We would like to offer two critical comments on the PNAS paper by Doose et al. entitled “MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells” (1).
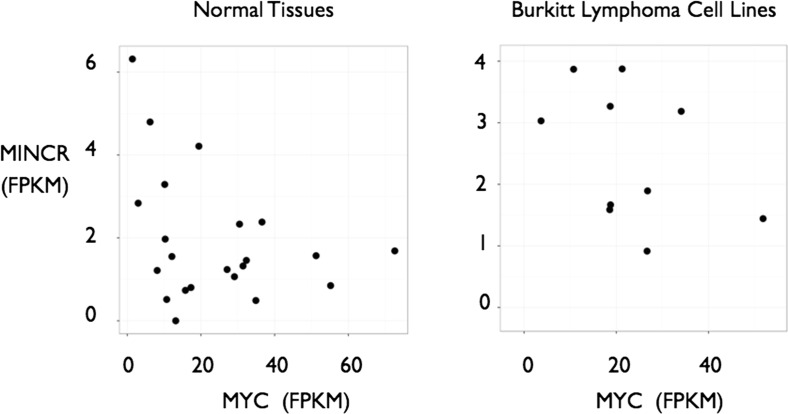
Doose et al.’s (1) paper claims that MINCR is a MYC-induced long noncoding (lnc) RNA. The authors base this claim upon three pieces of evidence, correlation of expression in P493-6 cells, in hT-RPE-MycER cells, and in Burkitt lymphoma cells versus germinal center B-cell lymphoma cells. However, an examination of publicly available data (some of which were also used by the authors) shows that this correlation between MINCR and MYC is not universal, nor is it reproducible using independent Burkitt lymphoma data. An analysis of the normal tissue data (2) from which figure S3C in ref. 1 is derived reveals no correlation between MYC and MINCR expression in the various tissue types, as demonstrated in Fig. 1, Left. Additionally, independent RNAseq data on Burkitt lymphoma cell lines obtained from the Cancer Cell Line Encyclopedia also fail to show such a correlation (3). Here, both MYC and MINCR levels vary substantially, but do not correlate with each other, as shown in Fig. 1, Right. Although such a correlation is seen in the particular dataset presented by Doose et al. (1), this dataset appears not to be broadly representative of the transcriptional regulation of MINCR. Because the expression of MYC and MINCR do not generally correlate, nor do they correlate in Burkitt lymphoma, we consider the term “MINCR” as “MYC-induced noncoding RNA” a misnomer.
Fig. 1.
Expression levels of MINCR plotted against levels of MYC taken from normal tissues and cell lines (Left, data from ref. 2) and Burkitt lymphoma cell lines (Right, data from ref. 3) show a lack of significant positive correlation between MYC and MINCR. FPKM, fragments per kilobase of transcript per million mapped reads.
Doose et al. (1) claim that they are the first to analyze the MYC-regulated noncoding transcriptome with RNAseq. As a matter of fact, at least three publications that use RNAseq for the analysis of MYC-regulated lncRNAs preceded their publication (4–6), in addition to numerous studies using microarray and quantitative PCR. This would be a minor breach of scientific etiquette, except for the fact that the paper by Doose et al. (1) relies extensively on the lncRNA expression data from two of these previously published studies (5, 6). In the first section of the Results and in figure 1 of ref. 1, Doose et al. use RNAseq data from the lymphoblastic cell line P493-6 as one of the selection criteria for MYC-regulated lncRNAs, and cite two early papers that merely document the origin of P493-6 cells, but do not deal with lncRNAs. The ordinary reader would then assume that the P496-3 lncRNA data are generated by Doose et al. Only in the SI Materials and Methods of ref. 1 is it revealed that the data are taken from a published study, but no appropriate attribution is given. We consider the listing of Gene Expression Omnibus identifiers here as not adequate. It is not in accordance with currently accepted norms of data reuse and data reanalysis and is particularly problematic in conjunction with claims of priority.
Footnotes
The authors declare no conflict of interest.
References
- 1.Doose G, et al. MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 2015;112(38):E5261–E5270. doi: 10.1073/pnas.1505753112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng GX, Do BT, Webster DE, Khavari PA, Chang HY. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol. 2014;21(7):585–590. doi: 10.1038/nsmb.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart JR, et al. Inhibitor of MYC identified in a Kröhnke pyridine library. Proc Natl Acad Sci USA. 2014;111(34):12556–12561. doi: 10.1073/pnas.1319488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart JR, Roberts TC, Weinberg MS, Morris KV, Vogt PK. MYC regulates the non-coding transcriptome. Oncotarget. 2014;5(24):12543–12554. doi: 10.18632/oncotarget.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]