODS
Study Sample
The study population consists of participants in the Research Program on Genes, Environment, and Health (RPGEH). This cohort was recruited from 3.3 million patients in the Kaiser Permanente Northern California (KPNC) health plan. The RPGEH includes a cohort with baseline survey data obtained in 2007–2008 from 400 000 adult patients in KPNC who had ≥2 years membership prior to the survey (average time, 23.5 years). More than 200 000 patients who had completed surveys were selected to receive Oragene saliva collection kits. Genotyping was conducted using returned saliva specimens from 110 266 RPGEH subjects (of whom 89 341 self-identified as white). The RPGEH imputed >25 million SNPs not covered in the original genotyping arrays, as well as HLA major histocompatibility complex (MHC) serotypes. Informed consent was obtained from all RPGEH subjects. Questionnaire data included demographic characteristics (eg, age and sex), as well as self-reports of disease history (eg, diabetes). The RPGEH study and the current study were approved by the KPNC institutional review board.
KPNC maintains electronic databases that include data on hospitalizations, clinic visits, laboratory testing results, and pharmacy dispensing records. These databases were used to create the case-control sample in the present study.
Identification of Cases
The study's phenotypes included culture-confirmed diagnoses of S. aureus infections identified using standard methods in the clinical microbiology laboratory of KPNC [12]. For repeated testing, we adapted the Clinical and Laboratory Standards Institute guideline and recommendations in the literature, including only the first isolate per person in a 365-day period [13, 14]. To identify isolates likely to be related to clinically relevant infections, we restricted our analyses to blood, bone, cerebrospinal fluid, body fluid, urine, tissue, respiratory, and miscellaneous bacterial specimens (eg, abscesses). Screening tests (nares) and cultures of genital specimens, feces/stool specimens, catheter tips, and throat specimens were excluded. The primary study phenotype included all S. aureus infections diagnosed in white subjects of the RPGEH cohort between 1995 and 2011. The secondary phenotype was the subset of community-acquired SSTIs diagnosed during the same interval. SSTIs were ascertained via linking to diagnostic data in the electronic databases (International Classification of Diseases, Ninth Revision, Clinical Modification [ICD-9]). SSTI diagnostic codes included erysipelas (035), cellulitis and abscess (566, 67510–67514, 6810–6829, 6850), mastitis (67520–67524), carbuncle and furuncle (6800–6809), acute lymphadenitis (683), impetigo (684), other skin infections (6860–6861, 6868–6869), folliculitis (7048), and hidradenitis (70583). An infection that was diagnosed ≥48 hours after admission to the hospital was classified as a hospital-onset case; all other infections were classified as community acquired.
Study controls included RPGEH subjects with no evidence of culture-confirmed S. aureus infection. Controls lacking a positive isolate but having records of an ICD-9 code for S. aureus infection and subsequent treatment with antibiotics were excluded. Our study used a frequency matched case-control design. Cases were matched on age (5-year age groups) at the time of specimen collection and sex to approximately 10 controls.
DNA Isolation, Genotyping, and Quality Control
DNA was extracted from saliva specimens, using an Agencourt AMPure XP kit (Beckman Coulter) in a high-throughput process. Genotyping was accomplished using the Affymetrix Axiom Genotyping Solution (available at: http://media.affymetrix.com/support/technical/datasheets/axiom_genotypingsolution_datasheet.pdf). The Axiom genotyping platform is a 2-color, ligation-based assay that uses 30-mer oligonucleotide probes synthesized in situ on a microarray substrate, with 96 samples per plate. A maximum of approximately 690 000 SNPs may be accommodated by this format. Performance of the array was assessed by assaying the white and Yoruban HapMap2 [15] populations. Call rates, sample concordance, reproducibility, and Mendelian consistency were extremely high. A large majority of SNPs have overall call rates of ≥97%. Genotyping of 89 341 saliva samples from the European (EUR) ancestry study subjects was completed using 3 Affymetrix Gene Titan systems and 3 Beckman Biomek Systems. The Affymetrix Powertools Package, version 1.12.0 (available at: http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx?hightlight=true&rootCategoryId=34002#13), was used to make genotype calls. The Affymetrix Axiom EUR array included 674 112 SNPs: 116 were mitochondrial, 289 were on the Y chromosome, 388 were in pseudo-autosomal regions of the X and Y chromosomes, and the remaining 660 989 SNPs were autosomal.
Examination of graphics from principal components (PC) analysis [16] (see the “GWAS Data Analysis” section, below) led to the identification of some individuals whose genetic ancestry appeared to be discordant from their self-report on the RPGEH survey (available at: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/GetPdf.cgi?id=phd004309). Some individuals with data on the African (AFR) array were estimated to have 100% European ancestry. Investigation of so-called discordant individuals revealed a discrepancy between the survey form and the computerized records from optical scanning. About 2% of surveys had been mis-scanned for race/ethnicity/nationality. This led to the systematic reassignment of these individuals to their original survey responses, supplemented by race/ethnicity information in the KPNC databases.
Our quality control (QC) filtering excluded SNPs with genotyping call rates of <95%, a minor allele frequency (MAF) of <0.4% (primary analysis) and <0.9% (secondary analysis), or a statistically significant (P < 10−3) departure from Hardy–Weinberg equilibrium (HWE) in the controls. Subjects with missing genotype rates of >5% or mismatch between reported and genetically determined sex were excluded. Samples were evaluated for cryptic relatedness through estimation of kinship coefficient, using KING [17] software. One sample was randomly excluded from sample pairs exhibiting a kinship coefficient of ≥0.0625.
Genotype Imputation Method
To increase coverage of common variants in the genotyped platform, particularly in the region where our most significant genotyped SNP GWAS results were found, imputation was done on the EUR array (for a comparison of SNP imputation vs other multiple imputation techniques, see Supplementary 1). This approach is now a well-established method to increase marker density in GWAS [18] and takes advantage of known multi-SNP haplotype structures that have been determined by sequencing large diversity panels, such as the 1000 Genomes Project [19]. Therefore, untyped SNPs can be inferred from the genotyped markers included on a genome-wide array with high degrees of confidence and used in association analysis. The data were prephased (inferring haplotypes) with SHAPE-IT v2.r644 [20], using the family structure of first-degree cryptic related individuals as available. With 1000 Genomes (phase I; March 2012) as a reference panel, SNP data were imputed using IMPUTE2 v2.2.2 [10, 21].
HLA Serotype Imputation Method
The HLA serotype imputation method used in our study was originally developed and described in detail by Jia et al [11]. Briefly, the complex genetic structure of MHC makes it difficult to collect high-resolution haplotype data in large cohorts. Long-range linkage disequilibrium (LD) between HLA loci and SNPs across the MHC region offers an alternative approach through imputation to interrogate HLA. We used the SNP2HLA program to impute individual amino acid–changing polymorphisms and classical 4-digit haplotypes at class I and II loci. SNP2HLA was developed using the following reference panels: (1) HapMap-CEPH (individuals of European ancestry) [22] and (2) Type 1 Diabetes Genetics Consortium reference panel (5225 unrelated individuals) [23]. Beagle software [18] was used to phase genotype data into individual haplotypes, taking into account familial relationships. Genotyped SNPs were extracted from within the MHC region (chromosome 6: 29–34 Mb on build 36/hg18); SNPs with a MAF of <2.5% were removed. Beagle imputed all missing SNPs, classical HLA alleles, and amino acid polymorphisms. Output included posterior probabilities, allelic dosages, and phased haplotypes for each individual.
Statistical Power
The original statistical power calculations showed that, under an additive logistic model with a 2-sided test, an α of 5 × 10−8, and a MAF of > 0.10, there was sufficient power to detect odds ratios (ORs) of ≥1.20, given a (pre-QC filtering) sample size of 56 100 subjects (5100 cases and 51 000 controls).
GWAS Data Analysis
Our study conducted case-control analyses for each S. aureus phenotype, using an additive genetic model of inheritance. Unconditional logistic regression was used for both primary outcome (all S. aureus infections) and secondary outcome (community-acquired SSTIs), testing the association with each genotyped and each imputed SNP separately. Logistic regression was also used in association testing of the imputed HLA serotype data in relation to the primary phenotype, as well as in further analysis, in which the 3 most significant SNPs were conditioned on an imputed HLA serotype variant. To account for population stratification and admixture, our regression models included adjustment for the first 10 eigenvectors from a PC analysis [24], using Eigensoft 4.2. To reflect the study sampling scheme, our regression models included age (5-year intervals) at time of specimen collection and sex covariates. PLINK software [25] was used in the association testing analyses of the genotyped data for both phenotypes [25]. PLINK and R software were used to analyze the imputed SNP and imputed HLA serotype data.
RESULTS
The initial study sample for the primary phenotype analysis (ie, all culture-confirmed S. aureus infections as outcome) included 53 322 subjects (4997 S. aureus cases and 48 325 controls). During QC filtering, 16 513 SNPs with genotyping call rates of <95%, 6958 with a MAF of <0.4%, and 66 106 showing significant departure from HWE in controls were excluded, leaving 584 535 SNPs for data analysis. Removal from the data set prior to analysis occurred for 9 subjects whose monozygotic twin was also in the study sample, 3178 subjects whose sample pair had a kinship coefficient of ≥0.0625, 54 controls with ICD-9 diagnostic coding for S. aureus infection followed by coding for appropriate treatment, 24 subjects who had a mismatch between their reported and genetically determined sex, and 12 subjects (of South Asian ancestry) who did not have estimates for European PC. Thus, our final sample for the primary phenotype analysis consisted of 50 045 unique subjects (4701 S. aureus cases and 45 344 controls).
Table 1 shows the distribution of study characteristics among 50 045 subjects. Approximately 51% of cases and 49% of controls were male; 73% of cases and 74% of controls were ≥60 years of age at the time of specimen collection. Fifteen percent of cases and 13% of controls had a history of diabetes. Of the 4701 S. aureus cases, 28% were methicillin-resistant S. aureus (MRSA) infections, 50% were SSTIs, and 96% were community-acquired infections. Laboratory order codes for isolates from the 4701 confirmed cases included blood culture for 3%, cerebral spinal fluid/other body fluid for 3%, and miscellaneous bacterial culture for 61%.
Table 1.
Distribution of Study Characteristics for 50 045 Subjects With (Cases) or Without (Controls) Staphylococcus aureus Infection in the Primary Phenotype Analysis Data Set
| Characteristic | Cases, No. (%) | Controls, No. (%) |
|---|---|---|
| Sex | ||
| Female | 2315 (49) | 23 322 (51) |
| Male | 2386 (51) | 22 022 (49) |
| Age at specimen collection date, y | ||
| 20–24 | 16 (<1) | 171 (<1) |
| 25–29 | 19 (<1) | 214 (<1) |
| 30–34 | 44 (1) | 446 (1) |
| 35–39 | 82 (2) | 822 (2) |
| 40–44 | 112 (2) | 1067 (2) |
| 45–49 | 194 (4) | 1892 (4) |
| 50–54 | 308 (7) | 3069 (7) |
| 55–59 | 461 (10) | 4625 (10) |
| 60–64 | 715 (15) | 7079 (16) |
| 65–69 | 666 (14) | 6654 (15) |
| 70–74 | 607 (13) | 6134 (14) |
| 75–79 | 629 (13) | 6133 (14) |
| ≥80 | 848 (18) | 7038 (15) |
| History of diabetes | ||
| Yes | 705 (15) | 5895 (13) |
| No | 3996 (85) | 39 449 (87) |
| History of cancer | ||
| Yes | 799 (17) | 7255 (16) |
| No | 3902 (83) | 38 089 (84) |
| HIV infected | ||
| Yes | 66 (1.4) | 136 (0.3) |
| No | 4635 (98.6) | 45 208 (99.7) |
| Source of S. aureus | ||
| Blood culture | 141 (3) | … |
| Cerebrospinal fluid or other fluid | 141 (3) | … |
| Urine culture | 282 (6) | … |
| Respiratory culture | 329 (7) | … |
| Tissue/biopsy culture | 940 (20) | … |
| Miscellaneous bacterial culture | 2868 (61) | … |
| Overall | 4701 (100) | 45 344 (100) |
Abbreviation: HIV, human immunodeficiency virus.
Prior to QC filtering, 50 576 subjects (2251 cases and 48 325 controls) were available for analysis of the secondary phenotype, community-acquired SSTI; 16 499 SNPs with call rates of <95%, 13 752 with a MAF of <0.9%, and 46 730 showing departure from HWE in controls were excluded, leaving 597 131 SNPs for data analysis. Eight subjects whose monozygotic twin was also in the study sample, 2915 subjects whose sample pair had a kinship coefficient of ≥0.0625, 54 controls with an ICD-9 code for S. aureus infection, 23 subjects who had a mismatch between self-reported and genetically determined sex, and 12 subjects who did not have estimates for European PC were removed. Our final analysis sample for the secondary phenotype consisted of 47 564 subjects (2130 cases and 45 434 controls).
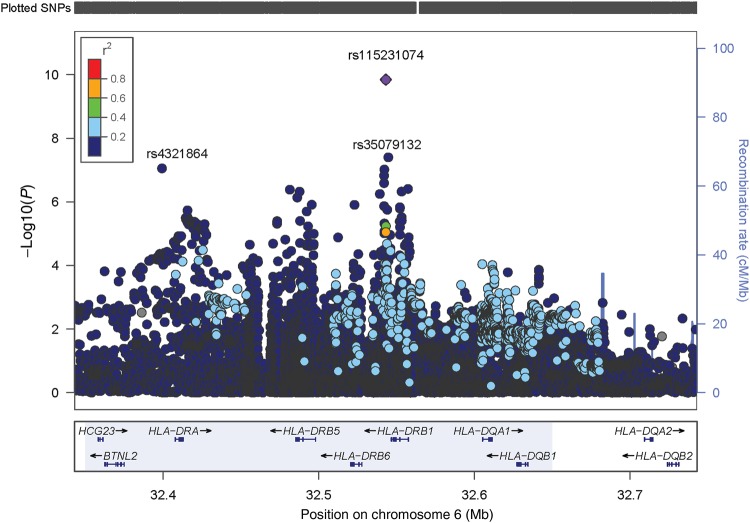
A Q-Q plot of the expected distribution of association test statistics across all genotyped SNPs in the primary phenotype analysis in comparison to observed P values is presented in Supplementary Figure 1; the genomic inflation factor was estimated to be 1.01, indicating adequate control of population stratification. Table 2 presents results of logistic regression analysis for both genotyped and imputed SNPs in relation to the primary and the secondary phenotype outcomes. The primary phenotype model revealed 1 genotyped SNP, rs4321864, that approached genome-wide significance (OR, 1.13; P = 8.85 × 10−8; Supplementary Figure 2). This variant is located in the HLA class II region of chromosome 6 (position: 32 399 187-32 399 187; band: 6p21.32). SNP rs4321864 appears to be at the 5′ terminus of the HLA-DRA gene. Given that rs4321864 approached the threshold (P ≤ 5.0 × 10−8) of genome-wide significance, we conducted additional regression analyses examining imputed SNPs on chromosome 6. Two imputed HLA class II variants were significantly associated with the primary phenotype (Table 2): imputed SNP rs115231074 (OR, 1.22; P = 1.3 × 10−10) and rs35079132 (OR, 1.24; P = 3.8 × 10−8); imputed SNP rs189516143 (OR, 1.21; P = 9.2 × 10−8) approached genome-wide significance. All 3 SNPs are intergenic near the HLA-DRB1 gene locus. Another imputed SNP, rs17210959 (OR, 1.23; P = 3.5 × 10−7), is located within the HLA-DRB1 gene. A focused view of the region in chromosome 6 where these SNPs are located is shown in Figure 1. All of these SNPs had a MAF of >0.10. A subanalysis in which MRSA infection was the outcome did not show any SNPs with P values that approached genome-wide significance (not shown). Also, analyses stratified by 3 large age groups did not show differences in SNP effect estimates across the age strata.
Table 2.
Final Results of Logistic Regression Analysis of Primary and Secondary Phenotypes Versus Genotyped and Imputed Single-Nucleotide Polymorphisms (SNPs), and Primary Phenotype vs Imputed HLA Serotype
| Model, SNP | Chromosome | Gene | Location | A1 | A2 | OR | P Valuea |
|---|---|---|---|---|---|---|---|
| Primary phenotype (all S. aureus infections) SNP association model | |||||||
| Genotyped | |||||||
| rs4321864 | 6 | HLA-DRA | 5′ end of gene | A | C | 1.13 | 8.8 × 10−8 |
| Imputed | |||||||
| rs115231074 | 6 | HLA-DRB1 | Intergenic: 3′ downstream of –DRB1 | T | C | 1.22 | 1.3 × 10−10 |
| rs35079132 | 6 | HLA-DRB1 | Intergenic | C | T | 1.24 | 3.8 × 10−8 |
| rs189516143 | 6 | HLA-DRB1 | Intergenic | A | T | 1.21 | 9.2 × 10−8 |
| rs190073676 | 6 | HLA-DRB1 | Intergenic | T | A | 1.18 | 1.6 × 10−7 |
| rs184932624 | 6 | HLA-DRB1 | Intergenic | T | C | 1.23 | 2.5 × 10−7 |
| rs17210959 | 6 | HLA-DRB1 | Within locus | A | G | 1.23 | 3.5 × 10−7 |
| Secondary phenotype (community-acquired SSTc S. aureus infections) SNP association model | |||||||
| Genotyped | |||||||
| rs4321864 | 6 | HLA-DRA | 5′ end of gene | A | C | 1.14 | 8.1 × 10−5 |
| Imputed | |||||||
| rs115231074 | 6 | HLA-DRB1 | Intergenic: 3′ downstream of –DRB1 | T | C | 1.28 | 5.2 × 10−8 |
| rs12526396 | 6 | … | Intergenic | G | A | 0.80 | 4.3 × 10−7 |
| Primary phenotype (all S. aureus infections) imputed HLA serotype association model | |||||||
| Imputed HLA serotype | … | … | … | … | |||
| HLA_DRB1_04 | … | … | … | Present | … | 1.08 | .01 |
| HLA_DRB1_0401 | … | … | … | Present | … | 1.08 | .04 |
| HLA_DRB1_0402 | … | … | … | Present | … | 1.36 | .002 |
Analyses were adjusted for age, sex, and the first 10 eigenvectors from a principal components analysis.
Abbreviations: A1, minor allele; A2, major allele; OR, odds ratio; S. aureus, Staphylococcus aureus; SST, skin and soft tissue.
a Threshold for genome-wide significance: P ≤ 5 × 10−8.
Figure 1.
Primary (all Staphylococcus aureus infections) phenotype: focused view of the locus of genotyped and imputed single-nucleotide polymorphisms (SNPs) between 32.3 and 32.5 Mb on chromosome 6. The plot also shows the SNP location in relation to known genes in that locus. Linkage disequilibrium of index SNP rs115231074 with other plotted SNPs is shown as a range of r2 values, in which red = 0.81–1.0, orange = 0.61–0.80, green = 0.41–0.60, light blue = 0.21–0.40, and dark blue = 0.01–0.20.
Secondary Analysis: Community-acquired SSTI
Results from the secondary phenotype analyses (community-acquired SSTIs) demonstrated that genotyped SNP rs4321864 did not approach genome-wide significance (P = 8.1 × 10−5). However, imputed SNP rs115231074 closely approached genome-wide significance (OR, 1.28; P = 5.2 × 10−8). No other genotyped or imputed SNPs approached genome-wide significance.
Sensitivity analysis was conducted for both the primary and secondary phenotypes by excluding subjects who had a history of diabetes diagnosis. Results were essentially the same as those for the analyses that included patients with diabetes.
HLA Serotype Association and Conditional Association Analysis
Table 2 also presents selected significant results from association testing for the imputed HLA serotypes. Given the significant association from the genotyped and imputed SNP data analysis in the HLA class II region, we examined the association of imputed HLA-DR serotype variants to primary phenotype to determine whether the imputed SNP association results could be attributed to the effect of 1 or more classical HLA haplotypes. Among several HLA-DRB1 serotypes significantly associated with the primary phenotype, HLA-DRB1*04 variants showed the largest effect estimates (eg, HLA-DRB1*0402: OR, 1.36 [P = .002]). We then conducted a conditional association analysis by fitting separate logistic regression models based on the most significant results from the genotyped and imputed SNP GWAS analyses conditioning on imputed HLA-DRB1*04 serotype variant (eg, logit [primary phenotype] = β1 rs4321864 + β2 HLA-DRB1*04 + β3 age-sex group + β4 PC1+ … + β14 PC10; Table 3). Results showed that, while the P value for genotyped SNP rs4321864 had increased, it still remained significant even after conditioning on imputed HLA-DRB1*04 serotype; the effect estimate was unchanged (OR, 1.14; 95% confidence interval, 1.07–1.20). Moreover, the P value and effect estimate for imputed SNP rs115231074 were virtually unchanged after adjustment for HLA-DRB1*04. These results indicate that the individual SNP associations are not due to LD with HLA class II haplotypes and provide further support for the association between both SNPs and the primary phenotype.
Table 3.
Results of Logistic Regression Analysis of Primary Phenotype vs Genotyped and Imputed Single-Nucleotide Polymorphisms (SNP) Conditioned on Imputed HLA-DRB1*04 Serotype
| HLA SNP | Chromosome | Gene | Location | OR | P |
|---|---|---|---|---|---|
| Genotyped | |||||
| rs4321864 | 6 | HLA-DRA | 5′ end of gene | 1.14 | 1.38 × 10−5 |
| Imputed | |||||
| rs115231074 | 6 | HLA-DRB1 | Intergenic: 3′ downstream of –DRB1 | 1.22 | 5.45 × 10−9 |
| rs35079132 | 6 | HLA-DRB1 | Intergenic | 1.24 | 1.72 × 10−6 |
Analyses were adjusted for imputed serotype HLA-DRB1*04, age, sex, and the first 10 eigenvectors from a principal components analysis.
Abbreviation: OR, odds ratio.
DISCUSSION
In the current study, we used a sample of >50 000 unique white subjects to identify 2 imputed SNPs (rs115231074 and rs35079132) located in the HLA class II region of chromosome 6 that achieved genome-wide significance in the primary phenotype GWAS analysis. These results strengthened our finding of genotyped SNP rs4321864, located on chromosome 6 near the 5′ terminus of the HLA-DRA gene, in the primary phenotype association testing. The secondary phenotype analysis also revealed a significant finding for imputed SNP rs115231074.
HLA-DRA encodes the sole α chain for the β chains encoded by HLA-DRB-1 and HLA-DRB-3, HLA-DRB-4, and HLA-DRB-5. Together, the proteins encoded by HLA-DRA and HLA-DRB genes form an antigen binding heterodimer that presents foreign peptides to trigger the immune response. A significant body of evidence supports the possibility that HLA class II haplotypes may influence human susceptibility to S. aureus infection. First, specific HLA haplotypes (HLAII DR14/DQ5) are associated with susceptibility to invasive Streptococcus pyogenes infection in patients [26] and determine the severity of response to bacterial superantigens from both S. pyogenes [27] and S. aureus [28]. Second, S. aureus superantigens, including toxic shock syndrome toxin (TSST-1), bind to the HLA II DR1 molecule [29, 30] and are critical in the development of S. aureus bacteremia and endocarditis [31, 32]. Third, nasal carriage of S. aureus is associated with the HLA-DR3 and HLA-DR7 class II serotypes [33]. Finally, polymorphisms in HLA-DRB1 are strongly associated with rheumatoid arthritis, an inflammatory disease characterized by a high risk of S. aureus infection [34–36]. The HLA-DR α chain is a relatively nonpolymorphic gene containing 5 exons and spanning approximately 33–35 kDa. Because of its relative stability, the positions in the HLA-DR β chain (HLA-DRB) appear to play a major role in binding to and presenting different antigens for recognition by T cells. Recognition of staphylococcal toxins and the subsequent immunologic response may be critical elements in determining infection. The intergenic location of the most significant imputed SNPs (rs115231074 and rs35079132) suggests that functional elements (eg, regulatory element) may exist in the region. Such variants might change the quantity of a specific variable chain produced, rather than its sequence. Alternatively these SNPs could be in LD with multiple variants that collectively constitute a functional haplotype. Finally, it is also possible that there is LD with genic variation that was not genotyped or imputed. Further study is therefore warranted.
Refining our primary end point to include only community-acquired SSTI did not result in more-significant findings. This may be due in part to the emergence of the highly virulent S. aureus USA300 clone [37] during the case ascertainment period. Unfortunately, bacterial genotyping was not available in this study.
Both our primary and secondary phenotype sample sizes were adequate to power GWAS testing that could detect relatively small risk-ratio estimates for common variants (MAF, >1%) at genome-wide significance. Because a GWAS relies on LD between common genotyped markers and relatively common causative variants, it generally has inadequate power to detect significant associations with rare causative variants. Much of the genetic component to common infectious disease may be attributable to the cumulative effects of many rare mutations with limited penetrance [38]. While rare mutations could account for much of the genetic component, our imputed SNP results support a consensus that common variants are a part of the genetic component for which there is evidence in other diseases/conditions [39–41].
Previously, we reported a case-control study of 361 white individuals with a diagnosis of S. aureus bacteremia who were matched to 699 controls [8]. That study did not find any genetic associations that reached genome-wide significance (P ≤ 5 × 10−8). Ye et al conducted a case-control design in which the outcome included all S. aureus infections (309 cases and 2925 controls) from a cohort of approximately 20 000 individuals of northern European ancestry [9]. That study also failed to identify any SNPs reaching genome-wide significance. More-targeted approaches, including murine sepsis models, have identified candidate genes associated with susceptibility to S. aureus infection in murine chromosomes [1, 2, 42]. Thus, the current study is the first to identify candidate polymorphisms associated with susceptibility to S. aureus infection at a genome-wide significant level.
Our study had limitations. First, the genotype of the infecting S. aureus isolates is not known. Specific S. aureus clones possess different combinations of virulence genes (fnbpA and fnbpB [43] and sasX [44]), human immune evasion clusters (scn and chp) [45], and enterotoxins that influence their ability to cause and continue infection in humans. In addition, specific nonsynonymous SNPs in key virulence genes have been shown to enhance [46] and reduce [47] bacterial virulence. Second, we were unable to control for certain environmental factors (eg, nutrition) that may influence how S. aureus interacts with host gene variants. Third, it is possible that some of the S. aureus respiratory isolates may have reflected colonization instead of active infection. However, this possibility would have led to a reduction in the difference between cases and controls. Fourth, our study population was limited to white subjects (see Supplementary 2 for further discussion of this topic). Thus, our findings cannot be generalized to other racial groups. Finally, we were unable to perform additional subgroup analyses in potentially important populations, such as patients with recurrent S. aureus infections.
Despite these limitations, our fully powered GWAS identified both genotyped and imputed genetic variants in the HLA class II region that are associated with susceptibility to S. aureus infection. These findings are independent of classical haplotype associations. Future studies using whole-genome sequencing experiments in patients with complicated and uncomplicated S. aureus bacteremia and admixture mapping studies to evaluate susceptibility to S. aureus in African American patients are currently underway. Further knowledge of host genetic response to S. aureus infection will contribute to our understanding and eventually inform our management of this serious, common infection.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the Research Program on Genes, Environment, and Health research team (Cathy Schaefer and Neil Risch, codirectors) for providing the genotyped single-nucleotide polymorphisms (SNPs), imputed SNPs, imputed human leukocyte antigen serotypes, and survey data, as well as advice during our study activities.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the NIH (grant 2R01-AI068804 to V. G. F.) and the National Center for Advancing Translational Sciences, NIH (award UL1TR001117).
Potential conflicts of interest. V. G. F. was chair of Merck's V710 scientific advisory committee; received grant support and has grants pending from the NIH, MedImmune, Forest/Cerexa, Pfizer, Merck, Advanced Liquid Logics, Theravance, Novartis, and Cubist; was a paid consultant for Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines, Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy, and Contrafect; received royalties from UpToDate; received payment for development of educational presentations from Green Cross, Cubist, Cerexa, Durata, and Theravance; and has a patent pending on bacterial diagnostics. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed
References
- 1.Ahn SH, Deshmukh H, Johnson N et al. Two genes on A/J chromosome 18 are associated with susceptibility to Staphylococcus aureus infection by combined microarray and QTL analyses. PLoS Pathog 2010; 6:e1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson NV, Ahn SH, Deshmukh H et al. Haplotype association mapping identifies a candidate gene region in mice infected with Staphylococcus aureus. G3 (Bethesda) 2012; 2:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griesbeck-Zilch B, Osman M, Kuhn C et al. Analysis of key molecules of the innate immune system in mammary epithelial cells isolated from marker-assisted and conventionally selected cattle. J Dairy Sci 2009; 92:4621–33. [DOI] [PubMed] [Google Scholar]
- 4.Bonnefont CM, Rainard P, Cunha P et al. Genetic susceptibility to S. aureus mastitis in sheep: differential expression of mammary epithelial cells in response to live bacteria or supernatant. Physiol Genomics 2012; 44:403–16. [DOI] [PubMed] [Google Scholar]
- 5.Zimakoff J, Rosdahl VT, Petersen W, Scheibel J. Recurrent staphylococcal furunculosis in families. Scand J Infect Dis 1988; 20:403–5. [DOI] [PubMed] [Google Scholar]
- 6.Holland SM, DeLeo FR, Elloumi HZ et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 2007; 357:1608–19. [DOI] [PubMed] [Google Scholar]
- 7.Tanabe F, Kasai H, He L et al. Improvement of deficient natural killer activity and delayed bactericidal activity by a thiol proteinase inhibitor, E-64-d, in leukocytes from Chediak-Higashi syndrome patients in vitro. Int Immunopharmacol 2009; 9:366–70. [DOI] [PubMed] [Google Scholar]
- 8.Nelson CL, Pelak K, Podgoreanu MV et al. A genome-wide association study of variants associated with acquisition of Staphylococcus aureus bacteremia in a healthcare setting. BMC Infect Dis 2014; 14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Z, Vasco DA, Carter TC, Brilliant MH, Schrodi SJ, Shukla SK. Genome wide association study of SNP-, gene-, and pathway-based approaches to identify genes influencing susceptibility to Staphylococcus aureus infections. Front Genet 2014; 5:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44:955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia X, Han B, Onengut-Gumuscu S et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013; 8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray GT, Suaya JA, Baxter R. Trends and characteristics of culture-confirmed Staphylococcus aureus infections in a large U.S. integrated health care organization. J Clin Microbiol 2012; 50:1950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shannon KP, French GL. Validation of the NCCLS proposal to use results only from the first isolate of a species per patient in the calculation of susceptibility frequencies. J Antimicrob Chemother 2002; 50:965–9. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Graber CJ, Karr M et al. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004–2005. Clin Infect Dis 2008; 46:1637–46. [DOI] [PubMed] [Google Scholar]
- 15.Frazer KA, Ballinger DG, Cox DR et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007; 449:851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banda Y, Kvale MN, Hoffmann TJ et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics 2015; 200:1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics 2010; 26:2867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porcu E, Sanna S, Fuchsberger C, Fritsche LG. Genotype imputation in genome-wide association studies. Curr Protoc Hum Genet 2013; 78:1.25.1–1.25.14. [DOI] [PubMed] [Google Scholar]
- 19.Delaneau O, Marchini J. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun 2014; 5:3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connell J, Gurdasani D, Delaneau O et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet 2014; 10:e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Bakker PI, McVean G, Sabeti PC et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006; 38:1166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007; 447:661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–9. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotb M, Norrby-Teglund A, McGeer A et al. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med 2002; 8:1398–404. [DOI] [PubMed] [Google Scholar]
- 27.Nooh MM, El-Gengehi N, Kansal R, David CS, Kotb M. HLA transgenic mice provide evidence for a direct and dominant role of HLA class II variation in modulating the severity of streptococcal sepsis. J Immunol 2007; 178:3076–83. [DOI] [PubMed] [Google Scholar]
- 28.Llewelyn M, Sriskandan S, Peakman M et al. HLA class II polymorphisms determine responses to bacterial superantigens. J Immunol 2004; 172:1719–26. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Urban RG, Strominger JL, Wiley DC. Toxic shock syndrome toxin-1 complexed with a class II major histocompatibility molecule HLA-DR1. Science 1994; 266:1870–4. [DOI] [PubMed] [Google Scholar]
- 30.Lavoie PM, Thibodeau J, Cloutier I, Busch R, Sekaly RP. Selective binding of bacterial toxins to major histocompatibility complex class II-expressing cells is controlled by invariant chain and HLA-DM. Proc Natl Acad Sci U S A 1997; 94:6892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salgado-Pabon W, Breshears L, Spaulding AR et al. Superantigens are critical for Staphylococcus aureus Infective endocarditis, sepsis, and acute kidney injury. MBio 2013; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol 2004; 186:2430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinsman OS, McKenna R, Noble WC. Association between histocompatability antigens (HLA) and nasal carriage of Staphylococcus aureus. J Med Microbiol 1983; 16:215–20. [DOI] [PubMed] [Google Scholar]
- 34.Suarez-Almazor ME, Tao S, Moustarah F, Russell AS, Maksymowych W. HLA-DR1, DR4, and DRB1 disease related subtypes in rheumatoid arthritis. Association with susceptibility but not severity in a city wide community based study. J Rheumatol 1995; 22:2027–33. [PubMed] [Google Scholar]
- 35.Thomson W, Harrison B, Ollier B et al. Quantifying the exact role of HLA-DRB1 alleles in susceptibility to inflammatory polyarthritis: results from a large, population-based study. Arthritis Rheum 1999; 42:757–62. [DOI] [PubMed] [Google Scholar]
- 36.Rau R, Herborn G, Zueger S, Fenner H. The effect of HLA-DRB1 genes, rheumatoid factor, and treatment on radiographic disease progression in rheumatoid arthritis over 6 years. J Rheumatol 2000; 27:2566–75. [PubMed] [Google Scholar]
- 37.Diep BA, Gill SR, Chang RF et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 2006; 367:731–9. [DOI] [PubMed] [Google Scholar]
- 38.Gorlov IP, Gorlova OY, Frazier ML, Spitz MR, Amos CI. Evolutionary evidence of the effect of rare variants on disease etiology. Clin Genet 2011; 79:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imamura M, Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives [Review]. Endocr J 2011; 58:723–39. [DOI] [PubMed] [Google Scholar]
- 40.Ishak MB, Giri VN. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol Biomarkers Prev 2011; 20:1599–610. [DOI] [PubMed] [Google Scholar]
- 41.Rafiq S, Khan S, Tapper W et al. A genome wide meta-analysis study for identification of common variation associated with breast cancer prognosis. PLoS One 2014; 9:e101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Q, Sharma-Kuinkel BK, Deshmukh H et al. Dusp3 and Psme3 are associated with murine susceptibility to Staphylococcus aureus infection and human sepsis. PLoS Pathog 2014; 10:e1004149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nienaber JJ, Sharma Kuinkel BK, Clarke-Pearson M et al. Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J Infect Dis 2011; 204:704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Du X, Villaruz AE et al. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med 2012; 18:816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spaan AN, Surewaard BG, Nijland R, van Strijp JA. Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu Rev Microbiol 2013; 67:629–50. [DOI] [PubMed] [Google Scholar]
- 46.Lower SK, Lamlertthon S, Casillas-Ituarte NN et al. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci U S A 2011; 108:18372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLeo FR, Kennedy AD, Chen L et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci U S A 2011; 108:18091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.