TO THE EDITOR
Basal cell carcinomas (BCCs) are the most common cancers in the United States, with an annual national incidence of ~ 2 million (Lomas et al., 2012). Although the majority are localized to the skin and cured by surgery, in rare cases, they can progress to advanced and metastatic tumors that result in severe morbidity and death. BCCs are typically caused by activating mutations in the sonic hedgehog (HH) pathway, most commonly through loss of the receptor Patched1 (PTCH1) or activation of the G-protein-coupled receptor Smoothened (SMO; Epstein, 2008). Here, we report the presence of Kinastrin (kinetochore-localized astrin/SPAG5 binding protein (KNSTRN)) mutations in BCC. KNSTRN encodes a kinetochore-associated protein that is an essential component of the mitotic spindle and is required for faithful chromosomal segregation during mitosis (Fang et al., 2009). It is expressed in a broad range of tissues, including skin, and its mutations have been detected in both squamous-cell carcinoma (SCC) and melanoma, leading to its recent classification as an oncogene (Lee et al., 2014).
In order to elucidate the role of KNSTRN in BCC, we interrogated 18 advanced (inoperable and >3 cm in size) and 30 early stage (<2 cm in size) BCCs for mutations in KNSTRN (Stanford Human Subjects panel approval and subsequent written informed consent was obtained from patients for tumor sequencing (Protocol 18325)). Among the advanced BCCs, whole-exome sequencing revealed a mean of 1,428 somatic coding mutations per tumor, equivalent to a nonsynonymous single-nucleotide variant rate of 47.6 per Mb (range 5.5–125.6 per Mb), which is in the low range of previous reports (Jayaraman et al., 2014; Atwood et al., 2015). In addition, all tumors had a mutational profile characteristic of exposure to UV radiation, with C → T and A → T transitions present in 68% of alterations (SD 21%; Figure 1a). In all, 72% of samples harbored mutations in PTCH1 or SMO, consistent with the known pathogenesis of BCC (Figure 1a and b; Jayaraman et al., 2014; Atwood et al., 2015).
Figure 1. The spectrum of basal cell carcinomas (BCCs) which harbor kinetochore-localized astrin/SPAG5 binding protein (KNSTRN) mutations.
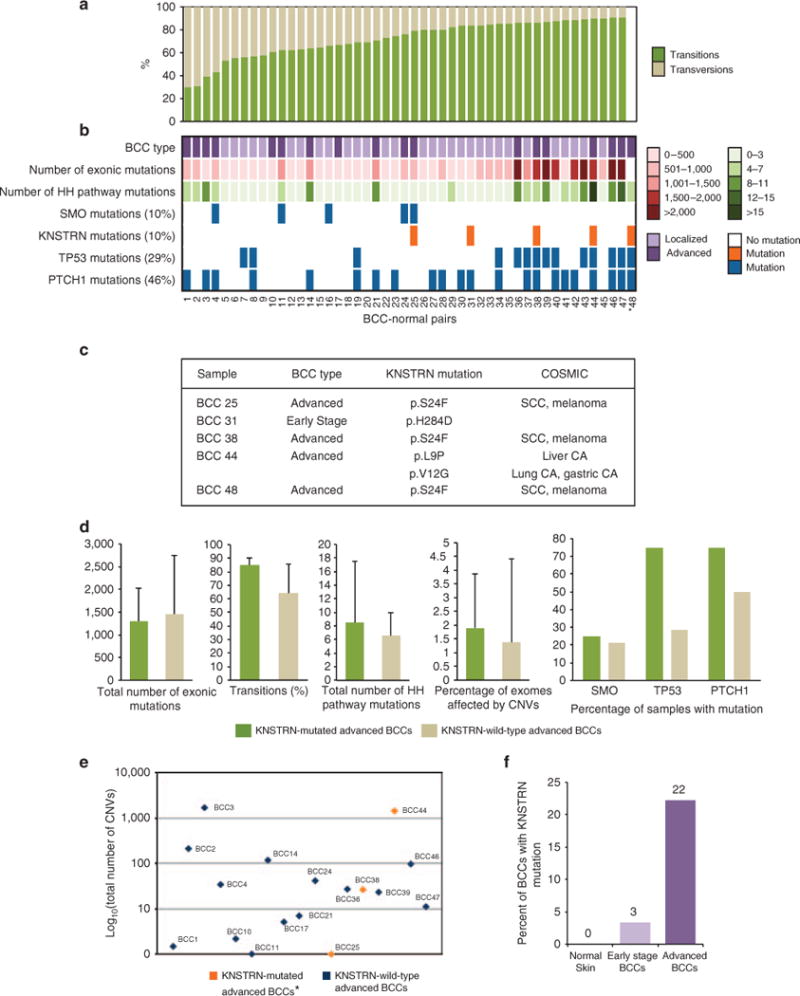
(a) The percentage of transitions and transversions in 48 early stage and advanced BCCs. (b) Characteristics of the 48 BCCs. (c) Table of KNSTRN point mutations identified in BCCs and Catalogue of Somatic Mutations in Cancer (COSMIC) database annotation. (d) Comparison of mean exonic mutations, sonic hedgehog (HH)-pathway mutations, transitions, copy-number variations (CNVs), and SMO, TP53, and PTCH1 mutations in KNSTRN-mutated advanced BCCs and KNSTRN-wild-type advanced BCCs. Error bars represent ± SD (e) Scatterplot showing the total number of CNVs detected in each advanced BCC. (f) Percent of samples with mutations in KNSTRN for normal skin (n = 48), early stage BCCs (n = 30) and advanced BCCs (n = 18). *BCC48 lacked a paired normal sample, so certain calculations could not be performed (see Supplementary Methods online).
Interestingly, 4 advanced BCCs (21%) contained somatic mutations in KNSTRN (Figure 1a and c). Three harbored the KNSTRN p.S24F mutation, which has previously been detected in both SCC and melanoma (Lee et al., 2014). The affected region of KNSTRN corresponds to a UV-signature hotspot and involves a C → T transition characteristic of UV-induced mutagenesis. KNSTRN p. S24F is predicted to be deleterious by SIFT (Ng and Henikoff, 2001) and Polyphen (Sunyaev et al., 2000), and disrupts chromatid cohesion, increases aneuploidy, and promotes Ras-driven tumorigenesis in cell-based assays of keratinocytes and in vivo mouse models (Lee et al., 2014). The fourth advanced BCC contained two KNSTRN mutations, p.L9P and p.V12G, both of which were predicted to be non-damaging by SIFT and Polyphen; however, identical mutations have been detected in liver, lung, and gastric cancer, supporting a role in tumorigenesis. KNSTRN p.S24F, p.L9P, and p.V12G have all been reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) and are absent from dbSNP137 and ESP6500 (Figure 1c).
Compared with KNSTRN-wild-type advanced BCCs, KNSTRN-mutated advanced BCCs did not have a significant difference in average mutational load or mean number of HH pathway mutations, suggesting that HH pathway-driven tumorigenesis is not diminished or replaced by downstream effects from mutant KNSTRN (Figure 1a and d; Supplementary Figure 1 and Supplementary Table 1 online). However, KNSTRN-mutated BCCs appeared to have a higher rate of transitions (85 vs. 64% transitions, P = 0.003), supporting a role for KNSTRN in UV-related mutagenesis (Figure 1d and Supplementary Table 1 online).
Mutant KNSTRN has been functionally shown to disrupt sister chromatid cohesion and chromosome segregation in keratinocytes; in KNSTRN-mutated SCCs, this results in enhanced tumor aneuploidy and increased genomic copy-number aberrations (Lee et al., 2014). To evaluate the functional relevance of mutated KNSTRN in BCC, we expressed wild-type- or S24F-mutated KNSTRN in a murine BCC cell line (Figure 2a) and assessed chromosome segregation during mitosis. Interestingly, we found that mutant KNSTRN expression disrupted sister chromatid cohesion, as demonstrated by a higher percentage of unpaired sister chromatids in these cells (Figure 2b and c). In conjunction with similar functional studies in primary human keratinocytes and SCCs (Lee et al., 2014), these data support a role for KNSTRN in maintaining chromosomal stability. To investigate large-scale genomic instability and tumor aneuploidy in our advanced BCCs, we employed a Bayesian-based algorithm, BIC-seq, to detect the copy-number variations (CNVs) in our samples (Xi et al., 2011). PTCH1 deletions were detected in 12% of tumors, consistent with previous reports (Jayaraman et al., 2014). There was a wide range in the number of CNVs among both KNSTRN-mutated and KNSTRN-wild-type samples (Figure 1e), suggesting that advanced BCCs have a large range of genetic instability. On average, 1.9% (SD 2.0%) of the exome was affected by CNVs in KNSTRN-mutated tumors versus 1.4% (SD 3.0%) in KNSTRN-wild-type tumors (P = 0.74). Although this was not statistically significant, our ability to detect differences may have been limited by a small sample size.
Figure 2. S24F kinetochore-localized astrin/SPAG5 binding protein (KNSTRN) disrupts sister chromatid cohesion in a murine basal cell carcinoma (BCC) cell model.
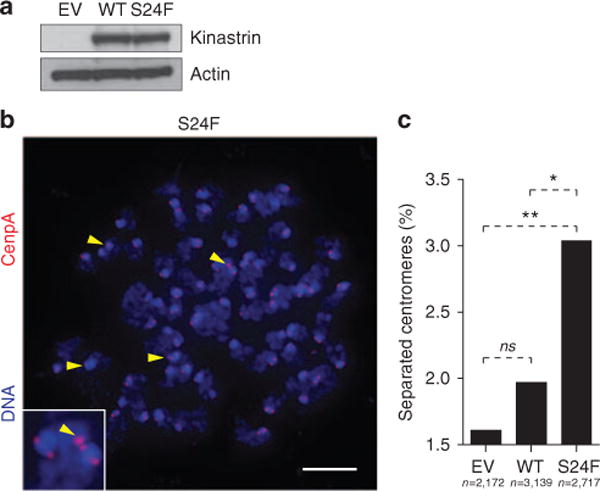
(a) Western blot analysis of ASZ001 cells following transduction with empty vector (EV), wild-type Kinastrin (WT), or S24F mutant Kinastrin (S24F). The level of enforced protein expression used to assess sister chromatid cohesion is shown. (b) Disrupted sister chromatid cohesion in ASZ001 cells transduced to express EV, WT, or S24F Kinastrin. Arrowheads mark unpaired chromatids. Scale bar = 5 μm. (c) Quantification of unpaired chromatids. *P = 0.04, **P = 0.005. ns, not significant.
In SCC, KNSTRN p.S24F is present in 19% of tumor precursors, suggesting that it arises early in disease progression (Lee et al., 2014). To determine whether KNSTRN mutagenesis is an early event in BCC development as well, we screened 30 early stage BCCs for KNSTRN mutations. We identified a nonsynonymous KNSTRN mutation in only 1/30 (3%) early stage BCCs, suggesting that, unlike in SCC, mutant KNSTRN in BCC appears to be acquired later in disease and is possibly a marker of aggressive behavior (Figure 1c and f). The mutation, pH284D, is absent from dbSNP137 and ESP6500 and is predicted to be deleterious by SIFT; however, it has not previously been reported in the COSMIC database and is not predicted to be damaging by Polyphen.
These findings are the first to implicate KNSTRN in BCC tumorigenesis. Alongside recent data offering a role for KNSTRN in SCC and melanoma, our work supports the classification of KNSTRN as an oncogene and an important contributor to the pathogenesis of malignancies related to UV-exposure. In both SCC and BCC, mutant KNSTRN disrupts sister chromatid cohesion and promotes genomic instability in functional assays. However, unlike in SCC, KNSTRN mutations in BCC appear to occur late in disease progression and are preferentially found in advanced tumors. Further exploration of the role of KNSTRN in skin cancer and genomic stability is warranted.
Supplementary Material
Acknowledgments
This work was funded by the V Foundation Translational Award, NIAMS (5ARO54780, 2AR046786), NIH Pathway to Independence Award 1K99CA176847 (SXA), the Damon Runyon Clinical Investigator Award (JYT), Stanford Medical Scholars Program (PDJ) and the Dermatology Foundation Career Development Award (KYS).
Abbreviations
- BCC
basal cell carcinoma
- CNV
copy-number variation
- COSMIC
Catalogue of Somatic Mutations in Cancer
- HH
sonic hedgehog pathway
- KNSTRN
kinetochore-localized astrin/SPAG5 binding protein
- PTCH1
patched 1
- SCC
squamous-cell carcinoma
- SMO
smoothened
- TP53
tumor protein 53
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
References
- Atwood SX, Sarin KY, Whitson RJ, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27:342–53. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer. 2008;8:743–54. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Seki A, Fang G. SKAP associates with kinetochores and promotes the metaphase-to-anaphase transition. Cell Cycle. 2009;8:2819–27. doi: 10.4161/cc.8.17.9514. [DOI] [PubMed] [Google Scholar]
- Jayaraman SS, Rayhan DJ, Hazany S, et al. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Investig Dermatol Symp Proc. 2014;134:213–20. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- Lee CS, Bhaduri A, Mah A, et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat Genet. 2014;46:1060–2. doi: 10.1038/ng.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012;166:1069–80. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- Xi R, Hadjipanayis AG, Luquette LJ, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci USA. 2011;108:E1128–36. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


