Significance
γ-Secretase is a conserved and ubiquitous intramembrane-cleaving protease (I-CLiP). Its normal function is required for proper notch signaling, and its processing of amyloid precursor protein is implicated in Alzheimer’s disease. Although γ-secretase has over 90 reported substrates, little is known about the mechanism by which γ-secretase recruits its substrates while at the same time distinguishing substrate from nonsubstrate within the protein-crowded environment of cellular membranes. In contrast to previous studies, our data demonstrate that substrate transmembrane domain drives its interaction with γ-secretase. We find that the γ-secretase component nicastrin acts to sterically block substrates with large ectodomains from interacting with γ-secretase, providing the mechanism by which γ-secretase selectively recruits ectodomain-shed substrates while also preventing cleavage of nonsubstrates.
Keywords: γ-secretase, nicastrin, notch, intramembrane-cleaving protease, Azheimer’s disease
Abstract
γ-Secretase is an intramembrane-cleaving protease that processes many type-I integral membrane proteins within the lipid bilayer, an event preceded by shedding of most of the substrate’s ectodomain by α- or β-secretases. The mechanism by which γ-secretase selectively recognizes and recruits ectodomain-shed substrates for catalysis remains unclear. In contrast to previous reports that substrate is actively recruited for catalysis when its remaining short ectodomain interacts with the nicastrin component of γ-secretase, we find that substrate ectodomain is entirely dispensable for cleavage. Instead, γ-secretase–substrate binding is driven by an apparent tight-binding interaction derived from substrate transmembrane domain, a mechanism in stark contrast to rhomboid—another family of intramembrane-cleaving proteases. Disruption of the nicastrin fold allows for more efficient cleavage of substrates retaining longer ectodomains, indicating that nicastrin actively excludes larger substrates through steric hindrance, thus serving as a molecular gatekeeper for substrate binding and catalysis.
Regulated intramembrane proteolysis (RIP) involves the cleavage of a wide variety of integral membrane proteins within their transmembrane domains (TMDs) by a highly diverse family of intramembrane-cleaving proteases (I-CLiPs) (1). I-CLiPs are found in all forms of life and govern many important biological functions, including but not limited to organism development (2), lipid homeostasis (3), the unfolded protein response (4), and bacterial quorum sensing (5). As the name implies, RIP must be tightly regulated to ensure that the resultant signaling events occur only when prompted by the cell and to prevent cleavage of the many nonsubstrate “bystander” proteins present within cellular membranes. Despite this, very little is known about the molecular mechanisms by which I-CLiPs achieve their exquisite specificity. Although traditional soluble proteases maintain substrate specificity by recognizing distinct amino acid sequences flanking the scissile bond, substrates for intramembrane proteases have little to no sequence similarity.
Recent work on rhomboid proteases has demonstrated that this family of I-CLiPs achieves substrate specificity via a mechanism that is dependent on the transmembrane dynamics of the substrate rather than its sequence of amino acids (6, 7). Here, rhomboid possesses a very weak binding affinity for substrate and, in a rate-driven reaction, only cleaves those substrates that have unstable TMD helices that have had time to unfold into the catalytic active site, where they are cleaved before they can dissociate from the enzyme–substrate complex. Although it may be tempting to speculate that this is a conserved mechanism for all I-CLiPs, rhomboid is the only family of I-CLiPs that does not require prior activation of substrate through an initial cleavage by another protease (8). Specifically, site-2 protease substrates must be first cleaved by site-1 protease (9), signal peptide peptidase substrates are first cleaved by signal peptidase (10), and ectodomain shedding by α- or β-secretase is required before γ-secretase cleavage of its substrates (11, 12). These facts suggest that the diverse families of I-CLiPs likely have evolved fundamentally different mechanisms by which they recognize and cleave their substrates.
Presenilin/γ-secretase is the founding member of the aspartyl family of I-CLiPs. The importance of γ-secretase function in biology and medicine is highlighted by its cleavage of the notch family of receptors, which is required for cell fate determination in all metazoans (2, 13–16), and of the amyloid precursor protein (APP), which is centrally implicated in Alzheimer’s disease (AD) (14, 17). In addition to APP and notch, γ-secretase has over 90 other reported substrates, many of which are involved in important signaling events (12, 18). Despite this, little is known about the mechanism by which γ-secretase binds and cleaves its substrates. Currently, the only known prerequisite for a substrate to be bound and hydrolyzed by γ-secretase is that it be a type-I integral membrane protein that first has most of its ectodomain removed by a sheddase, either α- or β-secretases (11, 12, 19). How γ-secretase selectively recognizes ectodomain-shed substrates and recruits them for catalysis while at the same time preventing cleavage of nonsubstrates remains unsettled.
γ-Secretase is a multimeric complex composed of four integral membrane proteins both necessary and sufficient for full activity: presenilin, nicastrin, Aph-1, and Pen-2 (20–24). Presenilin is the proteolytic component, housing catalytic aspartates on TMDs 6 and 7 of its nine TMDs (17, 25, 26). After initial complex formation, the mature proteolytically active complex is formed when presenilin undergoes auto-proteolysis, resulting in N- and C-terminal fragments (NTF and CTF, respectively) (17, 27, 28), a process thought to be stimulated by the three-TMD component Pen-2 (29). The seven-TMD protein Aph-1 is believed to play a scaffolding role in complex formation (30, 31). Nicastrin is a type-I integral membrane protein with a large, heavily glycosylated ectodomain (32–34) that contains multiple stabilizing disulfide bridges (24, 34).
The ectodomain of nicastrin is structurally homologous to a bacterial amino peptidase (34). Although nicastrin lacks the specific amino acids required for peptidase activity, it has been proposed to bind the N terminus of ectodomain-shed substrate, thereby directing substrate TMD to the γ-secretase active site for cleavage (35, 36). This mechanism has been suggested to depend on a key binding interaction between the free amine at the N terminus of the shortened substrate ectodomain and E333 of the vestigial amino peptidase domain of nicastrin (35, 36). However, the importance of nicastrin in substrate recognition has been questioned (37, 38), and although an initial high-resolution structure of γ-secretase suggested a role for nicastrin in substrate recognition (24), the most recent structures of the γ-secretase complex and the nicastrin ectodomain reveal that E333 is actually buried within the interior of nicastrin and resides on the opposite side of the complex relative to the active site (39, 40). Although this makes it unlikely that nicastrin is involved in direct substrate binding barring a large, energy-intensive conformational change, the basic mechanism of substrate recognition by γ-secretase remains controversial and requires resolution.
Here, we demonstrate that nicastrin functions to sterically exclude substrates based on ectodomain size rather than actively recruit them for catalysis. This blocking mechanism allows γ-secretase to distinguish substrate from nonsubstrate and explains why substrate ectodomain shedding by α- or β-secretases is a prerequisite for γ-secretase catalysis. In contrast to rhomboid, γ-secretase apparently binds substrate TMD tightly, making the nicastrin steric hindrance mechanism necessary to prevent cleavage of nonectodomain-shed substrates and nonsubstrates alike.
Results
The Neo-Ectodomain of Notch Is Not Required for Its Cleavage by γ-Secretase.
We chose to use a notch-based substrate for the majority of our γ-secretase–substrate interaction studies for two main reasons: (i) Relative to other substrates, we found notch to be more easily expressed and purified with different permutations (point mutations, deletions, etc.) from Escherichia coli, and (ii) notch signaling is essential for metazoan cellular identity and organism development (2, 16), thus making its proper cleavage a likely evolutionary driver of γ-secretase function. We would therefore expect the fundamental mechanisms by which γ-secretase interacts with notch to extend to many, if not all, of γ-secretase’s other substrates.
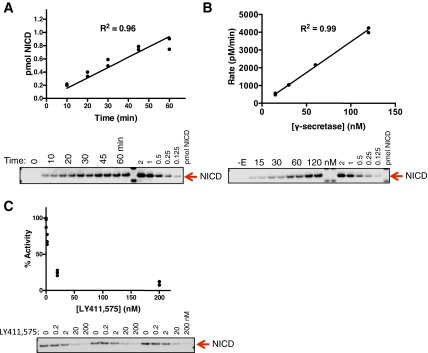
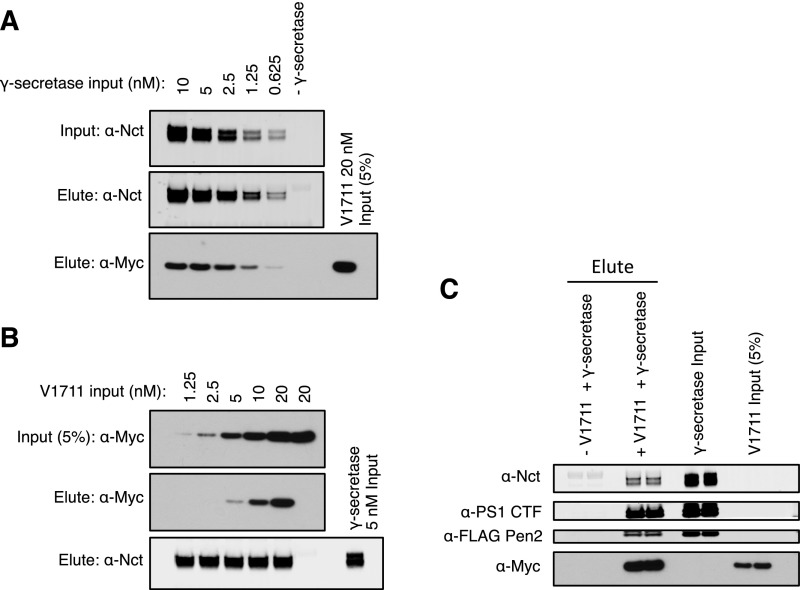
C-terminally truncated (at T1811) notch1 protein was produced in E. coli by expressing the substrate with an N-terminal tag, which was later cleaved off during purification to yield the native Val1711 at the α-secretase cleavage site (Materials and Methods). In a detergent-solubilized state, cleavage of this substrate (V1711) by γ-secretase purified from our S20 CHO cell line (stably overexpressing human PS1, Pen2, Aph1, and nicastrin) (41) was linear with respect to time for at least the first 45 min of the reaction (Fig. S1A). Generation of product was monitored in an end-point assay by Western blot using a cleavage-specific antibody raised to the neoepitope of the notch intracellular domain (NICD) created by γ-secretase. Turnover rate doubled with doubling enzyme concentration (Fig. S1B), suggesting this assay is well suited for studying γ-secretase kinetics. We determined the amount of active γ-secretase in our in vitro assays to be ∼30 nM (Fig. S1C) by titrating the active complex with the tight-binding inhibitor LY411,575.
Fig. S1.
Establishment of a detergent-solubilized γ-secretase assay with a notch-based substrate. (A) Cleavage of V1711 with γ-secretase was monitored by Western blot to cleaved NICD. Cleavage was linear with respect to time for the first 45 min. (B) The reaction rate doubled with doubling enzyme concentration (mean ± SD). (C) The amount of active γ-secretase enzyme used in the detergent-soluble assay was titrated with the highly potent inhibitor LY411,575.
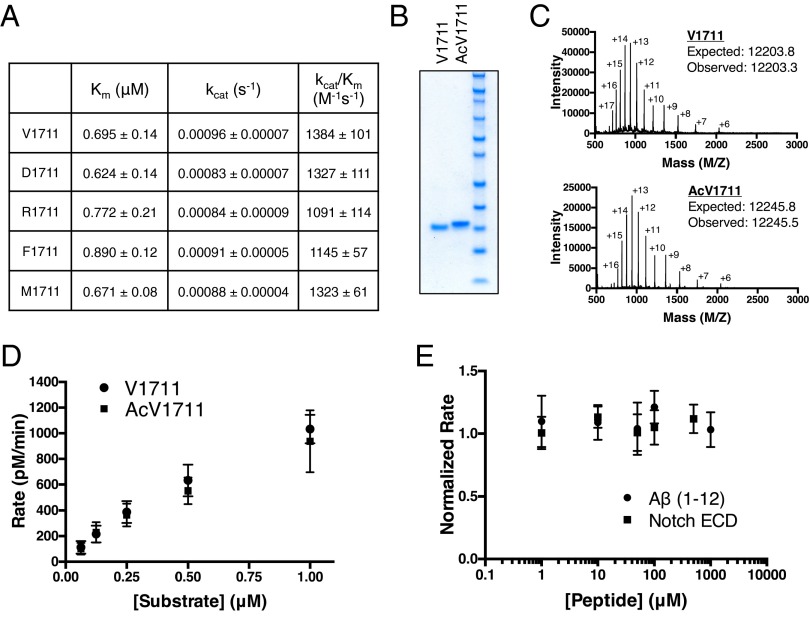
Given the reported importance of the free N terminus of substrate ectodomain in the proposed nicastrin–substrate binding mechanism (35, 36), we hypothesized that the identity of the N-terminal amino acid should play a role in the efficiency of γ-secretase cleavage of substrate. We therefore produced several notch substrates varying the N-terminal amino acid at the native α-secretase cleavage site. Mutating the naturally occurring Val at position 1711 to acidic (Asp), basic (Arg), and bulky hydrophobic (Phe, Met) amino acids had no effect on the ability of γ-secretase to cleave these substrates (Fig. 1A). To test the proposed key interaction between nicastrin and the positive charge of the N-terminal amine of substrate, we neutralized the positive charge by generating N-terminally acetylated V1711, along with an unacetylated control, by native chemical ligation (42). The ligation reactions were driven to >95% completion based on Coomassie staining and were of high purity (Fig. 1B). The correct mass of each protein was confirmed by electrospray ionization time-of-flight (ESI-TOF) mass spectrometry (Fig. 1C). Surprisingly, γ-secretase cleaved both acetylated and unacetylated V1711 with equal efficiency, indicating that a free amino group at the N terminus of notch is not important for its recognition (Fig. 1D).
Fig. 1.
γ-secretase activity toward N-terminally manipulated notch. (A) Summary of kinetic data from γ-secretase cleavage of notch substrates containing varying N-terminal amino acids. Cleavage was monitored by Western blot using a cleavage-specific antibody to NICD (mean ± SD, n = 3). (B) Coomassie-stained gel of semisynthetic native V1711 and N-terminally acetylated AcV1711 generated by native chemical ligation. (C) Electrospray ionization time-of-flight mass spectrometry of V1711 and AcV1711. Intact masses were obtained after deconvolution of the multiply charged states of each protein. (D) γ-secretase activity toward semisynthetic V1711 and AcV1711 (mean ± SD, n = 2). (E) In trans peptide inhibition of recombinant V1711 with notch ectodomain and Aβ (1–12) peptides. Data points are normalized to untreated V1711 cleavage (mean ± SD, n = 2).
Alhough the N-terminal identity of the short notch neo-ectodomain appeared to be unimportant for γ-secretase recognition and cleavage, we sought to identify if there were other determinants within the ectodomain that may be essential for its binding and proteolysis. If there were a binding site for the notch neo-ectodomain on nicastrin (or any other component of γ-secretase), then notch ectodomain peptide would be expected to compete with V1711 for this binding site and inhibit its cleavage. Incubating γ-secretase with up to 500 μM synthetic notch ectodomain peptide caused no inhibition of V1711 cleavage, even though the ectodomain peptide was in 1,000-fold excess over substrate (Fig. 1E). Similarly, Aβ (1–12) peptide had no inhibitory effect up to 1 mM concentrations (Fig. 1E), as had been previously suggested (43). Quite surprisingly, cleavage of a notch substrate having all but four amino acids of the ectodomain deleted (ΔEct) was nearly identical to that of native V1711 (Fig. 2 A and B), demonstrating that the ectodomain of notch is dispensable for its recognition and cleavage by γ-secretase.
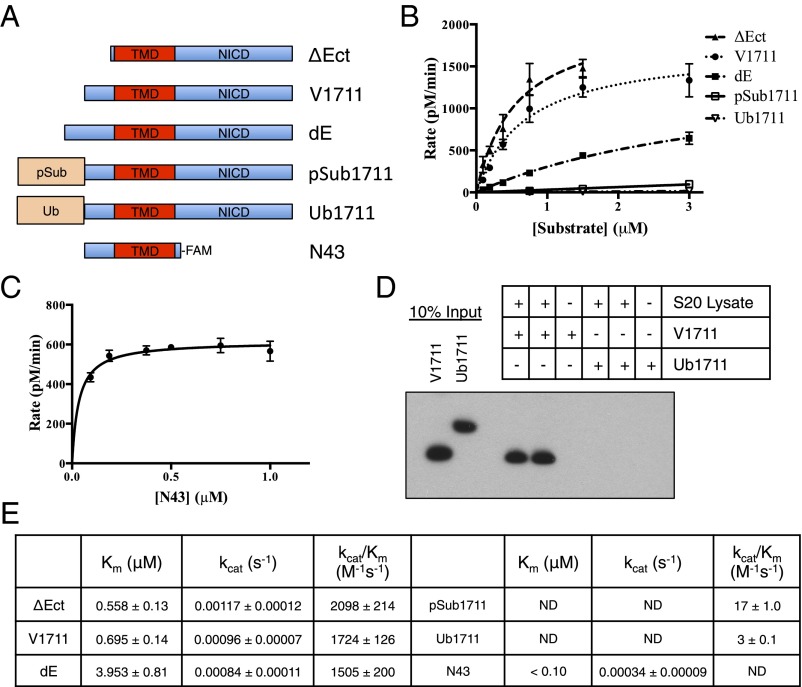
Fig. 2.
The effects of notch ectodomain length on its interaction with γ-secretase. (A) Schematic diagram of notch-based substrates. N43 is a synthetic peptide, whereas the other substrates were produced recombinantly. (B) Kinetic analysis of notch with varying ectodomain length (mean ± SD, n = 3) and (C) of N43 substrate (mean ± SD, n = 3). (D) Comparing the ability of γ-secretase to immunoprecipitate in complex with V1711 and Ub1711. γ-secretase was immunoprecipitated via its HA tag on Aph-1. Substrates were visualized by their C-terminal myc tag. (E) Summary of kinetic data from B and C (mean ± SD, n = 3; ND, not determined).
Notch TMD Drives Its Interaction with γ-Secretase.
In agreement with previous findings (44), we observed that extending the neo-ectodomain of V1711 notch by 13 amino acids (dE notch) (Fig. 2A) decreased its ability to be cleaved by γ-secretase (Fig. 2B). This effect is derived mostly from an increase in Km (suggesting a decrease in binding affinity) (Fig. 2E). Extending the ectodomain even farther by fusing either the 8 kDa prodomain of subtilisin protease (pSub1711) or ubiquitin (Ub1711) to the N terminus of V1711 nearly ablated NICD production (Fig. 2B). We were unable to approach saturation using pSub1711 or Ub1711, suggesting the Km values for these substrates were increased dramatically compared with the shorter ectodomain substrates (Fig. 2E). This trend of Km values decreasing with decreasing ectodomain length (ΔEct ≤ V1711 < dE < pSub1711/Ub1711) (Fig. 2E and Fig. S2) suggests that the binding affinity between γ-secretase and substrate is derived from the TMD of substrate rather than its ectodomain, which is increasingly inhibitory with increasing length. To eliminate the possibility that the NICD is involved in this γ-secretase–substrate interaction, a notch peptide (N43) was synthesized in which almost the entire NICD was omitted and replaced with fluorescein to monitor the cleavage reaction (Fig. 2A). This substrate was readily hydrolyzed by γ-secretase, with a very low Km (<100 nM) (Fig. 2 C and E), indicating NICD is not required for its interaction with γ-secretase. Together, these data suggest that the γ-secretase–substrate binding event is driven by the notch TMD and the intramembranous active site of γ-secretase, rather than through an interaction between the ectodomains of substrate and nicastrin, as deletion of the notch ectodomain did not cause an increase in Km (decreased binding affinity) or a decrease in the efficiency of its cleavage by γ-secretase compared with native V1711.
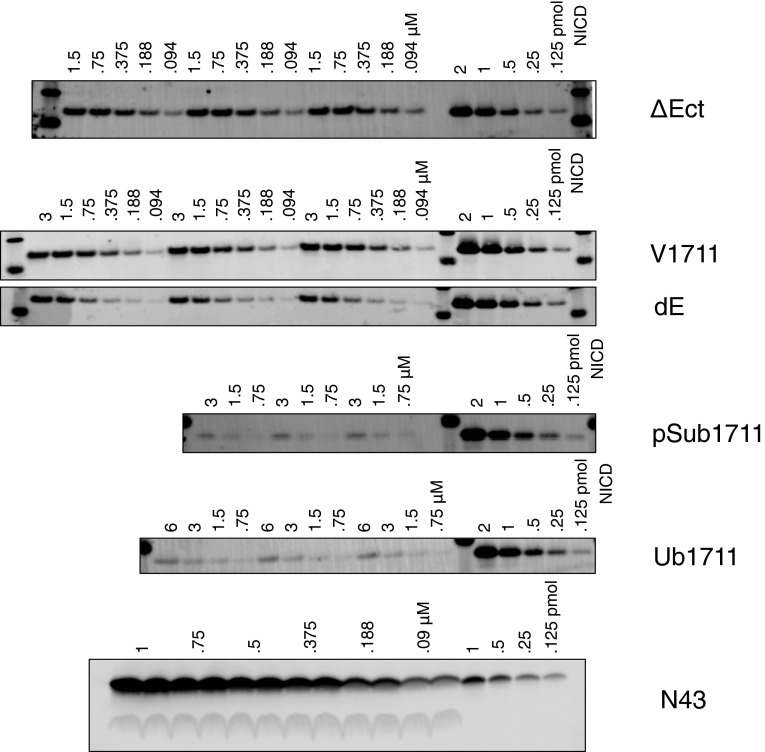
Fig. S2.
Representative Western blot images from kinetic assays in Fig. 2.
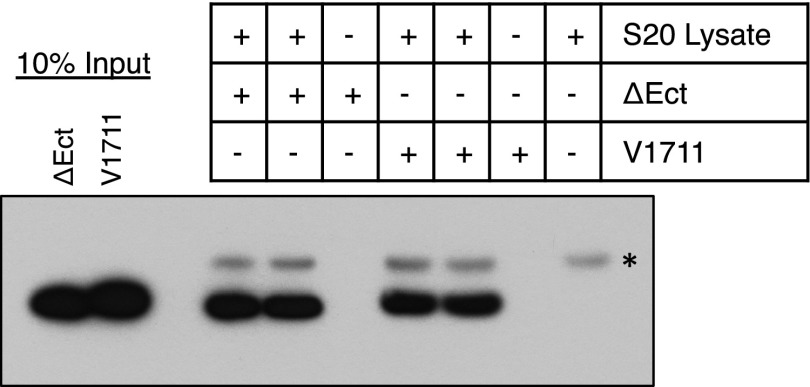
The sub-μM Km values calculated here suggest that γ-secretase possesses a strong binding affinity for substrate TMD. However, given that true binding affinity (Kd) can sometimes deviate significantly from Km, we performed a series of pull-down experiments at known concentrations of purified enzyme and substrate to get an estimate of the binding affinity between γ-secretase and substrate. Here we see that even at low nM concentrations of both γ-secretase and substrate, there is robust coimmunoprecipitation (co-IP) of V1711 bound to γ-secretase (Fig. S3). Quantification of the fraction of substrate bound (or fraction γ-secretase bound in the reciprocal pull-down) allowed us to estimate apparent Kd values averaging in the mid-nM range for the γ-secretase–V1711 complex, which are in good agreement with our Km measurements. To confirm that the observed increases in Km values caused by longer ectodomain length are a reflection of decreased binding affinity between substrate and γ-secretase, we compared the ability of γ-secretase from cell lysate to be immunoprecipitated with V1711 and the larger ectodomain-containing Ub1711. Although V1711 was again readily pulled down in complex with γ-secretase, no Ub1711 could be detected (Fig. 2D), indicating there is likely only a low affinity and transient interaction between γ-secretase and Ub1711. ΔEct was immunoprecipitated to the same extent as V1711 (Fig. S4), suggesting ΔEct and V1711 bind in a similar manner to γ-secretase.
Fig. S3.
Estimation of the binding affinity between γ-secretase and substrate. (A) γ-secretase concentrations ranging from 0.625–10 nM were incubated with 20 nM V1711 before co-IP via the HA tag on Aph-1. (B) Myc-tagged V1711 concentrations ranging from 1.25–20 nM were incubated with 5 nM γ-secretase before co-IP via the HA tag on Aph-1. (C) A total of 5 nM γ-secretase was incubated with 20 nM V1711 before pull-down of the complex via the myc tag of V1711 with anti-myc affinity resin. Quantification reveals apparent Kd values averaging in the mid-nM range.
Fig. S4.
Immunoprecipitation of ΔEct and V1711 with γ-secretase. Lysate from S20 cells was incubated with notch substrate before co-IP via the HA tag on γ-secretase. ΔEct and V1711 were visualized by Western blot (anti-myc).
Notch Is Efficiently Cleaved Within the Lipid Bilayer.
The apparently strong interaction between γ-secretase and notch TMD is of particular interest, as it is in stark contrast to the binding mechanism of rhomboid proteases, another family of I-CLiPs, which have no physiologically relevant binding affinity between enzyme and substrate within the lipid bilayer (7). To verify that the apparently high-affinity γ-secretase–substrate interaction that we observed in the detergent-solubilized assay also dominates in a more physiological context, we measured cleavage of notch by γ-secretase within the lipid bilayer of a proteoliposome using the elegant techniques recently established by Dickey et al. to study rhomboid intramembrane kinetics (7).
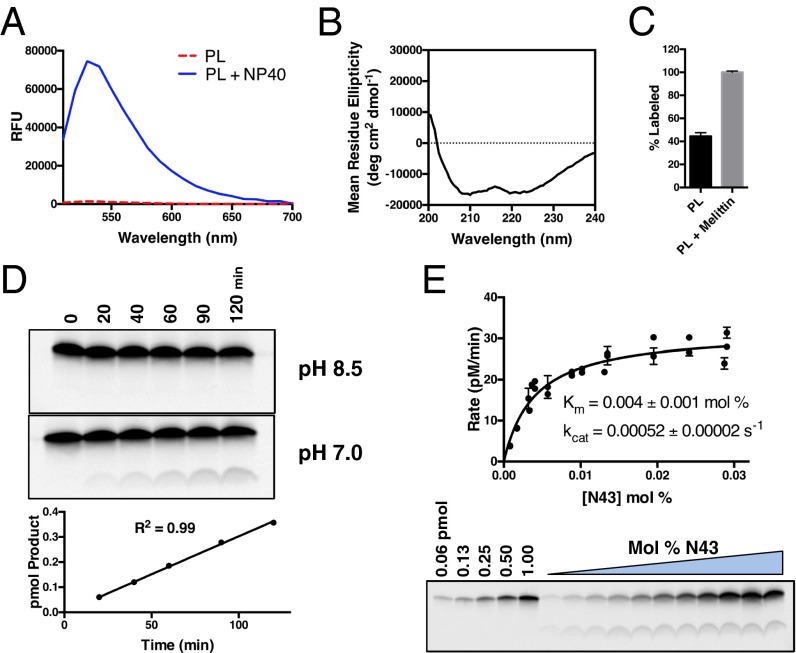
We found that incorporation of the synthetic, fluorescein-labeled notch substrate N43 (see Fig. 2A) into lipid vesicles led to efficient quenching of the fluorophore (Fig. 3A), as was reported for a fluorescein-labeled rhomboid substrate in vesicles (7). The notch substrate maintained its α-helical structure, as determined by circular dichroism after incorporation into the proteoliposome (Fig. 3B). The orientation of substrate within the proteoliposome was determined to be roughly 50% in (N → C), 50% out (C → N) by labeling a synthetic notch peptide containing a Cys on its N terminus with a Cys-reactive membrane-impermeable dye in the absence versus presence of melittin to allow passage of the dye through the membrane (Fig. 3C).
Fig. 3.
Intramembrane kinetics of γ-secretase within a proteoliposome. (A) Fluorescence quenching of N43 when incorporated into vesicles is alleviated after dissolving the proteoliposome with 0.25% Nonidet P-40 detergent. (B) Circular dichroism spectrum of N43 incorporated into a proteoliposome reveals an α-helical structure. (C) Labeling of a notch peptide with a Cys on its N terminus with a membrane impermeable dye in the presence and absence of melittin to allow dye passage into the interior of the proteoliposome (mean ± SD, n = 2). (D) After reconstitution of γ-secretase and N43 substrate into proteoliposomes, the reaction was initiated by resuspending the proteoliposomes in 50 mM Hepes pH 7.0 and 150 mM NaCl. Product was formed linearly with time. (E) Kinetics of N43 cleavage by γ-secretase within a proteoliposome (mean ± SD, n = 4).
The key to measuring the kinetics of an intramembrane protease within a proteoliposome is to prevent cleavage of substrate from occurring during the lengthy process of incorporating enzyme and substrate into the same vesicle and then being able to initiate the reaction when desired. Similar to rhomboid (7), a pH shift served as a suitable mechanism for enzyme inhibition and subsequent activation. With an optimal pH of 6.5 (45), we found that γ-secretase was essentially inactive in bicine buffer at pH 8.5 during the reconstitution procedure (Fig. 3D). Returning the proteoliposomes to a neutral pH of 7.0 initiated the reaction, and product was produced linearly with respect to time (Fig. 3D), allowing us to measure initial rates of catalysis with both γ-secretase and substrate incorporated into a lipid bilayer.
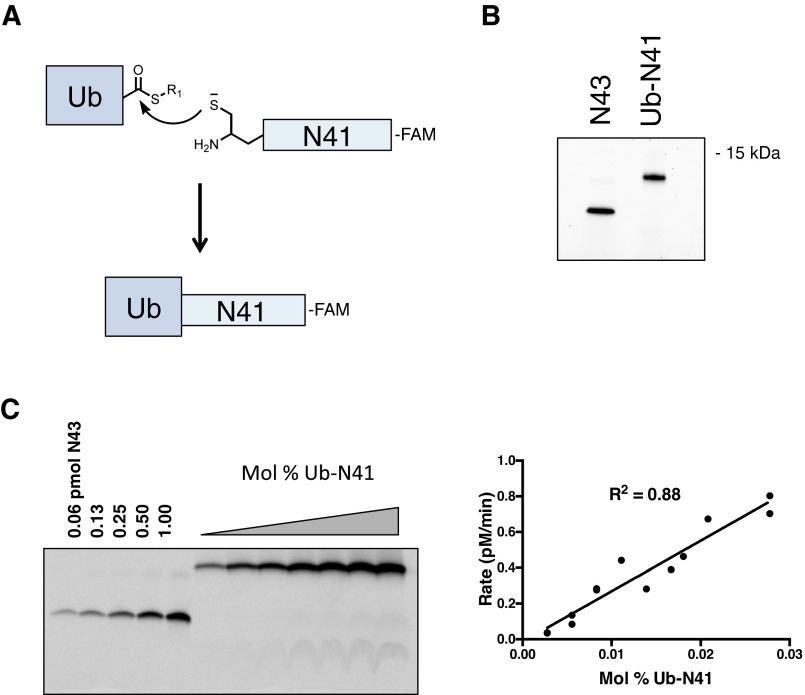
Using this method, we were able to measure an intramembrane Km of 0.004 mol% for N43 (Fig. 3E). This value is nearly two orders of magnitude lower than that of GlpG rhomboid (0.14–0.3 mol%) (7) for its substrate and indicates what is likely a tight-binding interaction between γ-secretase and notch substrate. To verify that substrates with extended ectodomains are inefficiently cleaved in the context of the lipid bilayer as they are in the detergent-solubilized assay, we used expressed protein ligation (46) to generate a semisynthetic substrate comprised of recombinant ubiquitin ligated to synthetic, fluorescently tagged notch TMD, termed Ub-N41 (Fig. S5 A and B). γ-Secretase cleaved Ub-N41 very inefficiently compared with N43, with overnight reaction times being required to produce measurable cleavage product (Fig. S5C), whereas product was observed after only 20 min for N43 (Fig. 3D). Cleavage of Ub-N41 was not saturable over the same concentration range as N43 (Fig. S5C), indicating a greatly increased intramembrane Km and reduced binding affinity for γ-secretase within the membrane. Based on these data, we estimate that the catalytic efficiency for the cleavage of Ub-N41 is reduced about 300-fold compared with N43. These results establish that the apparently tight-binding interaction observed between γ-secretase and notch TMD in the detergent-solubilized assay is also prevalent in the more physiologically relevant context of the lipid bilayer and that extending substrate ectodomain length similarly disrupts this interaction.
Fig. S5.
Kinetics of Ub-N41 cleavage in the proteoliposome assay. (A) Ligation of ubiquitin–thioester to N41 notch peptide. (B) Purified Ub-N41 and N43 visualized via their C-terminal fluorescein tag. (C) After 18 h, Ub-N41 cleavage product was monitored by running proteoliposome reactions on a 16.5% Tris–tricine gel and imaging with a fluorescence scanner. Cleavage was greatly reduced compared with N43 (compare with Fig. 3E), and saturating concentrations were not achieved (n = 2).
Reduction of Nicastrin Disulfide Bonds Allows for More Efficient Cleavage of Substrates Containing Longer Ectodomains.
The decreased binding and cleavage of substrates containing longer ectodomains suggests a mechanism by which these substrates are prevented from interacting with γ-secretase, possibly through steric hindrance between substrate ectodomain and a component of the γ-secretase complex. Recently, it was demonstrated that the archaeal presenilin homolog PSH—which retains activity in the absence of nicastrin, Pen2, and Aph1 cofactors (47)—was fully capable of hydrolyzing the APP-based substrate C99 containing a large ∼40 kDa N-terminal maltose binding protein tag, whereas γ-secretase could not (48). This result suggests a γ-secretase component other than the catalytic presenilin is responsible for this selective cleavage function. Several EM structures of γ-secretase dating back as far as a decade have demonstrated that the ectodomain of nicastrin sits on top of the extracellular side of the γ-secretase TMDs (24, 49–51). It would therefore stand to reason that nicastrin itself might act to sterically block substrates with large ectodomains from entering the γ-secretase complex for catalysis.
To test this hypothesis, we sought a method to selectively disrupt the fold and function of the nicastrin ectodomain without compromising the integrity of the other components of the complex. Because mutations of nicastrin can lead to reduced γ-secretase complex formation and stability (36, 37, 52) and have led to contradictory results in the literature (35–37), we hoped to find a way to disrupt nicastrin after mature wild-type γ-secretase had been formed and purified in its native state. Nicastrin is the only component of γ-secretase known to contain any disulfide bonds (24, 34). We therefore decided to take advantage of this and attempt to disrupt nicastrin function by reducing these highly conserved disulfide bridges.
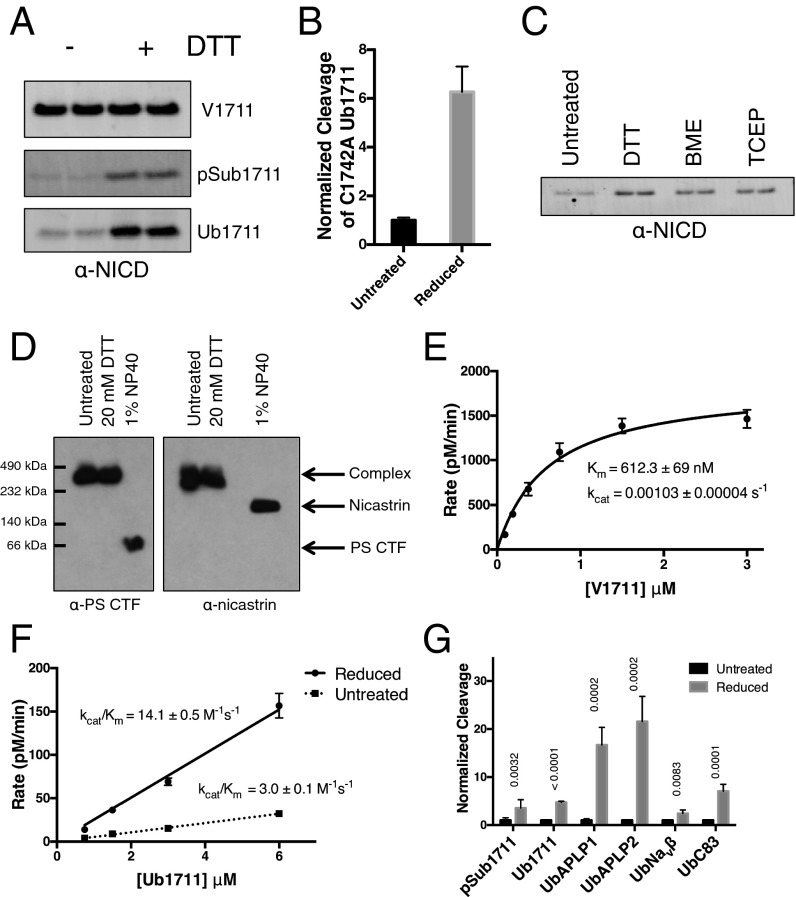
Pretreatment of γ-secretase with 10 mM dithiothreitol (DTT) had no apparent effect on cleavage of the short ectodomain-containing V1711 (Fig. 4A). However, cleavage of the large ectodomain-containing pSub1711 was increased roughly threefold under the same conditions (Fig. 4A). This observation was not specific to the pSub ectodomain, as Ub1711 was also cleaved around 5–6-fold more effectively after the addition of reducing reagent (Fig. 4A). To eliminate the possibility that reduction of substrate was causing this effect, we generated a Cys-free Ub1711 substrate by mutating the single notch Cys at residue 1742 to Ala. After reduction with DTT, cleavage of this Cys-free Ub1711 was still enhanced ∼sixfold (Fig. 4B), indicating that the reducing reagent is acting on enzyme rather than substrate.
Fig. 4.
Activity of γ-secretase after treatment with reducing reagents. (A) Western blot of NICD product after the cleavage of V1711, pSub1711, and Ub1711 with γ-secretase in the presence and absence of DTT. (B) Cleavage of cysteine-free Ub1711 with untreated and reduced γ-secretase (mean ± SD, n = 2). (C) Western blot of NICD product after γ-secretase cleavage of pSub1711 untreated and in the presence of DTT, BME, or TCEP. (D) Native blue gel electrophoresis of γ-secretase untreated or in the presence of DTT or Nonidet P-40 detergent. Nicastrin and presenilin CTFs were detected by Western blot. (E) Kinetic analysis of V1711 cleavage in the presence of DTT (mean ± SD, n = 3). (F) Kinetic analysis of Ub1711 cleavage in the presence and absence of DTT (mean ± SD, n = 3). (G) Cleavage of pSub1711 (n = 6) and N-terminally ubiquitin (Ub)-tagged notch (n = 3), APLP1 (n = 2), APLP2 (n = 2), Navβ (n = 2), and C83 (n = 2) (mean ± SD, t test; P values are shown above each substrate).
In addition to DTT, betamercaptoethanol (BME) and tris(2-carboxyethyl)phosphine (TCEP) both had similar stimulatory effects on γ-secretase cleavage of pSub1711 (Fig. 4C). Pretreatment of γ-secretase with up to 20 mM DTT for 2 h did not dissociate the complex, as determined by native gel electrophoresis, whereas Nonidet P-40 detergent readily broke down γ-secretase into its individual components, as expected (Fig. 4D). Kinetic analysis of the cleavage of native V1711 (short ectodomain) after reduction revealed no difference compared with untreated, with nearly identical Km and Vmax values in the presence of DTT relative to its absence (compare Fig. 4E to Fig. 2 B and E). This result suggests that the effects of reducing reagent do not significantly perturb the TMDs of γ-secretase and that the presenilin active site remains unchanged during catalysis in the presence of reducing reagent. In contrast to V1711, the catalytic efficiency of Ub1711 cleavage was enhanced about fivefold after reduction of γ-secretase (Fig. 4F). However, it is important to note that saturation is not achieved, and the rate of cleavage is still lower than that of V1711 even at the highest concentrations of Ub1711 used (Fig. 4F). This result indicates that some but not all of the inhibitory effects of the large substrate ectodomain of Ub1711 are alleviated by reduction of γ-secretase, which would be expected as the nicastrin ectodomain is likely changing conformation after the addition of reducing reagent but is not being removed.
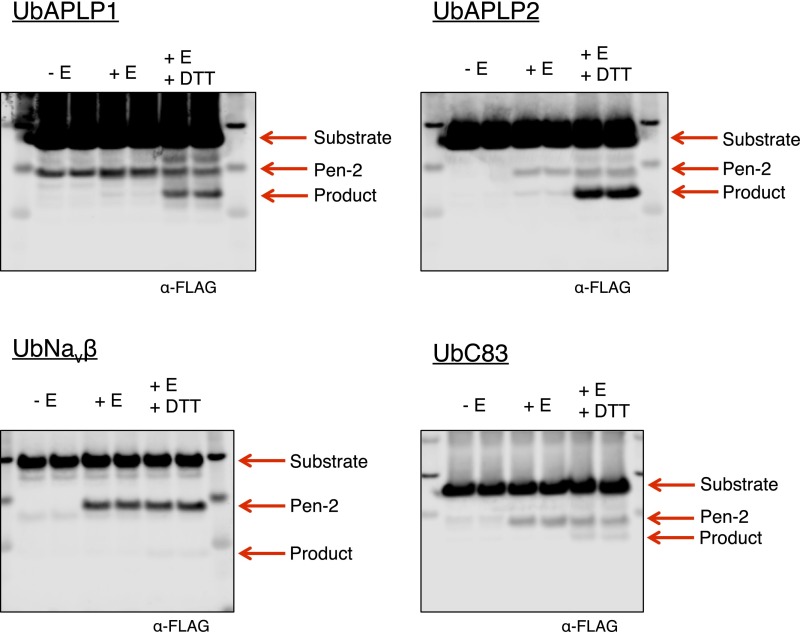
To determine if, in addition to notch, reduced γ-secretase could more effectively hydrolyze other known γ-secretase substrates having large ectodomains, we expressed and purified N-terminally ubiquitin-tagged forms of amyloid precursor-like protein (APLP)1, APLP2, the voltage-gated sodium channel beta subunit (Navβ), and the APP-based substrate C83. With each substrate, γ-secretase was able to produce significantly more cleavage product after its reduction (Fig. 4G and Fig. S6). In the cases of UbAPLP1 and UbAPLP2, which do not contain any cysteine residues, cleavage was enhanced a remarkable ∼20-fold by reducing agent. Although we were unable to detect any cleavage of substrates such as UbCD44 or UbC99 (possibly due to their poor solubility), for every longer ectodomain-containing substrate that we were able to detect basal levels of cleavage, we observed increased hydrolysis after the reduction of γ-secretase. These results suggest that, through nicastrin, the ability of γ-secretase to exclude substrates with large ectodomains likely extends to all of its substrates, not only notch.
Fig. S6.
Representative Western blot images of cleavage of N-terminally ubiquitin-tagged γ-secretase substrates APLP1, APLP2, Navβ, and C83 in the presence and absence of reducing reagents from Fig. 4H.
Reduction of Nicastrin Disrupts Its Normal Fold.
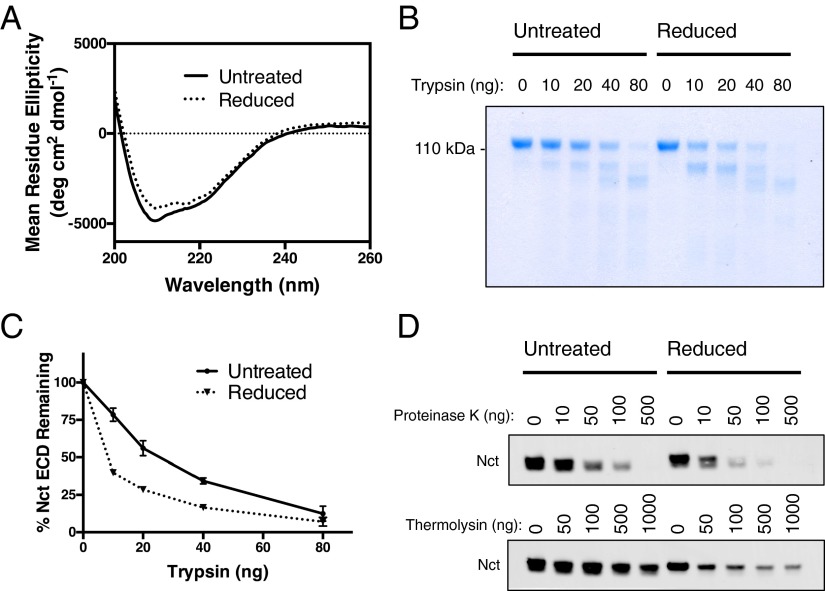
The above results are consistent with the idea that the fold of the nicastrin ectodomain is stabilized by its disulfide bonds and that, upon reduction, it undergoes a destabilizing conformational change, allowing large ectodomain-containing substrates to more easily enter the active site of γ-secretase where catalysis can occur. To verify biochemically that the nicastrin fold is indeed destabilized by reduction, we expressed the full ectodomain of human nicastrin as an Fc fusion protein (53) in HEK293 cells. We were able to obtain highly pure nicastrin ectodomain after removal of the Fc tag by factor Xa protease followed by anion exchange and size exclusion chromatography (Fig. S7). The circular dichroism spectra of the nicastrin ectodomain in the presence and absence of the TCEP reducing reagent reveals a subtle but consistent change in secondary structural elements, suggesting that reduction of nicastrin’s disulfide bonds causes a conformational change (Fig. 5A). To verify this, we used limited proteolysis, which is a well-characterized technique used to measure changes in the conformational stability and flexibility of proteins (54). Digestion of the purified ectodomain of nicastrin with varying amounts of trypsin in the presence or absence of reducing reagent revealed that reduced nicastrin was substantially more sensitive to proteolysis than the unreduced protein (Fig. 5B), with roughly half the amount of trypsin being required for its degradation (Fig. 5C).
Fig. S7.
Purification of human nicastrin ectodomain. The Fc tag on nicastrin–Fc was removed with factor Xa protease. Pure nicastrin ectodomain used in circular dichroism and limited proteolysis studies was obtained after anion exchange and size exclusion chromatography.
Fig. 5.
The effects of chemical reduction of disulfides on nicastrin structure. (A) Circular dichroism spectrum of purified nicastrin ectodomain with and without TCEP present. (B) Limited proteolysis of pure nicastrin ectodomain with varying amounts of trypsin in the presence and absence of DTT. (C) Quantification of intact nicastrin ectodomain from B (mean ± SD, n = 2). (D) Limited proteolysis of nicastrin in the γ-secretase complex with varying amounts of proteinase K and thermolysin proteases.
To confirm that nicastrin in the context of the formed γ-secretase complex is similarly destabilized by reduction, we again measured its susceptibility to proteolysis after the addition of reducing reagent. Although trypsin was nearly inactive in the detergent conditions required to keep the γ-secretase complex intact, proteinase K and thermolysin both retained activity. In each case, the addition of reducing reagent increased the rate of nicastrin degradation, as expected (Fig. 5D).
Discussion
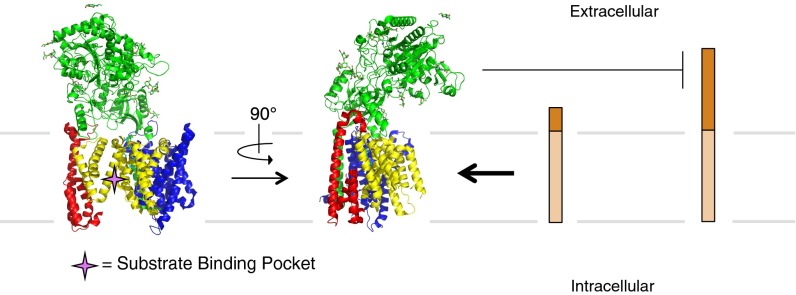
In this study, we provide multiple lines of evidence that nicastrin acts as a steric block, controlling substrate entry into the γ-secretase active site. This solves the long-standing question as to why ectodomain shedding by α- or β-secretases is required before γ-secretase cleavage of its substrates. A decade ago, nicastrin was first proposed to act as a receptor that bound the free N terminus of ectodomain-shed substrates via its aminopeptidase-like domain (around E333) to facilitate substrate cleavage by γ-secretase (35). More recently, an initial high-resolution cryo-EM structure of γ-secretase appeared to support this speculative role for the nicastrin ectodomain in substrate recognition, in that E333 of nicastrin was placed directly over the presenilin active site that was putatively assigned to the concave side of the horseshoe-shaped array of TMDs of γ-secretase (24, 55). However, while our present study was being completed, two higher resolution cryo-EM structures were solved in which all of the γ-secretase TMDs are now accurately assigned, revealing that the presenilin active site actually resides on the convex side of the horseshoe-shaped TMDs (39, 40). Based on the experimental data reported herein and with this new configuration available, we note that the bilobed nicastrin ectodomain is well positioned to block substrates with large ectodomains from entering the presenilin active site (Fig. 6). In contrast, the originally proposed substrate binding site in the vestigial amino peptidase “active site” of nicastrin is now located to the opposite side of the complex relative to the presenilin active site and is buried within the interior of nicastrin rather than being solvent-exposed (39, 40).
Fig. 6.
Model of the nicastrin-mediated substrate-gatekeeping mechanism of γ-secretase. The ectodomain of nicastrin sits on top of the extracellular side of the γ-secretase TMDs with its large lobe extending over the presenilin active site. This prevents substrates with large ectodomains from entering the active site and subsequently being cleaved. Substrates with small ectodomains are able to pass under the nicastrin ectodomain gatekeeper and be cleaved by presenilin. Front and side views of the atomic resolution structure of γ-secretase are depicted (Protein Data Bank ID code 5A63).
In the newest structure, the large lobe of the nicastrin ectodomain extends about 25 Å perpendicular from the TMDs of γ-secretase, directly over the substrate binding pocket of presenilin (Fig. 6). Hovering roughly 20 Å above the membrane, the large lobe provides a cutoff under which only substrates with small ectodomains can easily pass and subsequently be cleaved. Partial disruption of the fold of nicastrin through reduction of its known disulfide bridges likely allows more space for substrates to enter under this barrier. We speculate that the disulfide bond between C230 and C248 may be critical for this function, given its position at an apparent hinge region between the small and large lobes of nicastrin. Determining this definitively will be difficult, given that these highly conserved cysteines are important for the assembly and maturation of the γ-secretase complex within cells (52).
We have established that there is no functional interaction between the short neo-ectodomain of notch and the large ectodomain of nicastrin. We demonstrated that manipulation (e.g., N-terminal acetylation, mutation) and importantly even deletion of the notch ectodomain does not alter γ-secretase’s ability to hydrolyze the notch TMD. Struhl and Adachi had established early on that relatively short ectodomains were required for presenilin-dependent notch TMD cleavage (11), and recently Funamoto et al. showed that substrates with shorter ectodomains are cleaved better by γ-secretase (44). However, nearly complete deletion of substrate ectodomain had not been previously explored. Our study makes it clear that substrate neo-ectodomain is not required for its productive recruitment into the γ-secretase complex or its subsequent cleavage. In fact, in some cases the remaining short neo-ectodomains may even be inhibitory (43).
We also show that disrupting nicastrin’s natural function—by destabilizing its fold—has no effect on the efficiency of turnover of native V1711, indicating there is no functional connection between the ectodomain of nicastrin and the presenilin active site during catalysis. This finding seems to decisively exclude nicastrin acting as a receptor for the notch neo-ectodomain. Given that notch signaling was likely a key driver of γ-secretase function from an evolutionary standpoint, it is highly improbable that nicastrin would evolve to recognize the ectodomain of other γ-secretase substrates but not notch. Furthermore, with the apparently strong binding affinity derived from the interactions between substrate TMD and γ-secretase active site, it is not obvious why additional binding energy from substrate neo-ectodomain would be necessary for productive recruitment of substrate into the active site.
Our combined data suggest that the physiologically relevant Km values and apparently tight-binding interaction between notch TMD and the γ-secretase active site are common characteristics shared by other γ-secretase substrates. Indeed, Km values of ≤1 μM have been reported for APP-, APLP-, ErbB4-, and N-cadheren–based substrates as well as notch (44, 56, 57). This apparently strong interaction between substrate TMD and γ-secretase is in stark contrast to the binding between rhomboid and its substrate, where no physiologically relevant binding affinity between the two is observed. The intramembrane Km of γ-secretase (0.004 mol%) for substrate is roughly two orders of magnitude lower than that of GlpG rhomboid (0.14–0.3 mol%) (7). This extremely low intramembrane Km is even lower than the interfacial Km values of some of the most efficient lipid-metabolizing enzymes such as PTEN (0.04 mol%) (58). This apparently high affinity for substrate would not only allow γ-secretase to be highly effective at finding low concentrations of its substrates within the protein-crowded environment of the lipid bilayer but also provide the means by which γ-secretase catalyzes multiple successive cleavages of its substrates such as APP. If γ-secretase did not possess a reasonably high affinity for substrate, it would likely prematurely release longer cleavage products, which would remain embedded in the membrane rather than be secreted from the cell.
Our data demonstrate that γ-secretase and rhomboid clearly evolved different mechanisms for interacting with their respective substrates. The striking difference in Km values and apparent binding affinities for their substrates may have arisen due to the nicastrin ectodomain blocking mechanism of γ-secretase described in this study. Through nicastrin, γ-secretase is able to distinguish substrate from nonsubstrate based on the presence and size of its ectodomain. Rhomboid has no such mechanism, as it is responsible for direct shedding of large ectodomain-containing substrates (8, 59, 60), without the requirement of prior “activation” of substrate by another protease like γ-secretase substrates are. Thus, rhomboid requires a different mechanism to distinguish substrate from nonsubstrate and has apparently evolved to bind its substrates with very low affinity, only hydrolyzing those substrates whose unstable TMD helices have had time to unfold in the active site and subsequently be cleaved (7). Rhomboid cleavage is therefore dependent on differences in rate rather than binding affinity for distinguishing between substrates, in a mechanism similar to DNA glycosylases (61). In contrast, differences in binding affinity between preferred and nonpreferred substrate play a major role in γ-secretase’s cleavage of its substrates. In the absence of the nicastrin steric blocking function identified here, a strong interaction between γ-secretase and substrate TMD would allow γ-secretase to cleave type-I intramembrane proteins nonspecifically, causing improper and unregulated signaling.
The nicastrin-mediated steric blocking of large ectodomain-containing substrates of γ-secretase is not the only example of such a mechanism used by an intramembrane protease. The E. coli I-CLiP RseP of the site-2 protease family has evolved a similar size exclusion mechanism to prevent unregulated signaling of the periplasmic unfolded protein response (62). RseP achieves steric blocking of its substrate RseA through its two tandem periplasmic PDZ domains. Normally RseA must have its large periplasmic region removed by DegS protease before being cleaved by RseP. Both deletion and structurally destabilizing mutations of the PDZ domains of RseP allow for direct cleavage of RseA, bypassing the need for prior removal of its periplasmic domain (62). Ironically, similar to nicastrin, the PDZ domains of RseP were originally proposed to be involved in substrate binding (62). This was later ruled out after they were shown to be dispensable for catalysis, and high-resolution structures of the PDZ domains demonstrated that the putative substrate-binding pocket is both too narrow to accommodate substrate and physically covered by a helix (62, 63).
Although nicastrin does not actively recruit substrates as originally proposed, it appears to discriminate among different ectodomain-shed substrates based upon the size of their remaining ectodomain stubs, imparting substrate specificity in that manner. We find that γ-secretase preferentially cleaves V1711 over dE notch, which has a longer ectodomain by only 13 amino acids. Similarly, the shorter ectodomain α-secretase–generated C83 fragment of APP is reported to be a better substrate than the longer β-secretase–generated C99 fragment (44). Like APP, the ectodomains of many other γ-secretase substrates can be cleaved at distinct sites by either α- or β-secretase (12, 64, 65), resulting in different neo-ectodomain lengths. This may provide the cell with a mechanism by which it can temporally control signaling events from the same substrate through the preferential cleavage of substrates with smaller neo-ectodomains by γ-secretase.
It should be mentioned that nicastrin knockout cells show very low levels of endogenous notch processing that was shown to require ectodomain shedding and the other components of γ-secretase (38). However, notch signaling was so low in this case that it could not rescue the embryonic lethal phenotype of deficient notch signaling. Additionally, the authors could not rule out that changes in substrate subcellular localization postectodomain shedding were responsible for the ectodomain shedding dependency they observed. Indeed, α-secretase shedding of notch occurs at the plasma membrane, whereas in the absence of nicastrin, the other γ-secretase components are retained in earlier secretory compartments (66). The putative active tripartite complex lacking nicastrin could not be isolated from cells, but based on this report, we cannot rule out contributions from the other components to substrate gate-keeping.
Our findings help elucidate the mechanism by which one previously explored approach to therapeutic inhibition of C99 cleavage may work. Both antibodies and peptides targeting the N terminus of the C99 ectodomain have been found to be effective inhibitors of Aβ production (35, 44). Their mode of action was thought to function by blocking the specific nicastrin–substrate ectodomain binding interaction. Our results suggest that these agents act instead to increase the effective size of the ectodomain, which is then sterically blocked by nicastrin, rather than masking a specific recognition motif on the substrate itself.
The kcat values determined here for γ-secretase are in close agreement with the values recently published by Kamp et al. (67). These exceptionally slow rates are similar to rhomboid (7), suggesting slow rates of catalysis may be a common theme for all I-CLiPs. The reason for this is currently unknown, although it may be due to lateral substrate gating into the active site (7) or conformational changes in substrate and/or enzyme after initial substrate binding.
With this new model for nicastrin functioning as a steric gatekeeper for substrate entry into the γ-secretase complex, additional work is now required to define the nature of the substrate TMD interaction with presenilin and the catalytic steps that subsequently occur. This could further elucidate how familial AD mutations in presenilin contribute to pathogenesis. In light of the wealth of structural information from the recently solved γ-secretase structures, it should only be a matter of time before the remaining mysteries of γ-secretase function are resolved, with broad implications for biology and medicine.
Materials and Methods
Antibodies.
The following antibodies were used in this study: α-Myc (9E10 Pierce #20168), α-Flag (M2 Sigma #F3165), α-HA (3F10 Roche #11867423001), α-Nct (3632S Cell Signaling Technology #3632S), α-PS1–CTF (EP2000Y Cell Signaling Technology #ab76083), and α-cleaved notch (D3B8 Cell Signaling Technology #4147).
Expression and Purification of γ-Secretase.
We purified γ-secretase from the S20 CHO cell line overexpressing human presenilin-1, Pen2, Aph1, and nicastrin, as described previously (41).
Expression and Purification of γ-Secretase Substrates.
All notch-based substrates were expressed with an N-terminal tag, either the prodomain of subtilisin (pSub) protease or His-ubiquitin in E. coli (BL21) in the pPAL7 vector from Bio-Rad. pSub-tagged notch cells were grown to a density of 0.6 at 37 °C, at which point 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added and the temperature cooled to 16 °C for overnight expression. Ubiquitin-tagged substrates were grown at 37 °C until an OD of 0.8 was reached. We then added 1 mM IPTG, and expression was allowed to proceed for 4 h at 37 °C. All cells were lysed by French press. To obtain notch substrates with native N termini or N-terminal mutants, the subtilisin prodomain tag was cleaved off using the eXact tag purification system from Bio-Rad (#156–3000) according to the manufacturer’s protocol. After cleavage from the pSub tag, notch substrate was centrifuged at 100,000 × g for 30 min to remove aggregates. All substrates were stored in 50 mM Tris pH 7.0 with 0.25% Nonidet P-40 detergent at –80 °C. Purity was checked by SDS/PAGE.
All additional substrates were expressed with N-terminal His-ubiquitin tags positioned just before their native α-secretase cleavage sites. If the α-cleavage site is unknown, the Ub tag was arbitrarily placed 15 amino acids from the substrate’s TMD. These substrates were purified over a bed of Ni-NTA (Qiagen), washed, eluted, and stored the same as notch substrates.
Peptides.
The following peptides were synthesized and purified by Anaspec Inc.: N43, amine-VKSEPVEPPLPSQLHLMYVAAAAFVLLFFVGCGVLLSRRRAAK(5-FAM)-amide; N41, amine-CEPVEPPLPSQLHLMYVAAAAFVLLFFVGCGVLLSRRRAAK(5-FAM)-amide; Notch Ectodomain, amine-VKSEPVEPPLPSQ-acid; and Aβ (1–12), amine-DAEFRHDSGYEV-acid. The peptides for native chemical ligation were as follows: amine-VK-thiobenzyl ester and acetyl-VK-thiobenzyl ester.
Native Chemical Ligation.
N-terminally His-tagged notch was expressed in E. coli with a tobacco etch virus (TEV) protease cut site after the His tag but preceding the notch neo-ectodomain, which omits the N-terminal VK and contains a Cys in the place of Ser1713 to facilitate the native chemical ligation reaction. After nickel purification, the His tag was removed with TEV protease (Invitrogen) to yield an N-terminal Cys at a concentration of 0.2 mg/mL in 50 mM Tris pH 8.0 and 1% Triton-X 100 at 4 °C overnight during dialysis to remove imidazole retained from nickel column elution. The TEV protease (itself His-tagged) and the His tag removed from notch were then depleted with nickel beads.
The resulting notch protein containing a free Cys at its N terminus was ligated to either AcVK- or VK-thioester peptide under the following conditions: 2 mM peptide, 100 mM mercaptoethanesulfonate (MESNA), 100 mM Tris pH 7.0, and 500 μM TCEP were incubated with N-Cys notch (0.2 mg/mL) for 20 h at room temperature. Following completion of the ligation, the ligated product was dialyzed for 24 h at 4 °C to remove excess peptide and reducing reagents. After dialysis, the semisynthetic proteins were further purified and concentrated by the FLAG tag on its C terminus using M2 anti-FLAG beads from Sigma. After overnight binding at 4 °C, the immunoprecipitated substrates were washed thoroughly, eluted with 100 mM glycine pH 2.5 and 0.25% Nonidet P-40, and immediately neutralized with Tris buffer pH 7.0. The mass of each protein was verified using an Agilent 6220 ESI-TOF mass spectrometer after Nonidet P-40 detergent was replaced with 0.2% n-dodecyl -D-maltoside (DDM) detergent. The intact masses were identified using the MaxEnt deconvolution algorithm.
Expressed Protein Ligation.
Ubiquitin–thioester was generated as follows: human ubiquitin containing an N-terminal His-tag was cloned into the intein-containing vector pTXB1 from New England Biolabs. The resulting ubiquitin–intein fusion protein was expressed in E. coli by inducing BL21 cells at an OD of 0.7 with 1 mM IPTG for 4 h at 37 °C. Cell pellets were lysed by French press and the lysate passed over chitin beads (New England Biolabs). The ubiquitin–thioester was isolated by incubating the fusion protein bound to the chitin beads with 600 mM sodium MESNA (Sigma), 50 mM Tris pH 7.0, and 150 mM NaCl overnight at room temperature. Following cleavage from the intein, the ubiquitin–thioester was concentrated and used immediately in ligation reactions.
Purified ubiquitin–thioester was ligated to N41 peptide in the presence of 100 mM MESNA, 500 μM TCEP, 50 mM Tris pH 7.2, 150 mM NaCl, and 1% Sarkosyl detergent. Ligations proceeded for 24 h, after which the newly generated semisynthetic Ub-N41 was purified from unreacted N41 peptide by isolating His-tagged Ub-N41 with nickel beads. Ub-N41 was eluted from the nickel beads with 150 mM imidazole, 50 mM Tris pH 7.0, and 0.25% Nonidet P-40 detergent. Imidazole was subsequently removed in a final dialysis step.
Detergent-Solubilized Assay.
The γ-secretase was preincubated for 30 min at 37 °C in the presence of 50 mM Hepes pH 7.0, 150 mM NaCl, and 0.25% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO) detergent supplemented with 0.1% phosphatidylcholine (DOPC) and 0.025% phosphatidylethanolamine (DOPE). Reactions were initiated with the desired amounts of substrate and allowed to proceed at 37 °C for various lengths of time (usually 15–30 min for notch-based substrates) before being quenched by SDS. Cleavage product was visualized by Western blot using an antibody to either notch NICD using the notch cleavage product-specific antibody from Cell Signaling or an epitope tag (FLAG or Myc). In all kinetic assays, the amount of product produced was determined by running a standard curve of NICD on each gel. NICD containing the native Val at the cleavage site 1744 (for antibody recognition) was generated using the eXact tag purification system as described above.
For assays examining the effects of reducing reagent on γ-secretase activity, γ-secretase was preincubated in 50 mM Hepes pH 7.0, 150 mM NaCl, and 0.25% CHAPSO detergent with 0.1% DOPC and 0.025% DOPE in addition to reducing reagent (5 mM DTT unless otherwise stated) at room temperature for 2 h. Reactions were then initiated with substrate and allowed to proceed at 37 °C for 30 min (pSub1711) or 4 h (Ub1711) for notch-based substrates or overnight for nonnotch substrates. Reactions were quenched by SDS and visualized by Western blot using either the notch cleavage-specific antibody or anti-FLAG.
Proteoliposome Formation and Kinetic Assay.
DOPC and DOPE lipids from Avanti Polar Lipids were dried under a stream of nitrogen in glass test tubes before being hydrated at 50 °C with 50 mM Bicine pH 8.5 and 150 mM NaCl for 20 min. Following hydration, large multilamellar vesicles were generated by vigorous vortexing before extrusion through a filter with a pore size of 200 nm to obtain unilamelar vesicles of uniform size. All vesicles used in proteoliposome assays are composed of 90% DOPC and 10% DOPE.
Vesicles (3 mM total lipid) were incubated with γ-secretase enzyme and varying amounts of N43 or Ub-N41 notch substrates for 20 min at room temperature in 50 mM bicine pH 8.5 and 150 mM NaCl. Dilution of the mixture to below the critical micelle concentration (CMC) of detergent with 10 mM bicine pH 8.5 and 10 mM NaCl allowed for incorporation of enzyme and substrate into the vesicle. The proteoliposomes were then pelleted by ultracentrifugation at 100,000 × g for 20 min at 4 °C. To initiate proteolysis, the proteoliposome pellets were then resuspended in 50 mM Hepes pH 7.0 and 150 mM NaCl. Reactions were allowed to proceed for 2 h (N43) or overnight (Ub-N41) at 37 °C before being quenched by SDS. The quenched reactions were run on a 16.5% Tris–tricine gel and the fluorescein-tagged product visualized using a Storm imager from GE. Bands were quantified using a standard curve of fluorescein-labeled N43 substrate run concurrently on each gel. The fluorescent wavelength spectrum of labeled peptide incorporated into proteoliposome was taken on a Synergy H1 plate reader from BioTek.
Proteoliposome Labeling.
Proteoliposomes incorporated with the N41 notch substrate containing an N-terminal Cys were generated as described above. After ultracentrifugation, the proteoliposomes were resuspended in PBS containing the membrane-impermeable Cys reactive IRDye 800CW maleimide from Licor (929-80020) at a concentration of 1 μM with or without melittin peptide at a concentration of 1 mg/mL. Labeling was allowed to proceed for 2 h at room temperature followed by quenching with 50 mM DTT for 30 min. The labeled peptide was then separated from unreacted dye on 16.5% Tris–tricine gel and visualized with an Odyssey scanner from Licor.
Coimmunoprecipitation of γ-Secretase from Cell Lystate with Substrate.
One confluent 15-cm dish of S20 CHO cells stably overexpressing human γ-secretase components was lysed in 200 μL of 50 mM Hepes pH 7.0, 150 mM NaCl, and 1% CHAPSO detergent for 1 h on ice. Lysate was then centrifuged to remove cell debris and incubated in the presence of 10 μM of the noncompetitive γ-secretase inhibitor III-31C for 1 h at room temperature before the addition of 50 pmol myc-tagged ΔEct, V1711, or Ub1711 substrate, which was incubated for an additional 1 h at room temperature. The γ-secretase and substrate complex was then immunoprecipitated via HA-tagged Aph-1 using anti-HA affinity resin (Pierce #88836).
Coimmunoprecipitation of Purified γ-Secretase and Substrate for Kd Estimation.
Purified γ-secretase at the indicated concentration was preincubated in assay buffer (50 mM Hepes pH 7.0, 150 mM NaCl, 0.25% CHAPSO, 0.1% DOPC and 0.025% DOPE, 2% BSA) in the presence of 2 μM III-31C for 1 h at room temperature. Purified myc-tagged V1711 was then incubated with γ-secretase for 1 h before pull-down with either anti-HA or anti-myc magnetic affinity beads for 4 h with mixing at room temperature. The immunoprecipitated complex was then washed three times and eluted with SDS loading buffer before Western blot. The fraction of substrate or γ-secretase components bound was determined by densitometry.
Nicastrin Ectodomain Expression and Purification.
A HEK293 cell line stably overexpressing the ectodomain (aa 1–669) of nicastrin fused to human IgG1 Fc was selected for as previously reported (53). The fusion protein contained a factor Xa cleavage site for Fc tag removal. The cells were grown in shaker flasks in Freestyle 293 media from Invitrogen supplemented with 1% FBS and pen/strep antibiotic. After 24–72 h of growth, the cells were pelleted and the Nct–Fc containing supernatant was collected for purification. After filtration through a 0.2-μm filter to remove cellular debris, the supernatant was passed over a bed of protein A beads (GE Healthcare). After washing to remove contaminants, Nct-Fc was eluted from the column using 100 mM glycine pH 2.5 and immediately neutralized with Tris buffer pH 8.0. The fusion protein was then concentrated to 2 mg/mL.
The Fc tag was removed by factor Xa protease (50 μg/mL, Invitrogen) for 16 h at 10 °C. The nicastrin ectodomain was then purified from the mixture by anion exchange chromatography followed by size exclusion chromatography in 20 mM sodium phosphate buffer pH 7.4 on an AKTA FPLC. The pure protein was again concentrated to 2 mg/mL and subsequently used in proteolysis and CD experiments.
Native Blue Gel Electrophoresis.
The γ-secretase was preincubated in 50 mM Hepes pH 7.0, 150 mM NaCl, and 1% digitonin with the addition of either 20 mM DTT or 1% Nonidet P-40 detergent for 2 h at room temperature. Each sample was then prepared for blue native PAGE in a solution of 0.25% G250 Brilliant Blue (Sigma B0770) and NativePAGE Sample buffer (Invitrogen BN20032). Samples were loaded on a 2–16% bis–Tris NativePAGE gel (Life Technologies, BN2111), and electrophoresis was carried out with anode buffer and dark blue cathode buffer from the NativePAGE Running Buffer Kit (Life Technologies, BN2007) at 150 V until the dye front had run ∼1/3 of the gel length. Dark blue cathode buffer was substituted with light blue cathode buffer for the duration of electrophoresis. The γ-secretase components nicastrin and presenilin CTF were then visualized by Western blot.
Limited Proteolysis Experiments.
Purified nicastrin ectodomain (2 μg) was incubated with PBS in the presence or absence of 5 mM DTT for 2 h at room temperature. After that, varying amounts of trypsin protease were added and proteolysis was allowed to proceed for 15 min at 37 °C before being quenched by SDS. The digestion products were separated by SDS/PAGE and visualized by Coomassie stain.
We preincubated γ-secretase in 50 mM Hepes pH 7.0, 150 mM NaCl, and 1% CHAPSO in the presence or absence of 5 mM DTT for 2 h at room temperature before the addition of varying amounts of proteinase K or thermolysin proteases. Proteinase K and thermolysin digestions were incubated at 37 °C and 65 °C, respectively, for 1 h and then quenched with SDS. Nicastrin digestion was measured by Western blot.
Circular Dichroism.
All CD spectra were obtained using a Jasco J-815 spectropolarimeter in a cuvette with a path length of 0.2 cm. The CD spectra were averaged from five scans from an individual experiment. All experiments were repeated three times. The temperature within the cell was maintained using a Peltier temperature control unit. The spectra of the N43 notch peptide substrate was taken after the peptide was incorporated into a proteoliposome as described above but in a different buffer: 10 mM Tris pH 7.0. The proteoliposomes were diluted such that the final concentration of peptide was 0.2 mg/mL during the spectral scan. Pure nicastrin ectodomain stored at 2 mg/mL in 20 mM sodium phosphate buffer was diluted in water or in 1 mM TCEP in water to a final concentration of 0.1 mg/mL before spectral reading. Data are represented in terms of mean residue ellipticity.
Acknowledgments
We thank our colleagues Allen Chen, Tim Bartels, Oliver Holmes, Ulf Dettmer, and Andrew Newman for helpful discussions. We thank Sin Urban for helpful insights involving the establishment of the γ-secretase proteoliposome assay. The nicastrin–Fc construct was supplied by Regina Fluhrer. Mass spectrometry was performed by Sunia Trauger at the Small Molecule Mass Spectrometry core facility at Harvard. This work was supported by NIH Grant P01 AG15379.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 1112.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512952113/-/DCSupplemental.
References
- 1.Urban S. Mechanisms and cellular functions of intramembrane proteases. Biochim Biophys Acta. 2013;1828(12):2797–2800. doi: 10.1016/j.bbamem.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Huppert SS, et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405(6789):966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 3.Rawson RB, et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1(1):47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson LG, et al. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc Natl Acad Sci USA. 2007;104(3):1003–1008. doi: 10.1073/pnas.0608140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moin SM, Urban S. Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. eLife. 2012;1:e00173. doi: 10.7554/eLife.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickey SW, Baker RP, Cho S, Urban S. Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell. 2013;155(6):1270–1281. doi: 10.1016/j.cell.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman M. Proteolysis within the membrane: Rhomboids revealed. Nat Rev Mol Cell Biol. 2004;5(3):188–197. doi: 10.1038/nrm1334. [DOI] [PubMed] [Google Scholar]
- 9.Rawson RB. The site-2 protease. Biochim Biophys Acta. 2013;1828(12):2801–2807. doi: 10.1016/j.bbamem.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 10.Voss M, Schröder B, Fluhrer R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim Biophys Acta. 2013;1828(12):2828–2839. doi: 10.1016/j.bbamem.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 11.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6(3):625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 12.Haapasalo A, Kovacs DM. The many substrates of presenilin/γ-secretase. J Alzheimers Dis. 2011;25(1):3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398(6727):522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 14.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 15.Mumm JS, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 16.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe MS, et al. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398(6727):513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 18.Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65(9):1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopan R, Ilagan MXG. Gamma-secretase: Proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5(6):499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 20.Francis R, et al. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3(1):85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 21.Edbauer D, et al. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5(5):486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 22.Kimberly WT, et al. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci USA. 2003;100(11):6382–6387. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takasugi N, et al. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422(6930):438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 24.Lu P, et al. Three-dimensional structure of human γ-secretase. Nature. 2014;512(7513):166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YM, et al. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405(6787):689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 26.Esler WP, et al. Transition-state analogue inhibitors of gamma-secretase bind directly to presenilin-1. Nat Cell Biol. 2000;2(7):428–434. doi: 10.1038/35017062. [DOI] [PubMed] [Google Scholar]
- 27.Thinakaran G, et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17(1):181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 28.Fukumori A, Fluhrer R, Steiner H, Haass C. Three-amino acid spacing of presenilin endoproteolysis suggests a general stepwise cleavage of gamma-secretase-mediated intramembrane proteolysis. J Neurosci. 2010;30(23):7853–7862. doi: 10.1523/JNEUROSCI.1443-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn K, et al. Activation and intrinsic gamma-secretase activity of presenilin 1. Proc Natl Acad Sci USA. 2010;107(50):21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaVoie MJ, et al. Assembly of the gamma-secretase complex involves early formation of an intermediate subcomplex of Aph-1 and nicastrin. J Biol Chem. 2003;278(39):37213–37222. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- 31.Gu Y, et al. APH-1 interacts with mature and immature forms of presenilins and nicastrin and may play a role in maturation of presenilin.nicastrin complexes. J Biol Chem. 2003;278(9):7374–7380. doi: 10.1074/jbc.M209499200. [DOI] [PubMed] [Google Scholar]
- 32.Yu G, et al. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407(6800):48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 33.Kimberly WT, et al. Complex N-linked glycosylated nicastrin associates with active gamma-secretase and undergoes tight cellular regulation. J Biol Chem. 2002;277(38):35113–35117. doi: 10.1074/jbc.M204446200. [DOI] [PubMed] [Google Scholar]
- 34.Xie T, et al. Crystal structure of the γ-secretase component nicastrin. Proc Natl Acad Sci USA. 2014;111(37):13349–13354. doi: 10.1073/pnas.1414837111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah S, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122(3):435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Dries DR, et al. Glu-333 of nicastrin directly participates in gamma-secretase activity. J Biol Chem. 2009;284(43):29714–29724. doi: 10.1074/jbc.M109.038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chávez-Gutiérrez L, et al. Glu(332) in the Nicastrin ectodomain is essential for gamma-secretase complex maturation but not for its activity. J Biol Chem. 2008;283(29):20096–20105. doi: 10.1074/jbc.M803040200. [DOI] [PubMed] [Google Scholar]
- 38.Zhao G, Liu Z, Ilagan MXG, Kopan R. Gamma-secretase composed of PS1/Pen2/Aph1a can cleave notch and amyloid precursor protein in the absence of nicastrin. J Neurosci. 2010;30(5):1648–1656. doi: 10.1523/JNEUROSCI.3826-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai X-C, et al. An atomic structure of human γ-secretase. Nature. 2015;525(7568):212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L, et al. Structural basis of human γ-secretase assembly. Proc Natl Acad Sci USA. 2015;112(19):6003–6008. doi: 10.1073/pnas.1506242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraering PC, et al. Purification and characterization of the human gamma-secretase complex. Biochemistry. 2004;43(30):9774–9789. doi: 10.1021/bi0494976. [DOI] [PubMed] [Google Scholar]
- 42.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 43.Tian Y, Bassit B, Chau D, Li Y-M. An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat Struct Mol Biol. 2010;17(2):151–158. doi: 10.1038/nsmb.1743. [DOI] [PubMed] [Google Scholar]
- 44.Funamoto S, et al. Substrate ectodomain is critical for substrate preference and inhibition of γ-secretase. Nat Commun. 2013;4:2529. doi: 10.1038/ncomms3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quintero-Monzon O, et al. Dissociation between the processivity and total activity of γ-secretase: Implications for the mechanism of Alzheimer’s disease-causing presenilin mutations. Biochemistry. 2011;50(42):9023–9035. doi: 10.1021/bi2007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95(12):6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, et al. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493(7430):56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 48.Dang S, et al. Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH. Proc Natl Acad Sci USA. 2015;112(11):3344–3349. doi: 10.1073/pnas.1502150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarov VK, et al. Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci USA. 2006;103(18):6889–6894. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osenkowski P, et al. Cryoelectron microscopy structure of purified gamma-secretase at 12 A resolution. J Mol Biol. 2009;385(2):642–652. doi: 10.1016/j.jmb.2008.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, et al. Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes. Structure. 2014;22(1):125–135. doi: 10.1016/j.str.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pamrén A, et al. Mutations in nicastrin protein differentially affect amyloid beta-peptide production and Notch protein processing. J Biol Chem. 2011;286(36):31153–31158. doi: 10.1074/jbc.C111.235267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fluhrer R, et al. The Nicastrin ectodomain adopts a highly thermostable structure. Biol Chem. 2011;392(11):995–1001. doi: 10.1515/BC.2011.169. [DOI] [PubMed] [Google Scholar]
- 54.Bolduc D, et al. Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis. eLife. 2013;2:e00691. doi: 10.7554/eLife.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfe MS, Selkoe DJ. γ-secretase: A horseshoe structure brings good luck. Cell. 2014;158(2):247–249. doi: 10.1016/j.cell.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 56.Fernandez MA, Klutkowski JA, Freret T, Wolfe MS. Alzheimer presenilin-1 mutations dramatically reduce trimming of long amyloid β-peptides (Aβ) by γ-secretase to increase 42-to-40-residue Aβ. J Biol Chem. 2014;289(45):31043–31052. doi: 10.1074/jbc.M114.581165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chávez-Gutiérrez L, et al. The mechanism of γ-secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31(10):2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McConnachie G, Pass I, Walker SM, Downes CP. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: Evidence for activation by anionic phospholipids. Biochem J. 2003;371(Pt 3):947–955. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 2002;21(16):4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deas E, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20(5):867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49(24):4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kroos L, Akiyama Y. Biochemical and structural insights into intramembrane metalloprotease mechanisms. Biochim Biophys Acta. 2013;1828(12):2873–2885. doi: 10.1016/j.bbamem.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hizukuri Y, et al. A structure-based model of substrate discrimination by a noncanonical PDZ tandem in the intramembrane-cleaving protease RseP. Structure. 2014;22(2):326–336. doi: 10.1016/j.str.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Eggert S, et al. The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves alpha-, beta-, gamma-, and epsilon-like cleavages: Modulation of APLP-1 processing by n-glycosylation. J Biol Chem. 2004;279(18):18146–18156. doi: 10.1074/jbc.M311601200. [DOI] [PubMed] [Google Scholar]
- 65.Luo X, et al. Cleavage of neuregulin-1 by BACE1 or ADAM10 protein produces differential effects on myelination. J Biol Chem. 2011;286(27):23967–23974. doi: 10.1074/jbc.M111.251538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang YW, et al. Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J Biol Chem. 2005;280(17):17020–17026. doi: 10.1074/jbc.M409467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamp F, et al. Intramembrane proteolysis of β-amyloid precursor protein by γ-secretase is an unusually slow process. Biophys J. 2015;108(5):1229–1237. doi: 10.1016/j.bpj.2014.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]