Abstract
T lymphocyte development branches off from other lymphoid developmental programs through its requirement for sustained environmental signals through the Notch pathway. In the thymus, Notch signaling induces a succession of T-lineage regulatory factors that collectively create the T-cell identity through distinct steps. This process involves both the staged activation of T-cell identity genes and the staged repression of progenitor-cell-inherited regulatory genes once their roles in self-renewal and population expansion are no longer needed. With the recent characterization of Innate Lymphoid Cells (ILCs) that share transcriptional regulation programs extensively with T cell subsets, T-cell identity can increasingly be seen as defined in modular terms, as the processes selecting and actuating effector function are potentially detachable from the processes generating and selecting clonally unique T-cell receptor structures. The developmental pathways of different classes of T cells and ILCs are distinguished by the numbers of prerequisites of gene rearrangement, selection, and antigen contact before the cells gain access to nearly-common regulatory mechanisms for choosing effector function. Here, the major classes of transcription factors that interact with Notch signals during T-lineage specification are discussed in terms of their roles in these programs, the evidence for their spectra of target genes at different stages, and their cross-regulatory and cooperative actions with each other. Specific topics include Notch modulation of PU.1 and GATA-3, PU.1-Notch competition, the relationship between PU.1 and GATA-3, and the roles of E proteins, Bcl11b, and GATA-3 in guiding acquisition of T-cell identity while avoiding redirection to an ILC fate.
Keywords: T cell development, transcription factor, gene regulation, Notch, GATA-3, PU.1, E2A
I. Introduction: T-cell identity and processing of T-cell progenitors
A. T-cell development in the context of hematopoiesis
Hematopoietic stem and progenitor cells generate an exceptional diversity of cell types throughout life, and this poses a series of challenges for explanation of developmental dynamics, developmental choice hierarchies, and the mechanisms used to make each of the myriad differentiation programs remain coherent. T cell development is one of the most interesting and complex of the pathways in this system. Hematopoietic cells have traditionally been divided into erythroid/megakaryocytic, myeloid, and lymphoid branches, and T cells have been considered a subspecies of adaptive immune cells that splits off from B cells only once their joint lymphoid fate is confirmed. This classic view has been based on the unique antigen receptor generation strategy based on somatic mutation that T and B cells share, and on the early separation that seemed to occur between precursors that could generate lymphocytes and precursors that could generate all other hematopoietic cell types. However, a more subtle picture of cell type identity and of lineage relationships has emerged in the past decade as increasing knowledge has been gained about the dynamics of lineage-specific developmental processes and the transcriptional regulatory apparatus that drives them. T cell development has turned out to be a particularly revealing branch of hematopoiesis, because the intermediates in the process leading to T-cell commitment are readily isolated, characterized, and then monitored kinetically if their development is allowed to resume. The results from T-cell development have been important to catalyze new understandings of hematopoietic hierarchy in general, and the relationship between T cell fates and other lymphoid and myeloid fates alike.
T-cell development is distinctive within the hematopoietic families both because of its site and because of the nature of the cells produced. The inductive microenvironment needed for T-cell development is not available in the bone marrow or fetal liver. Virtually all known T-cell development occurs in the thymus, where multipotent progenitors immigrate at a low rate. Despite a low absolute cell input per day, the precursors begin to proliferate massively as they begin the T-cell program under the influence of thymic environmental signals, mostly interaction of Notch1 receptors on the surfaces of the hematopoietic cells with Delta-like 4 (DLL4) Notch ligands on the thymic stroma. The thymus then serves both as a development zone and as an immunological filtering zone, where the cells are tested for their newly-expressed antigen-recognition specificities. All are killed but those with the most useful specificities (defined below), and only the cells that have passed through this selection are exported to the periphery to serve in immune responses. Unlike many other types of hematopoietic cells, but like their B-lymphocyte cousins, surviving T cells preserve the potential for extensive cell division when they are triggered as well as the ability to remain quiescent for long periods of time between stimulation. They thus maintain features associated with stem-ness as well as the mature functional properties they acquire through differentiation.
The basic elements of T-cell development and the genes required for T-cell development to proceed have been characterized as summarized in multiple reviews (De Obaldia and Bhandoola, 2015; Miyazaki et al., 2014; Naito et al., 2011; Rothenberg, 2014; Thompson and Zúñiga-Pflücker, 2011; Yui and Rothenberg, 2014). However, for the most part specific regulatory genes have been studied as isolated influences that are characterized only in terms of “necessity” for the T-cell developmental pathway at one stage or another. In this chapter, we not only review what is known of the multiple transcriptional regulators that regulate this process, but also relate the ways that their roles intersect with each other, especially as subcircuits of specific factors can be viewed as elements of potential modules of functional programming.
B. Origins of T-cell precursors and the initial steps of T-cell development
In mammals, T cell precursors enter the thymus most prominently between the last half of fetal life and puberty. The epithelial structure of the thymus forms in mid-gestation by an outpocketing of branchial cleft endoderm and begins to be populated by hematopoietic progenitors that migrate through disorganized mesenchyme even before the thymus is vascularized. Once blood vessels supply the thymus, there are multiple successive waves of hematopoietic immigration. The cells coming into the thymus in the first wave may be highly biased toward a T-cell fate (Harman et al., 2005; Masuda et al., 2005; Ramond et al., 2014), but by later in gestation and through postnatal life, the thymic immigrants enter as multipotent cells (Ramond et al., 2014). Within the thymus, the strong environmental signals force the cells into the T-cell pathway or pathways, through the mechanisms that are the focus of this chapter. However, before reaching the thymus, the kinds of cells that enter have a constellation of alternative potentials that include lymphoid, myeloid, and dendritic-cell alternatives. There is considerable evidence that both lymphoid-biased precursors (Common Lymphoid Progenitors, CLP) and more balanced lymphomyeloid precursors (Lymphoid-primed MultiPotent Precursors, LMPP) can enter the thymus and give rise to T cells through a common pathway, although the LMPPs take somewhat longer (Saran et al., 2010; Serwold et al., 2009). An important confirmation that the cells enter the thymus as multipotent cells is that even after the initial stage(s) of T cell development, it is still possible to remove early T-cell precursors from the Notch-signaling environment of the thymus and show that they can still give rise to a variety of non-T cells (Bell and Bhandoola, 2008; Benz and Bleul, 2005; Lu et al., 2005; Luc et al., 2012; Sambandam et al., 2005; Tan et al., 2005; Wada et al., 2008). Intrathymically, such early T cell precursors are actively responding to the Notch signaling environment as they begin to turn on T-cell genes, but their status remains reversible at this point: they have not yet internalized mechanisms cutting off their alternative developmental potential. This plasticity is significant because there is a discrete point in T-cell development, a little later, at which they lose their intrinsic potential to access the alternative fates (Masuda et al., 2007; Yui et al., 2010). That transition is what is referred to in the following as “commitment”.
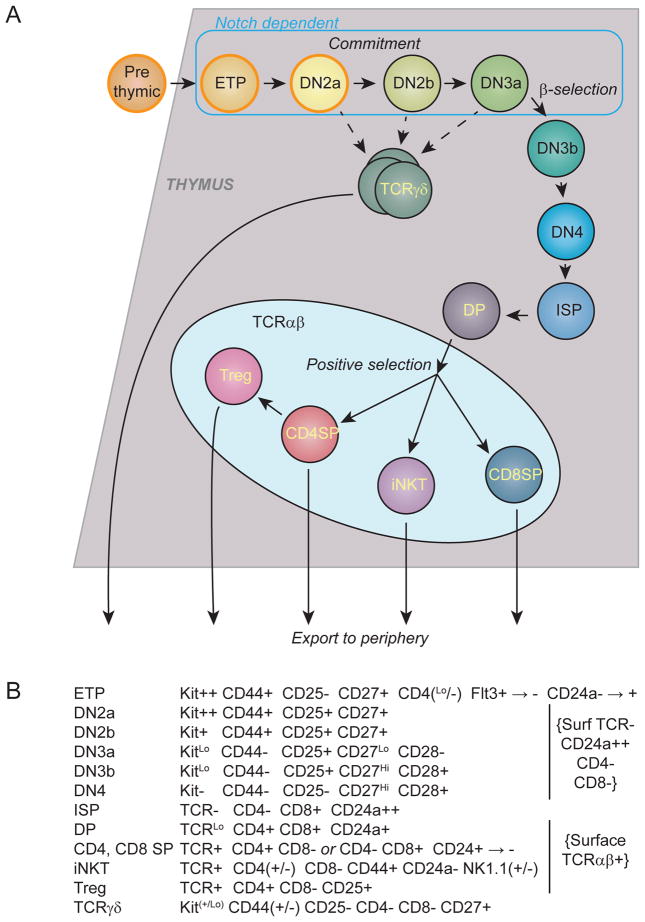
Fig. 1A shows the details of the pathway through which cells progress to expression of a T-cell receptor complex. The initial stages are called “DN” to signify that they are “double negative” for expression of either of the two T-cell coreceptors, CD4 and CD8. In mice, these stages are subdivided according to their expression of other surface molecules, the growth factor receptors Flt3 and Kit, the adhesion molecule CD44, and the solitary IL-2R α chain (CD25)(Fig. 1B). In humans, the subdivisions are made based on expression of different molecules: CD34, CD7, and CD1a (Vicente et al., 2010); however, most of this discussion will focus on the mouse where relationships and mechanisms have been easier to dissect using mouse genetics and in vivo cell transfers. Within the first population [Kit+ DN1, or “Early T-cell Precursors” (ETP)], the cells that enter the thymus in each wave, initially called thymus-settling progenitors, still express Flt3, but rapidly downregulate this while maintaining or upregulating Kit expression. The resulting cells appear to undergo multiple rounds of cell division under the influence of Notch pathway signaling before progressing to the next recognizable stage. CD25 expression is finally activated by the Notch signals, and this signals the beginning of the DN2 stage. DN2 cells initially proliferate even faster than ETPs, but then go through a transition, at first barely detectable, at which they downregulate Kit expression and start to slow down their proliferation. Although the phenotypic shift is subtle (called DN2a to DN2b), this turns out to signal a major regulatory transition that coincides with lineage commitment. The cells begin to shut off CD44 expression, downregulate Kit almost completely, and slow their proliferation to the point of G1 arrest, at the DN3a stage. At this point the enzymes required to rearrange the TCR genes are strongly upregulated, and the cells await the successful expression of either a TCRβ chain or the combination of TCRγ and TCRδ chains.
Figure 1. Stages of intrathymic T cell development.
A) Scheme of stages of mouse T-cell development discussed in the text, showing the stages affected by Notch-DLL4 signaling, intrathymic branchpoints in development, and the timing of three watershed events: commitment, β-selection, and positive selection.
B) Markers used to define the stages shown in A.
Unlike all other essential T cell genes, TCR genes require more than transcriptional regulation. Each chain is encoded in the genome in multiple segments and depends on success of highly error-prone somatic gene rearrangements to generate a full TCR coding sequence, ultimately yielding TCR heterodimers composed either of intact TCRβ and TCRα chains or of TCRγ and TCRδ chains. For the majority of DN3a cells, further survival depends on in-frame TCRβ rearrangements. When this occurs, TCRβ chains assemble with a pre-synthesized set of TCR complex components to trigger a strong burst of proliferation and phenotypic transformation called β-selection. The cells sweep through a succession of states distinguished as DN3b, DN4, and immature single positive as they proliferate on their way to acquire a CD4+ CD8+ Double Positive (DP) phenotype (Fig. 1A). This is a major change in cellular transcriptional and regulatory state, and the resulting cells, the DP cells, finally stop proliferating and accumulate in a G0 state once more to rearrange their TCRα genes.
DP cells are allowed about 3–4 days of survival in which to assemble a TCRαβ complex that successfully forms a low-affinity interaction with major histocompatibility complex (MHC) molecules on the surrounding stroma (positive selection). Most of the cells normally die unsuccessful. For those that survive, the TCR interaction specificity with class II or class I MHC, respectively, determines whether they will be positively selected to become mainstream CD4 cells or CD8 cells, or some form of innate-like T cells (e.g. iNKT cells) with a noncanonical specificity. These choices lead the selected cells to maturation in the thymic medulla, further negative selection against strongly self-reactive cells, and finally export of the surviving cells to the periphery.
If TCRγδ is successfully rearranged first in DN cells, a shorter burst of proliferation is triggered than by TCRβ rearrangement, but this too causes recombinase inhibition and sets in train a series of events that ultimately make the receptor choices final. Interestingly, the TCRγδ lineage opens the door to a variety of effector programs that differ in certain respects from those of TCRαβ subsets (see Section I.C.2). If γδ vs. αβ lineage choice were only determined stochastically, by the luck of rearrangement, then lineage fate should only be settled at the DN3a stage. However, both genetic and single-cell tests of differentiation in vitro show that by the time cells reach the DN3a stage, ready to carry out the definitive V-DJ rearrangements of their TCRβ genes, their ability to give rise to γδ cells has contracted to a small fraction of what it was earlier (Ciofani et al., 2006; Feng et al., 2011; Shibata et al., 2014). This implies that many cells with γδ potential leave the mainstream after the early DN2 stages, even before the peak of TCR gene rearrangement activity. If so, then transcriptional changes involving differential expression of key regulatory genes (Haks et al., 2005; Melichar et al., 2007; Narayan et al., 2012) may also contribute to bias the outcome of the γδ vs. αβ lineage choice even before completion of rearrangement.
C. Definition of T cell maturity in gene expression terms
1. Pan-T cell genes
The gene regulatory events that operate during T cell differentiation must endow T cells with a combination of pan-T characteristics and subset-specific characteristics. Pan-T characteristics confer the cells’ ability to recognize antigen with a T-cell receptor (TCR) complex and trigger a response. Thus, all T cells express either an αβ or γδ TCR, assembly partner proteins CD3γ, δ, and ε and TCRζ (CD247) that enable it to form a signaling-competent complex, and the specialized set of signaling adaptors, kinases and phosphatases that transduce activation signals when TCR ligands are engaged. Finally, all T cells grow and maintain viability throughout both resting and immune response phases by cytokine stimulation through receptors of the γc (Il2rg) family. Most T cells must therefore express γc robustly in order to survive, together with a variety of partner chains that assemble with it to complete the IL-7, IL-2, IL-4, IL-15, IL-9, or IL-21 receptors (Yamane and Paul, 2012). While γc expression and its signaling mediators are constitutive in T cells, the roles of the alternative partners for γc are more dynamic and subset-specific, as indicated below for subset-specific effector genes.
Because gene rearrangement is required to enable the cells to express a TCR, all T cells must go through a phase when they also express the lymphoid-specific recombinase enzymes RAG1 and RAG2, and usually terminal deoxynucleotidyl transferase (encoded by Dntt) to make the process more mutagenic as well. RAG1-RAG2 complexes are indispensable to catalyze the recombination of TCR V, sometimes D, and J segments to assemble TCR coding sequences. These enzymes collaborate with DNA-dependent protein kinase (encoded by Prkdc), Artemis (Dclre1c), and DNA ligase 4 (Lig4) and other canonical components of the nonhomologous end-joining pathway in order to complete the TCR gene segment joining process. For T cells, the phase of somatic mutation is transient and unique to their intrathymic development; mature T cells can no longer alter their TCR coding sequences. However, their existence depends on the earlier, precisely regulated hit-and-run activity of these mutagenic enzymes.
Signaling molecules required by all mature T cells include the Src-family kinase Lck, the ζ-associated tyrosine protein kinase Zap70, the adaptors LAT and Slp76 (Lcp2), Ca2+-activated TEC family kinase Itk, the tyrosine protein phosphatases CD45 and Csk, and additional adaptors including Grap2 (GADS). Most T cells also express the costimulatory receptor CD28 and coreceptors CD4 or CD8α and CD8β. These signaling components couple to more broadly expressed signaling partners such as 4, 5-phosphatidylinositide 3-kinase (PI3K), Akt, Ras, Raf, multiple MAP kinases, various isoforms of protein kinase C, Orai and CRAC Ca2+ channel components, Target of Rapamycin (TOR) complexes 1 and 2, and the transcription factors that these signaling molecules can trigger. Again, some subsets use additional signaling components, such as SLAM Associated Protein (SAP, Sh2d1), but these are specialized requirements. Finally, there are additional signaling molecules such as the multiple related GTPases of the IMAP family (Gimap1–9), most of which are strongly expressed in a mature T-cell restricted way and maintain cell viability through pathways that are less well understood (Schwartzberg et al., 2009; Wang et al., 2010; Yano et al., 2014).
To summarize, at a minimum, the core T-cell developmental program must do two things. It must induce the properly timed bursts of RAG1 and RAG2 expression and allow their silencing, and it must set up stable expression of the transcription factors and genomic accessibility states needed to induce and sustain expression of the broad set of pan-T cell genes. These are requirements that apply to essentially all T cells of all subsets.
2. Subset-specific genes
T cell maturity is actually a collection of diverse states, because the T cell developmental pathway is highly branched. T-cell precursors encounter a succession of developmental choice points both during their development in the thymus and after they leave the thymus The effector programs as they are modulated in the peripheral immune response are of extreme clinical interest and have become major fields in themselves. But some points about this effector specialization are important to note, even in simplified form. First, subset specialization choices are made at a variety of developmental stages (Fig. 2). Second, one large fate decision or group of fate decisions has to do with the specific form of the TCR that is to be expressed: γδ or αβ. The γδ vs. αβ decision is made without actually triggering the TCR; however, the relative outputs of cells that have taken these divergent pathways do depend on TCR engagement. Third, all subsequent choices within the T-cell pathway require TCR engagement as one component of the trigger to drive the cells to one fate or another. Fourth, effector subset choice involves a constellation of gene regulation alterations: the cells must express an appropriate γc-containing growth factor receptor; they must set up a poised state in which a specific set of effector genes becomes inducible; and they must do this by establishing stable expression of a network of subset-specific transcription factors.
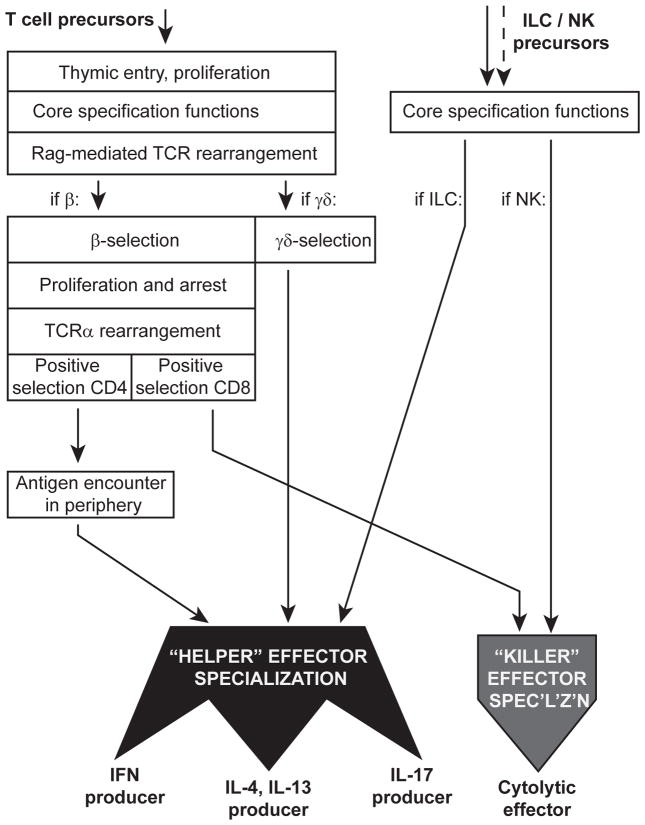
Figure 2. Modularity of the T-cell developmental program: differential access to effector specialization functions depending on choice of T-cell or innate-cell lineage.
The figure depicts a surprisingly common set of regulatory programming used to distinguish “killer” and different “helper” subtypes of effector T cells and innate lymphoid cells (ILC and natural killers, NK), and the layers of developmental programming that cells must undergo before gaining access to these common programs. Note that TCRαβ lineage T cells have a highly protracted, multistep pathway requirement before they can access the specialization functions, in contrast to ILC and NK cells. As described in the text, TCRγδ cells appear to be intermediate between the extreme of the longer path taken by CD4+ αβ T cells and the apparently short paths taken by ILCs.
Subset-specific effector genes often encode cytokines that are used to stimulate or inhibit other cells (IL-4, IFNγ, IL-17, IL-10), chemokine receptors to guide migration preferences, specific costimulatory molecules and auxiliary cytokine receptors, and sometimes cytolytic factors (granzyme B, perforin). Importantly, under normal conditions without antigen challenge, many effector genes remain silent in specific T-cell subsets despite being primed for expression. However, in simplest terms Th1 cells are defined by their ability to express IFNγ on stimulation; Th2 cells, by their ability to express IL-4 (and IL-13); Th17 cells, by their ability to express IL-17a; and cytolytic T cells, by their ability to express granzyme B and perforin.
Effector programs depend on an endogenous transcriptional regulatory state that sets the default gene expression preference of the cells, plus specific real-time interactions with STAT (signal transducers and activators of transcription) family factors that convey dynamic signals to the nucleus from most different kinds of cytokine receptors. STAT factors collaborate with stably expressed factors in the cells to define the set of enhancers across the genome that are available for activation at any given time (Vahedi et al., 2013; Vahedi et al., 2012). Actual transcription of the effector genes is then triggered by signals through the T-cell receptor, which are mediated by stereotypical signal transduction factors of the NF-κB, NFAT, and AP-1 families. The sustained regulatory states that distinguish Th cells of different classes from each other between bouts of stimulation are defined by specific “lineage-determining” factors (sometimes romantically called “master regulators”). Each effector subclass is characterized by at least one of these stably expressed lineage determining transcription factors and at least one specific type of growth factor receptor capable of mobilizing the right STATs to collaborate with the lineage determining factors (Murphy and Stockinger, 2010; O’Shea and Paul, 2010; Oestreich and Weinmann, 2012; Vahedi et al., 2013). Thus, very broadly, the Th1 state is characterized by T-bet collaborating with STAT1 and STAT4, the Th2 state is characterized by GATA-3 collaborating with STAT6, the Th17 state by RORγt (Rorc isoform) collaborating with STAT3, the Tfh state by Bcl6 collaborating with STAT3, and the Treg state by Foxp3 supported by STAT5. In CD8 cytolytic cells, T-bet or Eomes and Runx3 collaborate with STAT1 and STAT5; and there are also some alternative CD8 activation states which appear to be regulated roughly analogously to the CD4 states.
In depth, the alternative effector types in fact specialize through somewhat more complex regulatory programs, and different effector programs can antagonize each other’s operation to create fairly coherent alternative regulatory states (Ciofani et al., 2012; Naito and Taniuchi, 2010; O’Shea and Paul, 2010; Yosef et al., 2013). Importantly, the branching of these alternatives follows a similar pattern whenever cells become specialized for effector function, even though this can happen at a variety of developmental stages (Fig. 2; and section I.D). As described below, there are ways for developing lymphocytes to arm themselves with these effector functions even without their expressing a TCR at all. One key question, then, is what the threshold may be that cells need to cross before they are able to specialize for effector function.
D. Programming for T-cell effector function: intrathymic and postthymic
One notable feature of the transcriptional regulation of effector subset identity is that at least some of the “lineage determining factors” have more than one role in T cell development, working at different stages. RORγt in peripheral T cells is crucial for the Th17 fate, downregulated in the Th1 fate and excluded from the programs for the Th1 and Treg fates; yet it is earlier expressed by all TCRαβ cells during the DP stage. GATA-3 is not only the major regulator of the Th2 fate, but also critical for the choice of all CD4 cell fates during positive selection. It is also indispensable for all DN cell survival in the ETP stage, for differentiation, and for survival from DN2 to DN3 and during β-selection as discussed at length in Section III.C.1. The DN stages and the transition to the DP stage are the periods when pan-T cell features are programmed uniquely. Thus, most if not all regulatory gene activity in these early stages, including roles of factors like GATA-3 and RORγt, is likely to be conferring functions that the cell will use no matter what future effector function it chooses. The roles of the same factors in selection of effector functions are exerted later, through a series of asynchronous branchpoints.
For most TCRαβ T cells, there is a large divide between the functional programming of CD8 cells, future killers, and CD4 cells, mostly future helpers. This separation occurs in the thymus shortly after successful TCRα rearrangement (Fig. 2), as a result of the interaction of the TCR and CD8 or CD4 coreceptors on DP cells with class I or class II MHC in the thymic microenvironment. The CD4 cell fate is triggered by GATA-3 upregulation and then sealed by the action of a zinc finger-POZ domain factor, Th-POK (Zbtb7b), which antagonizes the CD8 cell fate defined by action of the Runx factor, Runx3. The CD4 (helper) vs. CD8 (killer) decision is generally irreversible after the cells leave the thymus, as a function of the robust gene regulatory network circuitry that enforces mutually exclusive regulatory states (Carpenter and Bosselut, 2010; He et al., 2010; Naito et al., 2011). However, most CD4 cells continue to make effector lineage decisions after CD4/CD8 lineage choice, long after TCR gene rearrangement, both within the thymus and much later, during responses to antigen stimulation. Two additional decisions that are now recognized to occur in the thymus are the specification of some DP cells to become iNKT cells (invariant TCR, natural killer-like T cells) or other innate-like cells (Gangadharan et al., 2006), and others as Tregs (regulatory T cells). DP cells are triggered to differentiate into iNKTs through their TCR’s strong interaction with non-classical class I MHC molecules expressed by their fellow DP thymocytes. Other CD4 cells are specified to become tTregs (thymic Tregs, or nTregs, natural Tregs), based on their interaction with self antigens in the thymic medulla, which programs the cells to act as suppressors of other T cells in their peripheral responses. Both of these intrathymic variant decisions are based on fairly strong TCR-ligand interaction (“agonist selection”). In the case of iNKT cells, the agonist interaction activates expression of the transcription factor Promyelocytic Leukemia Zinc Finger (PLZF) or Zbtb16, through a pathway involving Egr2 (Seiler et al., 2012). It is their expression of PLZF that enables iNKT cells to express effector genes with greatly reduced need for priming by TCR signaling (Savage et al., 2011). For the Tregs, the agonist interaction induces expression of the transcription factor Foxp3, through a pathway involving NF-κB and IL-2 (Hsieh et al., 2012; Moran et al., 2011; Nunes-Cabaco et al., 2011; Wirnsberger et al., 2011).
Like the programs induced in the agonist-selected iNKT and Treg cells, the later, post-thymic decisions for these TCRαβ cells all depend on TCR stimulation, which is now joined with stimulation by various polarized combinations of cytokines in the environment. These clinically important events are foundational to the generation of major Th1, Th2, Th17, and Treg subsets of T cells in the periphery, as well as other rarer types of effectors, and to the specific designation of cells as Tfh (follicular helpers) for intense collaboration with B cells in lymph node follicles.
Interestingly, the late lineage decisions made after the cells leave the thymus are not always entirely definitive. Careful analysis of these decisions at a single-cell level have revealed that the initial patterns of activation of the lineage-determining factors (e.g. GATA-3 and T-bet) can be far more overlapping than the effector functions of the cells that emerge, and even after polarization, these decisions have substantial plasticity, especially between Th1 and Th17 and between Th17 and iTreg (peripherally induced Treg)(Antebi et al., 2013; Murphy and Stockinger, 2010; Zhu and Paul, 2010). In some cases, even the CD4/CD8 lineage decision may be reversible (Mucida et al., 2013). In contrast, the decision to be a T cell and to express a core set of pan-T-cell genes appears to be irreversible.
E. Modularity of effector and recognition T cell identity elements
Because most effector subtype choices occur long after programming of multipotent progenitors to become T cells, it would be easy to assume that they are hierarchically subordinate to the prior events that generate a mature TCRαβ+ cell in the first place. Such an impression could seem to be supported by the fact that DP thymocytes are extremely poor at turning on any of the effector genes of mature T cell subsets even when they are stimulated with chemical proxies for TCR signaling that bypass the TCR entirely. However, multiple lines of research over the past 10 years have converged to show that in fact the mechanisms generating TCR and priming the cells for TCR-dependent selection events are functionally quite independent from the mechanisms controlling effector function, and that in certain lineages of cells these mechanisms can be uncoupled.
Early hints for this separability came from the responses to chemical activators of cells much more immature than DP cells. It emerged that although DP cells are unable to express effector cytokines, their precursors are much more competent. IL-2 activation, among other responses, is easy to elicit from DN cells that have not yet undergone β-selection, including cells from RAG-knockout animals that lack the ability to rearrange any kinds of TCR genes (Chen and Rothenberg, 1993; Diamond et al., 1997). By inference, in this respect DP cells are not so much immature as paralyzed. Bevan and coworkers first identified RORγt as one of the transcription factors expressed in DP cells that actively blunts access to Th1 and Th2-like response genes in essentially all TCRαβ lineage precursors throughout the period between β-selection and “positive selection” to a CD4 or CD8 cell fate (He et al., 2000). Positive selection is required to downregulate RORγt and to restore functional responsiveness at the end of the DP stages. If RORγt is ever expressed again, it is only in the context of Th17 differentiation.
The access to effector genes in DN pro-T cells is not necessarily exploited by these immature precursor cells in the thymic microenvironment in vivo. However, it is significant that those DN2 cells that split off to become TCRγδ cells may begin using their effector genes after a much shorter wait than those that become TCRαβ cells (Fig. 2). Recent detailed analyses of γδ cells have revealed that they are at least as diverse functionally as αβ cells, and they begin to be diversified within the thymus, before being exported to the periphery at all (Bonneville et al., 2010; Prinz et al., 2013; Vantourout and Hayday, 2013). Different TCRγδ cell functional subsets have characteristic biases in the gene segments they use for rearrangement to assemble their TCRγδ genes, and so it is easy to separate them and ascertain that their preference for different kinds of effector response are established while they are still in the thymus. The basis for different TCRγδ gene segment choice is determined at least in part by different transcription factor requirements to make these segments accessible: STAT5 activates the TCRγ locus in general, but certain Vγ’s are used more or less depending on the ratio of bHLH E protein activity to Id-family E protein inhibitors (Bain et al., 1999). However, there are other dramatic regulatory differences between TCRγδ cells of different subsets (Narayan et al., 2012). While many γδ cells express IFNγ, some do so exclusively like Th1 cells, some are IL-4 producers like Th2 cells, while others specialize in IL-17 production like Th17 cells. Thus, for such TCRγδ cells, the decisions that CD4+ TCRαβ cells defer till antigen response in the periphery are precociously shifted to a milieu more like the one in which TCRαβ cells choose CD4 vs. CD8 fates. Notably, this is not because they undergo more extensive programming than TCRαβ cells immediately after TCR expression; the number of cell divisions triggered in TCRγδ cells is significantly less. However, the maturation programs of these cells induce a strongly dynamic transcriptional cascade that yields at least three major types of effectors, apparently based on interaction with “self” microenvironmental molecules alone.
While most TCRαβ cells delay functional specialization much longer, the iNKT cells (and in human, MAIT cells)(Chandra and Kronenberg, 2015) show that rapid intrathymic effector differentiation is possible in principle for TCRαβ cells too. These cells were first noticed to be distinctive because of their rapidly triggered expression of various cytokines that normally would require priming and restimulation before activation, and it has turned out that they represent a discrete branch or set of branches of αβ T-cell development [rev. by (Alonzo and Sant’Angelo, 2011; Chandra and Kronenberg, 2015; D’Cruz et al., 2010b; MacDonald and Mycko, 2007; Yamagata et al., 2006)]. These cells rapidly express IL-4 (a Th2 signature cytokine) as well as IFNγ (a Th1 signature cytokine), and they can also activate the IL-2 gene. Their rapid response capability appears to be correlated with their expression of the zinc finger transcription factor PLZF (Zbtb16). The ability of one population to express all these cytokines initially appeared to be due to a special violation of conventional subset boundaries in the iNKT cell effector program. However, recent data have suggested a more interesting alternative: namely, that diverse types of iNKT cells that were classically thought to be stages in maturation might instead be end states of alternative iNKT cell maturation programs (Lee et al., 2013). All express the hallmark PLZF factor, but at different levels. Whether or not these are absolute end states, the cells in three different intrathymic iNKT subsets all exhibit functional competence, but with significant differences: some better as IFNγ producers, some better as IL-4 producers, and some better as IL-17 producers. Signature transcription factors of peripheral T cell effector subsets as well as PLZF are also differentially expressed in these subsets (Cohen et al., 2013): whereas all express GATA-3, there is more T-bet (and Runx3) expression in those that can express most IFNγ, more PLZF in those that express most IL-4, and more RORγt in those that express most IL-17 (Lee et al., 2013). In general, then, PLZF and GATA-3 can each be implicated as a rate-limiting factor for priming cells to express Il4 and/or its linked neighboring gene Il13. Thus, something very much like the same effector lineage choice involving similar transcription factors can be made either post-thymically, as a late response to repeated antigen stimulation in the periphery; or intrathymically shortly after positive selection, for certain αβ T cells that become iNKT cells; or earlier intrathymically for cells that branch off from the αβ mainstream even before β-selection, without passing through a DP stage at all, to become functionally specialized γδ cells (Fig. 2).
An exciting perspective on T cells that has recently emerged is that a very similar kind of effector lineage choice can be implemented by newly appreciated lymphoid cells that do not enter the T-cell pathway at all. Natural killer cells long offered a prototype of lymphocytes that lack TCR for recognition, yet exercise T cell-like function in the body: they mimic the effector program of activated cytolytic T cells. With the recent description of three other classes of Innate Lymphoid Cells, completely TCR-negative equivalents of Th1, Th2, and Th17 cells have been discovered as well: to simplify slightly, these are ILC1, ILC2, and ILC3, respectively (Di Santo, 2014; Diefenbach et al., 2014; Hazenberg and Spits, 2014; Spits et al., 2013). All of the innate lymphoid classes including NK cells are specified through a pathway that requires the bZIP transcription factor Nfil3 at least transiently (Geiger et al., 2014; Seillet et al., 2014), and among these all the helper-type ILCs are distinguished by origin from precursors with early expression of PLZF, GATA3, and TOX (Constantinides et al., 2015; Constantinides et al., 2014; Seehus et al., 2015). Notably familiar transcription factors are involved in the alternative effector programs of ILCs: T-bet for ILC1 (and NK) cells, GATA-3 and TCF-1 for ILC2 cells, and RORγt for ILC3 cells (Cherrier et al., 2012; Hoyler et al., 2012; Klose et al., 2014; Mjösberg et al., 2012; Serafini et al., 2014; Yagi et al., 2014; Yang et al., 2013). For ILC2 cells especially, there is a close correspondence between the regulatory requirements for generating these cells and for generating committed T-cell precursors, even though they lack TCR expression (Furusawa et al., 2013; Klein Wolterink et al., 2013; Serafini et al., 2014; Walker et al., 2015; Yagi et al., 2014; Yang et al., 2013; Yu et al., 2015). Some of them can even descend from Common Lymphoid Precursors that have had a history of transient RAG1-RAG2 expression (Karo et al., 2014; Yang et al., 2011b), as can some NK cells (Welner et al., 2009). However, these cells are not T cells, and express only scattered members of the pan-T cell gene battery (Robinette et al., 2015). While some ILCs express one or another of the CD3 genes used in the TCR complex, they do so neither as highly nor as coordinately as T cells. Ironically, the ILC2s that have the most extensive transcription factor overlap with T cell precursors express some of the lowest levels of these pan-T cell genes. Thus, it is not necessary to turn on a pan-T cell gene expression program at all to gain access to the choice point between cytolytic program, Th1-like effector function, Th2-like effector function, and Th17-like effector function (Fig. 2). These programs, and the gene network cross-regulation mechanisms enforcing the choices between them, are modular and deployable completely independently of the TCR assembly and selection programs. Making competent T cells thus requires both activation of the regulatory factors needed to drive expression of pan-T cell genes, and also negotiating the developmental conditions and activation thresholds that the cells will need to deploy one of these effector function modules.
II. Transcriptional dynamics of the T-cell program
A. Onset of T cell gene expression
Activation of the pan-T cell genes occurs in the thymus. The precursors of T cells enter the thymus with little if any detectable expression of the pan-T genes or major subset markers. As the cells first arrive, they are expressing chemokine receptors including Ccr9 and supported by the crucial growth factor receptors Flt3 and Kit. Some of the thymus-settling precursors are likely to have had a history of IL-7R and/or RAG gene expression as bone marrow Common Lymphoid Progenitors before arriving, but as the cells settle in the initial reception compartment and begin to proliferate as Early T cell Precursors (ETP), expression of these genes is silent. The earliest detectable phenotypic transitions within the thymic environment actually involve the repression of Ccr9 and Flt3 in response to Notch signals (Krishnamoorthy et al., 2015; Ramond et al., 2014; Sambandam et al., 2005), leaving Kit expressed at high levels, before substantial activation of pan-T cell genes is under way. The cells begin to turn on T-cell genes primarily as they make the transition from ETP to DN2a stage, a transition that may occur within just a day in the fetal thymus but may take over a week in the adult thymus. In the adult, based on population expansion kinetics and computational modeling, the cells are likely to undergo ten rounds of cell division on average before they reach this point. CD25 (Il2ra) and Il7ra are the earliest T-cell genes that reach full expression in murine T-cell precursors. As the cells cross the DN2a to DN2b transition and become committed, the expression of other T-cell genes increases substantially.
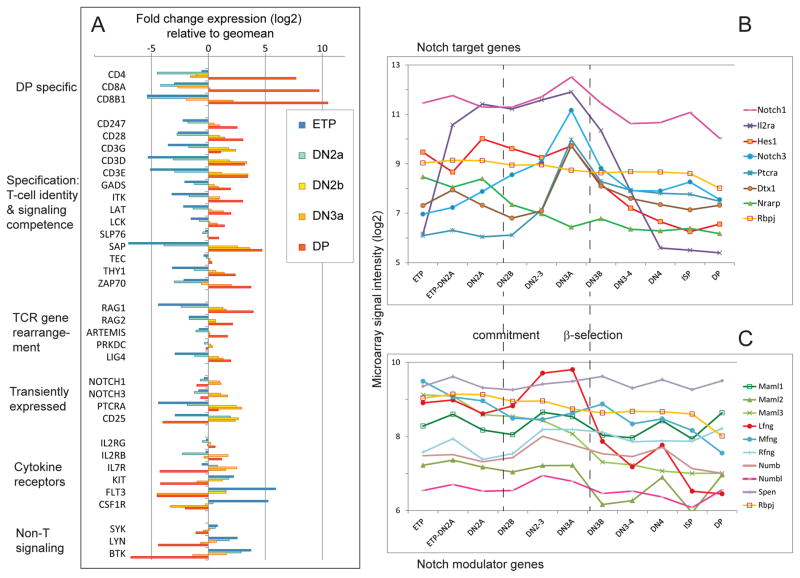
The gene expression changes in early T cells from ETP stage through β-selection are complex, with different sets of genes responding to different underlying regulatory state changes, as shown in Figure 3A [data from (Zhang et al., 2012b)]. In this chart, pre-commitment stages (ETP, DN2a) are represented in shades of blue, post commitment DN stages (DN2b, DN3a) in yellow and orange, and DP stage in red (transitional β-selection stages are not shown). To capture the great dynamic range of change for some genes, each gene’s expression pattern is shown in terms of differences at these stages from its own geometric mean value across all stages. While there are a few genes in this set that are turned on only in DP cells (top group), most of the T-cell identity and signaling genes undergo their greatest increases from DN2a to DN3a stages (Fig. 3A, light blue to orange). The DN3 and DP stages are the main stages when TCR gene rearrangement occurs, and RAG1 and RAG2, Artemis, and Ligase IV are also strongly upregulated following commitment as if they too were T-cell identity-specific. The CD3 genes and LAT achieve levels at or close to their maxima soon after commitment, by the DN3a stage, with ZAP70 and GADS only somewhat lagging behind. Thus, between the ETP and DN3a stages, the cells are substantially armed with T-cell specific response competence.
Figure 3. Trajectories of gene regulation in T-cell precursors through commitment and β-selection.
Data for (A) are RNA-seq analyses taken from (Zhang et al., 2012b), and data for (B, C) are highly curated microarray analyses taken from www.immgen.org (Mingueneau et al., 2013).
A) Patterns of expression of T-cell identity genes and signaling components through the transition from ETP to DP. Data are presented on a log2 scale, with increases and decreases in expression plotted as changes relative to the geometric mean of values for each gene. Thus, genes with stable expression at high or low levels have little fold change. Precommitment stages are indicated by blue bars, postcommitment stages by yellow to red bars. Note that T-cell identity and signaling competence genes are drastically upregulated during commitment, while cytokine receptor genes are more diversely regulated.
B) Notch target genes have developmentally distinct patterns of expression. Absolute microarray hybridization intensities at the indicated stages are presented on a log2 scale. All the genes shown are sharply affected by interruptions or increases in Notch signaling intensity in early T cells except the Notch signal-transducing regulatory gene Rbpj, which is shown to indicate the stability of the Notch response machinery.
C) Little developmental change in expression of known Notch signal modulating genes despite dynamic target gene expression. The indicated modifiers are plotted as for the samples in (B), but the scale is expanded for greater sensitivity to change, with Rbpj shown again for reference. Note that only Lfng is highly regulated across these stages.
This rapid, parallel increase in T-cell gene expression contrasts with the cytokine receptor genes, which behave very individually. As already noted, Il7ra, like Il2ra, is strongly upregulated during the DN2-DN3 stages, only to decline again in the DP cells. Flt3 and genes coding for other non-T growth factor receptors such as Csf1r (c-fms, M-CSF receptor) are active in the thymus-settling precursors but steeply repressed at the earliest stage transition. Kit expression, initially high, continues until after commitment, but is then silenced. Of these receptors, only Kit and IL-7R are functional in early T cells. The Il2ra gene product encoding CD25, although it can serve as an α chain for the IL-2 receptor, does not work that way here, for it is not accompanied in these cells by its obligate assembly partner IL-2Rβ. Il2rb expression instead serves as a marker for certain γδ cell lineages and cells developing into NK cells. Interestingly, the ETP and DN2a cells initially express a number of kinases that are normally considered specific to non-T cells, but these too are downregulated and silenced during the stages immediately following commitment. The T-cell differentiation program thus includes precisely timed silencing and transient up- and down-regulation activities as well as the steady increase in T cell identity gene expression. These features hint at the regulatory complexity that underlies the program.
B. Notch signaling: driver and modulator
1. Notch target genes
The indispensable exogenous trigger for T-cell development is the stimulation of the Notch pathway, by interaction of Notch1 transmembrane molecules on the lymphoid precursors with Delta-like 4 molecules on thymic epithelial cells (Fig. 1A). Notch signaling not only induces T-cell development, but also begins blocking access to the B-cell developmental pathway and induces an intrinsic loss of B-lineage potential shortly after precursors enter the thymus. Notch signaling also inhibits NK, myeloid, and dendritic cell alternative developmental pathways for ETP and DN2a cells, and is ultimately required to induce the mechanisms that close off these options by the DN2b stage. Thus, before the cells stop responding to Notch signals during β-selection, Notch-induced inherent regulatory changes render the cells’ commitment Notch-independent.
Notch signaling is well known to affect transcription directly. To simplify (Borggrefe and Oswald, 2009; Radtke et al., 2010), engagement of the extracellular domain of a Notch molecule causes a proteolytic cleavage that liberates the cytoplasmic domain from the plasma membrane; its nuclear localization sequence then enables it to enter the nucleus, where it works as a direct transcriptional coactivator. A simple expectation might therefore be that T-cell identity genes could include a large number of direct Notch target genes. Indeed, some T-cell identity-associated genes appear to be strongly dependent on Notch signaling throughout the early developmental stages, including Il2ra and the gene encoding the surrogate light chain that is expressed as a transient partner for TCRβ, Ptcra (Pre-TCRα). Interrupting the contact with Delta-like molecules or chemically inhibiting the protease-dependent cleavage of Notch causes sharp losses of expression of these genes over a 1–2 day period (Del Real and Rothenberg, 2013; Franco et al., 2006; Liu et al., 2010a; Maillard et al., 2006).
However, the T-cell program as a whole is not simply a Notch signaling response. First, Notch signaling in general is shut off at β-selection, as the cells transition from DN3a through proliferation and intermediate phenotypes to DP. By that time, any permanently expressed T-cell genes must be weaned from Notch-dependence in order to persist. Another complication is that different genes which are clearly Notch-dependent in T-cell development are not co-regulated. Instead, as shown in Fig. 3B, they are expressed in diverse patterns [data from (Mingueneau et al., 2013); note the extended log2 scale], some like Hes1 turned on early and sustained throughout the DN stages, some like Ptcra turned on only at the last DN stages before β-selection, while others like Nrarp are restricted to the earlier stages of T-cell development and paradoxically turned off as other Notch target genes are more strongly activated. Importantly, in the postnatal mouse thymus, ETPs receive Notch signals for an entire week before expressing the definitive T cell genes that define the DN2a stage (Porritt et al., 2003). Thus T cell development as a whole is not an immediate response to Notch signaling.
In fact, Notch plays a variety of distinct roles during the T-cell specification and commitment process, dependent on signal intensity and developmental stage. In mice and humans, in the early ETP stages, Notch signals enhance proliferation but are not essential for viability (Balciunaite et al., 2005; De Smedt et al., 2005; Garcia-Peydro et al., 2006; Schmitt et al., 2004; Taghon et al., 2005), whereas by the time the cells reach DN3a stage, Notch signals become indispensable for survival (Ciofani and Zúñiga-Pflücker, 2005). The intensified Notch dependence correlates with the fact that in mouse, differentiation to undergo β-selection requires a stronger and more prolonged period of Notch signaling than differentiation to undergo γδ selection (Ciofani et al., 2006; Garbe et al., 2006; Taghon et al., 2006); as noted above, in mouse precursors, some γδ cells split off from the DN stage progression before the DN3a stage. Interestingly, this relationship of Notch signaling to TCR choice is reversed in human T cells (Van de Walle et al., 2009; Van de Walle et al., 2013). The evolutionary reversal implies that Notch signaling is being used only as a switch and not as an integrally essential regulator to distinguish these classes of TCR genes themselves. To explain the shifting impacts of Notch signaling on the cells, clearly some additional regulatory influence is directing the impact of the Notch signal to different target genes.
2. Notch mediators
Notch signals are modulated by multiple other gene products, including Numb and its family members as negative regulators, Spen (also called Msx2-interacting protein MINT, SMRT/HDAC1 associated repressor protein SHARP, or RBM15c) as a competitive negative regulator, Mastermind-like (MAML) family proteins as obligate positive regulators, and modulators of Notch protein post-translational processing. Glycosyltransferases of the Fringe family (Lfng, Mfng, Rfng) also strongly modulate the preference of Notch proteins for interaction with Delta-like ligands as opposed to Jagged family ligands. There is indeed an important developmental change in Lfng expression as T-cell precursors differentiate, such that DN cells are generally Lfng high and thus competitively favored for interaction with the DLL4 presented in the thymic microenvironment, whereas DP cells sharply downregulate Lfng and become unable to compete (Visan et al., 2006). However, Lfng is the exception, not the rule: overall the patterns of expression of Notch signaling modulators are remarkably unaffected by progression through T-cell development (Fig. 3C). Thus, the shifting spectra of Notch target genes between ETP and DN3a stage are not readily explained by changes in the Notch signaling machinery itself.
The overall intensity of the Notch pathway signal received by the cells may increase as the cells progress through commitment, due to the downregulation of two antagonists. One of the transcription factors expressed early in the cells is the myeloid and multilineage regulator PU.1, and PU.1 activity levels apparently blunt Notch signaling in a dose-dependent way (see section V.B.1). This Notch-inhibitory effect seems to be general, because it applies similarly to “early” and “late” stage Notch-dependent targets (Champhekar et al., 2015; Del Real and Rothenberg, 2013). The fact that PU.1 is normally turned off during commitment could thus unleash stronger potential for Notch signaling by the DN3a stage. At least two Notch dependent genes also have the potential to act as feedback negative regulators of Notch signaling, i.e. Nrarp and Dtx1. As differentially regulated Notch target genes, Dtx1 and Nrarp themselves exemplify the sharp developmental stage-dependence of Notch effects. Dtx1 is particularly labile, and its dynamics are complex, but Nrarp expression is mostly confined to the precommitment period and thus its negative effects on Notch signaling are relieved by the DN3a stage. However, during β-selection most Notch signaling is downregulated for the rest of intrathymic T-cell development. It only re-emerges to be utilized, much later, to modulate mature T-cell functional responses (Osborne and Minter, 2007; Radtke et al., 2013). Thus all Notch induction of pan-T cell genes must be hit-and-run, and the genes that continue to be expressed transition to Notch-independence.
III. Dynamic transcription factor expression
Notch signaling in principle must continue throughout the 12–14 cell cycles that precursors probably undergo between entry into the thymus and arrest at the DN3a stage checkpoint in postnatal mice (Lu et al., 2005; Manesso et al., 2013; Porritt et al., 2003). In contrast, the transcription factors expressed by the cells shift dramatically across this interval. These changes are at least indirectly regulated by Notch signaling, but most of them do not reflect a close dependence of transcription factor gene expression on the immediate status of Notch signaling. They are instead the outcome of cascades of regulatory factor actions. An overview of the dynamics of the major factor groups is shown in Fig. 4A.
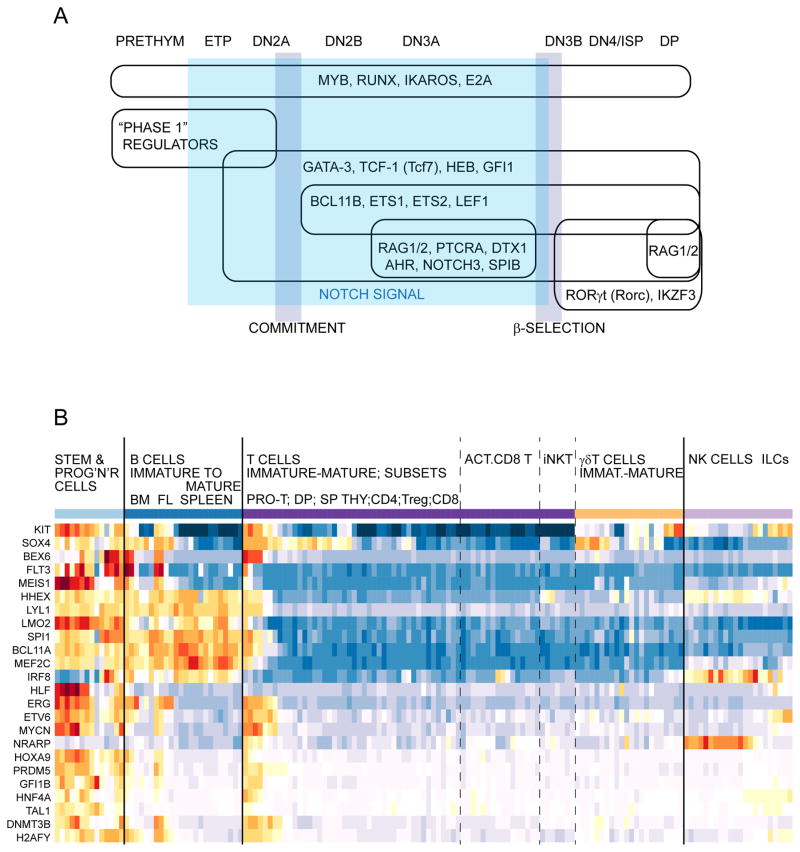
Figure 4. Dynamic regulatory stages in developing T-cell precursors.
A) Diagram representing the major groups of regulatory factors at the stages when they are deployed in early T-cell development. The period dominated by Notch signaling is shown by shading. All the factors shown are transcription factors or cofactors except RAG1/2 (recombinases), Ptcra (TCRα surrogate chain used in β-selection), and Dtx1 (Notch pathway target gene). “Phase 1” regulators are the stem/progenitor-associated factors that are highly expressed in thymic immigrants but then silenced during commitment and immediately afterwards. “RUNX” refers to the constant presence of members of the Runx family, although Runx2 and Runx3 are more highly expressed in the ETP stages and Runx1 is more highly expressed at the DN3a stage.
B) Phase 1 regulators have distinct patterns of usage in stem/progenitor cells and in other classes of lymphocytes. Patterns of expression are shown for two cytokine receptor genes (Kit and Flt3) and 22 phase 1 regulatory factor genes, using the ImmGen database (Heng et al., 2008; Mingueneau et al., 2013; Robinette et al., 2015) and interactive heatmap tool for “MyGeneSet” (http://rstats.immgen.org/MyGeneSet/)(July, 2015). Red and orange cells represent highly expressed genes in the indicated cells relative to their average, blue represents very low levels of expression relative to their average. Note that all these genes have their expression in T cells confined to the left end samples in the “T Cells” series, which correspond to the stages shown in Fig. 3B, C. However, they are all expressed in addition in at least some stem and progenitor cells (“Stem & Prog’n’r Cells”) and distinct groups are also shared with early B cells, B cells generally, and/or NK and ILC cells.
A. Stem-progenitor specific factors
The cells enter the thymus continuing expression of a set of transcription factors used in stem and progenitor cells. Some of these factors persist and even increase in expression during T-cell commitment: these include Myb, Gfi1, Runx1, Ikaros (Ikzf1) and the ubiquitous E protein E2A (E12/E47, “transcription factor 3”, Tcf3). Others play a role only in the earliest stages and are downregulated, and these can be termed stem-progenitor specific factors. We have also referred to these as “phase 1” transcription factors, and their patterns of expression are summarized in Fig. 4B, using data from the Immunological Genome Project (Heng et al., 2008; Mingueneau et al., 2013).
Developing T cells shut off the expression of some stem-progenitor specific factors relatively soon after entering the thymus, but others persist through at least one Notch-dependent transition and multiple rounds of division. The first round of silencing occurs before the cells leave the ETP compartment, probably in parallel with the downregulation of Flt3: this is when Hoxa9, Hlf, Lmo2, Meis1 and Mef2c are turned off (Fig. 4B). A second round of silencing occurs significantly later, during commitment, essentially in parallel with the downregulation of Kit. Here, PU.1 (Spi1, Sfpi1), Hhex, Gfi1b, N-Myc (Mycn), Tal1, and Bcl11a are turned off, with Lyl1 persisting only slightly longer. Clearance of the final progenitor-cell regulatory factors occurs only from DN3a stage to entry into β-selection, when Erg and Etv6 are turned off. Most of these regulatory genes once repressed are never activated in T-cells again (Fig. 4B), except in case of malignant transformation (Yui and Rothenberg, 2014). Any cross-regulatory stimulation that they provide to each other’s expression is lost as they are silenced. This is likely to be one of the features making T-cell specification irreversible.
Despite their temporary expression, the factors in the stem and progenitor set do make a positive contribution to T-cell generation. Elsewhere (Yui and Rothenberg, 2014) we have described substantial evidence that their engagement in self-renewal mechanisms can help to sustain the extensive proliferation that precedes lineage commitment, which is crucial to generate a large pool of T-cell precursors that can be winnowed stringently by TCR-dependent selection. Lyl1 and Bcl11a are both important for sustaining the expansion of normal prethymic and early pro-T cells up to the DN2 stages in vivo (Yu et al., 2012c; Zohren et al., 2012). Recently it has become clear that even PU.1, a factor with explicit ability to promote alternative cell fates (Anderson et al., 2002; Del Real and Rothenberg, 2013; Dionne et al., 2005; Franco et al., 2006; Laiosa et al., 2006; Lefebvre et al., 2005), is needed to contribute to optimal T-cell generation (Champhekar et al., 2015), by retarding the timing of T-cell lineage commitment. Special Notch-dependent mechanisms make it possible for these progenitor factors to work within the confines of the pro-T cell program until they are silenced, as described in Section V.B.2.
B. Factor families with shifting intrafamily dominance: Runx and ETS
T cell development involves roles for some transcription factors that are products of multigene families, such as the ETS family and the RUNX family. ETS and RUNX family transcription factors are likely to play important roles as participants in most lymphoid gene expression, based on the extreme enrichment of their binding motifs in enhancers of lymphoid genes with various patterns of activity. These families are always represented among the transcription factors expressed in the cells, from prethymic stages all the way through to mature T cell effector subsets. With their near-indistinguishable DNA binding affinities, this guarantees that some family member will always be available to occupy an appropriate target site. However, the early T-cell specification stages are times when there is particularly dynamic shifting among members of these two families for dominance in expression.
Runx1, Runx2, and Runx3 family members (depicted collectively as “RUNX” in Fig. 4A) are all expressed in the ETP stage. Later, their expression patterns diverge in different subsets of αβ and γδ T cells, but initially they are coexpressed. During commitment, Runx2 and Runx3 are downregulated while Runx1 rises to its highest level (David-Fung et al., 2009). After the DP stage, Runx3 is specifically re-induced in CD8 T cells, to match the high levels of expression it sustains in most ILCs. Runx activity is implicated in CD4 gene silencing during positive selection of CD8 cells and in the silencing of PU.1 earlier, during the DN2a to DN2b transition (Collins et al., 2009; Egawa et al., 2007; Grueter et al., 2005; Huang et al., 2008; Naito and Taniuchi, 2010; Zarnegar et al., 2010). However, a complete analysis of Runx factor roles is complicated by alternative splicing and promoter use isoforms of all three genes and by uncertainty about whether the family members act redundantly or competitively.
The ETS family is arguably the most complex, as more than a dozen family members are expressed during intrathymic T-cell development (Anderson et al., 1999; David-Fung et al., 2009; Mingueneau et al., 2013; Zhang et al., 2012b). The dominant ETS factor of mature T cells, Ets1, is poorly expressed when the cells enter the thymus, and its closest paralogue Ets2 is initially silent. However, the cells express multiple other ETS factors that undergo little change in expression during T-cell development (GABPα, Fli1, Elf1, and other Elf, Elk, and Etv subfamily members), and also Erg, Etv6 (TEL), Elk3, and PU.1 which are confined to the early DN stages. During commitment, not only is PU.1 turned off but Ets1 and Ets2 dramatically turn on, Ets2 rising to a summit at DP stage and then falling in more mature cells. A different family member, Etv5 (ERM), is turned on sharply in a γδ-specific way and acts as a consistent feature of most TCRγδ lineage cells thereafter (Narayan et al., 2012); unfortunately nothing is known yet about how this factor could contribute to the special qualities of TCRγδ cell functional maturation. The dominant members of the ETS family thus shift their relative expression levels by orders of magnitude during commitment and progression to the DP stage or to selection of a TCRγδ fate.
It is still uncertain how much these related ETS-family factors compete with each other for the same binding sites. Whereas most ETS factors bind similar DNA sequences containing a GGAA core in vitro, there are characteristic differences between them in their preferences for flanking sequences, so they are unlikely to bind to identical spectra of target sites in living cells (Wei et al., 2010). Nevertheless, for any given target gene it is unlikely that only a single unique ETS factor can provide a regulatory input: in closely studied cases there is clear evidence for redundancy [e.g. (Xue et al., 2004)]. Thus, the impact of ETS family transcription factors on T-cell development is probably vastly larger than has been revealed so far by any single-gene knockout phenotypes.
Of the ETS factors, one which is easiest to interrogate for a specific role is PU.1. It is the founding member of a “Spi” subfamily with a distinctive preference for purines flanking the GGAA core, so it binds to sites that may not be redundantly occupied (Wei et al., 2010), and it is the only member of the Spi subfamily expressed in the early ETP and DN2a/2b stages. PU.1 itself is silenced after commitment and never expressed again except in a rare subset of helper T cells (Th9 cells) (Chang et al., 2010; Chang et al., 2005), but it has a potent and well-studied role in myeloid cells, dendritic cells, and B cells. In fact, PU.1 appears to be directly responsible for the expression of some “myeloid-like” genes in the earliest stages of T-cell development and for the duration of access to dendritic-cell or myeloid fate options that the cells preserve in the stages before commitment. However, as described below, PU.1 is not expressed in early T cell precursors only to be a subverter of the T cell program; its role is illustrative of the interlocking between hematopoietic programs that is characteristic of the lineage commitment process.
C. “T cell” transcription factors
The factors that separate the T-cell program from other hematopoietic programs most clearly are strongly upregulated after the cells enter the thymus. They are members of four different families: GATA-3 (GATA family); TCF-1 (encoded by Tcf7, founding member of the TCF/LEF family); HEB (Tcf12) as a partner of E2A (Tcf3), a fellow member of the bHLH factor family; and Bcl11b, a zinc finger transcription factor whose closest relative, Bcl11a, is expressed in a mirror-image pattern. GATA-3, TCF-1, and Bcl11b are virtually T-lineage specific within hematopoiesis, and play roles in most if not all subsets of T cells. TCF-1 is also detectably expressed in NK cells, but the only other blood cells in which they are expressed substantially are ILCs, especially ILC2 cells. HEB as a partner for E2A is much more common in the T-cell lineage than in any other hematopoietic cell type, and the dimer plays a crucial role throughout specification and commitment, in particular separating the T-cell pathway from all the NK and ILC types. With essential support from progenitor-inherited factors like Myb and Runx1, Gfi1, and members of the Ikaros family, these four factors collaborate to create the T-cell identity.
1. GATA-3 and TCF-1
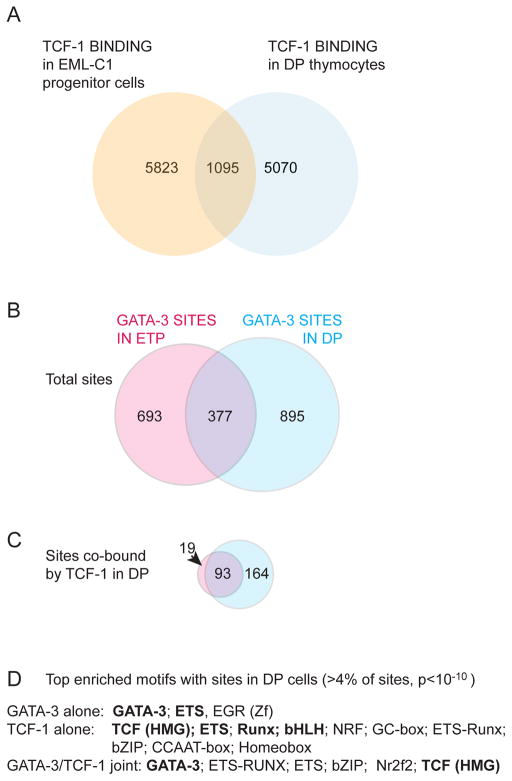
The GATA-3 and TCF-1 factors are activated first, beginning their expression within the ETP population. TCF-1 is most clearly a Notch target gene (Germar et al., 2011; Weber et al., 2011), and it both positively auto-regulates and positively regulates GATA-3 expression. Whether Gata3 has direct transcriptional input from Notch is currently uncertain. Later, Notch signaling inhibition can result in higher GATA-3 expression rather than lower, so that Gata3 at least has some Notch-independent modes of expression. In T-lineage cells, Gata3 is positively regulated by the progenitor-cell factor Myb as well as by TCF-1 (Del Real and Rothenberg, 2013; Gimferrer et al., 2011; Maurice et al., 2007). Long after lineage commitment, during positive selection and in the periphery, GATA-3 can be activated by other factors, but its association with TCF-1 expression is maintained (Yang et al., 2013; Yu et al., 2009).
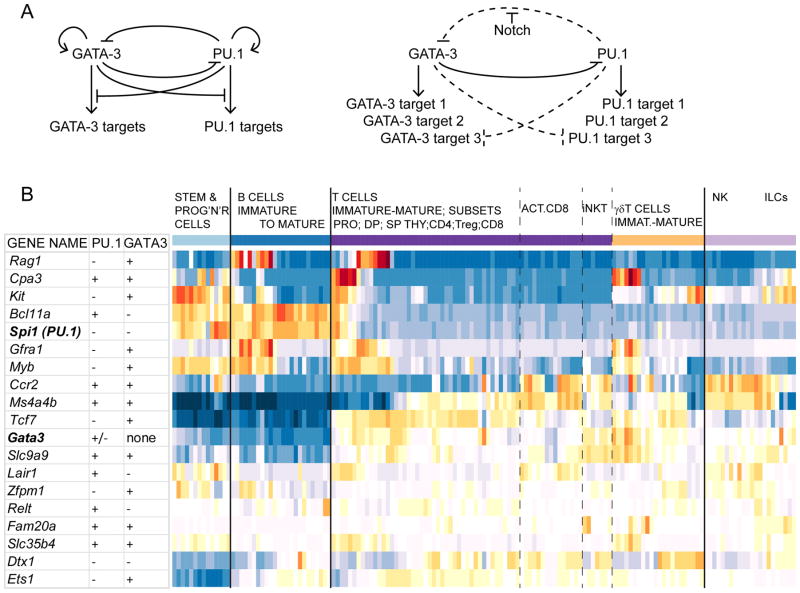
GATA-3 is essential for T-cell development from an early stage, and even partial dose reductions can lead to pro-T cell death especially at ETP, DN3, and later stages (García-Ojeda et al., 2013; Hattori et al., 1996; Hosoya et al., 2009; Hozumi et al., 2008; Scripture-Adams et al., 2014). GATA-3 deficient cells cannot progress beyond a stage resembling DN2 stage, although even the apparent DN2 cells are abnormal in gene regulation pattern. Consistent gene expression perturbations have been reported in GATA-3-deficient cells generated both in prolonged OP9-DL1 culture (García-Ojeda et al., 2013; Scripture-Adams et al., 2014) and in short-term acute deletion experiments from ETP-DN2a precursors (Scripture-Adams et al., 2014). For example, GATA-3-deficient “DN2” cells have reduced Cd3e, Rag1, Ets1, Bcl11b, Itk, Tcf7, Zfpm1, and Kit expression, while they overexpress SpiB, Bcl11a, Dtx1, Spi1 (PU.1), and possibly also Lmo2 and Nfil3. Some of these abnormalities could simply be indicators of a general developmental retardation, but others clearly indicate derangement of the normal developmental sequence. Later, GATA-3 deficiency causes failure of β-selection as well as failure to generate CD4 cells (Hernández-Hoyos et al., 2003; Pai et al., 2003).
TCF-1 is also crucial for T-cell development, with abnormal and reduced ETP populations, absent DN2 populations, and reduced, developmentally defective DN3 populations (Germar et al., 2011; Jeannet et al., 2008; Verbeek et al., 1995; Yu et al., 2012b). Although some of the residual DN3 cells in Tcf7−/− mice appear to express TCRβ chains, they are incompetent to go through β-selection normally (Yu et al., 2012b), and generate small and abnormal later thymocyte populations. The earliest precursors in the Tcf7−/− thymic populations are not true ETPs since they lack high Kit expression, and they already express lower levels of Cd3e and Cd3d than controls while expressing abnormally high Id2, Il7r, and Tcf3. The roles TCF-1 plays in the following stages are complicated by the mixture of activating and repressive isoforms in which it is normally expressed and by the potential for redundancy with its relative, LEF-1, which is turned on sharply starting in DN2b stage (Tiemessen et al., 2012; Yu et al., 2012b). The need for TCF-1 appears to be somewhat dependent on ontogeny’, with knockouts grossly defective in post-weaning adult T-cell development but allowing fetal thymopoiesis to occur apparently normally (Germar et al., 2011; Jeannet et al., 2008; Schilham et al., 1998; Weber et al., 2011). This difference could be due mostly to the augmented requirements for proliferation and survival in adult animals, because Tcf7 mutants generally show their sharpest defects in all proliferative stages of T cell development as well as in survival of DP cells (Germar et al., 2011; Schilham et al., 1998; Staal et al., 2004; Verbeek et al., 1995; Wang et al., 2011).
Although they are activated specifically in early T-cell development, neither factor has a simple relationship of DNA binding to T-cell specific target gene activation. TCF-1 levels become extremely high in early T cell development, with mRNA transcript levels rising to approach those for β-actin (Actb). By the DP stage, available TCF-1 binding data shows the protein engaged broadly across nearly all open chromatin throughout the genome, not limited to developmentally specific genes (Dose et al., 2014). TCF-1 family members are renowned for their roles as Wnt signaling pathway effectors, dependent on nuclearized β-catenin as a cofactor [rev. by (Staal and Sen, 2008)], but current evidence strongly disfavors any Wnt/β-catenin dependence of the functions that TCF-1 plays in the earliest stages of T-cell development (Jeannet et al., 2008; Weber et al., 2011). It is possible that TCF-1 is serving instead as a scaffolding or architectural component in the construction of T-lineage specific enhancer/promoter complexes in these cells (Giese et al., 1995). GATA-3 levels remain much lower than TCF-1 levels at all stages, and GATA-3 binding is much more selective, but the sites bound vary substantially from stage to stage (Wei et al., 2011; Zhang et al., 2012b). At a minimum, other factors must be strongly influencing GATA-3 site choice and outcomes of GATA-3 engagement.
2. E proteins
HEB and E2A are the main components of the “E protein” heterodimers in early T cells. Whereas E2A continues to be expressed at unchanging levels from prethymic precursors, HEB is strongly upregulated. HEB-E2A complexes are crucial for the growth arrest, recombinase gene expression, and fidelity of the β-selection checkpoint in DN3a cells, and are vital again for enabling TCRα rearrangement in DP cells (Barndt et al., 2000; D’Cruz et al., 2010a; Jones and Zhuang, 2007). DP cells deprived of these complexes progress prematurely through positive selection-associated changes and shift disproportionately to a CD8-like lineage fate (Jones-Mason et al., 2012). Unlike the situation in B cells, where E proteins are degraded in response to Notch signaling, HEB/E2A complexes are stable in the early T-cell milieu; whether the HEB components that are much more common in T cells play a role in this stability is not known. HEB is expressed in two major isoforms in early T cells: the “canonical” one uses the standard transcriptional start site in exon 1, and its regulation is not well understood. The second “alternative” isoform uses an internal promoter that encodes a truncated product with an alternative N-terminal sequence called “HEBalt”. HEBalt is expressed as a classic Notch target gene, paralleling the rise of Il2ra in the early T cells from ETP to DN2a and then being abruptly and permanently silenced after DN3a stage during β-selection (Wang et al., 2006a; Yui and Rothenberg, 2004). HEBalt can complex with E2A as well, and the further heterogeneity of E2A isoforms between E12 and E47 alternative splice variants increases the diversity of E protein heterodimers in the cells. Although mapping of E protein dimer binding sites across the genome is difficult in early T cells, the results available for cells in DN3 stage and β-selection (Miyazaki et al., 2011) and later (Zhang et al., 2012a) show strong binding to many sites of T-cell differentiation genes, and there is good evidence that these genes are in fact functionally regulated by E proteins (Ikawa et al., 2006; Miyazaki et al., 2011; Schwartz et al., 2006; Xu et al., 2013).
Note that in later T-cell development, E protein activity is often dynamically antagonized by induction of the antagonists, Id3 and Id2, which form non-DNA binding heterodimers with E2A and/or HEB (Bain et al., 2001; Kaech and Cui, 2012; Yang et al., 2011a). High-level expression of Id2 especially is characteristic and essential for ILC development, implying that the innate-cell fate depends on neutralizing E proteins [rev. by (De Obaldia and Bhandoola, 2015; Mjösberg et al., 2012; Serafini et al., 2015)]. However, in normal early T cell development from ETPs until DN3a stage, these Id factors are normally held to extremely low levels, thus leaving E2A and HEB free to form active DNA-binding complexes instead.
Complexes of E2A and HEB are crucial for numerous aspects of early T-cell specification, but it is not certain yet which responses are specifically mediated by which isoforms. Nevertheless, the rapid increase in HEB expression during early T-cell specification could have a strong effect on the deployment of the total E2A protein in the cells, because E2A would otherwise be engaged in a different kind of complex entirely. Two of the progenitor-associated factors, Tal1 and Lyl1 (see Fig. 4B), are also bHLH factors that have important roles in the gene regulatory networks in multipotent progenitors. These heterodimerize with E2A and make it possible to form super-complexes with GATA factors (usually GATA-2 in progenitors) and with the bridging components Lmo2 (or Lmo1) and Ldb1, binding as a coordinated unit to a characteristic set of composite GATA/Tal1 sites (Lahlil et al., 2004; Xu et al., 2003). The bridging functions depend on the Tal1 or Lyl1-specific structure of the bHLH heterodimers, and Lyl1 expression is especially high until just after commitment. Thus, by raising levels of HEB to the point that it can compete against Lyl1 for limiting amounts of E2A, the early T cells may be able to break up the progenitor-associated DNA binding complexes and reposition the E2A to different sites. If this should turn out to be the case, then the downregulation of Lmo2 within the ETP stage should collaborate with the initial upregulation of HEB to transform the spectrum of bHLH target genes expressed in the early T cells.
3. Bcl11b
The final member of the T-cell specific set of factors is Bcl11b. This zinc finger transcription factor is not expressed at all until the cells are in the mid-DN2a stage (Rothenberg et al., 2008; Tydell et al., 2007), and then its expression is strongly implicated in the repression of Kit and other changes that define the committed state. Bcl11b is still poorly characterized as a genome-wide regulatory agent, but it has two distinct, stage-dependent roles. When it is first turned on, it is needed to shift the cells from a program of progenitor-cell gene expression which can be sustained by growth in permissive cytokines, to a commitment program of limited growth, obligate progenitor-cell gene silencing, and continued differentiation to a DN3a climax state (Ikawa et al., 2010; Li et al., 2010a). If committed cells are deprived of Bcl11b, they lose viability and shift to a natural killer-like or ILC1-like gene expression profile (Avram and Califano, 2014; Liu et al., 2010b). Ironically, the cells that do not yet express Bcl11b express its highly related family member Bcl11a, which has roles in B cells, dendritic cells, and in the viability of lymphoid progenitors (Ippolito et al., 2014; Liu et al., 2003; Yu et al., 2012c). The extent to which Bcl11a and Bcl11b play overlapping or competing roles needs to be determined. However, Bcl11b itself has a unique status as the factor the expression of which seems most closely related with T-cell lineage commitment. The connection that may exist between its one-time lineage commitment roles and its roles in regulating access to cytolytic effector programs is discussed below (section IV).
Bcl11b knockouts fail to generate αβ T cells (Wakabayashi et al., 2003), while later deletion has particularly severe effects on DP cells, iNKT cells, and CD4 cells, especially Tregs (Albu et al., 2007; Albu et al., 2011; Avram and Califano, 2014; Kastner et al., 2010; Vanvalkenburgh et al., 2011). One could easily infer that an obligate lineage commitment regulator should be equally essential for all T cells. However, there has been a striking exception to this prediction in Bcl11b knockouts. There is a subset of TCRγδ cells that appears to be able to develop reliably from precursors lacking Bcl11b. Such TCRγδ cells are enriched for use of fetal-specific TCRγδ gene rearrangements that are typical of the first waves of T-cell development in the organism (Li et al., 2010a; Shibata et al., 2014), and they are selectively enriched for the ability to make IFNγ on stimulation. Such fetal-type thymocytes are normally characterized by their unusually high expression of Id2 and Il2/Il15rb, two genes usually associated with NK or ILC1 development. These may only be the progeny of TCRγδ precursors that branch off of the common TCRαβ/γδ pathway at the DN2 stage (Shibata et al., 2014), in harmony with the fact that Bcl11b may not be turned on yet in these precursors even in wildtype mice. Thus, the exception to the requirement of T-cell development for Bcl11b appears to be a very particular lineage of innate-like T cells.
D. Collaborating factors
“T cell factors” cannot complete the job of T-cell specification without collaborating with factors involved in growth factor receptor signaling, the repressor Gfi1, and the multifunctional zinc finger factors of the Ikaros family (Ikzf). Growth factor receptor signaling is not only permissive but also, to some extent, an instructive regulatory input. For example, in postnatal mouse T-cell precursors, signaling through Kit and IL-7R actually needs to be reduced substantially in order to permit arrival at the β-selection checkpoint (Balciunaite et al., 2005; Wang et al., 2006b). STAT5 is a major transducer of signals through the IL-7R, and can act directly to establish epigenetic accessibility for the TCRγδ locus (Schlissel et al., 2000; Ye et al., 2001). Thus, both viability support and STAT5 action as a transcription factor are likely to explain why IL-7/IL-7R signaling is crucial for TCRγδ cells. However, for progression to the αβ-biased, DN3a stage, a more important switch may be the PI 3-kinase cascade, also mobilized by IL-7R signaling, that inhibits activity of FoxO factors (Pallard et al., 1999). FoxO1 is crucial for RAG gene activation in B-cell precursors (Mansson et al., 2012; Welinder et al., 2011), and if the FoxO factors are similarly needed in DN3 pro-T cells, then excessive IL-7/IL-7R signals could retard competence for TCR rearrangement directly. However, once TCRβ is successfully expressed, remaining IL-7R expression becomes important again to promote the strong burst of proliferation during β-selection (Boudil et al., 2015).
Gfi1 has long been known to play a vital role in lymphocyte development, but its structure suggesting an obligate repressor activity has made it difficult to identify the specific targets that it needs to control in early T cells. There has been evidence that in peripheral T cells it can interact with GATA-3 to stabilize it, and that it can restrain excessive expression of the IL-7 receptor (Park et al., 2004; Shinnakasu et al., 2008). Recently, Gfi1 has been shown to be essential in LMPPs and ETPs to enable cells to be stimulated by Notch signals. Without Gfi1, Notch signaling is toxic, and the cells fail to activate a T-cell gene expression program (Phelan et al., 2013). The mechanism is still being dissected. Collaborative Gfi1-GATA3 interaction could be involved, since GATA-3 deletion or knockdown also prevents successful development under Notch signaling conditions (Hozumi et al., 2008; Scripture-Adams et al., 2014). Gfi1 is needed for granulocyte and B-cell fates as well, where it apparently protects the cells from powerful macrophage-promoting effects of PU.1 (Laslo et al., 2006; Spooner et al., 2009); conceivably a similar mechanism could be involved in early T cells. Finally, Gfi1 is activated by E proteins, and a positive feedback effect on E protein activities is also possible because Gfi1-deficient animals overexpress E protein antagonists Id1 and Id2 in their thymocytes (Hock et al., 2003; Yücel et al., 2003). This suggests that Gfi1 may help to enforce the E protein activity levels that are essential to prepare cells for β-selection. In T cell development as well as in ILC subsets, Gfi1 expression roughly parallels GATA-3 expression until positive selection, when it is turned off; thus, the expression pattern of Gfi1 is consistent with any of these possibilities.
Ikaros family members are crucial for the development of multiple lymphocyte types, and the main reason that any T-lineage cells are made after deletion of the founding member Ikzf1 (Ikaros) is that from the earliest stages they express Ikzf2 (Helios) as well (Hahm et al., 1998; Kelley et al., 1998). The Ikaros family zinc finger factors can multimerize and have complex roles in regulating differentiation (Schjerven et al., 2013), but one function is particularly interesting. They appear to compete for binding to Notch target gene sites, working to damp activation in response to Notch signals (Chari et al., 2010; Geimer Le Lay et al., 2014; Kathrein et al., 2008; Kleinmann et al., 2008). The role of Ikaros is indispensable for maintaining the integrity of the β-selection checkpoint especially. Loss of Ikaros function otherwise converges with gain of Notch signaling activity to drive malignant transformation of early T cells at the β-selection checkpoint. In addition, the implications for normal T-cell development are important. Although Notch signaling drives the T-cell program forward and maintains viability in DN2b and DN3a cells, the T cell program itself may still require Notch-dependent transcription to be restrained.
IV. Commitment: transfer of lineage fidelity from environment to intrinsic factors
Lineage commitment is defined as the point when the developing T cells give up the intrinsic competence to embark on any other developmental pathway. Left in the thymic environment, essentially all of the pro-T cells from ETP through β-selection will normally give rise to T-cell progeny because of the Notch signaling, but there are dramatic differences from one stage to the next in what they can do if they are removed from the Delta-rich thymic milieu. In fact, lineage commitment is a process rather than a single event, because adult pro-T cells lose access to the B-cell pathway soon after entering the thymus, but only lose access to myeloid, granulocyte, and NK fates significantly later, at DN2b stage [rev. by (Rothenberg, 2011)].
Close analysis of conditional knockouts that affect expression of certain transcription factors in the DN stages shows that the molecular basis of these alterations in response depends on at least three mechanisms. One is “commitment by subtraction” of PU.1: the silencing of PU.1 between DN2a and DN3a cuts a regulatory tie between the T-cell precursors and the granulocyte/monocyte lineage. Runx1, GATA-3, and possibly also TCF-1 are implicated in this repression (Huang et al., 2008; Rosenbauer et al., 2006; Scripture-Adams et al., 2014; Taghon et al., 2007; Zarnegar et al., 2010). A second mechanism is “commitment by addition” of GATA-3: from the earliest stage of GATA-3 expression, even at a low level, it completely blocks access to the B-cell fate through a still-uncharacterized mechanism (García-Ojeda et al., 2013; Scripture-Adams et al., 2014). A third is “commitment by addition” of Bcl11b (Ikawa et al., 2010; Li et al., 2010a; Li et al., 2010b).
A unique role for GATA-3 in the initial exclusion of B-cell potential is implied by the fact that GATA-3-deficient DN2 cells, unlike any normal DN2 cells, can still trans-differentiate into B cells if Notch signaling is withdrawn (García-Ojeda et al., 2013; Scripture-Adams et al., 2014). The direct targets of repression by GATA-3 that are relevant for B-cell development are not known but can be partially inferred from the genes that are upregulated in DN2 cells lacking GATA-3. Potential candidates include Bcl11a and possibly also PU.1 and SpiB, all of which play roles in B-cell precursor viability (Liu et al., 2003; Schweitzer and DeKoter, 2004; Yu et al., 2012c).
At the final commitment transition, as Bcl11b is upregulated, the cells become dependent on Notch signals and unable to survive long enough to give rise to anything else if Notch-Delta interaction is withdrawn. The lineage commitment role of Bcl11b also includes lineage-selective repressive effects. There are two effects of Bcl11b: a hit-and-run role in crossing the commitment threshold, and a permanent role in guarding against NK cell gene expression. Pro-T cells that have never expressed Bcl11b at all can become highly proliferative for weeks in culture, as long as strong Notch signals are continuously provided in the environment, and these cells continue to express multiple stem/progenitor genes. Thus, the absence of Bcl11b prevents them from triggering the repressive mechanisms that terminate the progenitor-gene-dominated state (Ikawa et al., 2010; Li et al., 2010a). However, if Bcl11b is deleted after commitment, the stem/progenitor regulatory genes and myeloid-associated genes like PU.1 do not resume expression; only the NK program is activated. This implies that the role of Bcl11b in the silencing of stem and progenitor genes can be a hit-and-run transitional one, unnecessary once the “phase 1” regulatory gene network is dismantled.
In contrast, the mechanism blocking NK and other ILC gene expression is a repressive effect that Bcl11b, once expressed, must exert continuously, especially if Notch signals are not sustained (Li et al., 2010a; Li et al., 2010b). The nature of the stimulus that would activate this substantial effector gene battery is not clear, but could reflect an ILC-like increased ability to use cytokine signals without TCR signals as priming activators for effector function. Removal of Bcl11b at any stage enables these genes to turn on within 48 hr if cytokine stimulation is present (Li et al., 2010b). Bcl11b-deficient DP cells also spontaneously activate genes that would normally only turn on as a result of positive selection, suggesting a reduced threshold for activation or reduced checkpoint enforcement at the DP stage (Kastner et al., 2010). Even a hypomorphic allele of Bcl11b that allows T cell development in general converts positively selected CD8 cells into “innate-like” cells, with many ILC-like genes upregulated (Hirose et al., 2015). The interactions of Bcl11b with the T-cell gene regulatory network thus link it both to the transitional process of T-cell identity generation and to the subtler differences in activation response that distinguish conventional from innate-like T cells.
V. From transcription factors to developmental process: glimpses of regulatory circuitry
A. Contextual modulation of transcription factor activity
The transcription factors that drive T cell development are all required, but this simple genetic observation leaves the core mechanistic questions about how they work in T-cell development unanswered. First, how are transcription factors deployed to serve different functions at different stages, and in different lineage contexts? In this context, what “placeholders” may keep a memory of lineage defining changes? Second, do transcription factor roles in early T-cell development operate in distinct subcircuits? What are the respective roles of repression and activation in the roles of key factors? Finally, how is modularity of different T-cell identity elements organized in terms of transcription factor action? The kinds of data that need to be marshalled to address these questions are becoming much more accessible, but are all imperfect. It is now possible to map the genome-wide distributions of many transcription factors’ binding in vivo by chromatin immune precipitation analyzed by deep sequencing (ChIP-seq), provided that good antibodies and large, well-characterized cell populations are available. However, sites of binding do not necessarily correspond to sites of specific function. Perturbation experiments are crucial, but because transcription factor effects are stage specific, gain or loss of function experiments must be carried out in a restricted developmental time frame in order to make comparisons with controls meaningful. This is often difficult because the kinetics of Cre deletion, RNA interference, and turnover of pre-existing RNA and protein can be slow compared to developmental progression in some important intervals. Thus current evidence has limitations that will need to be overcome with more sophisticated technology in the future. Nevertheless, it is already possible to glimpse some relationships between the activity spectra of key transcription factors in early T cell precursors that may point toward features of the fundamental modules of T-lineage developmental programming. In this final section, we review available evidence for the ways combinations of transcription factors coordinate and cross-regulate each other’s effects during the pivotal DN2 stages when T-cell identity is being selected.
1. Preferential recruitment by pioneer and placeholder factors
As ChIP-seq has been used increasingly to document transcription factor binding genome-wide, it has become clear that the binding patterns of transcription factors depend on developmental context. Lineage-specific factor combinations and often the presence or absence of particular stimulus-dependent factors cause redeployment of a given transcription factor to different genomic sites. For example, PU.1 is used in B cells, macrophages, progenitors, and early T cells, and occupies largely different sets of genomic sites in these different hematopoietic cells (Ghisletti et al., 2010; Heinz et al., 2010; Heinz et al., 2013; Ostuni et al., 2013; Zhang et al., 2012b). In turn, however, any transcription factors that depend on interaction with PU.1-bound DNA to stabilize their own binding will be recruited preferentially to the appropriate lineage-specific target sites, where PU.1 already resides (Ostuni and Natoli, 2013): thus, genomic sites already rich in transcription factor occupancy can get richer. Stabilization can involve direct factor-factor interactions or simply a displacement of nucleosomes from the neighborhood of DNA sites where transcription factors are engaged (Barozzi et al., 2014). A striking demonstration of the collective binding that can result is the pattern of multiplex transcription factor occupancy seen across the genome in hematopoietic progenitor cells (Beck et al., 2013; Wilson et al., 2010). Active enhancers, whether “super” or compact, are marked in these cells by clusters of 5–10 different transcription factors binding all together. GATA, ETS family, and bHLH family transcription factors with known roles in hematopoiesis are repeatedly seen engaged together, with Runx factors as well. The power of the interaction can be inferred because a significant fraction of the binding of individual factors appears to be occurring with grossly relaxed criteria for recognition of these factors’ normal target motifs in the DNA.
What kinds of mechanisms determine how hematopoietic factors get recruited to lineage-specific sites as T-cell specification begins? In embryonic stem cell differentiation, pioneer factors have been defined that can enter closed chromatin and render it accessible (Zaret and Carroll, 2011). There are several candidates that could provide pioneering-like activity in early T cell precursors. Runx1 is expressed in prethymic cells and throughout early T-cell development, and has target motifs highly enriched in lymphoid gene enhancers. It could at least maintain a hematopoietic identity state. Some data are available for Runx binding in DP cells (Yu et al., 2012a). However, it is not clear yet which sites Runx1 is binding in the early stages of T-cell development. Another potential candidate as an identifier of “starter” hematopoietic cis-regulatory sites is PU.1, even though its role is restricted to the early stages of T-cell specification. PU.1 protein is very stable (Kueh et al., 2013) and can continue to occupy a large fraction of its target sites even as its own RNA levels begin to drop. Initially, PU.1 is found binding to the majority of active enhancers and promoters in the ETP and DN2a cells. Although it selects different sites for engagement in T-lineage cells than it does in B or myeloid cells, this binding pattern is very stable within the first stages of T-cell development (Zhang et al., 2012b). There is a correlation between PU.1 binding sites and gene activity in these early T cells, implying that it is most often contributing to positive regulation, although only a small fraction of the genes near where it binds are PU.1 dependent for their expression (Champhekar et al., 2015). Interestingly, in ETP cells PU.1 binds to some sites that may become active in positive gene regulation only one or two stages later, such as the Il7r and Bcl11b cis-regulatory elements (Li et al., 2013; Zhang et al., 2012b). In these cases, PU.1 cannot be directing the majority of the transcriptional activity, but it may be helping to give access to the factors that will drive expression later. However, functionally important sites where PU.1 would bind in multipotent progenitors or B cells can be efficiently occluded from PU.1 binding in pro-T cells if they are engaged in repressed chromatin structures, marked by H3K27me3 (Zhang et al., 2012b): examples include the intronic enhancer for the B-cell lineage determining gene Pax5 (Decker et al., 2009). Thus, conceivably PU.1 could contribute to T-lineage specific gene expression too, both an early placeholder and a sensor to discriminate permissive from nonpermissive chromatin states at regulatory sequences of genes for early T cells.
To open up T-cell genes de novo, of course, a T-cell specific input is required. The most abundant candidate among the first wave of factors induced by Notch signaling is TCF-1 (Tcf7 gene product). In later T cells, TCF-1 is found binding to a very large fraction of all open cis-regulatory elements (Dose et al., 2014), a distribution similar to that of the powerful lineage-determining factor Pax5 in B cells (Cobaleda et al., 2007). However, TCF-1 again is not sufficient to determine all of its own target sites; it also respects context. TCF-1 is normally T-cell specific but is also found expressed in an immortal hematolymphoid progenitor cell line, EML-c1 (Wu et al., 2012). Despite approximately equivalent numbers of binding sites in EML-c1 and in total normal thymocytes (Dose et al., 2014), the occupied sites in the two cell types are largely non-overlapping (Fig. 5A). Thus TCF-1, despite its high abundance, binds only a subset of its possible sites, forming selective complexes at distinctive cell type-specific cis-regulatory elements in the two different cellular contexts. Where it does bind, TCF-1 can be a powerful indicator of sites that are likely to be functionally important for expression. TCF-1 engages its own regulatory elements, and TCF-1 occupancy sites are major elements of the far-distal enhancer recently described for the Bcl11b gene (Germar et al., 2011; Li et al., 2013; Weber et al., 2011). Interestingly, TCF-1 binding can also distinguish between different classes of sites engaged by other T-cell specific factors, for example GATA-3.
Figure 5. GATA-3 and TCF-1 DNA binding sites across the genome in different cellular contexts: higher developmental stability in sites of co-occupancy.
The figure summarizes ChIP-seq data from EML-c1 progenitor cells (A), DP T-lineage cells (A–C), and ETP (DN1) pro-T cells (B–C). The Venn diagrams show the extent of overlap between the sequences recovered as binding sites for the two factors, or the same factors in different contexts. Data for GATA-3 in ETP and DP cells were taken from (Zhang et al., 2012b), data for TCF-1 in EML-c1 cells were from (Wu et al., 2012), and data for TCF-1 in DP cells were from (Dose et al., 2014). Peak calling of mm9 aligned sequences was performed with the HOMER package (Heinz et al., 2010) and filtered for a peak score ≥ 15. Peaks were identified using findPeaks.pl with the –style factor parameter and normalized to sequencing inputs; overlapping peaks were identified and Venn-diagram parameters were retrieved using mergePeaks.pl (default parameters with the Venn-diagram option).
A) TCF-1 overlapping and non-overlapping ChIP-seq peaks in EML-C1 progenitor cells (Gene Expression Omnibus accession number GSE31221) and DP thymocytes (GSE46662).
B) GATA-3 overlapping and non-overlapping ChIP-seq peaks in ETP (DN1) T-cell precursors and DP thymocytes (GSE31235). The poor overlap is also noted with a different graphical presentation in (Zhang et al., 2012b).
C) Overlap between Gata3 sites in DP cells shared with TCF-1 bound sites in DP cells and the Gata3 sites in ETP cells shared with TCF-1 bound sites in DP cells. Note that these sites are far more likely to overlap than total GATA-3 sites.
D) Evidence that occupancy involves recurrent partner factors: enrichment of motifs for GATA-3, TCF-1 [TCF (HMG)], and additional factors at GATA-3 and TCF-1 binding sites in DP cells. HOMER was used for de novo motif analysis within a 200 bp window of the sites of GATA-3 and/or TCF-1 occupancy in DP cells. Bold type identifies the most common co-enriched motifs, present at >15% of occupancy sites. Note the recurrent co-enrichment of ETS and Runx family motifs.
As noted above, GATA-3 can be a problematic factor to study by ChIP-seq target mapping. In some respects, GATA-3 could play a role as a pioneer, because it does not appear to be excluded by Polycomb-repressed chromatin, and it binds to some future enhancer sites at least one to two stages before they become active at all. For example, its binding to functionally important cis-regulatory sites of Cd3d and the Rag1,2 genes anticipates the activation of these genes by at least one stage (Zhang et al., 2012b). However, as early T-cell development progresses, the sites bound by GATA-3 shift substantially from stage to stage, suggesting that its interactions are highly dependent on partner factors. Fig. 5B shows that only a minority of the sites bound by GATA-3 in the DN1 (ETP) stage overlap with sites bound by GATA-3 in DP cells, and vice versa. However, if one focuses on GATA-3 sites adjacent to sites where TCF-1 binds [sample mostly consisting of DP cells (Dose et al., 2014)], there is a far more consistent pattern of GATA-3 occupancy. In these cases, over 85% of the sites where GATA-3 starts binding in the ETP stage still retain GATA-3 binding by the DP stage (Fig. 5C). Note that these are also predicted to be sites for collaborating factors of the ETS and Runx family (Fig. 5D). Many of these sites are associated with functionally important genes for T-cell development, including Cd247 (TCRζ), Itk, Jak1, the positive selection gene Tespa1 (Themis, GASP), the TCRα and TCRβ enhancers (near Dad1 and Prss2, respectively), Il9r, and transcription factor genes Zfpm1 (FOG1), Tox2, and Tcf7 itself [data from (Dose et al., 2014; Zhang et al., 2012b)], among many others. Intriguingly, again, although the local GATA-3 binding is already established by ETP stage, some of these genes like Tespa1 and the elements rearranging to generate TCRα are activated only in later stages of T-cell development, long after the ETP stage: additional cases where GATA-3 occupancy greatly precedes activation. This set of composite sites might thus mediate unusually stable assemblies of GATA-3 and TCF-1 together that guide the construction of a T-cell identity.
2. Notch signaling: modulator as well as independent input
The transcription factors that work in the cell during the earliest phases of T-cell specification have their effects influenced by the ongoing level of Notch signaling. In part this is due to Notch-activated target genes like Hes1, which encode transcription factors that themselves add to the regulatory mix. As a repressor, Hes1 can silence the expression of other regulatory genes that could hold back T cell development or promote alternative fates: for example, it can repress the myeloid factor C/EBPα (De Obaldia et al., 2013). There may also be additional mechanisms through which Notch signaling impacts the activities of other factors in the cells so as to change their roles. This can be seen when other transcription factor levels are manipulated in T cell precursors in the presence or absence of Notch signaling. If T cell precursors are forced to express PU.1 at elevated levels, this can shut off the T cell regulatory gene program and rapidly activate a myeloid program if Notch signaling is absent (Del Real and Rothenberg, 2013; Franco et al., 2006; Laiosa et al., 2006). However, if Notch signaling continues, it can provide a buffer against repression of T-cell genes by PU.1, directly or indirectly protecting a wide spectrum of T-cell regulatory genes including Tcf7, Ets1, Zfpm1, Myb, and even Gata3 (Del Real and Rothenberg, 2013). Also, in the presence of Notch signals, overexpressed PU.1 activates a select spectrum of target genes, with less activation of myeloid-specific genes like Csf1r, while continuing to activate progenitor-associated target genes like Bcl11a (Del Real and Rothenberg, 2013). The selectivity may directly reflect the specific repression of C/EBPα factor expression by Hes1 (De Obaldia et al., 2013). Where PU.1 drives myeloid-specific gene expression, C/EBP family factors very frequently need to be co-recruited (Heinz et al., 2013), but not at PU.1 targets in B cells and progenitors (Heinz et al., 2010). Thus, when Notch induces expression of Hes1, silencing of Cebpa may redirect PU.1 to a more progenitor-cell like spectrum of target genes.
Interestingly, Notch signaling can also modulate the responses to transcription factors that are more conventionally T-cell specific, like GATA-3. Despite its crucial function in T-cell development, GATA-3 activity to support the T-cell program is acutely dose-dependent, with inhibitory effects at high doses as well as failures to support survival or developmental progression at low doses (Scripture-Adams et al., 2014; Taghon et al., 2007; Xu et al., 2013). In both respects, GATA-3 effects depend on interaction with Notch signaling status. High-level GATA-3 itself is antagonistic to the T-cell program (Taghon et al., 2007; Xu et al., 2013), and this is a complex function of its tendency to drive alternative lineage fates that are antagonized by Notch as well as its tendency to promote death when Notch signaling is high. Of particular interest, high-level GATA-3 can become repressive for TCF-1 (Tcf7) expression itself, but only when Notch signals are removed (Taghon et al., 2007). This contrasts dramatically with the effects of normal levels of GATA-3 which enhance Tcf7 in the presence of Notch signals. Conversely, only when Notch signaling is present do hematopoietic precursors become dependent on a basic threshold level of GATA-3 for survival (Scripture-Adams et al., 2014). This is well established in the T-cell system, but interestingly it may also be true of the most Notch-dependent ILC lineage (ILC2), which is GATA-3 dependent (Hoyler et al., 2012; Radtke et al., 2013; Serafini et al., 2014; Yagi et al., 2014), and has recently been shown to require TCF-1 as well (Yang et al., 2013). A well-balanced Notch-GATA-3 collaboration may thus be a fundamental regulatory circuit element deployable in both T and ILC contexts.
B. Competitive circuitries pacing the onset of T-cell gene expression
1. PU.1-Notch mutual antagonism
Whereas Notch signaling modulates the effects of PU.1 on its targets, especially protecting T-cell genes from repression, the relationship between Notch and PU.1 in pro-T cells is actually a bistable, competitive switch. Recent data imply that PU.1 itself works dose-dependently to blunt the efficiency of Notch signal-induced target gene activation (Champhekar et al., 2015; Del Real and Rothenberg, 2013). PU.1 seems to accomplish this by activating a gene or genes that inhibit Notch transcriptional activity indirectly (Champhekar et al., 2015). The identities of these intermediates are still uncertain. However, even in the context of sustained access to Notch ligands, both early and later-stage Notch-activated genes alike increase expression in ETP and DN2 cells when normal endogenous PU.1 is reduced or neutralized (Champhekar et al., 2015). This implies that PU.1 and Notch are competitively restraining each other throughout the early stages of T-cell development, when PU.1 levels are high. The competition is biased against PU.1 as long as Notch signaling persists, because eventually Notch-activated transcription factors including GATA-3 and also Runx1 will ultimately turn Spi1 off (Huang et al., 2008; Scripture-Adams et al., 2014; Zarnegar et al., 2010); but the mutual antagonism exacerbates the power of PU.1 as a driver of lineage diversion if the Notch signal is interrupted. This could therefore mean that PU.1 has a role as a threshold setter for Notch signal intensity, such that only above a certain level of Delta in the thymic environment can an adequate level of Notch signal be induced to trigger as well as to protect T-cell gene expression (Champhekar et al., 2015; Del Real and Rothenberg, 2013).
2. GATA-3 vs. PU.1
An influential guiding paradigm in the systems biology of hematopoiesis is that GATA family factors are mutually antagonistic with PU.1, and that this antagonism splits erythro-megakaryocytic precursors from granulocyte-macrophage precursors (Huang et al., 2007)(Fig. 6A, left panel). There is abundant evidence that protein-protein interactions between PU.1 and GATA factors in a cell can blunt the transactivation activity of each on its specific target genes, and also evidence that PU.1 and GATA factors can mutually repress each other’s expression at the RNA level. Different “common myeloid progenitor” subsets with skewed potential for erythroid vs. macrophage/neutrophil fates can be distinguished accordingly by their divergent expression of GATA-1 vs. PU.1 (Arinobu et al., 2007; Månsson et al., 2007; Nutt et al., 2005). Yet under certain circumstances, GATA factors and PU.1 can also collaborate, such as in certain zebrafish hematopoietic progenitors (Monteiro et al., 2011) and more stably in mammalian mast cells (Walsh et al., 2002). It is therefore clear that the antagonism depends on several elements, including not only the doses of the two factors but also on the presence of certain exclusionary cofactors, which may or may not be expressed in given contexts.
Figure 6. GATA-3 and PU.1 relationship in early T cells: not a simple bistable antagonism.
A) Two diagrams contrasting the classic view of GATA/PU.1 interactions in hematopoiesis (left) with the relationship actually seen in early T-cell precursors (right). Note that the actual relationship is missing the positive autoregulation loops for GATA-3 and PU.1 and the categorical antagonism at the protein level. Instead, Notch signaling enables an asymmetric relationship between GATA-3 and PU.1 at the transcriptional level, and effects of PU.1 and GATA-3 on each other’s activities are specific for particular target genes, a small subset of their total functional targets.
B) Genes subject to regulation by both PU.1 and GATA-3 in early T-cell precursors, shown with the direction of the effects of PU.1 and GATA-3 inferred from perturbation experiments in DN2 cells developed in vitro from fetal liver precursors (Champhekar et al., 2015; Scripture-Adams et al., 2014). Note that all combinations of effects are seen. The patterns of expression of these genes in stem/progenitor cells, T cells, and other lymphocytes, mostly from adult animals (Heng et al., 2008; Mingueneau et al., 2013; Robinette et al., 2015), are also shown as in Fig. 4B to indicate the diversity of developmental regulation patterns involved, not simply correlated with expression of PU.1 and GATA-3 themselves.
In early T-cell development, GATA-3 and PU.1 can act as antagonists. Overexpressed GATA-3 sharply downregulates PU.1, and GATA-3 deletion or inhibition with shRNA causes PU.1 mRNA levels to rise, implying that GATA-3 normally exerts at least some restraint on Spi1 (PU.1) transcription (Scripture-Adams et al., 2014; Taghon et al., 2007). GATA-3 levels, in protein and RNA expression, reach their height as PU.1 begins to be silenced. However, PU.1 has a more complex effect on GATA-3: it can repress Gata3, but this effect depends on current Notch signaling status. When Notch signals are withdrawn, PU.1 can repress Gata3 considerably, yet in the presence of strong Notch signaling, it may even slightly enhance Gata3 expression (Del Real and Rothenberg, 2013). As a result, in early T cell development under normal conditions the antagonism is not exclusive (Fig. 6A, right panel). Both factors are expressed together through most of the ETP stage and at equally high or higher levels in DN2a stage, before PU.1 expression is turned off. During this period, neither factor is completely inhibiting the other at the level of protein interference either, because both factors have significant normal roles in target gene expression throughout these precise stages (Champhekar et al., 2015; Scripture-Adams et al., 2014).
Genome-wide surveys of the functional targets of both factors imply that they have largely independent functions but some significant competitive interaction at the DN2 stage (Fig. 6B). Provisional relationships can be illustrated by the small number of genes that can be specifically affected both by PU.1 and by GATA-3 perturbations within short timespans in DN2 cells. Of ~240 genes affected by interference with PU.1 activity in DN2 cells, and ~100 genes altered by acute deletion of Gata3, only 19 genes were consistently affected by both, showing that these factors are not only global antagonists in DN2 gene expression. Furthermore, the directions of the effects on these genes were not always reciprocal. For three of the genes, i.e. transcription factor Bcl11a, tumor necrosis factor receptor family member Relt, and immunoglobulin superfamily member Lair1, PU.1 appeared to be an activator and GATA-3 a repressor. For seven other genes, Tcf7, Ets1, Myb, Gfra1 (glial-cell derived neurotrophic factor family receptor α1), Zfpm1 (FOG-1), Rag1, and Kit, GATA-3 appeared to be an activator and PU.1 a repressor. However, seven additional genes appeared positively regulated by both factors, and two important genes, Dtx1 and Spi1 (PU.1) itself, appeared to receive at least partially negative inputs from both. In general, PU.1 positively regulated genes with their major phases of expression in progenitors and non-T cells, whereas GATA-3 positively regulated T-cell genes and genes used by other GATA-expressing lineages, such as mast cells (Cpa3, Kit); but the expression patterns were quite varied. Thus, both factors contributed largely independent regulatory activities to the cells, using the full range of possible combinations of interactions.
Importantly, the kind of negative regulation that is seen in such experiments is weak “damping repression” rather than “silencing repression”. For targets like Myb and Zfpm1, PU.1 has the potential to extinguish expression completely, but it does not do so as long as Notch signaling is sustained (Del Real and Rothenberg, 2013). Thus, these factors reduce the maximum levels of expression of target genes but allow considerable expression to continue. In marked contrast to the classic bistability models in which GATA factors and PU.1 are each positively autoregulatory (Fig. 6A left) (Chickarmane et al., 2009; Huang et al., 2007), in pro-T cells these factors even exert level-stabilizing damping repression upon their own regulatory elements (Champhekar et al., 2015; Scripture-Adams et al., 2014; Taghon et al., 2007).
In this context, GATA-3 and PU.1 can provide positive and negative inputs in all combinations during a sustained period of cohabitation, and their partial, graduated effects on target genes can work additively over a wide dynamic range. PU.1 is free to work to promote expression of multiple genes that can contribute to the viability, stimulation responsiveness, migration behavior, and cytoskeletal dynamics of the developing thymocytes (Champhekar et al., 2015), even while GATA-3 is being turned on and for multiple cell divisions thereafter. However, the select set of genes on which PU.1 and GATA-3 do exert opposing effects include regulatory genes of great significance for the control of T-cell developmental progression. Because of the effects on these specific genes, the tipping of the PU.1/GATA-3 balance has the potential to act as an important T-cell developmental control point.
C. GATA-3, E protein, and Bcl11b circuitry
Although Notch triggers feed-forward positive circuitry to activate the T-cell program, direct control by Notch-dependent transcription does not account for the levels of expression of most T-cell genes. Thus, the Notch-triggered T-cell factors must be involved in the sequential activation of T-cell genes from ETP to DN3a stage. For example, when prethymic precursor cells are forced to express Tcf7 in gain of function experiments, they can activate a near-complete spectrum of DN3-like genes, except for particular Notch targets, without apparent input from the Notch pathway (Weber et al., 2011). Although GATA-3 is essential for pro-T cell viability and development, it alone does not account for the activation of the T-cell program. The most target genes regulated sensitively by GATA-3 in DN2 cells include a few with known or likely T-cell functions: positive targets Kit, Zfpm1 (FOG-1, a cofactor of GATA factors), Tcf7, Ikzf2 (Helios, a member of the Ikaros family expressed early in T progenitors and strongly expressed later in Tregs), and transcripts from TCRγ-C1; and negatively regulated targets Bcl11a, Spi1 (PU.1), and a few others. Some of the GATA-3 activated genes indeed are being turned on in DN2 stage for sustained use in the T-cell program or being primed for future activation, whereas others like Kit, Cpa3, Fgf3, Cd93, and Akr1c13 are expressed only transiently in early stages and are actually shut off during commitment. To account for the expression patterns of these genes, and to make the cells turn on additional T-cell genes, additional positive inputs are needed.
One of these inputs is from E proteins. E proteins are needed to activate many genes that distinguish the T-cell gene expression program from progenitors, NK cells, and ILCs alike, mostly during during DN2b and DN3a stages (Fig. 3A). Comparing the transcriptomes of wildtype and E2A-knockout DN2 cells (Xu et al., 2013) shows that Cd3g, Cd3e, Lat, Rag1, and genes coding for the transcription factors Bcl11b and Gfi1 are all E2A-dependent. At least by the DN3a stage, E protein activity is also important to maintain maximal levels of Notch1 itself (Yashiro-Ohtani et al., 2009). E proteins act both positively and negatively, and it is significant that in developing DN2 cells they show repressive effects on regulatory and effector genes Rora, Zbtb16, Ahr, Fcer1g, Id2, Gzmb, Zfp105 and Lmo4, all of which are normally most expressed in innate lymphoid cells. Thus, establishing a strong E protein activity level is a major contributor to the gene expression identity of the developing T cells.
Interestingly, two of the regulatory genes that are subject to damping repression by E2A are those coding for GATA-3 itself and its target and cofactor, Zfpm1 (FOG-1). This result groups the essential T-cell factor GATA-3 and Zfpm1 together with the innate-cell genes which need to be kept repressed, and are only released from repression when E2A is deleted. Kee and colleagues have shown that the 2–3 fold increase in GATA-3 activity that occurs with loss of E2A is a major contributor to the developmental arrest that E2A mutant cells undergo, and the cells can be rescued by using shRNA to knock down the GATA-3 to more normal levels (Xu et al., 2013). In other words, the excess of GATA-3 is more deleterious to T cell development than the reduction of E2A itself.
The effects of E2A have some arresting similarities to those of Bcl11b in the DN2-DN3 stages. Bcl11b also is needed to keep many ILC-specific genes off, and especially to limit activation of the ILC1 and NK cell genes. Although genome-wide analysis of Bcl11b knockout cells has only been published so far for later-stage thymocytes (Hirose et al., 2015; Kastner et al., 2010; Li et al., 2010b), Bcl11b knockout DN2 cells also upregulate Zbtb16, Nfil3, Id2, Fcer1g, Zfp105, Gzmb, and Prf1 (perforin) as well as turning on expression of numerous NK receptors (Li et al., 2010a) (unpublished results, J. A. Zhang, L. Li, & E.V.R.). Despite the fact that they work together in normal T-cell development to exclude alternative lineage fates, Bcl11b probably also exerts damping repression on GATA-3, very much like E2A. The roles of Bcl11b and E2A are so similar that they can be provisionally considered as part of one pathway. Furthermore, GATA-3 is also linked to Bcl11b, in a classic incoherent feed-forward circuit. GATA-3 is needed to activate Bcl11b itself (García-Ojeda et al., 2013; Scripture-Adams et al., 2014), only to be damped down by Bcl11b activity as it comes on. Thus although GATA-3 and E proteins may collaborate in the activation of crucial pan-T cell genes such as those coding for CD3 proteins, another of the effective collaborators of E proteins is Bcl11b, and its role may be to prevent the effects of high-level GATA-3.
The relationships between direct and indirect action remain to be clarified in this process. One of the key targets of Bcl11b repression is Id2, in the sense that any reduction in Bcl11b dosage rapidly enables cells to upregulate Id2. This enables high-level Bcl11b to collaborate strongly with E2A/HEB to maintain the characteristics of an E protein-dominant state. However, lower levels of Bcl11b can be compatible with Id2 expression, for example in effector CD8 T cells where both are expressed (Kaech and Cui, 2012; Masson et al., 2013; Yang et al., 2011a). It has even been found that Bcl11b at moderate levels is actually required for ILC2 cell development, despite these innate lymphocytes’ constitutive requirement for Id2 expression (Califano et al., 2015; Walker et al., 2015; Yu et al., 2015). There, Bcl11b collaborates with GATA-3, TCF-1, and Gfi1 to favor ILC2 as opposed to ILC3, ILC1, or NK development. Thus, Bcl11b itself is probably dose-dependent, playing an integral role in a network circuit to guide effector subtype specialization at low levels, even while it can support the roles of E proteins in enforcing activation thresholds and disfavoring any innate-like pathway when it is expressed at high levels.
VI. Conclusions: T cell development as an emergent process
The elements identified here reveal ways that Notch-dependent progression through the early stages of T cell development not only turns on key regulators like GATA-3, TCF-1, and increased levels of HEB, but also engages them in a succession of circuit interactions. A feature of most of these circuits is their ability to channel the impacts of the component transcription factors either by level control or by ensuring that they are directed to a particular subset of target genes. This is most obvious with the interaction between Notch signaling and PU.1, but it is also a feature of the circuitry connecting GATA-3 and Notch signaling, and GATA-3 with the commitment climax factors Bcl11b and E proteins. Recurrent features are the dose-dependence of transcription factor effects in this system, policed normally by incoherent feed-forward gene network circuits that constrain factor activities within an appropriate range, and the stage-dependence of factor actions that implies that each successive stage creates new “facts on the ground” as a point of departure for the next stage.
We can speculate that the earliest stages of T-cell development, represented in ETP cells, bring PU.1 activities under the control of Notch modulation. A key component is likely to be the early activation of Hes1, which deprives PU.1 of its pro-myeloid partner C/EBPα and forces it to regulate selective, pro-T-cell compatible genes. The situation can change when Notch signaling can activate GATA-3 and TCF-1, which collaborate with E proteins already present to begin to activate T-cell identity genes. There is still much to learn about the roles of the other “phase 1” factors, but the cells probably cannot experience a maximal intensity of Notch signal as long as PU.1 levels are high. The collaboration of GATA-3, TCF-1, and pre-existing factors like Runx1 can finally tip the balance to a new state, when PU.1 with Lyl1 and other progenitor factors is downregulated. As the restraints on Notch signal strength are lifted during commitment, new regulators like Bcl11b are activated by the convergence of T-lineage regulators, and the cells can become Notch-dependent for viability, thus ending lineage plasticity. Dilution of the competitor Lyl1 as HEB levels rise may now unleash high E protein activity as well. Strong RAG expression can begin, and selection checkpoints are imposed on the cells. Although some of the factors they express could give them access to effector function directly, the cells can no longer take advantage of this. They can now begin a future as T cells in which all the other effector capabilities they may acquire will be linked to a permanent dependence on TCR signals.
Acknowledgments
The authors thank members of the Rothenberg group for discussion of the ideas presented here and for sharing relevant results before publication. Research in the Rothenberg lab was supported by grants from the USPHS, AI083514, AI095943, HD076915, and HL119102. J. U. was supported by the Swedish Research Council (Linköping University) and E. V. R. was supported by the Albert Billings Ruddock Professorship of Biology.
References
- Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu DI, VanValkenburgh J, Morin N, Califano D, Jenkins NA, Copeland NG, Liu P, Avram D. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci U S A. 2011;108:6211–6216. doi: 10.1073/pnas.1014304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23:220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- Antebi YE, Reich-Zeliger S, Hart Y, Mayo A, Eizenberg I, Rimer J, Putheti P, Pe’er D, Friedman N. Mapping differentiation under mixed culture conditions reveals a tunable continuum of T cell fates. PLoS Biol. 2013;11:e1001616. doi: 10.1371/journal.pbio.1001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Avram D, Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J Immunol. 2014;193:2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- Bain G, Romanow WJ, Albers K, Havran WL, Murre C. Positive and negative regulation of V(D)J recombination by the E2A proteins. J Exp Med. 1999;189:289–300. doi: 10.1084/jem.189.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunaite G, Ceredig R, Fehling HJ, Zúñiga-Pflücker JC, Rolink AG. The role of Notch and IL-7 signaling in early thymocyte proliferation and differentiation. Eur J Immunol. 2005;35:1292–1300. doi: 10.1002/eji.200425822. [DOI] [PubMed] [Google Scholar]
- Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D, Thoms JA, Perera D, Schutte J, Unnikrishnan A, Knezevic K, Kinston SJ, Wilson NK, O’Brien TA, Gottgens B, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122:e12–22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville M, O’Brien RL, Born WK. γδ T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudil A, Matei IR, Shih HY, Bogdanoski G, Yuan JS, Chang SG, Montpellier B, Kowalski PE, Voisin V, Bashir S, et al. IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte β-selection. Nat Immunol. 2015;16:397–405. doi: 10.1038/ni.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano D, Cho JJ, Uddin MN, Lorentsen KJ, Yang Q, Bhandoola A, Li H, Avram D. Transcription factor Bcl11b controls identity and function of mature Type 2 innate lymphoid cells. Immunity. 2015 doi: 10.1016/j.immuni.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champhekar A, Damle SS, Freedman G, Carotta S, Nutt SL, Rothenberg EV. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev. 2015;29:832–848. doi: 10.1101/gad.259879.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Kronenberg M. Activation and Function of iNKT and MAIT Cells. Adv Immunol. 2015;127:145–201. doi: 10.1016/bs.ai.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Chari S, Umetsu SE, Winandy S. Notch target gene deregulation and maintenance of the leukemogenic phenotype do not require RBP-Jκ in Ikaros null mice. J Immunol. 2010;185:410–417. doi: 10.4049/jimmunol.0903688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Rothenberg EV. Molecular basis for developmental changes in interleukin-2 gene inducibility. Mol Cell Biol. 1993;13:228–237. doi: 10.1128/mcb.13.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V, Enver T, Peterson C. Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput Biol. 2009;5:e1000268. doi: 10.1371/journal.pcbi.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential Notch dependency at the αβ and γδ T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB, Monach P, Shinton SA, et al. ImmGen Project C. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–99. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, Bendelac A. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci U S A. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010a;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz LM, Yang CY, Goldrath AW. Transcriptional regulation of NKT cell development and homeostasis. Curr Opin Immunol. 2010b;22:199–205. doi: 10.1016/j.coi.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Obaldia ME, Bell JJ, Wang X, Harly C, Yashiro-Ohtani Y, Delong JH, Zlotoff DA, Sultana DA, Pear WS, Bhandoola A. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 2005;106:3498–3506. doi: 10.1182/blood-2005-02-0496. [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–1219. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Santo JP. Staying innate: transcription factor maintenance of innate lymphoid cell identity. Immunol Rev. 2014;261:169–176. doi: 10.1111/imr.12202. [DOI] [PubMed] [Google Scholar]
- Diamond RA, Ward SB, Owada-Makabe K, Wang H, Rothenberg EV. Different developmental arrest points in RAG2−/− and scid thymocytes on two genetic backgrounds: developmental choices and cell death mechanisms before TCR gene rearrangement. J Immunol. 1997;158:4052–4064. [PubMed] [Google Scholar]
- Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Dose M, Emmanuel AO, Chaumeil J, Zhang J, Sun T, Germar K, Aghajani K, Davis EM, Keerthivasan S, Bredemeyer AL, et al. β-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci U S A. 2014;111:391–396. doi: 10.1073/pnas.1315752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng N, Vegh P, Rothenberg EV, Yui MA. Lineage divergence at the first TCR-dependent checkpoint: preferential γδ and impaired αβ T cell development in nonobese diabetic mice. J Immunol. 2011;186:826–837. doi: 10.4049/jimmunol.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa J, Moro K, Motomura Y, Okamoto K, Zhu J, Takayanagi H, Kubo M, Koyasu S. Critical role of p38 and GATA3 in natural helper cell function. J Immunol. 2013;191:1818–1826. doi: 10.4049/jimmunol.1300379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCRαβ+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines αβ versus γδ lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ojeda ME, Klein Wolterink RG, Lemaître F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, Di Santo JP. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells. Blood. 2013;121:1749–1759. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- Garcia-Peydro M, de Yebenes VG, Toribio ML. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J Immunol. 2006;177:3711–3720. doi: 10.4049/jimmunol.177.6.3711. [DOI] [PubMed] [Google Scholar]
- Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer Le Lay AS, Oravecz A, Mastio J, Jung C, Marchal P, Ebel C, Dembélé D, Jost B, Le Gras S, Thibault C, et al. The tumor suppressor Ikaros shapes the repertoire of Notch target genes in T cells. Sci Signal. 2014;7:ra28. doi: 10.1126/scisignal.2004545. [DOI] [PubMed] [Google Scholar]
- Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108:20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Gimferrer I, Hu T, Simmons A, Wang C, Souabni A, Busslinger M, Bender TP, Hernandez-Hoyos G, Alberola-Ila J. Regulation of GATA-3 expression during CD4 lineage differentiation. J Immunol. 2011;186:3892–3898. doi: 10.4049/jimmunol.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, et al. Runx3 regulates Integrin αE/CD103 and CD4 expression during development of CD4 −/CD8 + T cells. J Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, Akashi K, Weissman IL, Fisher AG, Smale ST. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of γδTCR signaling efficiently diverts thymocytes to the αβ lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Harman BC, Jenkinson WE, Parnell SM, Rossi SW, Jenkinson EJ, Anderson G. T/B lineage choice occurs prior to intrathymic Notch signalling. Blood. 2005;106:886–892. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazenberg MD, Spits H. Human innate lymphoid cells. Blood. 2014;124:700–709. doi: 10.1182/blood-2013-11-427781. [DOI] [PubMed] [Google Scholar]
- He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu Rev Immunol. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- He YW, Beers C, Deftos ML, Ojala EW, Forbush KA, Bevan MJ. Down-regulation of the orphan nuclear receptor RORγt is essential for T lymphocyte maturation. J Immunol. 2000;164:5668–5674. doi: 10.4049/jimmunol.164.11.5668. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TSP, Painter MW Consortium TIGP. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hernández-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- Hirose S, Touma M, Go R, Katsuragi Y, Sakuraba Y, Gondo Y, Abe M, Sakimura K, Mishima Y, Kominami R. Bcl11b prevents the intrathymic development of innate CD8 T cells in a cell intrinsic manner. Int Immunol. 2015;27:205–215. doi: 10.1093/intimm/dxu104. [DOI] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi K, Negishi N, Tsuchiya I, Abe N, Hirano K, Suzuki D, Yamamoto M, Engel JD, Habu S. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur J Immunol. 2008;38:977–985. doi: 10.1002/eji.200737688. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12:157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito GC, Dekker JD, Wang YH, Lee BK, Shaffer AL, 3rd, Lin J, Wall JK, Lee BS, Staudt LM, Liu YJ, et al. Dendritic cell fate is determined by BCL11A. Proc Natl Acad Sci U S A. 2014;111:E998–1006. doi: 10.1073/pnas.1319228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of β-catenin and γ-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. E protein transcription factors are required for the development of CD4+ lineage T cells. Immunity. 2012;36:348–361. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo JM, Schatz DG, Sun JC. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 2014;159:94–107. doi: 10.1016/j.cell.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur J Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathrein KL, Chari S, Winandy S. Ikaros directly represses the Notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem. 2008;283:10476–10484. doi: 10.1074/jbc.M709643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, Morgan BA. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- Klein Wolterink RGJ, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, Veiga Fernandes H, Hendriks RW, Di Santo JP. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci U S A. 2013;110:10240–10245. doi: 10.1073/pnas.1217158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinmann E, Geimer Le Lay AS, Sellars M, Kastner P, Chan S. Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol. 2008;28:7465–7475. doi: 10.1128/MCB.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy V, Carr T, de Pooter RF, Akinola EO, Gounari F, Kee BL. Repression of Ccr9 transcription in mouse T lymphocyte progenitors by the Notch signaling pathway. J Immunol. 2015;194:3191–3200. doi: 10.4049/jimmunol.1402443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 2013;341:670–673. doi: 10.1126/science.1240831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol. 2005;174:6184–6194. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010a;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, Gounari F, Rothenberg EV. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010b;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chi AW, Arnett KL, Chiang MY, Xu L, Shestova O, Wang H, Li YM, Bhandoola A, Aster JC, et al. Notch dimerization is required for leukemogenesis and T-cell development. Genes Dev. 2010a;24:2395–2407. doi: 10.1101/gad.1975210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- Liu P, Li P, Burke S. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol Rev. 2010b;238:138–149. doi: 10.1111/j.1600-065X.2010.00953.x. [DOI] [PubMed] [Google Scholar]
- Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, Kawamoto H, Katsura Y. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- Luc S, Luis TC, Boukarabila H, Macaulay IC, Buza-Vidas N, Bouriez-Jones T, Lutteropp M, Woll PS, Loughran SJ, Mead AJ, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald HR, Mycko MP. Development and selection of Vα l4i NKT cells. Curr Top Microbiol Immunol. 2007;314:195–212. [PubMed] [Google Scholar]
- Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al Hashmi S, Liuba K, Thorén L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Mansson R, Welinder E, Åhsberg J, Lin YC, Benner C, Glass CK, Lucas JS, Sigvardsson M, Murre C. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci U S A. 2012;109:21028–21033. doi: 10.1073/pnas.1211427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol. 2013;190:4585–4594. doi: 10.4049/jimmunol.1300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- Masuda K, Kubagawa H, Ikawa T, Chen CC, Kakugawa K, Hattori M, Kageyama R, Cooper MD, Minato N, Katsura Y, et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26:3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Miyazaki M, Murre C. The establishment of B versus T cell identity. Trends Immunol. 2014;35:205–210. doi: 10.1016/j.it.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjösberg J, Bernink J, Peters C, Spits H. Transcriptional control of innate lymphoid cells. Eur J Immunol. 2012;42:1916–1923. doi: 10.1002/eji.201242639. [DOI] [PubMed] [Google Scholar]
- Monteiro R, Pouget C, Patient R. The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. EMBO J. 2011;30:1093–1103. doi: 10.1038/emboj.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, et al. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. Int Immunol. 2011;23:661–668. doi: 10.1093/intimm/dxr078. [DOI] [PubMed] [Google Scholar]
- Naito T, Taniuchi I. The network of transcription factors that underlie the CD4 versus CD8 lineage decision. Int Immunol. 2010;22:791–796. doi: 10.1093/intimm/dxq436. [DOI] [PubMed] [Google Scholar]
- Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, Kornfeld H, Xiong N, Cohen NR, Brenner MB, et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol. 2012;13:511–518. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Cabaco H, Caramalho I, Sepulveda N, Sousa AE. Differentiation of human thymic regulatory T cells at the double positive stage. Eur J Immunol. 2011;41:3604–3614. doi: 10.1002/eji.201141614. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat Rev Immunol. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Natoli G. Lineages, cell types and functional states: a genomic view. Curr Opin Cell Biol. 2013;25:759–764. doi: 10.1016/j.ceb.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Pallard C, Stegmann AP, van Kleffens T, Smart F, Venkitaraman A, Spits H. Distinct roles of the phosphatidylinositol 3-kinase and STAT5 pathways in IL-7-mediated development of human thymocyte precursors. Immunity. 1999;10:525–535. doi: 10.1016/s1074-7613(00)80052-7. [DOI] [PubMed] [Google Scholar]
- Park JH, Yu Q, Erman B, Appelbaum JS, Montoya-Durango D, Grimes HL, Singer A. Suppression of IL7Rα transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Phelan JD, Saba I, Zeng H, Kosan C, Messer MS, Olsson HA, Fraszczak J, Hildeman DA, Aronow BJ, Möröy T, et al. Growth factor independent-1 maintains Notch1-dependent transcriptional programming of lymphoid precursors. PLoS Genet. 2013;9:e1003713. doi: 10.1371/journal.pgen.1003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J Exp Med. 2003;198:957–962. doi: 10.1084/jem.20030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz I, Silva-Santos B, Pennington DJ. Functional development of γδ T cells. Eur J Immunol. 2013;43:1988–1994. doi: 10.1002/eji.201343759. [DOI] [PubMed] [Google Scholar]
- Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Radtke F, Macdonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- Ramond C, Berthault C, Burlen-Defranoux O, de Sousa AP, Guy-Grand D, Vieira P, Pereira P, Cumano A. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol. 2014;15:27–35. doi: 10.1038/ni.2782. [DOI] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M Immunological Genome C. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. T cell lineage commitment: identity and renunciation. J Immunol. 2011;186:6649–6655. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Annu Rev Immunol. 2014;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Saran N, Lyszkiewicz M, Pommerencke J, Witzlau K, Vakilzadeh R, Ballmaier M, von Boehmer H, Krueger A. Multiple extrathymic precursors contribute to T-cell development with different kinetics. Blood. 2010;115:1137–1144. doi: 10.1182/blood-2009-07-230821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Bendelac A. Promyelocytic leukemia zinc finger turns on the effector T cell program without requirement for agonist TCR signaling. J Immunol. 2011;186:5801–5806. doi: 10.4049/jimmunol.1100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilham MW, Wilson A, Moerer P, Benaissa-Trouw BJ, Cumano A, Clevers HC. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, Lawson GW, Bensinger SJ, Farnham PJ, Witte ON, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14:1073–1083. doi: 10.1038/ni.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS, Durum SD, Muegge K. The interleukin 7 receptor is required for T cell receptor γ locus accessibility to the V(D)J recombinase. J Exp Med. 2000;191:1045–1050. doi: 10.1084/jem.191.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J Exp Med. 2004;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9:39–46. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- Schweitzer BL, DeKoter RP. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J Immunol. 2004;172:144–154. doi: 10.4049/jimmunol.172.1.144. [DOI] [PubMed] [Google Scholar]
- Scripture-Adams DD, Damle SS, Li L, Elihu KJ, Qin S, Arias AM, Butler RR, 3rd, Champhekar A, Zhang JA, Rothenberg EV. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol. 2014;193:3470–3491. doi: 10.4049/jimmunol.1301663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehus CR, Aliahmad P, de la Torre B, Iliev ID, Spurka L, Funari VA, Kaye J. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat Immunol. 2015;16:599–608. doi: 10.1038/ni.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler MP, Mathew R, Liszewski MK, Spooner C, Barr K, Meng F, Singh H, Bendelac A. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz GT, Carotta S. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N, Klein Wolterink RG, Satoh-Takayama N, Xu W, Vosshenrich CA, Hendriks RW, Di Santo JP. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113:807–815. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K, Yamada H, Nakamura M, Hatano S, Katsuragi Y, Kominami R, Yoshikai Y. IFN-γ-producing and IL-17-producing γδ T cells differentiate at distinct developmental stages in murine fetal thymus. J Immunol. 2014;192:2210–2218. doi: 10.4049/jimmunol.1302145. [DOI] [PubMed] [Google Scholar]
- Shinnakasu R, Yamashita M, Kuwahara M, Hosokawa H, Hasegawa A, Motohashi S, Nakayama T. Gfi1-mediated stabilization of GATA3 protein is required for Th2 cell differentiation. J Biol Chem. 2008;283:28216–28225. doi: 10.1074/jbc.M804174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells - a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal FJT, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol. 2008;38:1788–1794. doi: 10.1002/eji.200738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal FJT, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, van Dongen JJ. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J Immunol. 2004;172:1099–1108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon TN, David ES, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- Thompson PK, Zúñiga-Pflücker JC. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin Immunol. 2011;23:350–359. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Tiemessen MM, Baert MR, Schonewille T, Brugman MH, Famili F, Salvatori DC, Meijerink JP, Ozbek U, Clevers H, van Dongen JJ, et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol. 2012;10:e1001430. doi: 10.1371/journal.pbio.1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- Vahedi G, Kanno Y, Sartorelli V, O’Shea JJ. Transcription factors and CD4 T cells seeking identity: masters, minions, setters and spikers. Immunology. 2013;139:294–298. doi: 10.1111/imm.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Walle I, De Smet G, De Smedt M, Vandekerckhove B, Leclercq G, Plum J, Taghon T. An early decrease in Notch activation is required for human TCR-αβ lineage differentiation at the expense of TCR-γδ T cells. Blood. 2009;113:2988–2998. doi: 10.1182/blood-2008-06-164871. [DOI] [PubMed] [Google Scholar]
- Van de Walle I, Waegemans E, De Medts J, De Smet G, De Smedt M, Snauwaert S, Vandekerckhove B, Kerre T, Leclercq G, Plum J, et al. Specific Notch receptor-ligand interactions control human TCR-αβ/γδ development by inducing differential Notch signal strength. J Exp Med. 2013;210:683–697. doi: 10.1084/jem.20121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanvalkenburgh J, Albu DI, Bapanpally C, Casanova S, Califano D, Jones DM, Ignatowicz L, Kawamoto S, Fagarasan S, Jenkins NA, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208:2069–2081. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, Van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- Vicente R, Swainson L, Marty-Gres S, De Barros SC, Kinet S, Zimmermann VS, Taylor N. Molecular and cellular basis of T cell lineage commitment. Semin Immunol. 2010;22:270–275. doi: 10.1016/j.smim.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visan I, Tan JB, Yuan JS, Harper JA, Koch U, Guidos CJ. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat Immunol. 2006;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Walker JA, Oliphant CJ, Englezakis A, Yu Y, Clare S, Rodewald HR, Belz G, Liu P, Fallon PG, McKenzie ANJ. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med. 2015;212:875–882. doi: 10.1084/jem.20142224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Anderson MK. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006a;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- Wang H, Pierce LJ, Spangrude GJ. Distinct roles of IL-7 and stem cell factor in the OP9-DL1 T-cell differentiation culture system. Exp Hematol. 2006b;34:1730–1740. doi: 10.1016/j.exphem.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xiong Y, Bosselut R. Tenuous paths in unexplored territory: From T cell receptor signaling to effector gene expression during thymocyte selection. Semin Immunol. 2010;22:294–302. doi: 10.1016/j.smim.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xie H, Huang Z, Ma J, Fang X, Ding Y, Sun Z. T cell factor 1 regulates thymocyte survival via a RORγt-dependent pathway. J Immunol. 2011;187:5964–5973. doi: 10.4049/jimmunol.1101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder E, Mansson R, Mercer EM, Bryder D, Sigvardsson M, Murre C. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci U S A. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner RS, Esplin BL, Garrett KP, Pelayo R, Luche H, Fehling HJ, Kincade PW. Asynchronous RAG-1 expression during B lymphopoiesis. J Immunol. 2009;183:7768–7777. doi: 10.4049/jimmunol.0902333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wirnsberger G, Hinterberger M, Klein L. Regulatory T-cell differentiation versus clonal deletion of autoreactive thymocytes. Immunol Cell Biol. 2011;89:45–53. doi: 10.1038/icb.2010.123. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Seay M, Schulz VP, Hariharan M, Tuck D, Lian J, Du J, Shi M, Ye Z, Gerstein M, et al. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS Genet. 2012;8:e1002565. doi: 10.1371/journal.pgen.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Carr T, Ramirez K, McGregor S, Sigvardsson M, Kee BL. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood. 2013;121:1534–1542. doi: 10.1182/blood-2012-08-449447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nat Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, et al. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- Yamane H, Paul WE. Cytokines of the γc family control CD4+ T cell differentiation and function. Nat Immunol. 2012;13:1037–1044. doi: 10.1038/ni.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011a;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Saenz SA, Zlotoff DA, Artis D, Bhandoola A. Cutting edge: natural helper cells derive from lymphoid progenitors. J Immunol. 2011b;187:5505–5509. doi: 10.4049/jimmunol.1102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Carter C, Yoshida N, Abe T, Yamada A, Nitta T, Ishimaru N, Takada K, Butcher GW, Takahama Y. Gimap3 and Gimap5 cooperate to maintain T-cell numbers in the mouse. Eur J Immunol. 2014;44:561–572. doi: 10.1002/eji.201343750. [DOI] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye SK, Agata Y, Lee HC, Kurooka H, Kitamura T, Shimizu A, Honjo T, Ikuta K. The IL-7 Receptor controls the accessibility of the TCRγ Locus by Stat5 and histone acetylation. Immunity. 2001;15:813–823. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012a;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, Salay TM, Leeds KE, Du H, Wu B, Waterman ML, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-γ. Nat Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012b;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang C, Clare S, Wang J, Lee SC, Brandt C, Burke S, Lu L, He D, Jenkins NA, et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med. 2015;212:865–874. doi: 10.1084/jem.20142318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang S, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012c;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yücel R, Karsunky H, Klein-Hitpass L, Möröy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J Immunol. 2004;173:5381–5391. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14:529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar MA, Chen J, Rothenberg EV. Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012a;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012b;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohren F, Souroullas GP, Luo M, Gerdemann U, Imperato MR, Wilson NK, Göttgens B, Lukov GL, Goodell MA. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–769. doi: 10.1038/ni.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]