Significance
The DNA in eukaryotic organisms is packaged in nucleosomes, the fundamental repeating unit of chromatin. The ability of essential transcription regulatory factors to bind genomic recognition sites is contingent on the sequence-dependent conformation of DNA bound in nucleosomes. Our knowledge of the structure of nucleosome DNA is severely limited. The crystal structure of the nucleosome core particle reported here incorporating mouse mammary tumor virus promoter DNA provides new insight into the sequence-dependent structure of DNA as it exists in chromatin.
Keywords: chromatin, nucleosome, DNA, X-ray structure, MMTV
Abstract
The conformation of DNA bound in nucleosomes depends on the DNA sequence. Questions such as how nucleosomes are positioned and how they potentially bind sequence-dependent nuclear factors require near-atomic resolution structures of the nucleosome core containing different DNA sequences; despite this, only the DNA for two similar α-satellite sequences and a sequence (601) selected in vitro have been visualized bound in the nucleosome core. Here we report the 2.6-Å resolution X-ray structure of a nucleosome core particle containing the DNA sequence of nucleosome A of the 3′-LTR of the mouse mammary tumor virus (147 bp MMTV-A). To our knowledge, this is the first nucleosome core particle structure containing a promoter sequence and crystallized from Mg2+ ions. It reveals sequence-dependent DNA conformations not seen previously, including kinking into the DNA major groove.
DNA in eukaryotic cells is wrapped repeatedly in nucleosomes to form chromatin, the substrate engaged by the nuclear machinery to carry out repair, replication, recombination, and transcription of genomes. Nucleosome mapping in situ combined with biochemical studies has revealed that nucleosome positions determine access to DNA regulatory sequences essential to these processes (1, 2). Nucleosome position is determined chiefly by DNA sequence and ATP-dependent chromatin remodeling factors (3–5). The nucleosome includes a linker DNA of variable length and a nucleosome core containing a histone octamer and 147 bp of DNA (6). Many high-resolution structures of nucleosome cores with differing DNA sequences are required to see how the details of DNA conformation could affect nucleosome positioning and dynamics, as well as nuclear factor binding. The DNA studied would most interestingly represent natural sequences of transcription promoters and enhancer elements.
Our knowledge of sequence-dependent structure of DNA bound in the nucleosome core relative to the amount of DNA bound in genomes is extremely limited. A resolution of at least 2.6 Å is necessary to evaluate differences in DNA conformations and assess solvent interactions adequately. To date, this highly reliable “library” of DNA structural information pertinent to the nucleosome core consists primarily of two similar sequences of half α-satellite repeats and half the artificially “evolved” sequence 601 (7–10). Further investigations have been limited to substitution of short sequence elements in one of the α-satellite sequences (11, 12). These high-resolution structures have hinged on using palindromic sequences to avoid twofold averaging imposed by crystal packing. The lack of twofold symmetry in the full 601 sequence, for example, resulted in superposition of the electron density of the two different half-sequences (9).
We describe here the X-ray structure of a nucleosome core particle (NCP) containing a DNA sequence from mouse mammary tumor virus (MMTV) determined at 2.6 Å resolution. To our knowledge, this is the first NCP structure containing a sequence from a transcription promoter, and it reveals new sequence and nucleosome-dependent DNA conformations. This NCP represents the MMTV-A nucleosome and includes the first 143 bp of the MMTV transcript, and although the entire asymmetric sequence was used, the usual twofold averaging of nucleosome core halves did not occur. A palindromic sequence was not required to obtain a clear image of the entire DNA. An engineered variant of the histone H4 tail used appears to be important for this lack of averaging. This variant also allowed the structure to be crystallized using magnesium rather than manganese divalent cation, yielding, to our knowledge, the first look at an NCP under conditions closer to physiological than before.
Results
The MMTV-A NCP structure (MMTVA) was determined by molecular replacement using the 1.9-Å structure of the α-satellite palindromic (ASP) NCP [Protein Data Bank (PDB) ID code 1kx5] (7). MMTVA was rebuilt and refined using multiple rounds of energy refinement and simulated annealing (Table S1). The electron density for the DNA allowed the entire 147-bp sequence to be built unambiguously in the B-form. Over 900 water molecules could be placed in the structure. However, it was not possible to distinguish Mg2+ ions from water reliably. In one case, the octahedral ligand geometry permit Mg2+ ion assignment, and coincided with the interparticle bridging Mn2+ ion seen in the ASP structure (6).
Table S1.
MMTVA data collection and refinement statistics (molecular replacement)
| Data collection | MMTVA |
| Wavelength | 0.99988 Å |
| Space group | P212121 |
| a, b, c, Å | 107.87, 178.92, 109.06 |
| α, β, γ, ° | 90.0, 90.0, 90.0 |
| Resolution, Å | 30.0–2.63 (2.72–2.63)* |
| No. of unique hkl used | 59,566 (4,360) |
| Multiplicity | 6.8 |
| Completeness, % | 94.0 (73.6) |
| Rsym | 0.083 (0.33) |
| I/σI | 14.2 (1.8) |
| Refinement (all data) | |
| Rwork | 0.175 (0.300) |
| Rfree (4.7% of hkl) | 0.252 (0.367) |
| No. of atoms | 13,349 |
| Protein | 6,379 |
| DNA | 6,029 |
| Water | 936 |
| Ions | 5 |
| B-factors | |
| Protein | 75.9 |
| DNA | 138.1 |
| Water | 76.4 |
| Ions | 53.5 |
| Deviation from ideality | |
| Bond lengths, Å | 0.0105 |
| Bond angles, ° | 1.292 |
Values in parentheses are for the highest-resolution shell.
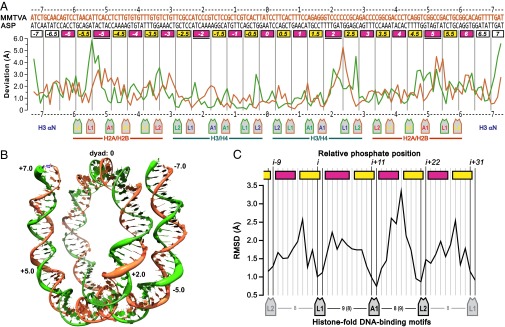
The DNA-backbone path deviations between MMTVA vs. ASP were assessed by aligning only the H3–H4 tetramer components of the two structures yielding rmsd of 1.83 and 1.77 Å for backbone phosphate groups and C4′ atoms, respectively. By comparison, the rmsd for Cα of the H3–H4 tetramer and the H2A–H2B dimers were only 0.27 and 0.40 Å, respectively. The deviations between the two different DNA sequences are generally elevated along each strand between phosphate groups not bound directly by the histone-fold DNA-binding motifs L1, L2, and A1 (8, 13) (Fig. 1A and Fig. S1). The largest distortions of the MMTVA DNA double helix relative to that for ASP are located in the major groove blocks at SHL +2, SHL ±5 and SHL ±7 (Fig. 1B). Superhelix location (SHL) and major and minor groove blocks have been defined previously (8). The rmsd of the phosphate groups bound to the H2A–H2B dimers is 2.08 Å, significantly larger than for the H3–H4 tetramer at 1.42 Å. Neither MMTVA nor ASP contain overwound, stretched DNA within the confines of the four histone-fold pairs, and therefore both DNA structures have the same registration of phosphate groups along the histone octamer surface.
Fig. 1.
Comparison of MMTVA and ASP DNA backbone paths. (A) Rmsd of the phosphate groups between the aligned structures. The I (gold) and J (green) strands are plotted 5′ to 3′ and 3′ to 5′, respectively (I and J designations correspond to the PDB files). The I-strand sequences are written above the assigned major (magenta) and minor (yellow) groove blocks (8). All blocks contain 5 bp, except where noted as containing 6 bp (underlined). The two primary phosphate groups bound by each L1, L2, and A1 DNA-binding motif are pointed to by the motif-label boxes. (B) MMTVA DNA superhelix. The I- and J-strand backbone representation has a diameter equal to the rmsd between the MMTVA and ASP phosphate groups. The SHL +2, ±5, and ±7 that show the largest deviation, as well as SHL 0, are labeled. (C) Rmsd between MMTVA and ASP at each phosphate position for all of the histone-fold motives combined. All eight DNA single-strand regions used were aligned in the 5′ to 3′ direction and registered with respect to each other based on L1 and L2 contacts. The major and minor groove blocks and primary phosphates are labeled as for A. The number of phosphate groups between primary phosphates is shown between motif labels.
Fig. S1.
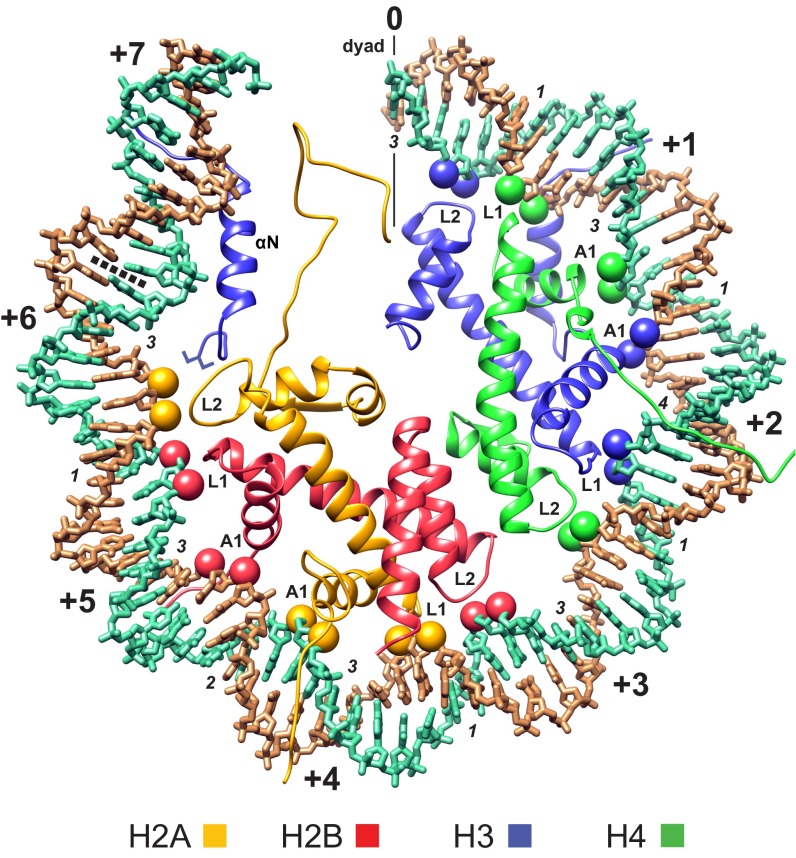
Superhelix locations and primary phosphates bound by the histone-fold motifs. One-half of the MMTVA DNA superhelix is shown (PDB file I chain 0 to +73, gold; J chain −73 to 0, green). The superhelix locations proceed from SHL 0 (0) at the DNA center (dyad) to SHL +7 (+7) near the DNA terminus. SHLs are shown at each place the DNA minor groove faces outward (every 10–11 bp). SHLs for the superhelix half not shown run from SHL −7 to SHL 0. The primary phosphate groups making interactions with the L1, L2, and A1 histone-fold DNA-binding motifs are shown as spheres colored according to the associated histone. The number of base pairs that do not contribute a primary phosphate is indicated for each major and minor groove block (italics). The terminal turn of DNA beyond the boundary for the central 128-bp steps (dotted line) is bound principally by the αN helix of the dyad-related H3, has approximately half the radius of curvature of histone fold-bound DNA, and may be affected by the end-to-end stacking of DNA between particles in the crystals.
The histone-fold DNA-binding motifs, L1, L2, and A1, interact primarily with two adjacent phosphate groups on each strand for each turn of the double helix, thereby guiding the form of the superhelix and placing constraints on the local conformation of the double helix (8, 14) (Fig. S1). The backbone deviation between the two structures is an indicator of the conformational flexibility of the DNA bound in the NCP and can be analyzed at each phosphate position by calculating the rmsd summed over all eight single-strand regions bound to the histone folds (Fig. 1C). Overall, a single histone fold spans 23 phosphate groups of a single strand from the L1 5′ to the L2 3′ primary phosphate group (i through i+22) (8). The center of the DNA strand bound between the L1 and L2 motifs is bound by the A1 motif of the paired histone fold. Three base pairs are directly bound via either or both of their 5′ and 3′ phosphate groups to the paired L1L2 sites and A1A1 sites, leaving only 1 or 3 bp not directly constrained by primary phosphate groups in minor groove or major groove blocks, respectively. The L1 and A1 motifs along a single strand are generally separated by nine phosphate groups, except for H4–L1 to H3–A1, where this separation is eight. This arrangement results in two special locations. The minor groove block at SHL ±4.5 and the major groove block at SHL ±2 have, respectively, 2 and 4 bp instead of 1 or 3 bp not directly constrained by primary phosphate groups (Fig. S1). Many of the phosphates not designated as primary binding groups also make histone interactions with, e.g., the H3 αN helix and the arginine side chains associate with each minor groove block. However, these interactions do not appear to constrain the DNA conformation as effectively as those made by the histone-fold motifs. The overall rmsd of the DNA backbone at the primary phosphate positions is limited to ∼1 Å, whereas the remaining positions between DNA-binding motifs show considerable variation between MMTVA and ASP (Fig. 1C).
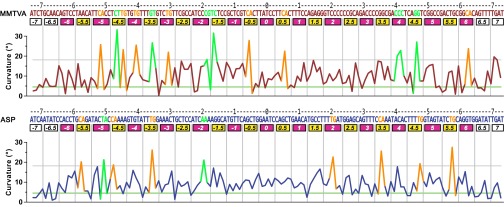
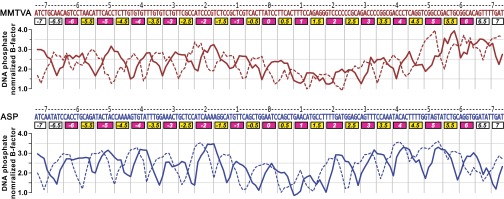
The greater propensity of MMTVA DNA to contort compared with ASP DNA is indicated by the mean curvature of 10.8° (σ = 6.8°) vs. 9.8° (σ = 5.1°) for the curvature of the DNA superhelix over the central 128 bp steps, the region bound by the histone-fold pairs. This difference stems from the greater degree of kinking, both in number and magnitude, for MMTVA vs. ASP (Fig. 2). The phosphodiester backbone segments in and adjacent to SHL −5, SHL +2, SHL +2.5, and SHL +5 show the largest deviations between MMTVA and ASP (Fig. 1A). These differences do not result from disorder or imprecise chain placement because the temperature factors in these regions are among the lowest in the DNA structure (Fig. S2). The major groove block at MMTVA SHL −5 is dominated by a CA=TG kink (−52, −51) at the central base-pair step, whereas for ASP there is a TA kink (−51, −50). In contrast, the major groove block at MMTVA SHL +5 is dominated by an undertwisted (18.2°) GG=CC step (+50, +51) that is correlated with an overtwisted (51.0°) and kinked GG=CC step (+46, +47) in the adjacent minor groove block at SHL +4.5 (Fig. S3). This undertwisting appears to affect the conformation of the adjacent base pairs causing the large deviations seen. Magnesium ions are possibly bound to the major groove in this region due to the high GC content (+46 to +55: GGTCGGCCGA) and may contribute to the DNA conformation here. The sequence of MMTVA SHL +2 through SHL +2.5 consists mainly of poly-C (TCCCCCCGCA) and shows large deviations from the equivalent region in ASP (TTGATGGAGC). The GATGG sequence for ASP is also found at MMTVA SHL −2 to SHL −2.5 and shows a substantial deviation from the pseudo twofold-related MMTVA SHL +2 to SHL +2.5. This difference is likely important for the asymmetry found in the crystal packing.
Fig. 2.
Curvature of the MMTVA and ASP DNA double-helix axes. The kinked base-pair steps are highlighted (orange: CA=TG; others: green). The horizontal lines show the values for ideal (green, 4.53°) and kink threshold (gray, 18.0°) curvatures. The horizontal axis labels are as for Fig. 1.
Fig. S2.
B-factors for MMTVA and ASP DNA phosphates. The plotted, normalized temperature factors were calculated by dividing each phosphate atom B-factor by the corresponding mean B-factor for either the Cα atoms of the MMTVA (62.4 Å2) or ASP (27.6 Å2) histone octamer core (H2A 17–117, H2B 36–123, H3 45–132, H4 27–100). The solid (sequence shown) and dotted lines represent the PDB file I chain and J chain, respectively. The horizontal axis labels are as for Fig. 1.
Fig. S3.
Base-pair step parameters. (A) MMTVA. The curvature components of roll and tilt contributing to superhelix formation are shown at each base-pair step, as are the values for shift, slide, and twist. The base-pair steps at which kinks are denoted in Fig. 2 have their roll and tilt contributions highlighted (orange, CA=TG; other, green). The slide and twist parameters are also highlighted for base-pair steps with roll/slide/twist correlation. The base-pair steps in minor groove blocks showing shift-assisted bending are denoted (pink). The horizontal axis labels are as for Fig. 1. The transcription start-site for the MMTV 3′-LTR containing the MMTV-A nucleosome is shown (red arrow). (B) ASP. As for A.
The large deviations between the MMTVA and ASP backbones at SHL −5 to SHL −5.5 (J-strand +53 to +56), SHL +5 to SHL +5.5 (I-strand +53 to +55), and SHL +2 (I-strand +23, +24) consist not only of lateral displacements of the entire double helix, but also of differences in the phosphate position along the backbone by approximately one-half base-pair step, including phosphates directly bound by histone-fold DNA-binding motifs (Fig. S4). These major groove blocks contain 6 bp compared with the others, which have 5 bp (Fig. 1), and are the principal sites for DNA stretching previously observed for ASP (PDB ID code 1kx4, 146 bp; PDB ID code 2nzd, 145 bp), NCP146b (PDB ID code 1kx3, 3utb, 146 bp) and nucleosome cores containing three versions of the 601 sequence (e.g., PDB ID code 3lz1, 145 bp) (7, 9, 14, 15).
Fig. S4.
Superposition of the MMTVA and ASP DNA structures (stereo). (A) The “negative” half of the MMTVA (red) and ASP (blue) DNA superhelices (PDB file I chain −73 to 0 and J chain +73 to 0). In addition to SHL 0, the superhelix locations are labeled at each place the DNA minor groove faces inward. The MMTVA phosphate groups that are approximately one-half base pair out of step with their ASP counterpart are marked (green dot). (B) The “positive” half of the MMTVA DNA superhelix (PDB file I chain 0 to +73 and J chain −73 to 0). This half corresponds to that shown in Fig. S1 and is labeled as for A. (C) The SHL −5.0 to SHL −5.5 region. Three of the four MMTVA phosphate groups that are out of step with those of ASP are labeled (J chain +53, +54, +55). The two primary phosphate groups that make hydrogen bonds (green) directly with the H2B–L1 motif are indicated (I chain −53, −54).
Three primary modes of DNA bending contributing to the form of the nucleosome core superhelix are apparent: (i) kinking in minor and major groove blocks, (ii) shift-assisted bending in minor groove blocks, and (iii) smooth bending in major groove blocks. The number of kinked base-pair steps based on total curvature is 14 for MMTVA and 9 for ASP (Table 1 and Methods, Analysis). Kinks in minor groove blocks typically comprise an extreme value for the roll base-pair step parameter and a roll/slide/twist correlation (8). The CA=TG kink is by far the most frequent, but others such as the GG=CC step at MMTVA SHL +4.5 also occur and can display the same roll/slide/twist correlation. The preference for CA=TG steps at kinks is consistent with the bendability exhibited in oligonucleotide structures (16). Nevertheless, the GG=CC steps at SHL +4.5 kinks strongly despite the adjacent CA=TG step (not kinked) in the same block. A GG=CC step was also noted to kink in a minor groove block in the 145-bp ASP structure containing stretched DNA (15). The TC=GA (−17, −16) step in the minor groove block MMTVA SHL −1.5 kinks predominantly via tilt instead of roll and is combined with shift-assisted bending. Kinks also occur in major groove blocks (Fig. 2). For MMTVA, these kinks occur at CA=TG (SHL −5, −4, −3, +6), CG (SHL −2) and CC=GG (SHL +4; Fig. 3A). On reexamining ASP using total curvature to define DNA bending, major groove blocks are seen to be kinked at CA=TG (SHL +2), TA (SHL −5), and AA=TT (SHL −2). Major groove kinks do not reveal any obvious correlation between base-pair step parameters such as the roll/slide/twist correlation seen for minor groove kinks. Of the 10 unique base-pair steps, only AT and GC steps do not show any kinking (Table 1). The AT step may be particularly resistant to kinking because it does not occur in any minor groove block bound to a histone-fold pair in either MMTVA or ASP.
Table 1.
Count of base-pair steps for MMTVA and ASP
| Type | Base-pair step | Kinked | Shift-assisted | ||||
| MMTVA | ASP | Total | MMTVA | ASP | MMTVA | ASP | |
| AA=TT | 14 | 25 | 39 | 1 | |||
| AC=GT | 21 | 14 | 35 | 1 | 1 | ||
| AG=CT | 18 | 18 | 36 | 1 | 3 | 5 | |
| AT | 5 | 15 | 20 | ||||
| CA=TG | 21 | 28 | 49 | 7 | 7 | 2 | 2 |
| CC=GG | 25 | 14 | 39 | 3 | 4 | 3 | |
| CG | 10 | 10 | 1 | 2 | |||
| GA=TC | 22 | 16 | 38 | 1 | 5 | 2 | |
| GC | 8 | 8 | 16 | 1 | 6 | ||
| TA | 2 | 8 | 10 | 1 | |||
| Total | 146 | 146 | 292 | 14 | 9 | 18 | 18 |
Only genomic sequences are counted to avoid biases from unnaturally selected sequences. The kinked CA=TG and GA=TC steps at SHL +7 were not counted due to possible effects from DNA end-to-end stacking between particles.
Fig. 3.
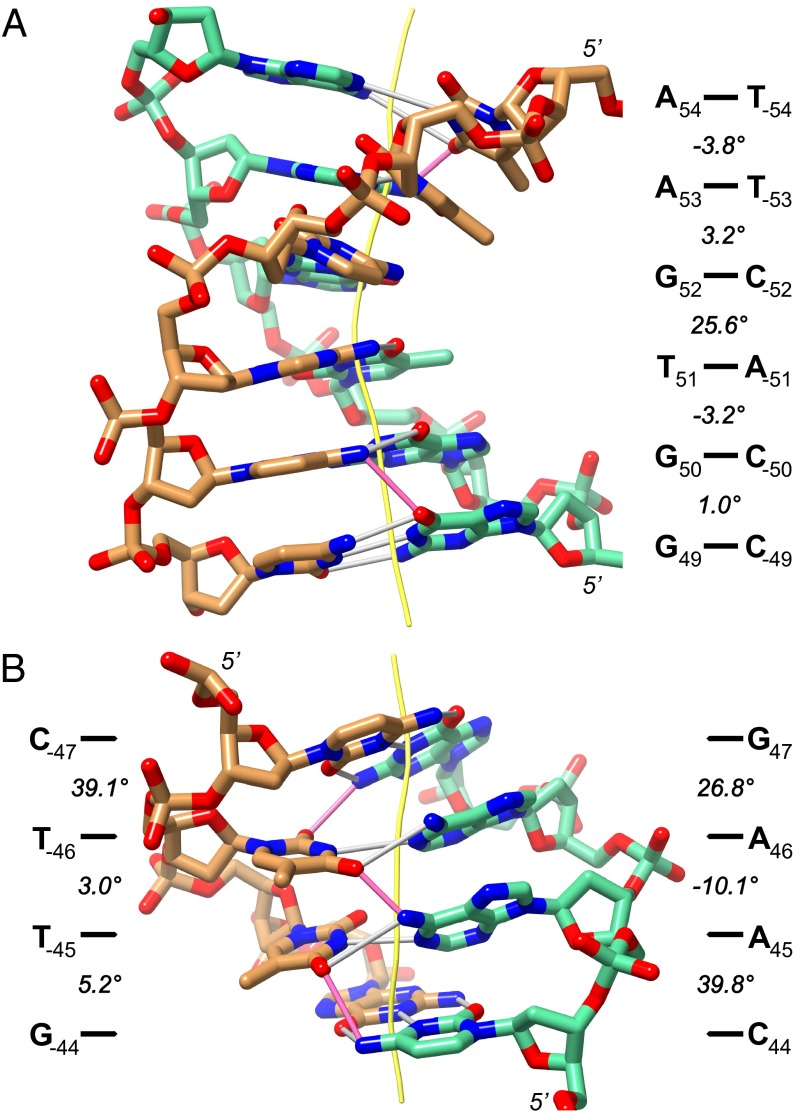
Examples of DNA kinks in MMTVA DNA. (A) SHL −5 is kinked into the major groove at the central CA=TG step. The curvature component forming the DNA superhelix is shown for each base-pair step. (B) SHL −4.5 displays a strand-distributed kink around an AA=TT step. The curvature components shown are calculated for both strands separately. The DNA double-helix axes (yellow) and cross-chain (pink) and Watson–Crick (white) hydrogen bonds are shown.
The sequence CTTG in the SHL −4.5 minor groove block appears to have both its AG=CT and CA=TG steps kinked sufficiently to bend the block into the superhelix (Fig. 2). However, the bending in this block is complex and better described using strand-specific parameters (Fig. 3B). In this case, the CTTG strand is extensively kinked (39.1°) at the CT step, whereas the CAAG strand is kinked at both the CA (39.8°) and AG (26.8°) steps. The AA step between them bends opposite (−10.1°) to the superhelical curvature. Both AT base pairs in this step have large propeller twist with T-45A45 at −43°, the most extreme in either MMTVA or ASP. The conformation of these three steps is stabilized by three cross-chain, bifurcated hydrogen bonds, one for each step (T−46–O2 and G47–N2; T−46-O4 and A45–N6; and T−45–O4 and C44–N4). Cross-chain hydrogen bonds occurring in AA=TT steps have been implicated in chain stiffening and should play a similar role here (16). There are five further locations in MMTVA (−26, −25; −15, −14; −6, −5; +15, +16; +37, +38) and two locations in ASP (−16, −15; +5, +6) where the curvature into the superhelix in one strand of a base-pair step exceeds the opposite strand by more than 34° as for the CA=TG (−45, −44) step. All of these locations are in minor groove blocks. However, except for SHL −4.5 containing the AA=TT step central to this strand-distributed kink, all other kinks are immediately compensated for by similar curvature in the opposite strand in an adjacent base-pair step.
The second primary mode of DNA bending occurs by alternation of the shift base-pair step parameter, described previously for ASP as smooth bending in minor groove blocks (8). Alternation of shift in minor groove blocks facilitates bending by reducing the steric interference between the edges of base pairs. The total number of shift-assisted base-pair steps is 18 both for MMTVA and for ASP (Table 1, Fig. S3, and Methods, Analysis).
The third primary mode of DNA bending is smooth bending in major groove blocks. There are 9 blocks in this category for MMTV and 12 for ASP. Unlike bending in minor groove blocks, bending into the major groove is less sterically encumbered (17). Consequently, the DNA sequence is less constrained, as indicated by rmsd curvatures for major and minor groove base-pair steps of 12.2° and 14.7° for MMTVA, and 10.9° and 12.0° for ASP, respectively. Indeed, consensus sequences extracted from positioned nucleosomes in systematic evolution of ligands by exponential enrichment (SELEX) studies demonstrate minor groove blocks dominate sequence dependency in the nucleosome core (10, 18).
Unlike minor groove blocks, each with an arginine side-chain inserted, bending in major groove blocks must be governed purely by intrinsic sequence-dependent flexibility because the only contacts with histones are with the primary phosphate groups at the ends of the blocks. The only exceptions are at major groove blocks SHL ±1, where the guanidinium group of H3 R40 interacts with the minor groove on the opposite face of the DNA. Coincidentally, the base-pair steps at −9, −8, and +8, +9 in these blocks display an out-of-phase preference for AA/TT/AT/TA steps (1). To explain this preference, the proximity H3 R40 was suggested to favor AT-rich vs. GC-rich steps on electrostatic grounds (19). However, in MMTVA, R40 makes a hydrogen bond with G−8–N3 of step C−9G−8 in SHL −1 (2.8 Å) and with T9–O2 of step T8T9 in SHL +1 (3.1 Å). In ASP, R40 makes a hydrogen bond with A9–N3 of step G8A9 in SHL −1 (3.0 Å) and again symmetrically in SHL +1. These interactions suggest a direct readout of sequence does not determine the sequence preference here. Likewise, steric interference due to a G–N2 group is unlikely because it can be accommodated in any of the four possible base positions without interfering with H3 R40. A larger, sequence-dependent context may play a role at this site.
A histone H4 variant, H18R, was used to improve the diffraction quality of MMTVA crystals. The H18R substitution was made to potentially strengthen crystal interparticle interactions based on the observation that several proteins bind the acidic patch of the nucleosome core analogously to the H4 tail, but have an arginine side-chain at the position homologous to H18 (20–22). In the crystal packing of ASP, the DNA at SHL +2 is bound by an H4 tail (16–26), whereas at the twofold symmetric SHL −2 it is not (6, 23). ASP H4 R17H18R19, localized by the interactions of the adjacent K20, V21, and R23 side chains with the H2A–H2B acidic patch of the neighboring NCP, binds the backbone of the SHL +2 TC step. This interaction apparently results in the displacement of the kink occurring at AA=TT in SHL −2 to the adjacent CA=TG step in SHL +2. For MMTVA, the H4 sequence 17–20 has reoriented due to H18R binding the acidic patch, extending the interaction with the neighboring NCP. The orientation of H4 variant R18 is similar to wild-type H4 H18 bound to a negatively charged patch of the BAH domain in the BAH domain/NCP structures (20, 24). As a consequence, the R17 and K20 side chains are in close proximity to the opposite side of the phosphate group at SHL +2 position −21 (Fig. 4). The DNA backbone here is displaced by 2.4 Å relative to the histone octamer compared with its position at MMTVA SHL −2 (or to ASP SHL ±2). In the context of the H4 H18R variant, potential steric interference of the R17 and K20 side chains with the DNA backbone may result in discrimination between the SHL +2 and SHL −2 sequences. Notably, SHL +1 through SHL +2.5 inclusive was the region used to determine the orientation of the MMTVA DNA initially because it had the lowest temperature factors (Figs. S2 and S5). Therefore, it is unlikely that this region is more flexible than the twofold-related region, and that differential flexibility accounts for the selective binding. Other aspects of the crystal packing may, however, contribute to the asymmetric arrangement. For example, the other histone–tail pairs also make asymmetric interparticle DNA interactions. Moreover, the DNA superhelices themselves make contacts between particles and may be an influence.
Fig. 4.
Interaction of the DNA backbone with the H4 N-terminal tail (green) variant H18R. The DNA backbone at SHL +2 (blue with P−21 B-factor of 72) is centered between side chains R17 and K20 with H18R anchoring the H4 tail to the acidic patch of the neighboring NCP in the crystal (solvent-excluded surface with coulombic coloring: red, negative; blue, positive). The backbone at SHL −2 (red with P−21 B-factor of 184) is superimposed based on twofold rotation around the NCP pseudo twofold axis.
Fig. S5.
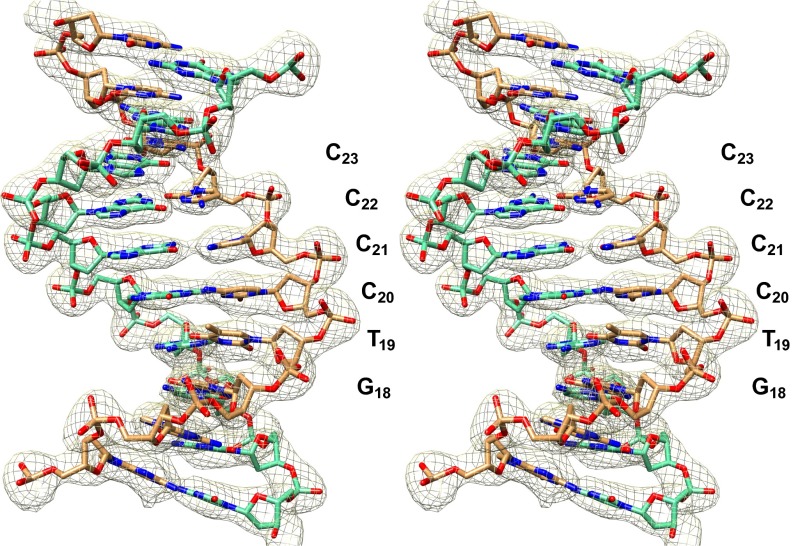
Electron density and model for the MMTVA SHL +2 DNA (stereo). The electron density map is contoured at 1.4× its overall SD.
Discussion
This study furthers our knowledge of DNA conformation for natural DNA sequences bound in the nucleosome core by over twofold. MMTVA is the first NCP structure containing a RNA Pol II transcription start-site and represents the +1 nucleosome in the MMTV 3′-LTR. Although the overall path of the DNA double helix in the NCP superhelix is highly similar for MMTVA and ASP with the overall backbone rmsd less than 2 Å, the local deviation can be much larger, exceeding 5 Å (Fig. 1 A and B). The combined deviations for the DNA backbone over all histone folds show that the DNA-binding elements L1, A1, and L2 suppress sequence-dependent conformation locally on the bound strand (Fig. 1C). The intervening positions, however, have substantial sequence-dependent variation, at the level observed for nuclear factors recognizing DNA sequence specifically.
The previous analysis of ASP at 1.9 Å revealed that NCP DNA adopts its superhelical form by kinking and shift-assisted bending in minor groove blocks and by smooth bending in major groove blocks. The current analysis of MMTVA and reanalysis of ASP newly reveals kinking in major groove blocks occurring at MMTVA SHL −5, −4, −3, +4, and +6, and ASP SHL −5 and ±2. Both major and minor groove kinks occur predominately at CA=TG steps (Table 1). However, kinked base-pair steps also occur in the major groove at CC=GG, TA, and AA=TT and in the minor groove at AC=GT, AG=CT, GA=TC, and CC=GG. The latter three of these were also seen to be kinked when coupled with DNA stretching in a 145-bp version of the ASP structure and an NCP containing a variant of the 601 DNA sequence (9, 15). The two most flexible base-pair steps are CA=TG and TA, and the preference for CA=TG vs. TA kinking in the minor groove has been discussed (8). It is notable that CA=TG steps are also important for kinking in major groove blocks, emphasizing their biflexibility. At MMTVA SHL −3.5, a kinked GT=AC step is uniquely combined with shift-assisted bending. Notably for shift-assisted bending, the base pair shared by shifted base-pair steps is always a GC base pair (Fig. S3), presumably due to the requirement to relieve steric interference from the extracyclic guanine N2 atom projecting into the minor groove. Conversely, neither MMTVA nor ASP have shifted-assisted bending involving AA=TT, AT, or TA steps.
The AA=TT step has been frequently implicated in nucleosome positioning by virtue of its lack of bending and stiffness (25, 26). Indeed, it has been found as a positioning signal in the middle of minor groove blocks by in situ nucleosome mapping (1). The strand-dislocated kink at MMTVA SHL −4.5 displays a conformation that can explain this positional propensity. The large propeller twists of the AA=TT step base pairs at the center of minor groove blocks evidently favors kinking in the adjacent base-pair steps. Although the SHL −4.5 strand-distributed kink is unique, the ASP SHL ±3.5 and SHL ±3.5 minor groove blocks also contain the sequence TTG in which the TG step is kinked.
DNA stretching occurs variably at SHL ±2 and SHL ±5 for MMTVA, ASP, NCP146b, and NCP containing 601-based sequences. The importance of DNA stretching for formation of a compact higher-order structure of nucleosomes and its likely ubiquitous presence in nucleosomes containing genomic DNA, based on sequence periodicity considerations, has been discussed previously (8, 27).
Sequence-dependent recognition of DNA wrapped on the nucleosome core may play a functional role for pioneer transcription factors such as FoxA, Oct4, Sox2, and Klf4 (28, 29). Unfortunately, there are no structures of pioneer factors bound to a nucleosome, and the existing structures of nucleosome core-binding proteins bound to a NCP are nonspecific complexes that make little contact with NCP DNA (20, 22, 30). The recent structure of the prototype foamy virus integrase bound to NCP shows that this enzyme makes substantial contact with SHL ±3.5, but the low resolution does not permit a deeper analysis (31). The integrase binds preferentially to SHL +3.5, and though the dyad position (origin) of the D02 DNA fragment most thoroughly studied is not known to base-pair accuracy, AG′GTAG,TT [prime sign (′) and comma (,) denote integration cleavage sites] is the sequence mapped at this location. The SHL +3.5 major groove block (minor groove facing outward) contains AGGT, and this sequence is coincidentally found at MMTVA SHL +4.5. In MMTVA, the GG step is kinked and the most highly overtwisted (51°) base-pair step in the structure. Although the H2A N-terminal tail was found to be crucial for complex formation and is most probably the NCP feature most selective for integration at SHL ±3.5, sequence-dependent DNA structure as seen in MMTVA may also be important.
The transcription factor NF-Y is a histone-fold dimer that mimics the H2A–H2B dimer structurally and in DNA binding (32). Superposition of NF-Y bound to a 25-bp cognate DNA with MMTVA H2A–H2B bound to SHL −5.5, −3.5 inclusive shows that the CA=TG step in the recognition-site sequence CAATT and SHL −5 align and that both of these base-pair steps are kinked. The NF-YA subunit, which is additional to the NF-Y histone-fold dimer, contributes to the specificity of binding by inserting a phenylalanine side-chain into a kinked CA=TG step from an α-helix bound in the DNA minor groove. Analogously, kinks in major groove blocks in nucleosome core DNA could contribute to sequence-specific minor groove recognition by transcription factors.
Methods
Crystal Preparation.
DNA for MMTVA was chemically synthesized (Life Technologies) and cloned to make milligram quantities of the 147-bp sequence (6). MMTVA was prepared from recombinant Xenopus laevis histones and DNA fragments (6). MMTVA samples [10 mM Tris⋅Cl (pH 7.5), 0.1 mM EDTA, 10 mM KCl] were concentrated to 4–6 mg/mL using Vivaspin500 spin concentrators (Sartorius) and filtered through 0.1-µM spin filters (Millipore). Crystals were grown by vapor diffusion using sitting drops in 24-well Cryschem plates (Hampton). A drop was 1:1 sample and 10 mM K-cacodylate (pH 6.0), 180 mM MgCl2, 50 mM KCl and equilibrated against a 1:4 dilution of the same solution. Plates were sealed and placed in 22 °C incubators (Rumed). Crystals were transferred from growth plates into a 50-µL drop of 2% (vol/vol) 2-methyl-2,4-pentanediol (MPD), 10 mM K-cacodylate (pH 6.0), 40 mM MgCl2, 12.5 mM KCl and allowed to equilibrate for 10 min. Crystals were prepared for cryocooling by stepwise addition (five steps of 2–3 min) of 40% (vol/vol) MPD to a final concentration of 26% (vol/vol) MPD. Trehalose was included in the final step at 2% (vol/vol) (12-h minimum, 22 °C). Crystals were fished onto nylon loops and flash cooled in liquid propane maintained at −120 °C before transferring to liquid nitrogen.
Structure Determination.
Diffraction datasets were collected from two crystals (300 0.4-s frames of 0.1° at 10 positions each, 100 K, wavelength 0.99988 Å) at the Swiss Light Source (X06SA) using the Pilatus 6M detector. Data were indexed using XDS and CCP4 REINDEX, and scaled with SCALA (33–35). Molecular replacement was performed with PHASER using ASP (PDB ID code 1kx5) with the histone N-terminal tails removed (7, 36). Refinement and model-building were performed using CNS and COOT, respectively (37, 38). The best PHASER solution was rigid-body refined, and then successive iterations of simulated annealing (2,500 K initial, −25 K steps), manual model building (sigmaA-weighted, solvent-flattened 2Fo–Fc and Fo–Fc maps), B-factor refinement, and energy minimization (maximum-likelihood targets on amplitudes) were carried out (39). Restraints were applied to the DNA to preserve hydrogen bonding within base pairs (NOE), to maintain purine and pyrimidine base planes (planarity) and to fix deoxyribose rings in the C2′-endo conformation (dihedral). Composite omit maps were calculated by systematic exclusion of 5% of the map sections coupled with simulated annealing of the model (300 K initial, −50 K steps) (40). These maps showed that the DNA is B-form and its sequence is distinct throughout. Notably, crystals incorporating wild-type H4 or divalent cations other than Mg2+ (e.g., Mn2+) yielded, at best, 3.0-Å resolution and were averaged by molecular twofold symmetry.
Water molecules were assigned exhaustively in rounds of selection and refinement of unassigned positive peaks in Fo–Fc difference maps using CNS. A peak was selected for refinement if there was a neighboring atom within 4.0 Å (center to center), and the distance between it and other atoms was greater than 2.6 Å (2.0 Å for O or N). After each round, assignments with occupancy less than 0.7 (final 0.65), B-factor greater than 150, or residue R-factor larger than 0.45 (final 0.50) were deleted. A water molecule was reassigned as a candidate Mg2+ ion if it had a distance to an O atom less than 2.4 Å or more than four O atoms were within 3.4 Å (K+ assignments were not made). Categorization based on occupancy, B-factor, residue R-factor, and ligand geometry yielded no significant effect on Rfree. The final refinement included 936 water molecules, 1 Mg2+ ion, and 4 Cl− ions.
Analysis.
DNA parameters were calculated with CURVES5.3, and DNA curvatures were calculated as previously described (8, 41). For the range of roll and tilt angles found in NCP, the curvature can be expressed as the root of the sum of the squares of these two base-pair step parameters. Kinked base-pair steps were identified previously using the component of DNA (magnitude ≥ 18.0°) directed into superhelix formation (8). A kink indicated that the DNA curvature required to maintain a superhelical path at a minor groove block occurred predominately in a single base-pair step. In this study, a total curvature ≥ 18.0° is used to define kinked base-pair steps (Fig. S6A). Using this simple definition, several kinks occur in major groove blocks and two MMTVA blocks contain two kinks. Shift-assisted bending occurs in minor groove blocks and requires that a base-pair step with a positive shift value be followed by one with a negative value. This criterion indicates that a base pair is shifted toward the major groove relative to its two adjacent neighbors. The threshold for the shift magnitude used is 0.9 Å (Fig. S6B). Molecular alignments and figures were made with CHIMERA (42).
Fig. S6.
Definitions for kinked and shift-assisted bending. (A) Kinked bending. The number of MMTVA and ASP base-pair steps binned in 1° increments of curvature (e.g., bin 18: 18.0° ≤ curvature < 19.0°) is plotted. Kinked base-pair steps are defined as those with curvature ≥ 18.0° (green) based on the decrease in number from bin 17–18. This choice corresponds to 8.6% of all base-pair steps being kinked. (B) Shift-assisted bending. The number of MMTVA and ASP base-pair steps contributing to pairs of consecutive base-pair steps having the first step above a slide magnitude threshold and the second step below the negated threshold is plotted vs. 0.1-Å increments of slide magnitude threshold. The threshold at 0.9 Å (pink) was selected to define shift-assisted bending based on the relatively constant number of base-pair steps for values from 0.7 to 0.9 Å. This choice corresponds to 30.4% of the base-pair steps in minor groove blocks aiding shift-assisted bending.
Acknowledgments
This work was support by Swiss National Fund Grant 310030B_138742 and European Research Council Grant FP/2007-2013/Agreement 322778.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5F99).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524607113/-/DCSupplemental.
References
- 1.Brogaard K, Xi L, Wang J-P, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486(7404):496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet. 2009;10(3):161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25(8):335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20(3):267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332(6032):977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 8.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 9.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J Mol Biol. 2010;403(1):1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 10.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 11.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361(4):617–624. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 12.Wu B, Mohideen K, Vasudevan D, Davey CA. Structural insight into the sequence dependence of nucleosome positioning. Structure. 2010;18(4):528–536. doi: 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 13.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115(6):2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua EYD, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012;40(13):6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol. 2007;368(4):1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 16.el Hassan MA, Calladine CR. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J Mol Biol. 1996;259(1):95–103. doi: 10.1006/jmbi.1996.0304. [DOI] [PubMed] [Google Scholar]
- 17.Calladine CR. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- 18.Thåström A, et al. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288(2):213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 19.Davey CA. Does the nucleosome break its own rules? Curr Opin Struct Biol. 2013;23(2):311–313. doi: 10.1016/j.sbi.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Armache K-J, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334(6058):977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbera AJ, et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311(5762):856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 22.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467(7315):562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306(5701):1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, et al. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc Natl Acad Sci USA. 2013;110(21):8495–8500. doi: 10.1073/pnas.1300126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: Structure-based computational analysis of nucleosome-binding sequences. Biophys J. 2009;96(6):2245–2260. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morozov AV, et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37(14):4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436(7047):138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 28.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 29.Soufi A, et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161(3):555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514(7524):591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maskell DP, et al. Structural basis for retroviral integration into nucleosomes. Nature. 2015;523(7560):366–369. doi: 10.1038/nature14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nardini M, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152(1-2):132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, et al. Iterative-build OMIT maps: Map improvement by iterative model building and refinement without model bias. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 5):515–524. doi: 10.1107/S0907444908004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: Conventions and principles. J Biomol Struct Dyn. 1989;6(4):655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]