Alzheimer’s disease is characterized by protein deposits of amyloid-β as plaques in the brain (1). The suite of enzymes that produce amyloid-β by cutting it out of the amyloid-β precursor protein (APP) have long been considered to be prime targets for therapeutic intervention. Converting this promise into reality, however, continues to be stalled by a series of obstacles, including an inaccurate and incomplete understanding of the underlying enzyme mechanisms (2). This challenge has been especially daunting for the multisubunit γ-secretase enzyme that lies embedded in the cell membrane, and ultimately delivers the final intramembrane cut that liberates amyloid-β. In PNAS, Bolduc et al. (3) have now added an unexpected new piece to the γ-secretase puzzle: Nicastrin, the largest component of the γ-secretase complex, functions as a “gatekeeper” that physically blocks large proteins from entering the enzyme’s catalytic center and being cut inappropriately.
Background
γ-Secretase was originally defined by its role in APP processing, but deeper study revealed it to be an enzymatic “hub” that processes scores of different cell surface proteins (4, 5). The most consequential is the Notch receptor: The effect of blocking γ-secretase activity is accounted for by genetic KO of Notch signaling alone (6). In fact, cleavage of Notch by γ-secretase is itself the trigger that completes a communication circuit between cells that is important during embryonic development and in rapidly renewing adult tissues. Specificity of this communication cascade, therefore, absolutely relies on γ-secretase reliably distinguishing activated from resting Notch receptors. The medical repercussions of this deceptively simple choice loom large: Loss of Notch cleavage is incompatible with life, whereas excess cleavage is implicated in cancer (7).
Ultimately, ligand binding to the Notch receptor leads to a series of rearrangements that result in cutting away of the extracellular portion (“ectodomain”) of Notch (8, 9). The short extracellular stub, transmembrane segment, and cytoplasmic signaling domain remain at the membrane and become a substrate for γ-secretase (6). In fact, APP, Notch, and dozens of other γ-secretase substrates share no apparent sequence identity, but all require this “priming” ectodomain shedding to convert them into γ-secretase substrates (10).
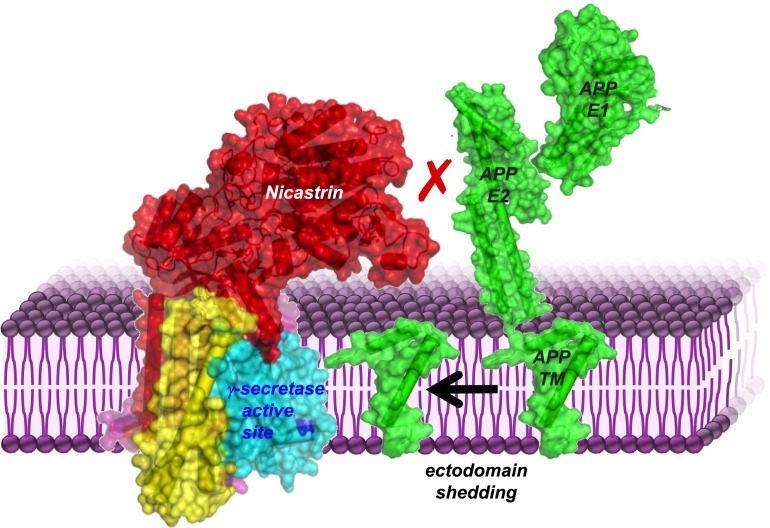
Fig. 1.
Nicastrin (red) physically blocks large substrates (APP in green) from entering the γ-secretase active site (dark blue) until their ectodomains are removed. Illustration uses coordinates 5A63 (γ-secretase, left complex), 4PWQ (APP E1), 3UMH (APP E2), 2LP1 (APP TM). The three APP domains are separate structures that would be connected through linkers that are not illustrated.
A lingering question has been precisely how γ-secretase keeps from cleaving full-length substrates when the sequence surrounding the cleavage of both “primed” and resting proteins is identical. One enticing model sprouted from an apparent motif hidden within one of the four essential γ-secretase subunits. Nicastrin, the only subunit with a large ectodomain, itself contains a short sequence that distantly resembles aminopeptidases, or enzymes that evolved to bind amino termini. Early evidence clearly supported this view (11), and nicastrin is now widely considered to be a substrate receptor that binds the new amino terminus as a prerequisite for efficient substrate processing by γ-secretase. Quantitative data to detail the mechanism underlying this attractive and prevailing model, however, have remained incomplete despite a decade of study.
A Mystery Unfolds
Bolduc et al. (3) confronted this challenge head on by judiciously developing a series of chemically defined substrates and quantitative enzyme assays to interrogate nicastrin function. Among their arsenal of innovative tools were proteins that they joined from synthetic and bacterially produced fragments to generate primed Notch substrates with either a natural amino terminus or one that had its amine character masked by acetylation. To their surprise, presenting either substrate to γ-secretase revealed no requirement for the amino terminus in substrate cleavage. Other
Bolduc et al. have now added an unexpected new piece to the γ-secretase puzzle: Nicastrin, the largest component of the γ-secretase complex, functions as a “gatekeeper” that physically blocks large proteins from entering the enzyme's catalytic center and being cut inappropriately.
modifications, including mutating the aminoterminal residue to various charged or bulky amino acids or flooding the system with 1,000-fold excess of “decoy” peptide, failed to hinder γ-secretase processing. These alarming results cast serious doubt that the newly generated amino terminus of Notch acts positively to enhance γ-secretase catalysis.
If binding between substrate and γ-secretase apparently is not mediated through the new amino terminus, then what part drives association with γ-secretase? Bolduc et al. (3) next beautifully adapted a new technology that allows measuring enzyme kinetics directly inside the cell membrane (12). They ultimately found that just the Notch transmembrane segment alone acted as an efficient substrate for γ-secretase (3). Although there is limited precedent for interpreting binding affinities within the 2D membrane bilayer system, the Km parameter they measured (3) was 100-fold lower than the Km parameter measured using the same assay with rhomboid proteases, which also cut inside the membrane (12). On at least a relative scale, therefore, it is the transmembrane segment of Notch alone that drives the “tight” association with γ-secretase.
If the new amino terminus does not stimulate γ-secretase processing, and association is driven by the Notch transmembrane segment, what is the role of nicastrin? A final clue came from progressively extending the Notch amino terminus: These modifications compromised catalysis, as expected, but did so by appearing to “block” the transmembrane interaction. This clue led Bolduc et al. (3) back to nicastrin: Does nicastrin ensure substrate specificity by physically blocking substrates with long ectodomains from entering the γ-secretase active site?
The incisive experiment to test this new model was conceptually simple but logistically complicated: Disrupting the nicastrin ectodomain should allow processing of full-length substrates, but disruptive mutations in nicastrin could confound γ-secretase complex assembly, stability, and/or transport. Bolduc et al. (3) thus developed a clever nonmutagenic means to disrupt the nicastrin ectodomain after the natural γ-secretase complex is already assembled and has matured.
Because nicastrin is the only γ-secretase subunit that has disulfide bridges to help stabilize its fold (13), Bolduc et al. (3) used reducing agents to destabilize its ectodomain. Remarkably, this treatment increased cleavage of diverse substrates with longer or even full ectodomains by as much as 20-fold, yet had no effect on the processing of primed substrates with short ectodomains. Reduction did not fully overcome the need for ectodomain shedding in every case, perhaps because this treatment destabilized, but did not remove, the bulky nicastrin ectodomain. Therefore, the nicastrin ectodomain functions negatively in substrate discrimination without affecting processing efficiency.
The steric block mechanism for nicastrin fits well with several recent observations that initially seemed paradoxical. The latest atomic structures of γ-secretase revealed the aminopeptidase-like region of nicastrin is positioned on the opposite side relative to the enzyme active site, and thus would require a large conformation change to move into position for productive aminoterminal binding during catalysis (13). The bulky face of nicastrin, in contrast, hovers reassuringly above the γ-secretase active site. Finally, a simple archaeal homolog of γ-secretase that lacks nicastrin (or other subunits) is able to process APP with a large ectodomain (14).
Implications and Future Directions
In addition to shedding new light on an old mystery, the work of Bolduc et al. (3) offers a different perspective on enzyme function inside the membrane. Methods to quantify the kinetics of protein cutting inside the membrane have been developed only recently (12), and, until now, the characterized examples revealed low affinity between enzymes and their transmembrane substrates. Bolduc et al. (3) now show that γ-secretase displays an ∼100-fold lower Km for Notch than any other enzyme/substrate pair analyzed to date (12, 15). Although this disparity likely reflects, at least partially, functional differences between γ-secretase and rhomboid proteases, perhaps even more exciting is the possibility that Notch is itself the root of the exception.
Bolduc et al. (3) deliberately chose to focus on Notch because it is the key γ-secretase substrate evolutionarily. By the same token, however, Notch could have experienced unique selective pressure to evolve a tight association (6). On a biochemical level, it also seems unlikely that γ-secretase could process scores of substrates with little regard for their transmembrane sequence but with a high binding affinity for most of these highly diverse substrates. In support of this speculative view, γ-secretase processing of APP inside the membrane was recently found to be remarkably similar to rhomboid catalysis, although it should be noted that APP was analyzed with a different assay system (15).
The inducible reconstitution assay that was developed with rhomboid (12) and now adapted for γ-secretase (3) provides an exciting common platform for more accurate and direct comparison of diverse membrane enzymes and substrates. This analysis should now be repeated with APP in parallel because of its clinical importance. In fact, the key challenge facing treating Alzheimer’s disease by targeting γ-secretase directly is the inability to lower pathological processing of APP while sparing the processing of Notch (2). Such nuanced intervention requires a complete understanding of how γ-secretase functions, and the work of Bolduc et al. (3) now provides both deeper insight and new methods that could find important but unexpected clinical implications.
Acknowledgments
Research in the laboratory of S.U. is funded by Grants 2R01AI066025 and R01AI110925 from the NIH. S.U. is a Fellow of the David & Lucile Packard Foundation and a Scholar of the Blavatnik Foundation.
Footnotes
The author declares no conflict of interest.
See companion article on page E509.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B, Chávez Gutiérrez L. Learning by failing: Ideas and concepts to tackle γ-secretases in Alzheimer’s disease and beyond. Annu Rev Pharmacol Toxicol. 2015;55:419–437. doi: 10.1146/annurev-pharmtox-010814-124309. [DOI] [PubMed] [Google Scholar]
- 3.Bolduc DM, Montagna DR, Gu Y, Selkoe DJ, Wolfe MS. Nicastrin functions to sterically hinder γ-secretase–substrate interactions driven by substrate transmembrane domain. Proc Natl Acad Sci USA. 2016;113:E509–E518. doi: 10.1073/pnas.1512952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopan R, Ilagan MX. Gamma-secretase: Proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5(6):499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 5.Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Proteomic profiling of gamma-secretase substrates and mapping of substrate requirements. PLoS Biol. 2008;6(10):e257. doi: 10.1371/journal.pbio.0060257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 7.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17(1):52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93(4):649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 9.Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- 10.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6(3):625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 11.Shah S, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122(3):435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Dickey SW, Baker RP, Cho S, Urban S. Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell. 2013;155(6):1270–1281. doi: 10.1016/j.cell.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai XC, et al. An atomic structure of human γ-secretase. Nature. 2015;525(7568):212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang S, et al. Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH. Proc Natl Acad Sci USA. 2015;112(11):3344–3349. doi: 10.1073/pnas.1502150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamp F, et al. Intramembrane proteolysis of β-amyloid precursor protein by γ-secretase is an unusually slow process. Biophys J. 2015;108(5):1229–1237. doi: 10.1016/j.bpj.2014.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]