Significance
Isocitrate dehydrogenase 1 (IDH1) mutations are drivers of hematological malignancy. Although these mutations are most often associated with myeloid disease, they are also found in lymphoid malignancies, including T-cell acute lymphoblastic leukemia (T-ALL). Treatment strategies targeting these mutations are currently being devised, including small-molecule inhibitors of the mutant IDH1 enzyme. A better understanding of the role of these mutations in tumorigenesis and their effects on tumor cells will allow these treatment strategies to be effectively translated to the clinic. Here we show that Idh1 mutations can contribute to the development of T-cell malignancies, including T-ALL, using a conditional knock-in mouse model. These mouse models provide a platform for further evaluation of treatment strategies for T-cell malignancy.
Keywords: isocitrate dehydrogenase, T-ALL, lymphoma, p53
Abstract
Gain-of-function mutations in isocitrate dehydrogenase 1 (IDH1) are key drivers of hematopoietic malignancies. Although these mutations are most commonly associated with myeloid diseases, they also occur in malignancies of the T-cell lineage. To investigate their role in these diseases and provide tractable disease models for further investigation, we analyzed the T-cell compartment in a conditional knock-in (KI) mouse model of mutant Idh1. We observed the development of a spontaneous T-cell acute lymphoblastic leukemia (T-ALL) in these animals. The disease was transplantable and maintained expression of mutant IDH1. Whole-exome sequencing revealed the presence of a spontaneous activating mutation in Notch1, one of the most common mutations in human T-ALL, suggesting Idh1 mutations may have the capacity to cooperate with Notch1 to drive T-ALL. To further investigate the Idh1 mutation as an oncogenic driver in the T-cell lineage, we crossed Idh1-KI mice with conditional Trp53 null mice, a well-characterized model of T-cell malignancy, and found that T-cell lymphomagenesis was accelerated in mice bearing both mutations. Because both IDH1 and p53 are known to affect cellular metabolism, we compared the requirements for glucose and glutamine in cells derived from these tumors and found that cells bearing the Idh1 mutation have an increased dependence on both glucose and glutamine. These data suggest that mutant IDH1 contributes to malignancy in the T-cell lineage and may alter the metabolic profile of malignant T cells.
Somatic mutations in isocitrate dehydrogenase 1 (IDH1) are frequently observed in a number of malignancies, including glioma, cholangiocarcinoma, chondrosarcoma, and several hematological malignancies (1). IDH1 is a cytoplasmic enzyme that catalyzes the NADP-dependent conversion of isocitrate to α-ketoglutarate (αKG). Mutations in IDH1 at arginine 132 (R132) cause an enzymatic gain of function that results in the NADPH-dependent conversion of αKG to d-2-hydroxyglutarate (2HG) (2). This metabolite is normally maintained at very low levels in cells and tissues and is not part of any known productive metabolic pathway. However, in cells and tissues of patients with IDH1 mutant tumors, 2HG builds up to high levels and is thought to contribute to tumorigenesis by inhibiting a class of αKG-dependent enzymes (1). The precise effects important for driving tumorigenesis downstream of IDH1 mutations are not fully understood and may differ between disease states.
In the hematopoietic system, IDH1 mutations are most often associated with myeloid diseases, where they are commonly found in myelodysplastic syndrome and acute myeloid leukemia (3). However, IDH1 mutations are also found in a small proportion of adult T-cell acute lymphoblastic leukemia (T-ALL) (4, 5). T-ALL is an aggressive malignancy of developing T cells and is responsible for ∼25% of adult ALL (6, 7). T-ALL is thought to arise via a multistep process of oncogenic mutation that leads to the transformation of immature T cells. The genetic landscape of the disease has been characterized, and a large number of driver mutations have been identified (6). The most common genetic feature of T-ALL is the presence of activating mutations in Notch1, which are present in more than 50% of patients (8). Interestingly, IDH1 mutations seem to be confined to a subset of adult patients with T-ALL bearing an immature T-cell gene expression signature and harboring other oncogenic mutations in genes more commonly associated with myeloid malignancy, including Flt3 and DNMT3A (4, 9). This subset of T-ALL has recently been recognized as a distinct disease entity called early T-cell precursor T-ALL and is associated with therapy resistance and a particularly poor outcome (10). The role of IDH1 mutations in this subset of T-ALL is not understood.
Using a myeloid lineage-specific conditional Idh1-R132-KI mouse model, we previously showed that mutant IDH1 partially blocks differentiation and produces a hematopoietic phenotype similar to human myelodysplastic syndrome (11). In this study, we crossed the Idh1-R132-KI mouse with Vav-cre animals to introduce the IDH1 R132 mutation into the entire hematopoietic system to investigate the role of Idh1 mutations in T-cell malignancy.
Results
Development of T-ALL in a Vav-Idh1-KI Mouse.
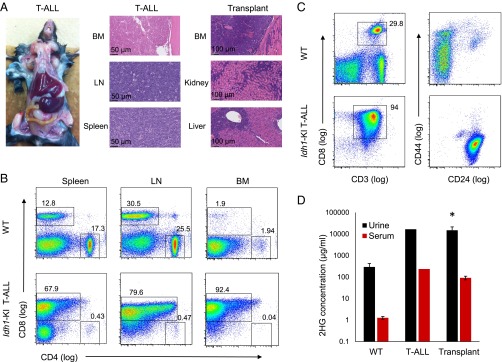
To investigate effects of Idh1 mutations in the lymphoid system, Idh1-R132-KI mice were crossed with Vav-cre mice to produce Vav-Idh1-KI animals that express the mutant protein throughout the hematopoietic system (12). As expected, these mice develop the myeloid phenotypes previously described (11). However, in addition, we observed the development of an aggressive, peripheral lymphadenopathy accompanied by splenomegaly in a Vav-Idh1-KI mouse (Fig. 1A). Histologically, neoplastic lymphocytes effaced more than 80% of the bone marrow, and diffuse sheets of these cells completely obliterated normal splenic and lymph node architecture (Fig. 1A). Flow cytometry showed that the malignant lymphocytes in the spleen, lymph nodes, and bone marrow were CD8-positive T cells (Fig. 1B). On the basis of expression of the surface markers CD44 and CD24, these cells resembled an abnormal population of early, immature T lymphocytes (Fig. 1C). As a result of these findings, the neoplastic disease in this mouse was characterized as T-ALL.
Fig. 1.
Development of T-ALL in a Vav-Idh1-KI mouse. (A) Gross pathology and H&E-stained histological sections of the indicated tissues from a Vav-Idh1-KI mouse diagnosed with T-ALL, and from a T-ALL-bearing animal after serial transplantation of the disease (fourth serial passage). (D) 2HG concentration in urine and serum from the indicated mice (wild-type, n = 4; T-ALL, n = 1; Transplant, n = 4). Error bars represent SD. *Significant difference compared with wild-type (P < 0.01). (B) Flow cytometry characterizing the cell surface phenotype of the malignant T cells isolated from spleen, lymph node (LN), and bone marrow (BM) of the T-ALL mouse compared with cells isolated from a wild-type animal. (C) Flow cytometry characterizing the malignant T-ALL cells relative to T cells obtained from the lymph node of a wild-type animal.
To further define this disease, T-ALL cells and tissue fragments isolated from the mutant mouse were transplanted into syngeneic C57BL6/J mice s.c. or into the mammary fat pad. Recipient animals developed a similar aggressive T-ALL phenotype within 3–6 wk that involved the bone marrow, secondary lymphoid organs, and other tissues, including the kidney and liver (Fig. 1A). The disease could be serially passaged (five passages have been performed), indicating a fully transformed phenotype. Interestingly, malignant cells could not be grown in suspension culture under the media conditions tested, which is consistent with previous difficulties encountered when attempting to maintain Idh1 mutant cells in culture. To determine whether the activity of the mutant IDH1 enzyme was retained in tumor cells on serial passage, 2HG levels were measured in serum and urine. In both cases, levels of 2HG were increased between 10- and 100-fold compared with wild-type animals, and the concentrations observed in transplanted animals were similar to those observed in the initial T-ALL mouse (Fig. 1D).
Whole-Exome Sequencing Identifies a Spontaneous Notch1 Mutation in Idh1-KI T-ALL.
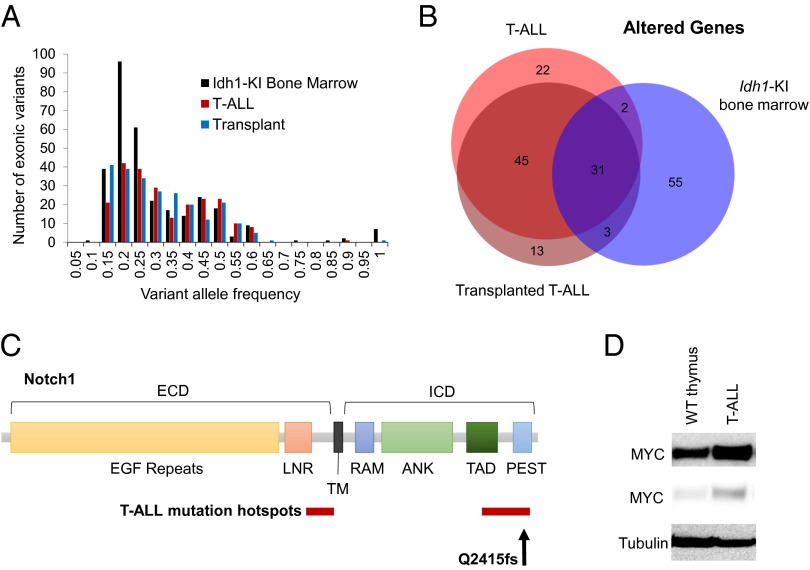
Because the T-ALL phenotype is not normally observed in these animals (penetrance of 1/110 mutant mice analyzed), we hypothesized that a spontaneous cooperating mutation contributed to this malignancy. To investigate this possibility, we conducted whole-exome sequencing on cells derived from the initial tumor and a tumor after the third serial passage. Bone marrow samples from a wild-type C57BL/6J mouse and a healthy age-matched Vav-Idh1-KI mouse were sequenced as controls. Genomic variants (single nucleotide variants and short indels) relative to the mm9 reference genome were identified using Haplotype caller. We next removed variants present in dbSNP or in the Sanger Mouse Genome Project SNP database (13). This step eliminates many false-positive calls resulting from strain variation, as well as deficiencies in the reference sequence. To further focus on likely somatic mutations, we eliminated variants identified in the wild-type C57BL/6J mouse sample or present in samples from two other C57BL/6J mice from our breeding colony that had been previously subjected to whole-exome sequencing, using the same analysis pipeline to eliminate alignment artifacts and strain background differences. Finally, we restricted our analysis to nonsynonymous exonic variants (Table 1). Interestingly, the number of exonic variants and the variant allele frequency distribution in the healthy Vav-Idh1-KI bone marrow sample was similar to that found in the two T-ALL tumor samples (Table 1; Fig. 2A). This suggests that the development of this T-ALL was not associated with a dramatic increase in genomic instability, which is consistent with observations in human T-ALL. There was a high concordance in the identities of the altered genes in the initial T-ALL sample and the third passage tumor, further suggesting the genome of this tumor was relatively stable during serial passage (Fig. 2B). There was also a significant overlap between the genes altered in the tumor samples and those found altered in the healthy Vav-Idh1-KI control bone marrow (Fig. 2B). This may be a result of false-positive identification of variants caused by misalignment of short-read sequences in highly repetitive genes.
Table 1.
Identification of coding exonic variants from whole-exome sequencing
| Sample | All variants* | Excluding SNPs† | Excluding wild-type variants | Exonic variants | Unique genes altered |
| Wild-type B6/J bone marrow | 5,379 | 3,744 | 0 | 0 | 0 |
| Idh1-KI bone marrow | 17,806 | 5,748 | 2,483 | 315 | 91 |
| T-ALL | 21,454 | 6,123 | 2,773 | 229 | 92 |
| T-ALL transplant | 21,080 | 6,061 | 26,84 | 237 | 100 |
Relative to the mm9 reference genome.
Compared with dbSNP128 and the Sanger Mouse Genomes Project SNP database.
Fig. 2.
Exome sequencing identifies a T-ALL-associated Notch1 mutation in Vav-Idh1-KI T-ALL cells. (A) The allele frequency distribution of exonic variants identified by whole-exome sequencing of malignant cells from the initial T-ALL-bearing mouse, an animal bearing T-ALL after serial transplant of the disease (fourth passage), and bone marrow from a healthy, age-matched Vav-Idh1-KI mouse. (B) A Venn diagram illustrating the unique altered genes in each sample. (C) Schematic of the Notch1 protein (ECD, extracellular domain; ICD, intracellular domain) illustrating the major protein domains. ANK, ankyrin repeats; LNR, LIN12/Notch repeats; PEST, PEST domain; RAM, RAM domain; TAD, transactivation domain; TM, transmembrane domain. Hotspots for T-ALL-associated mutations are shown in red, and the position of the Q2415fs mutation identified in the Vav-KI-IDH1 T-ALL is indicated. (D) Western blot showing levels of MYC in cells isolated from wild-type thymus, and Vav-Idh1-KI T-ALL cells (two exposures are shown). Tubulin is shown as a loading control.
Upon inspection of the list of altered genes (Dataset S1), a mutation in Notch1 was identified. This mutation was present in both the initial T-ALL tumor and the transplanted tumor, where the variant allele frequencies were 0.46 (51/95 reads) and 0.49 (37/72 reads), respectively, suggesting a heterozygous mutation. This mutation was not present in the healthy Vav-Idh1-KI bone marrow sample. The mutation is an 11-base pair deletion causing a frameshift at amino acid Q2415 (Q2415fs), in the PEST domain of the Notch1 protein, which is one of two regions commonly found mutated in human T-ALL (Fig. 2C). Mutations in the proline, glutamate, serine, and threonine rich (PEST) domain cause increased stability and increased nuclear retention of the intracellular domain of Notch, and thereby increase the transcription of a number of Notch target genes, including Myc, which is thought to contribute to T-ALL tumorigenesis (14, 15). Consistent with this mechanism of action, MYC levels were increased in T-ALL cells relative to thymic T cells from wild-type mice (Fig. 2D), suggesting the Q2415fs mutation identified in this tumor causes Notch pathway activation and may have contributed to the development of T-ALL in this animal.
Idh1 Mutations Accelerate Development of Thymic Lymphoma in Vav-p53fl/fl Mice.
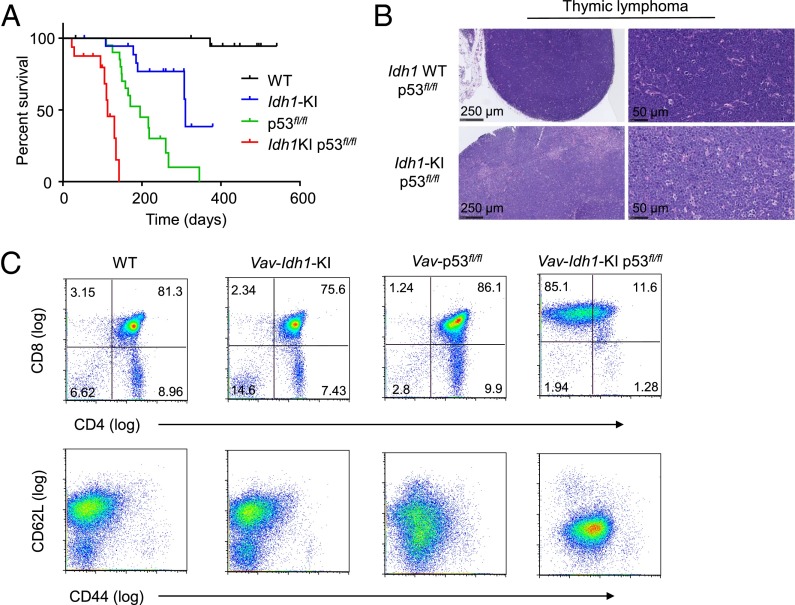
To further investigate the effect of Idh1 mutations on tumorigenesis in the T-cell lineage, we crossed Vav-Idh1-KI mice with conditional Trp53 null mice (p53fl/fl). Loss of p53 in this context is known to cause thymic T-cell lymphoma (16, 17). As expected, Vav-p53fl/fl mice developed thymic lymphoma with a median latency of ∼200 d (Fig. 3A). The addition of the Idh1 mutation significantly accelerated the development of thymic lymphoma and decreased the overall survival of double-mutant animals (Fig. 3A). Vav-Idh1-KI mice also displayed a shortened life span compared with wild-type animals, but this was not a result of abnormalities in the lymphoid system (Fig. 3A). The precise cause of the shortened life span in these animals is not fully understood, but appears to be related to the myelodysplastic syndrome-like phenotype, which includes splenomegaly and anemia.
Fig. 3.
Mutant Idh1 accelerates development of thymic lymphoma in Vav-p53fl/fl mice. (A) Kaplan-Meier curve showing overall survival of the indicated genotypes. All animals carry the Vav-cre allele. All curves are significantly different (P < 0.01, log-rank test). (B) Histology of representative thymic lymphomas in Vav-p53fl/fl and Vav-p53fl/fl Idh1-KI animals at two levels of magnification. (C) Representative flow cytometry characterization of the cell surface marker profiles of cells derived from the thymus (wild-type and Vav-Idh1-KI) or thymic T-cell lymphomas (Vav-p53fl/fl and Vav-Idh1-KI p53fl/fl) of the indicated genotypes.
Histological examination of the tumors that arose in Vav-p53fl/fl and Vav-Idh1-KI p53fl/fl animals showed no differences between the two diseases. Both sets of animals developed large localized thymic masses composed of sheets of neoplastic lymphocytes (Fig. 3B). Flow cytometric analysis confirmed that these were T-cell lymphomas, although the cells from double-mutant animals displayed a cell surface phenotype more similar to activated CD8 T-cells, with higher expression of CD44 and lower expression of CD62L compared with cells from Vav-p53fl/fl lymphomas (Fig. 3B). Thus, introduction of an Idh1 mutation accelerated the development of T-cell malignancy in this well-characterized mouse model.
Idh1 Mutations Render p53 Null Lymphoma Cells More Sensitive to Nutrient Withdrawal in Vitro.
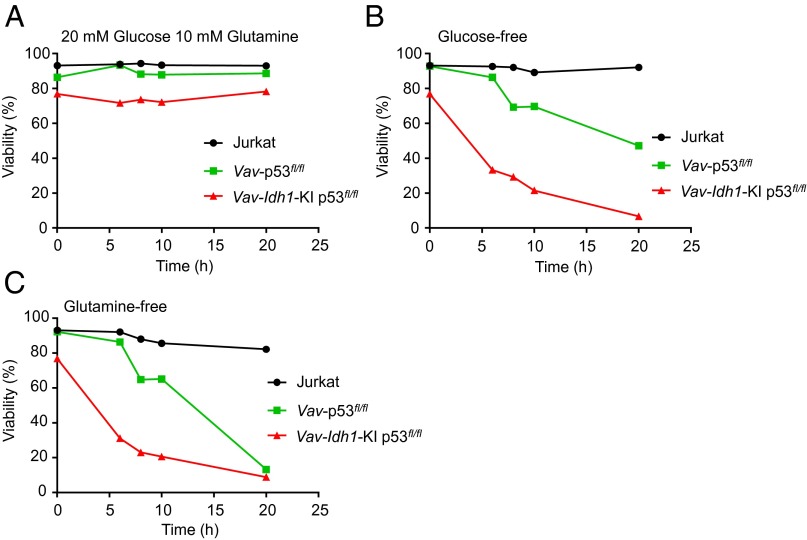
Both Vav-p53fl/fl and Vav-Idh1-KI p53fl/fl double-mutant lymphoma cells could be cultured in vitro. Because both IDH1 and p53 are known to affect cellular metabolism, we took this opportunity to determine whether the introduction of mutant IDH1 altered the nutrient requirements of these T-cell lymphoma cells. In a short-term nutrient deprivation assay, we found that double-mutant cells were more sensitive to withdrawal of glucose or glutamine from the growth media (Fig. 4). These data suggest that although the presence of mutant IDH1 accelerates tumorigenesis in p53 null T-cell precursors, these cells may be more dependent on both glucose and glutamine for maintenance of viability.
Fig. 4.
Mutant Idh1 increases sensitivity to in vitro nutrient deprivation in p53 null T-cell lymphoma cells. For all experiments, cells were resuspended in glucose and glutamine-free RPMI 1640 media containing 10% (vol/vol) dialyzed FBS, 1% FBS, and the indicated nutrient conditions, and viability was measured by flow cytometry, using annexin V/PI at the indicated times. (A) 20 mM glucose, 10 mM glutamine; (B) glucose-free, 10 mM glutamine; (C) glutamine-free, 20 mM glucose.
Discussion
Taken together, these data support the concept that Idh1 mutations can cooperate with other oncogenic events to drive malignant transformation in the T-cell lineage. Although this study focused exclusively on mutations in Idh1, it should be noted that IDH2 mutations are also observed in T-cell malignancies, including T-ALL. IDH2 is the mitochondrial counterpart of IDH1, and similar mutations in critical arginine residues of IDH2 lead to the production of 2HG. Although mutations in both genes result in gain of enzymatic function and production of 2HG, there are significant differences in the frequencies of IDH1 and IDH2 mutations in different diseases (1). Even among T-cell malignancies, this is the case. Whereas both IDH1 and IDH2 mutations are observed in adult early T-cell precursor T-ALL, a significant proportion of angioimmunoblastic T-cell lymphomas harbor exclusively IDH2 mutations (18). A better understanding of how these mutations affect T-cell development and malignant transformation may lead to an understanding of the different effects caused by mutations in these two very similar genes.
The development of a T-ALL-like disease in a Vav-Idh1-KI mouse is intriguing. The identification of a spontaneous, canonical T-ALL-associated Notch1 mutation in this tumor suggests Idh1 mutations can cooperate with Notch1 mutations to drive the development of this disease. Previous work using mouse models of common human Notch1 mutations, including those in the PEST domain similar to the mutation characterized here, have shown that similar to mutant Idh1, these Notch1 mutations alone are not sufficient to give rise to T-ALL (19). However, these mutations can cooperate with other oncogenic mutations, including those in Kras, to accelerate the development of lymphoid malignancy (19). A number of studies have identified Myc as the Notch target most likely to be responsible for its effects on the development of T-ALL (14, 15, 20). Given that MYC levels were elevated in the Vav-Idh1-KI T-ALL cells, it will be important to investigate whether mutant IDH1 can cooperate with MYC in driving tumorigenesis in the hematopoietic system. Future work will seek to formally investigate the contribution of Idh1 and Idh2 mutations to the development of T-ALL, in combination with other T-ALL-associated mutations. This may be particularly relevant for the newly recognized subset of early T-cell precursor T-ALL that appears to harbor other mutations more often associated with myeloid neoplasia.
The fact that Idh1 mutations accelerate the development of T-cell lymphoma in a well-characterized model driven by loss of p53 further illustrates the capacity of mutant IDH1 to contribute to tumorigenesis in the T-cell lineage. In this context, Idh1 mutations result in accelerated thymic T-cell lymphomagenesis, rather than disseminated T-ALL, or a more severe myeloid phenotype, reinforcing the concept that Idh1 mutations can contribute to different disease states, depending on the identity of the cooperating mutations.
Metabolic pathways and nutrient requirements of T cells are remarkably dynamic and can be rapidly remodeled during differentiation and activation and in response to extracellular conditions (21). Because mutant IDH1 alters core metabolic pathways involving the NADP:NADPH redox couple and αKG, and p53 has been shown to have effects on metabolic regulation (22), we investigated the nutrient requirements of malignant T cells isolated from thymic lymphomas, taking advantage of the opportunity to examine the effects of mutant Idh1 in a controlled in vitro system. The finding that mutant IDH1 renders cells more sensitive to nutrient deprivation suggests core metabolic pathways are altered by the presence of this mutant enzyme. Further work will be required to define the precise mechanisms responsible for these effects and to determine whether they may represent vulnerabilities that may be exploited for therapy.
There is very little known regarding the effects of IDH mutations on tumorigenesis in the T-cell lineage. First, it is unclear whether the same mechanisms responsible for malignant transformation in the myeloid lineage, or in other tissues, are also important for development and progression of T-cell malignancies. Second, it is unclear whether IDH mutations are early initiating events or act later in the multistep process of tumorigenesis in the lymphoid system. Finally, it is unclear whether the presence of IDH mutations in T-cell diseases may offer opportunities to personalize therapy. The mouse models described here will provide platforms to answer some of these questions and provide opportunities to evaluate treatment strategies in these disease states.
Materials and Methods
Animals.
Conditional Idh1-R132-KI (11, 23) and conditional Trp53 knockout (p53fl/fl) (24) mice have been described previously. Idh1-KI mice and p53fl/fl mice were bred with Vav-cre mice (Jackson Laboratories; cat. no. 008610) to produce Vav-Idh1-KI, Vav-p53fl/fl, and Vav-Idh1-KI p53fl/fl double-mutant animals. For the data shown in Fig. 3, the strain background was C57BL/6 × 129Ola9; F3, whereas for all other experiments, the strain background was C57BL/6 (F10). To serially transplant tumors, 2–3 mm3 tumor fragments, or single-cell suspensions in 50% PBS:50% (vol/vol) Matrigel (Corning) were surgically implanted s.c. or in the mammary fat pad, as previously described (25). All animal experiments were approved by the University Health Network Animal Care Committee.
Histology.
Tissues were fixed in 10% (wt/vol) formalin for 24 h, embedded in paraffin, and sectioned for histological evaluation. Sternums were rinsed and neutralized in an ammonium hydroxide solution, washed in PBS, and then fixed in 10% (wt/vol) formalin overnight. Sternums were then treated as other tissues for processing and embedding in paraffin. Sections were stained with H&E, using a standard protocol. Histological sections were examined by a board-certified veterinary anatomic pathologist.
Flow Cytometry.
Flow cytometry was performed as previously described (26). Single-cell suspensions were prepared from spleen, lymph nodes, thymus, or bone marrow preparations treated to lyse red blood cells. Cells (1–2 × 106) were preincubated with Fc block for 15 min at 4 °C and immunostained with antibodies recognizing the following: CD3 (145-2C-11), CD4 (GK1.5), CD5 (53-7.3), CD8 (53-6.7), CD13 (R3-242), CD16/CD32 (2.4G2), CD19 (1D3), CD24 (30F1), CD25 (PC61), CD44 (IM7), CD137 (BST2, PDCA-1), CD62L (MEL-14), and TCRb (H57-597), all from BioLegend, BD Biosciences, or eBioscience unless otherwise specified. Protein detection by intracellular staining was performed as described previously (26). Briefly, cells were fixed with 1.6% (wt/vol) paraformaldehyde and incubated for 30 min at room temperature. Data were acquired using BD FCMCanto flow cytometer and analyzed with the FlowJo analysis program (Tree Star Inc.).
Cell Culture.
Jurkat cells (ATCC) and cells derived from Vav-p53fl/fl and Vav-Idh1-KI p53fl/fl T-cell thymic lymphomas were cultured in nonadherent tissue culture plates in RPMI 1640 supplemented with 10% (vol/vol) FBS. Cells were maintained at 37 °C and 5% partial pressure CO2 in a humidified incubator. For nutrient deprivation experiments, cells were grown in glucose and glutamine-free RPMI 1640 media (Life Technologies) supplemented with the indicated concentrations of glucose and glutamine, 10% (vol/vol) dialyzed FBS, and 1% (vol/vol) FBS. Cell viability was measured by flow cytometry using Annexin V and propidium iodide, as previously described (27).
2HG Measurement.
For 2HG measurement, metabolites were extracted using 80% (vol/vol) aqueous methanol, as previously described (28). Extracts were subjected to ion-paired reverse-phase LC coupled to negative mode electrospray triple-quadrupole mass spectrometry, using multiple reaction monitoring. Integrated elution peaks were compared with metabolite standard curves for absolute quantification.
Exome Sequencing.
Genomic DNA was isolated using DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol. Whole-exome sequencing and data analysis were performed at the University Health Network Genomics Centre. Two hundred nanograms DNA was used to generate libraries after Agilent SureSelect XT target enrichment kit, according to the manufacturer’s protocol. A 750-ng library from each sample was hybridized for 24 h for mouse all-exon capture. Captured enriched libraries were size validated, using Agilent Bioanalyzer, and concentration validated by quantitative PCR. All libraries were normalized to 10 nM and pooled. Next, 10 pM of pooled libraries were loaded onto Illumina cBot for cluster generation. The clustered flow cell was then sequenced using paired-end 100 cycles on an Illumina HighSEq. 2000. Read quality was checked using FASTQC (v. 0.11.2), and raw sequence data were aligned to the mouse genome (mm9), using BWA-MEM (v. 0.7.10). Alignment quality was assessed using Qualimap (v. 2.0). Bam files were preprocessed using Picard (v. 1.124) to mark duplicates and GATK (v. 3.2–2) to perform local realignment around indels and to perform base quality score recalibration. HaplotypeCaller (Broad Institute) was used to identify variants relative to the mm9 reference genome. To eliminate variants that have a low likelihood of being somatic events, we removed variants present in the mouse dbSNP database (v128), and eliminated common variants identified by the Sanger Institute Mouse Genomes Project (13). To further identify potential somatic variants of interest, we eliminated variants present in the control wild-type C57BL/6J mouse from Jackson Laboratories, or in DNA from two C57BL/6J mice from our breeding colony. We further focused on exonic coding mutations. This approach should eliminate most variants called as a result of misalignment artifacts, sequencing artifacts, and errors in the underlying reference sequence.
Supplementary Material
Acknowledgments
We are grateful for the administrative assistance of Ms. Irene Ng and for technical assistance from the Princess Margaret Cancer Center flow cytometry facility, genotyping facility, and animal resource center, and the University Health Network Genomics Centre. This work was supported by a grant from the Canadian Institutes of Health Research (to T.W.M, R.A.C, and Z.H.) and by a grant from the Leukemia and Lymphoma Society (to T.W.M.). F.L. received a grant from the Institut National du Cancer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525354113/-/DCSupplemental.
References
- 1.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 2.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molenaar RJ, et al. Clinical and biological implications of ancestral and non-ancestral IDH1 and IDH2 mutations in myeloid neoplasms. Leukemia. 2015;29(11):2134–2142. doi: 10.1038/leu.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Vlierberghe P, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208(13):2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Vlierberghe P, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122(1):74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durinck K, et al. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp Hematol. 2015;43(8):625–639. doi: 10.1016/j.exphem.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 7.You MJ, Medeiros LJ, Hsi ED. T-lymphoblastic leukemia/lymphoma. Am J Clin Pathol. 2015;144(3):411–422. doi: 10.1309/AJCPMF03LVSBLHPJ. [DOI] [PubMed] [Google Scholar]
- 8.Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology (Am Soc Hematol Educ Program) 2009:353–361. doi: 10.1182/asheducation-2009.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haydu JE, Ferrando AA. Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol. 2013;20(4):369–373. doi: 10.1097/MOH.0b013e3283623c61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coustan-Smith E, et al. Early T-cell precursor leukaemia: A subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgiades P, et al. VavCre transgenic mice: A tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34(4):251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- 13.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 17.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4(1):1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 18.Cairns RA, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26(21):8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buck MD, O’Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212(9):1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 23.Hirata M, et al. Mutant IDH is sufficient to initiate enchondromatosis in mice. Proc Natl Acad Sci USA. 2015;112(9):2829–2834. doi: 10.1073/pnas.1424400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 25.Gorrini C, et al. Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc Natl Acad Sci USA. 2014;111(12):4472–4477. doi: 10.1073/pnas.1324136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Z, et al. Fas receptor expression in germinal-center B cells is essential for T and B lymphocyte homeostasis. Immunity. 2008;29(4):615–627. doi: 10.1016/j.immuni.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao Z, et al. The E3 ubiquitin ligase Mule acts through the ATM-p53 axis to maintain B lymphocyte homeostasis. J Exp Med. 2012;209(1):173–186. doi: 10.1084/jem.20111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27(18):1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.