Significance
Acyl-CoA thioesters are key substrates for energy conversion. Related ATP/GTP-producing synthetases form a large superfamily with members in all kingdoms of life. In contrast to their general importance, the underlying reaction mechanism of these enzymes is still not understood in all steps. Here, we describe various structures of a nucleoside diphosphate-forming acetyl--CoA synthetase from an evolutionary very old archaeon. A large conformational rearrangement within the enzyme is observed. The structures reveal a partial unwinding and reorientation by 120° of a phosphohistidine-containing segment. This conformational rearrangement couples the acyl-CoA binding site with the nucleoside diphosphate binding site. The presented structures prove a long-standing hypothesis and provide insight into the determinants for substrate selectivity.
Keywords: X-ray structure, metabolic energy conversion, protein dynamics, acyl-coenzyme A thioester, superfamily
Abstract
The NDP-forming acyl-CoA synthetases (ACDs) catalyze the conversion of various CoA thioesters to the corresponding acids, conserving their chemical energy in form of ATP. The ACDs are the major energy-conserving enzymes in sugar and peptide fermentation of hyperthermophilic archaea. They are considered to be primordial enzymes of ATP synthesis in the early evolution of life. We present the first crystal structures, to our knowledge, of an ACD from the hyperthermophilic archaeon Candidatus Korachaeum cryptofilum. These structures reveal a unique arrangement of the ACD subunits alpha and beta within an α2β2-heterotetrameric complex. This arrangement significantly differs from other members of the superfamily. To transmit an activated phosphoryl moiety from the Ac-CoA binding site (within the alpha subunit) to the NDP-binding site (within the beta subunit), a distance of 51 Å has to be bridged. This transmission requires a larger rearrangement within the protein complex involving a 21-aa-long phosphohistidine-containing segment of the alpha subunit. Spatial restraints of the interaction of this segment with the beta subunit explain the necessity for a second highly conserved His residue within the beta subunit. The data support the proposed four-step reaction mechanism of ACDs, coupling acyl-CoA thioesters with ATP synthesis. Furthermore, the determined crystal structure of the complex with bound Ac-CoA allows first insight, to our knowledge, into the determinants for acyl-CoA substrate specificity. The composition and size of loops protruding into the binding pocket of acyl-CoA are determined by the individual arrangement of the characteristic subdomains.
NDP-forming acyl-CoA synthetases (ACDs) catalyze the conversion of acyl-CoA thioesters to the corresponding acids and couple this reaction with the synthesis of ATP via the mechanism of substrate-level phosphorylation. ACDs have been studied in detail in hyperthermophilic archaea, where they function as the major energy-conserving enzymes in the course of anaerobic sugar and peptide fermentation (1–4). It is believed that ACDs represent a primordial mechanism of ATP synthesis in the early evolution of life. ACDs were found in all acetate (acid)-forming archaea (5, 6) and in the eukaryotic parasitic protists Entamoeba histolytica (7) and Giardia lamblia (8), but they have not been found in acetate-forming bacteria. In bacteria, with the exception of Chloroflexus (9), the conversion of inorganic phosphate and the thioester acetyl (Ac)-CoA to acetate and ATP is catalyzed by two enzymes, phosphate Ac-transferase and acetate kinase (10). Following the identification of ACD genes (5, 11) a novel protein superfamily of NDP-forming ACDs was proposed by a bioinformatics analysis (8). In addition to ACDs, this superfamily contains the well-characterized succinyl-CoA synthetases (SCSs) and ATP-citrate lyases (ACLYs) (8). Each ACD is composed of at least five subdomains with variable sequential arrangement (8). This phenomenon, termed “domain shuffling,” is one of the key features of this superfamily (8). Superposition of several structures of SCS from Escherichia coli (ecSCS) (12–14), Thermus aquaticus (15), the mammalian GTP-specific SCS from pig (16), and a truncated form of human ACLY (17, 18) revealed that subdomains 1–5 share a common arrangement in these enzymes. From detailed studies of the reaction mechanism of ecSCS, a crucial enzyme tightly connected to the TCA cycle, a three-step mechanism was proposed, which involves the phosphorylation of a highly conserved His residue at the first active site (site I) as an intermediate step (12, 13). Subsequently, the phosphoryl moiety is transferred onto an NDP that is bound at the second active site (site II). The distance of around 36 Å between site I and site II is assumed to be bridged by a so-called “swinging loop” (12, 14, 19). A mechanism comparable to the mechanism of the SCSs was presumed for the ACDs (20). A study based on sequence, biochemical, and mutational analyses identified a highly conserved and functional relevant His residue within the beta subunit of the ACDs (20). Of note, the SCS enzymes do not contain a comparable His residue. Thus, an extension of the mechanism by a fourth step was suggested for the ACD1 from Pyrococcus furiosus (20). Here, the second His residue is thought to serve as an additional intermediate, which gets phosphorylated transiently during the enzymatic reaction and subsequently transmits the phosphoryl moiety onto the bound NDP. The necessity for a four-step mechanism may originate from a shortening of the proposed swinging loop, which facilitates the phosphoryl transfer between the subunits (20). However, due to the completely different arrangement of the subdomains within ACD [alpha(1-2-5)/beta(3-4)] in comparison to ecSCS [alpha(1-2)/beta(3-4-5)], a different 3D arrangement of the subdomains in ACD and, as a consequence, a variation of the proposed reaction steps has to be considered.
ACDs are versatile enzymes that are capable of metabolizing a variety of CoA thioesters generated in the course of sugar and peptide fermentation (3, 4, 21). In contrast, the ecSCS specifically binds succinyl-CoA, accepting only small aliphatic CoA thioesters as additional substrates (22). The ACLY is even more specific for Ac-CoA (23). Because ACDs are involved in sugar and amino acid metabolism, they are ambivalent regarding their converted substrates (3, 4, 24, 25). In addition, domain shuffling creates a very diverse pattern of subdomain organization within members of the ACD family (8). Some ACD enzymes are even built up as single-chain proteins with fused alpha-beta or beta-alpha subunits. The linker region between the fused subunits is usually very short (8). Therefore, a structural model for ACDs based on the structure of ecSCS as presented by Bräsen et al. (20) cannot be fully compatible with those single-chain ACDs, because distances of more than 60 Å between the termini of individual alpha and beta subunits must be bridged.
So far, there are no structural data to explain these described peculiarities (necessity of a second His, different arrangement of the subdomains, and broader substrate selectivity). In this study, we present a comprehensive analysis of various crystal structures of the functional complex of the ACD isoform 1 from the hyperthermophilic archaeon Candidatus Korarchaeum cryptofilum (ckcACD1). Ca. K. cryptofilum belongs to the most ancestral archaea (26). Based on genome analyses, Ca. K. cryptofilum has been proposed to ferment amino acids using ACDs as major energy-conserving enzymes (26). Three ACD isoenzymes with different substrate specificities have been characterized (27). ckcACD1 converts small aliphatic CoA thioesters. It is composed of four protein chains (two alpha subunits and two beta subunits) that form an α2β2-heterotetramer. Each alpha subunit has one active site for acyl-CoA binding, and each beta subunit has one active site for NDP binding. Due to its similar molecular composition and kinetic properties as ACD1 from the hyperthermophilic archaeon P. furiosus (pfACD1), it is likely that ckcACD1 follows the same four-step catalytic reaction mechanism as has been proposed for the homologous pfACD1 (20). In particular, the structural rearrangements accompanying the phosphate shuttle process between alpha and beta subunits of ACD will be described for the first time to our knowledge. This work provides direct evidence for the proposed loop swinging, which, in addition, can explain the necessity for a second active-site His located in the beta subunit near site II. Further detailed analysis of the binding mode of Ac-CoA to ckcACD1 provides insight into substrate specificity of the enzyme.
Results
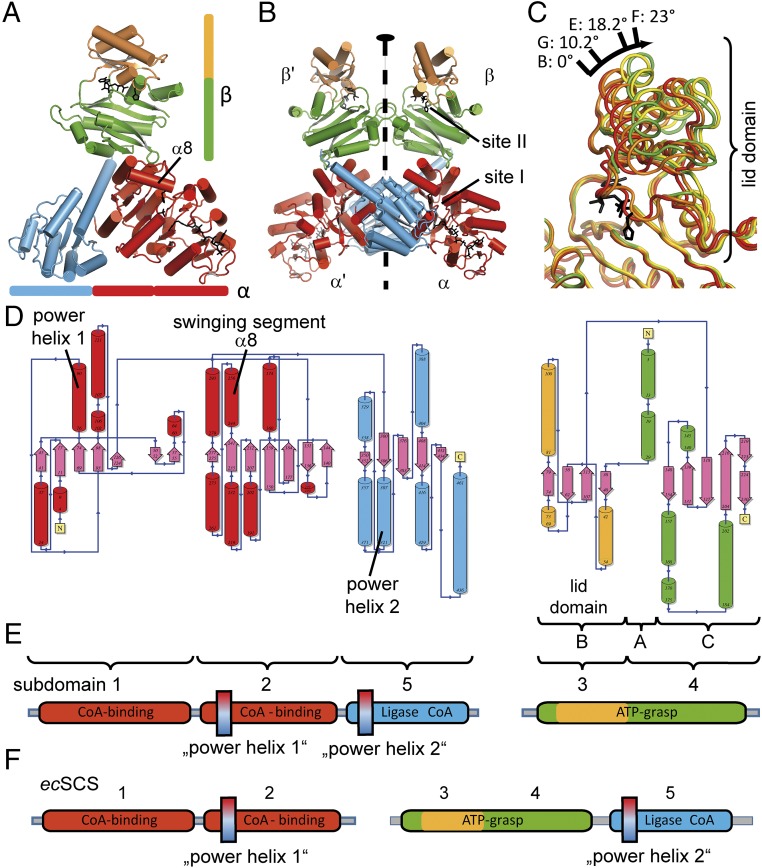
The 3D assembly of the two alpha and two beta subunits of the ckcACD1 complex revealed a unique α2β2-heterotetramer (Fig. 1). The protomer is composed of one alpha subunit and one beta subunit (Fig. 1 A and E and SI Appendix, Fig. S1). Two protomers are arranged with a twofold symmetry (Fig. 1B). The alpha subunit comprises three subdomains (1-2-5), and the beta subunit comprises two subdomains (3-4). The numbering is according to the nomenclature established for ecSCS (12, 19, 20) (Fig. 1F). All subdomains have an α/β secondary structure topology. With an interface of ∼2,000 Å2 between both alpha subunits, 740–770 Å2 between alpha and beta subunits, and 540 Å2 between both beta subunits, the assessment of the ckcACD1 complex was assigned as stable based on the free energy of assembly dissociation (ΔGdiss) as calculated by the Proteins, Interfaces, Structures and Assemblies (PISA) server (28, 29).
Fig. 1.
Overall structure and domain arrangement of ckcACD1. Ribbon representation of the protomer composed of one alpha subunit and one beta subunit (A) and the complete α2β2-heterotetramer (B). The twofold symmetry is indicated with a black oval. (C) Conformational differences of the beta subunit in various crystal structures (ckcACD1-B, ckcACD1-E, ckcACD1-F, and ckcACD1-G) (Table 1). The relative movement of subdomain 3 compared with subdomain 4 is noted in degrees (“opening angle”). Because motion of subdomain 3 is required for catalysis, this region is also called the “lid” (32). (D) Topology diagram generated with the program PDBsum (51). The colors of the helixes are in accordance with the color scheme used for the individual subdomains as outlined in E. (F) Domain arrangement for the heterodimeric ecSCS (12). The positions of the two power helices are indicated.
In the alpha subunit, the N-terminal subdomain (subdomain 1: Met1α–Phe131α) is responsible for CoA binding and contains a Rossmann-like fold (30), which is succeeded by two domains with a flavodoxin-like fold (31). The middle domain (subdomain 2: Gly132α–Asn293α) contains the conserved His (His254α), which is phosphorylated during the first part of the enzymatic reaction cycle. His254α is located within an array rich in small amino acids, providing this region an extraordinary mobility. The C-terminal domain of the alpha subunit (subdomain 5) consists of the amino acids Arg300α–Arg464α. According to the subdomain nomenclature for SCS and ACD enzymes, the beta subunit of ckcACD1 can be described as being built up of two different structural domains, subdomains 3 and 4 (8). Alternatively, the structure of the beta subunit can be referred to as an ATP-grasp fold, consisting of three subdomains A, B, and C (32). In ckcACD1 the subdomain A (Met1β–Pro33β) consists only of two helices and interacts tightly with subdomain C (Gly117β–Arg230β). Considering their similar atomic displacement parameters, both subdomains A and C can be regarded as one structural unit (subdomain 4). The substrate binding site (site II) is usually formed by the assembled subdomains A and C (32). In the context of ACD, this substrate is the phosphate-carrying moiety, as will be described later on. Residues Thr35β–Phe113β form subdomain B (subdomain 3). It harbors the conserved phosphate-binding T-loop (33) (residues Lys60β–Val75β), as well as a highly conserved HK(S/T)(D/E) motif (residues His68β–Asp71β) found in nearly all archaeal and bacterial ACDs (8). Interaction of subdomain 3 and subdomain 4 creates the nucleotide binding site.
ATP-grasp enzymes are known to carry out opening and closure motions to allow binding of the nucleotide (“open”) and to orient residues involved in catalysis properly (“closed”) (34). In the crystal structures of ckcACD1, the beta subunit was found in different states of “opening” (Fig. 1C). The most closed state was observed with bound Ca2+/ado-5′-β,γ-methylene triphosphate (AMPPCP; ckcACD1-B), and the most opened conformation was found in the structure ckcACD1-I with a rotation angle of nearly 25° (Fig. 1C and SI Appendix, Table S2).
ckcACD1 Binds its Acyl-CoA Substrates Comparable to Other Members of the ACD Superfamily: Binding Mode for CoA.
To understand the functionality of the ckcACD1 complex, we prepared crystals of the enzyme in complex with its substrates CoA, Ac-CoA, phosphate, and ADP, as well as the nonhydrolyzable ADP analog ado-5′-α,β-methylene diphosphate (AMPCP) and the nonhydrolyzable ATP analog AMPPCP. In all cases, CoA was bound to subdomain 1 in an elongated form, pointing its sulfur atom toward the site of the His phosphorylation (His254α) (Figs. 2 and 3 and SI Appendix, Fig. S2), in accordance with ecSCS referred to as site I. The Ade moiety is inserted into a hydrophobic cleft, and its position is additionally adjusted by two water molecules interacting with either the N6A or N1A and the carbonyl oxygen atoms of the residues Lys79α or Pro59α, respectively (SI Appendix, Fig. S2E). Upon CoA binding the highly flexible side chain of Lys24α becomes fixed via an interaction of its side-chain amino group with the oxygen atoms of the 3′ and 5′ alpha-phosphate groups (SI Appendix, Fig. S2D). Interestingly, the Sus scrofa SCS (ssSCS) features the Lys residue Lys26α at a structurally equivalent position. The side chain of this residue, however, is not involved in CoA binding (35).
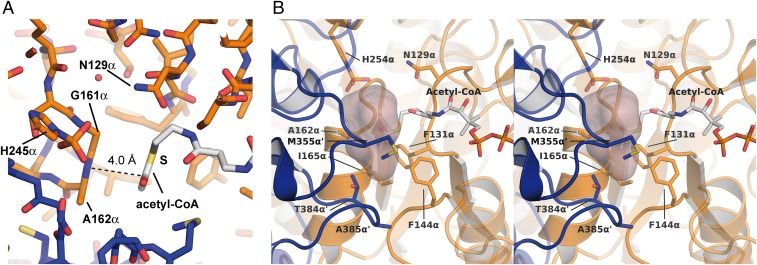
Fig. 2.
Binding site for the activated acyl moiety. (A) Orientation of the Ac group of Ac-CoA. A dashed line indicates its distance of 4.0 Å to the peptide bond between residues Gly161α and Ala162α. (B) Stereo representations of the estimated binding pocket for larger acyl groups. The space available for the acyl moiety was calculated with the program HOLLOW (52) using the crystal structure ckcACD1-E, a sphere radius of 1.4 Å, and CoA derived from Ac-CoA. Amino acid residues shaping the potential binding pocket are represented as sticks and are labeled. Carbon atoms in Ac-CoA are shown in white, nitrogen in blue, oxygen in red, and sulfur in yellow. The carbon atoms of the surrounding protein are colored in orange (alpha subunit) and blue (alpha subunit of the symmetry-related protomer), respectively.
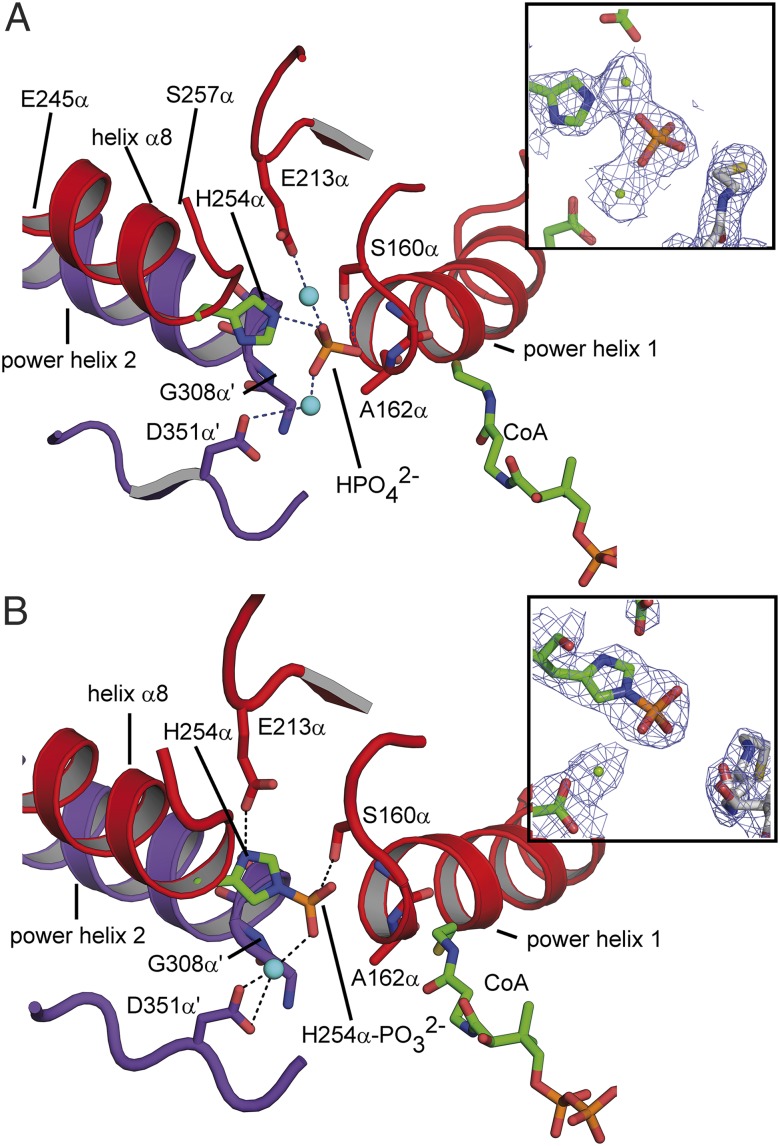
Fig. 3.
Phosphate binding site of ckcACD1. Environment of bound inorganic phosphate (A) and of the phosphohistidine residue His254α-P (B) within site I of the alpha subunit. The two so-called power helices (ckcACD1 162α–173α/308α′–319α′) and helix α8 of the phosphohistidine segment are displayed in ribbon representation, and the phosphate group and His254α, as well as interacting residues, are represented as sticks. Carbon atoms of His254α and the CoA are colored in green, and other carbon atoms of the surrounding protein are colored in red (alpha subunit, subdomain 2) and blue (alpha subunit of the symmetry-related protomer, subdomain 5), respectively. Magnesium ions are shown as cyan spheres. (Insets) Electron density distribution around the phosphate group with a contouring level of 1σ (electron density distribution is shown as blue mesh; carbon atoms of protein are shown in green, carbon atoms of CoA in white, nitrogen atoms in blue, oxygen atoms in red, sulfur atoms in yellow, and phosphor atoms in orange).
Binding Mode of the Ac Moiety Within Ac-CoA.
In one structure of ckcACD1 (ckcACD1-E), cocrystallized with Ac-CoA, the electron density allowed us to model the Ac group connected to CoA (SI Appendix, Fig. S2B). Thus, for the first time to our knowledge, an ACD with its CoA thioester substrate bound can be described. The Ac moiety appears to be nearly coplanar to the peptide bond between Gly161α and Ala162α, with a distance of 4.0 Å between the nitrogen atom of Gly161α and the carboxyl carbon of the Ac group (Fig. 2A). This distance is in the range of typical π-π interactions (36). Based on the chemical environment, the Ac group was arranged such that the methyl group was positioned to be accommodated by a more hydrophobic environment built up from the side chains of the residues Phe131α, Phe144α, Ala162α, Ile165α, Met355α, and Thr384α′ (the prime symbol indicates the “other” subunit) (Fig. 2B and SI Appendix, Fig. S2C). The phosphohistidine pocket at site I also comprises a highly conserved Asn residue (Asn173α). In the case of Ac-CoA, the distance to the thiocarboxyl carbon at 6.6 Å is rather long; however, due to its position, it might be involved in the cleavage of the thioester bond by inorganic phosphate.
Binding Mode of Phosphate Within Site I of the Alpha Subunit.
Crystals of ckcACD1 soaked with 50 mM Na2HPO4 revealed the phosphate ion positioned between both alpha subunits at the amino termini of the so-called “power helices” α5 and α11′ (ckcACD1-G and ckcACD1-I; Fig. 3A). Interestingly, interactions of the phosphate anion with the protein were mainly facilitated by the amino functions of the peptide bonds of four residues: Gly161α and Ala162α from one alpha subunit and Gly308α′ and Gly309α′ from the other alpha subunit. In addition, a strong hydrogen bond was formed to the side-chain hydroxyl group of Ser160α. The distance between the carbonyl carbon of the Ac group of Ac-CoA and the closest oxygen atom of the phosphate is ∼6 Å. The electron density surrounding the phosphate was interpreted as two metal ions. In ecSCS and ssSCS, phosphate ions are located in a very similar position (14, 16). As for the described SCS, the N termini of the two equivalent power helices participate in the compensation of the negative charge of the phosphate ion; however, no additional metal ions have been observed in SCS.
Environment of the Phosphorylated His Residue His254α.
We obtained crystals of the ckcACD1 complex with phosphorylated His254α (ckcACD1-E and ckcACD1-H; Fig. 3B). The conformation of the phosphorylated His was stabilized by ionic interaction with the side-chain carboxyl group of Glu213α. A hydrogen bridge (distance of 2.7 Å) provides an ideal geometry for the phosphorylation reaction by favoring the protonation state of the imidazole group. In addition, the phosphoryl moiety forms a hydrogen bond to the hydroxyl group of Ser160α. In the crystal structure ckcACD1-H, the phosphate moiety also interacts with a bound metal ion, which is coordinated by the side chain of Asp351α′. The interaction with the metal ion results in a further movement of the phosphohistidine and adjoining residues toward site I, and subsequent stabilization of the phosphorylation of His254α by forming several hydrogen bridges to the amino groups of the peptide bonds between the amino acids Ser160α–Gly161α, Gly161α–Ala162α, Gly307α′–Gly308α′, and Gly308α′–Gly309α′. Interestingly, the residues Ser160α and Asp351α′ are highly conserved throughout the ACD superfamily. Only Ser160α is sometimes exchanged to Thr, an amino acid with comparable side-chain characteristics.
Binding Mode of Ado Nucleotides Within Site II Located in the Beta Subunit.
The binding site for the nucleotide substrates of ckcACD1 is within the beta subunit (ATP-grasp domain). In ckcACD1, the Ade moiety is sandwiched in a hydrophobic environment created by residues of subdomain 3 and subdomain 4, with additional hydrogen bridges to the side chains of Gln111β and Lys60β, as well as the protein backbone (SI Appendix, Fig. S3). Furthermore, Lys60β, a residue conserved in nearly all ACDs, interacts with the alpha-phosphate group of ADP. A dual role of this particular Lys residue was also observed for the corresponding Lys46β of ecSCS (14). Furthermore, the phosphate groups of ADP in the crystal structures ckcACD1-B, ckcACD1-D, ckcACD1-G, and ckcACD1-H are in contact with the side-chain amine of Lys69β. Additionally, several interactions facilitate tight binding of the beta-phosphate moiety of the nucleotide to the protein. Hydrogen bridges were observed between the phosphate and the T-loop residues involving the side-chain hydroxyl group of Ser70β and the backbone nitrogens of Lys69 and Ser70β, respectively (SI Appendix, Fig. S3E). A magnesium ion was complexed by both alpha- and beta-phosphates in the structures ckcACD1-D and ckcACD1-G. In the case of the ckcACD1 structure with a closed ATP-grasp conformation (ckcACD1-B), a calcium ion is complexed by oxygen atoms of both alpha-phosphate and beta-phosphate, as well as the side-chain carboxyl group of Asp224β. All mentioned residues belong to the key fingerprint motif of ATP-grasp enzymes (32). The beta-phosphate additionally interacts with the guanidinyl group of Arg226β. However, this interaction is not observed for all beta subunits, which suggests that the side chain of Arg226β is flexible.
Due to the strong interactions of the T-loop with the phosphate groups of ADP, the opening and closing of the lid domain causes a stretching of the bound nucleotide (SI Appendix, Fig. S3D). Therefore, the movement of the lid domain appears to be involved in moving in and pulling out of the nucleotide substrate from the ATP-grasp domain. During this process, the magnesium ion remains complexed to the phosphate groups and is withdrawn from an interaction with the side-chain carboxyl group of Asp224β.
We also obtained one crystal structure with the nonhydrolyzable ATP analog AMPPCP (ckcACD1-C). In contrast to ADP, the Ade and sugar moiety of AMPPCP were shifted by 1–2 Å out of the nucleotide binding pocket. The beta-phosphate is rotated toward the surrounding solvent, with a concomitant change of the dihedral angle C5′-O5′-PA-O3A of ∼130° (SI Appendix, Fig. S3 C and E). Interestingly, the angle between the three phosphor atoms constitutes nearly 90°. A magnesium ion is complexed by oxygen atoms of all three phosphate groups, as well as the carboxyl group of Asp224β. There is also an interaction of the gamma-phosphate with the guanidinyl group of Arg226β. At the corresponding sequence position, other members of the ACD family feature either Arg or Lys residues. In the case of GTP bound to ssSCS, the amino function of the side chain of Lys222β interacts with the gamma-phosphate in a similar manner (16), which might indicate a specific role for this residue.
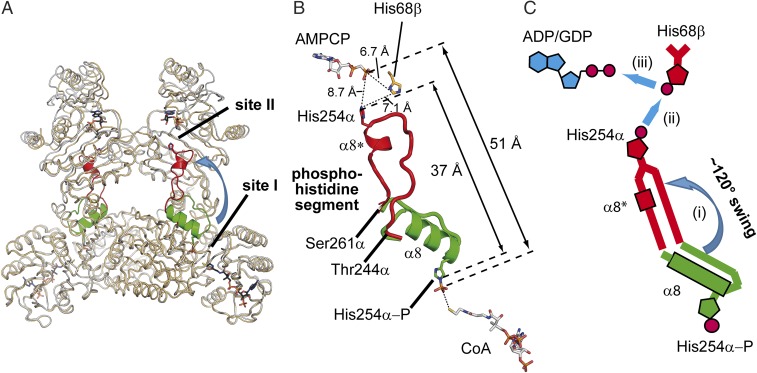
ckcACD1 Bridges a 51-Å Gap During its Enzymatic Cycle.
The beta subunit harbors two binding sites, one for the nucleotide and one for the activated phosphate group. Whereas the catalytic site within the alpha subunit is referred to as site I (binding of CoA and inorganic phosphate), the catalytic site within the beta subunit (binding pocket for NDP and activated phosphate) is referred to as site II (13). The distance between these two sites is very large. In ckcACD1, the space between the phosphor atom of the phosphate ion (bound to His254α) and the beta-phosphate of AMPCP (bound within the beta subunit) constitutes around 51 Å. Therefore, the transfer of the phosphoryl moiety between the alpha and beta subunits requires large rearrangements. For ecSCS, a swinging loop mechanism for the phosphohistidine-containing loop has been proposed (14). Here, we present, for the first time to our knowledge, detailed structural data of the “phosphohistidine segment” pointing toward the nucleotide-binding site of the beta subunit (crystal structure ckcACD1-C) (Fig. 4). This segment consists of the amino acids Gly242α to Val262α of the alpha subunit, and also contains the His residue His254α, which is phosphorylated during the enzymatic cycle (SI Appendix, Fig. S1). These residues undergo a major rearrangement in space, as well as in secondary structure, during movement from site I to site II (Fig. 4). Hence, we henceforth use the term phosphohistidine segment instead of the usual expression, swinging loop. This segment in its site I orientation is composed of an α-helix from residue Glu245α to Thr255α (helix α8), directing His254α toward the CoA binding site (Fig. 3). The amino acid residues Gly256α to Gly260α act as a flexible loop connecting the helix α8 with the subsequent helix α9. By passaging the (phosphorylated) His254α to the beta subunit of ckcACD1, the phosphohistidine segment loses most of its helical structure. Accompanying the partial unwinding of helix α8, the residues Thr244α to Thr255α are positioned in a nearly straight array bridging a distance of more than 10 Å. The upstream residues are assembled into a new short helix α8* (the asterisk indicates the rearrangement of helix α8) formed by the amino acids His254α–Ile258α. After the rearrangement of this segment, the τ-nitrogen of His254α travels around 37 Å between sites I and II (Fig. 4B). Interaction of the phosphohistidine segment with the beta domain also causes substantial changes in nearby residues. The formation of a hydrophobic pocket comprising residues Ile258α, Ala259α, Ile121β, Met135β, Val143β, Val149β, and Phe151β results in partial unwinding and reorientation of helix α5 (residues Gly140β–Phe146β), which, in turn, alters the arrangement of the hydrophobic interface between both beta domains (SI Appendix, Fig. S4A). As a further consequence, the residues between α6 and α7 are shifted in their position. The largest movement observed is 5.9 Å (maximal for Gly179α′) (SI Appendix, Fig. S4B).
Fig. 4.
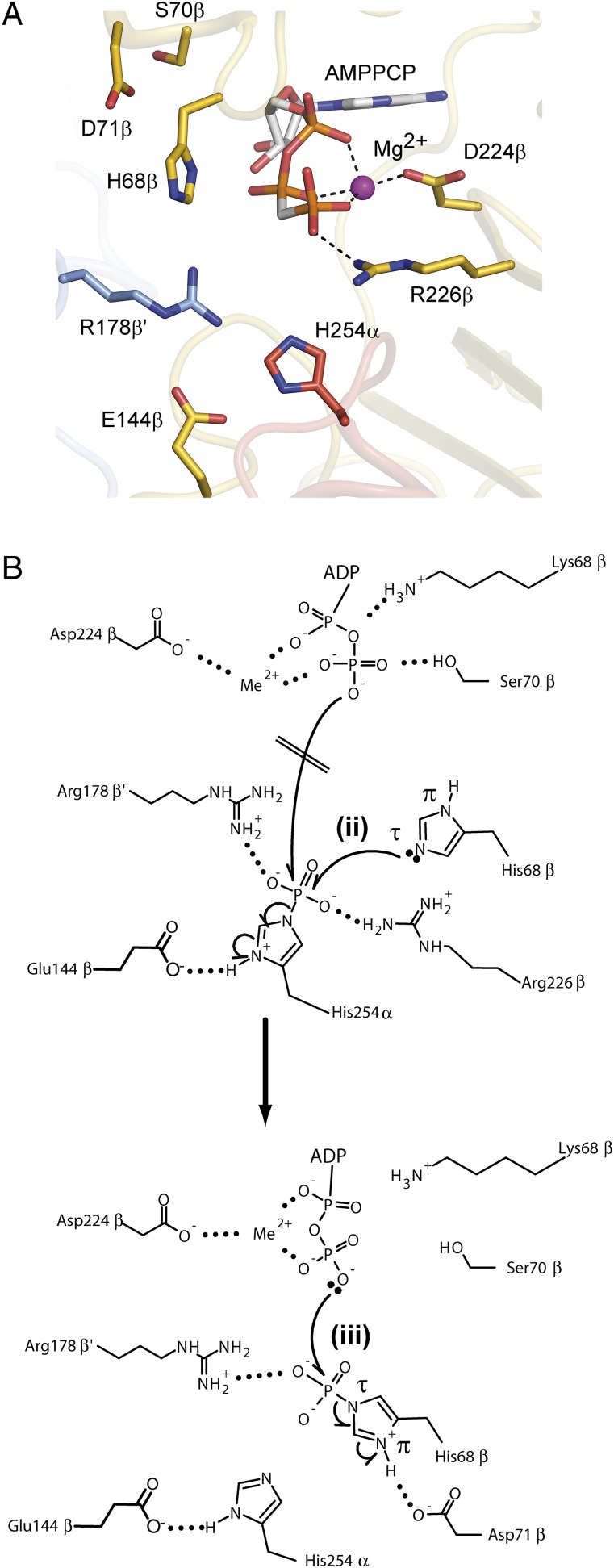
Rearrangement of the His254α-P containing segment upon phosphorylation. (A) Composition representation of two ckcACD1 structures to visualize the movement (blue arrow) of the phosphohistidine segment from site I to site II. Superimposed are the crystal structures ckcACD1-H (gray ribbons) and ckcACD1-C (orange ribbons). The phosphohistidine segment is colored in green if oriented toward site I (structure ckcACD1-H) and in red if oriented toward site II (ckcACD1-C). (B) Distances between site I and site II to highlight the gap that the activated phosphoryl moiety has to travel when attached to His254α of the phosphohistidine segment. Carbon atoms of CoA and AMPCP are colored in white, nitrogen atoms in blue, oxygen atoms in red, phosphor atoms in orange, and sulfur atoms in yellow. (C) Schematic representation of the reorientation of the phosphohistidine segment. After phosphorylation of His254α the phosphoryl moiety is transferred to the nucleotide binding site located in the beta subunit. For this step, reorientation and reorganization of the phosphohistine segment are needed (i) for transferring the phosphoryl moiety to His68β-forming phospho-His68β (ii), which, in turn, phosphorylates bound ADP/GDP (iii).
Discussion
Substrate Specificity.
The ecSCS has a high specificity for succinyl-CoA (22) but converts small aliphatic CoA esters (22) like ckcACD1 and other ACDs (2, 3, 5–7, 20, 21, 24, 37–39) as well. Interestingly, members of the ACD family are also able to transform aromatic CoA thioesters (3, 24, 39), although catalytic activity toward succinyl-CoA has only been described a few times (3, 25, 40). The high specificity of SCS enzymes for succinyl-CoA does make a lot of sense because this enzyme family is a central and, as such, an optimized element within the TCA cycle. In contrast, ckcACD1 and other members of this family are not integrated into a well-tuned metabolic pathway but, instead, have to conserve energy [e.g., during glycolysis or the catabolic degradation of amino acids (3, 7, 20, 24)]. In this respect, a broader substrate spectrum of ACDs appears plausible.
ckcACD1 and ecSCS Display Very Similar Features for the Catalytic Active Site I.
In the crystal structure of ckcACD1-E, Ac-CoA was bound to the enzyme. However, the resolution of this crystal was not good enough to allow the differentiation between its methyl and carbonyl moieties unambiguously. Therefore, we compared the binding pocket for the Ac moiety within ckcADC1 with the putative succinyl binding pocket of ecSCS to understand how the aliphatic moiety of the acid component within acyl-CoA should be positioned in ACDs. In both, ckcACD1 and SCS, the available space is large enough to harbor even bulkier substrates. The capacious volume is somewhat surprising, given the limited spectrum of transformed substances. In the case of ckcACD1, there is a small hydrophobic pocket next to the sulfhydryl group of CoA, which can be used to explain the substrate specificity of ckcACD1 (Fig. 2B). The size of this pocket is restricted by the side chains of the amino acids Ala162α, Ile165α, Phe144α, Met355α′, Thr384α′, and Ala385α′. Due to this limited size, this pocket permits only short-chained acyl-CoA thioesters to fit. The same region in ecSCS features a larger pocket surrounded by polar amino acids, which could accept the carboxy group of succinate of succinyl-CoA (SI Appendix, Fig. S5). The enzyme ACLY converts Ac-CoA (23). A comparable mode of interaction of the Ac group of Ac-CoA interacting with ACLY or ckcACD1, respectively, might be assumed. Hence, we superimposed ckcACD1 onto the crystal structure of human ACLY with bound citrate (17). The resulting distance between the carbonyl carbon of Ac-CoA of ckcACD1 and the carbon C1 of citrate in ACLY is ∼2.8 Å. Thus, we expect the binding site for the aliphatic moiety of acyl-CoA for ACDs in a similar location as observed in ACLY. Based on this comparison, the orientation of the Ac group of Ac-CoA is as shown in Fig. 2A. The thioate function of succinyl-CoA bound to ecSCS might, by analogy with ckcACD1, be situated coplanar to the peptide bond between the corresponding amino acids Cys123α and Pro124α. Interestingly, these residues were significantly distorted in the crystal structure of an ecSCS mutant with absent activity (13). The Cys residue itself, however, was shown not to be part of the catalytic mechanism (41). Furthermore, in the same study, it was shown that variations of the size of the side chain at the position of Cys123α deteriorate the enzyme activity. It appears that slight differences in the orientation of the Cys-Pro peptide bond may result in misalignment of the thioester moiety, and therefore a decrease in the enzymatic function.
Site I, Acyl-CoA Binding Site.
The CoA binding site I is situated at a highly conserved interface formed by residues of subdomains 1, 2, and 5. These subdomains are packed in a similar way in SCS, ACLY, and ckcACD1. A sequence alignment of several ACD proteins revealed that the second ligase-CoA domain (subdomain 5) has only moderate conservation in its primary structure (8), which is in strong contrast to the situation observed for SCS enzymes. Most interestingly, the sequence alignment revealed that ckcACD1 and pfACD1 not only share a similar substrate spectrum but also feature identical amino acids lining the potential substrate pocket (50% overall sequence identity). Those residues were dissimilar in other P. furiosus isoenzymes (35–42% overall sequence identity) with a different substrate spectrum (SI Appendix, Fig. S6). Hence, we propose that alterations of this cleft are responsible for different substrate specificity.
Site II, Purine Nucleotide Specificity.
ACD and SCS enzymes are able to bind and convert all purine nucleotides at site II, but some enzymes exhibit specificity for either the Ado or Guo nucleotide (2, 21, 24, 38, 40). Kinetic characterization of ckcACD1 revealed that both nucleotides are accepted as substrates (SI Appendix, Fig. S7 A and B). In this work, we crystallized the ckcACD1 complex solely with the Ado nucleotide ADP and its nonhydrolyzable analog AMPCP, as well as with the nonhydrolyzable ATP analog AMPPCP. The purine moiety is situated in a hydrophobic environment maintaining hydrogen bonds to the backbone nitrogen of Ala114β, the backbone oxygen of Glu112β, the side-chain carbonyl group of Gln111β, and the side-chain amino moiety of Lys60β. To gain insight into the potential binding mode of Gua nucleotides, we superimposed ckcACD1 with the ATP-grasp domain containing enzyme CK2 from Zea mays, which is known to metabolize Ade and Gua nucleotides (42–44). We found that (i) the Ade moiety of ATP bound to CK2 [Protein Data Bank (PDB) ID code 1DAW] is oriented comparable to ckcACD1 and (ii) the Gua moiety of GTP bound to CK2 (PDB ID code 1DAY) is shifted in its position and compares quite well with the binding mode found in the structure of GTP-specific ssSCS (16). Hence, we expect a comparable binding mode for Guo nucleotides in ckcACD1.
Consequences of Different Domain Arrangements.
The signature motifs for the superfamily of ACDs (NDP-forming) have been extensively studied by comparison of the sequences of bacterial, archaeal, and eukaryotic members (8). The same study highlights the different domain arrangements and connectivities observed in the various kingdoms of life. Based on the structure of the ecSCS and sequence comparison with related enzymes, it can be derived that a functional ACD is composed of five subdomains (subdomains 1–5), with ecSCS displaying the arrangement [alpha(1-2)/beta(3-4-5)]. In the context of domain shuffling, it has to be considered that the respective subdomains 3 and 4 should not be regarded as individual domains. The sequence region for these two subdomains is best described as an ATP-grasp domain, defining the binding site for the NDP and the binding site for the phosphate-carrying substrate (site II). In case of ACLY, the subdomain composition within a single protein chain is 3-4-5-1-2-CS with an additional domain (CS) fused to its carboxyl terminus. Due to the domain shuffling, the overall arrangement of the subdomains is significantly different between ecSCS and ckcACD1 (Fig. 1 and SI Appendix, Fig. S1). The existence of ACD as a α2β2-heterotetramer is a direct consequence of its specific domain arrangement [alpha(1-2-5)/beta(3-4)]. In contrast, SCS enzymes with the domain arrangement alpha(1-2)/beta(3-4-5) have representatives with an α2β2-heterotetrameric structure, as well as with αβ-heterodimeric structures, as observed for the SCS from E. coli (19), T. aquaticus (15), pig (16), and human (45). For ecSCS, two very important α-helices have been highlighted and termed power helices because they stabilize the phosphoryl moiety within the catalytic site I by their helix dipole moment (19). Whereas in ecSCS, the two power helices of one active site I are contributed from one alpha subunit and one beta subunit, in ckcACD1, the two power helices for each active site I are positioned on two different alpha subunits. To form a functional unit in ckcACD1, two alpha subunits have to come together, thus generating a heterotetramer, whereas a heterodimer is already the minimal composition for an active SCS (SI Appendix, Fig. S8).
As a further consequence of the domain arrangement, the distance between the phosphorylated His residue His254α and ADP as the final acceptor of the phosphoryl moiety bound to the beta subunit is significantly increased from around 35 Å (ecSCS) (SI Appendix, Fig. S9) to around 51 Å as observed in the presented complexes of ckcACD1 (Fig. 4). In one of our structures (ckcACD1-C), we observed an orientation of the phosphohistidine segment with the nonphosphorylated His254α pointing toward site II. This movement of the His254α necessitates a partial unwinding of helix α8 (site I), reorientation of the extended loop, and rewinding to helix α8* (site II) (Fig. 4 B and C). In this new orientation, His254α is in close proximity to the beta-phosphate group of ADP (∼9 Å) and His68β (∼7 Å), the second, highly conserved His residue in the ACD subfamily. The distance of His254α to ADP appears to be too large for a direct transfer of the activated phosphate but requires an intermediate mediator. Because His68β was indeed observed to be functionally relevant in an in vitro experiment with an only marginally active ckcACD1 variant (SI Appendix, Fig. S7C), we postulate that His68β is an essential part of the overall reaction. In accordance to ckcACD1, exchange of the corresponding residue of pfACD1 results in inactivation of the enzyme (20). Thus, we propose an additional, fourth reaction step for the overall reaction with phosphorylated His68β as an intermediate.
Postulated Reaction Steps.
Based on our various crystal structures and supplemental biochemical studies, we are able to explain the necessity of the additional fourth reaction step as suggested by Bräsen et al. (20) in comparison to the three-step reaction mechanism as formulated for SCS enzymes (12, 13, 19, 46). The first part of the overall reaction starts at active site I with the binding of Ac-CoA (structure ckcACD1-E) (Fig. 2A) and inorganic phosphate (structure ckcACD1-G and ckcACD1-I) (Fig. 3A). In the first reaction step, the cleavage of Ac-CoA into Ac-phosphate and CoA is coupled to the phosphorylation of His254α (structures ckcACD1-E and ckcACD1-H) (Fig. 3B) in the second step. These first two reaction steps are similar to those steps in pfACD-I and ecSCS (20). The activated phosphoryl moiety is transferred onto the NDP bound at the active site II. To facilitate this transfer, Bailey et al. (47) and Joyce et al. (14) postulated that the loop containing the phosphorylated His swings from site I to site II (SI Appendix, Fig. S9). With the structure of ckcACD1-C, we could prove this postulated rearrangement for the first time to our knowledge (Fig. 4). However, in contrast to the three partial reactions proposed for SCS, the ACD enzymes do not directly transfer the phosphoryl moiety from the phosphorylated His of the rearranged loop onto the NDP bound within the ATP-grasp domain [structures ckcACD1-B, ckcACD1-C, ckcACD1-D, ckcACD1-G, and ckcACD1-H (20); SI Appendix, Fig. S3]. After the rearrangement of the phosphohistidine-containing segment (step i in Fig. 4C), the ACD enzymes transfer in an additional step the phosphoryl moiety from phospho-His254α onto His68β to form phospho-His68β (step ii in Fig. 4C). In the fourth reaction step, the phosphoryl moiety is used to phosphorylate ADP or GDP (step iii in Figs. 4C and 5). The crucial role of His68β is underlined by the observation that mutation of His68β causes loss of activity (SI Appendix, Fig. S7C).
Fig. 5.
Rearrangement of the phosphohistidine segment and proposed involvement of His68β for phosphorylation of ADP. (A) Representation of the region around the phosphate-carrying His residue His254α pointing toward site II within the beta subunit (ATP-grasp domain) after delivery of the phosphate moiety (crystal structure ckcACD1-C). Residues involved in reaction steps ii and iii in Fig. 4C are labeled. Side-chain atoms are shown as carbon in magenta (alpha subunit), carbon in yellow (beta subunit), carbon in light blue (beta′ subunit), nitrogen in blue, and oxygen in red. Carbon atoms in AMPPCP are shown in white, nitrogen in blue, oxygen in red, and phosphor in orange. (B) Scheme for the transfer of the phosphoryl moiety from His254α to ADP via His68β within site II. Phosphorylated His254α is positioned in an ideal orientation for transfer of the phosphoryl moiety to His68β (reaction step ii in Fig. 4C). The negative charge from the phosphoryl moiety is neutralized by interaction with the guanidinyl group of Arg226β and Arg178β′. This makes phospho-His254α more susceptible to nucleophilic attack by His68β. Only the nitrogen atom Nε is in the correct position for an in-line attack on the phosphor atom. Phosphorylation of His68β must result in a displacement of the beta-phosphate group of ADP to bring one of its oxygen atoms into the optimal position for nucleophilic attack (reaction step iii in Fig. 4C). This reorientation needs rotation of the bond O5′-PA of ADP, as well as rotation of phospho-His68β toward the nucleotide.
We could not trap the phosphorylated His68β in a structure of ckcACD1. Based on the spatial proximity of His254α positioned toward site II and His68β (structure ckcACD1-C) (Fig. 5), we propose a nucleophilic attack by nitrogen Nε (τ-position) of the imidazole group of the His68β via an in-line mechanism. Due to steric reasons, the reaction would not be possible for the Nδ atom (π-position). His68β is situated in the T-loop of the ATP-grasp domain. For the final reaction step, the transfer of the phosphoryl moiety from phospho-His68β onto the NDP, a relocation of the diphosphate chain of the nucleotide will be necessary. In the crystal structure of ckcACD1-C with bound AMPPCP at site II, we observed such a conformation for the arrangement of the alpha-phosphate and beta-phosphate moieties of AMPPCP (SI Appendix, Fig. S3). In ecSCS, Glu197β is thought to mediate the optimal protonation state for His246α to facilitate direct phosphorylation of ADP (12, 14). In ckcACD1, the structurally equivalent residue Asp209β interacts with the main chain nitrogens of His254α and Thr255α and the hydroxyl group of the side chain of Thr255α (Fig. 5 and SI Appendix, Fig. S8C). Thus, it stabilizes completely the phosphohistidine segment in its optimal position for phosphoryl transfer onto the second His, His68β. In addition, from mutational studies of pfACD1, it is known that beside its charge, the length of this Asp residue (Asp212β in P. furiosus) is essential for enzymatic activity (20). On the contrary, the length of the side chain of the corresponding residue in ecSCS, Glu197β, only marginally influences the properties of the enzyme (13). Therefore, the role of ecSCS-Glu197β is different compared with the corresponding residue Asp209β in ckcACD1. In the crystal structure of ckcACD1-C, the side chain of the Glu residue Glu144β was found in a hydrogen bridge with Nδ of His His254α (SI Appendix, Fig. S4C). Interestingly, sequences of other ACDs feature a Glu residue corresponding to Glu144β in ckcACD1 (8). We believe that this residue maintains the ideal protonation state of phospho-His254α during the phosphoryl transfer to His68β in ckcACD1 and likely in other ACDs. The proposed mechanism for the unique fourth partial reaction is depicted in Fig. 5C.
ckcACD1 Domain Assembly Seems to be Prototypic for ACDs.
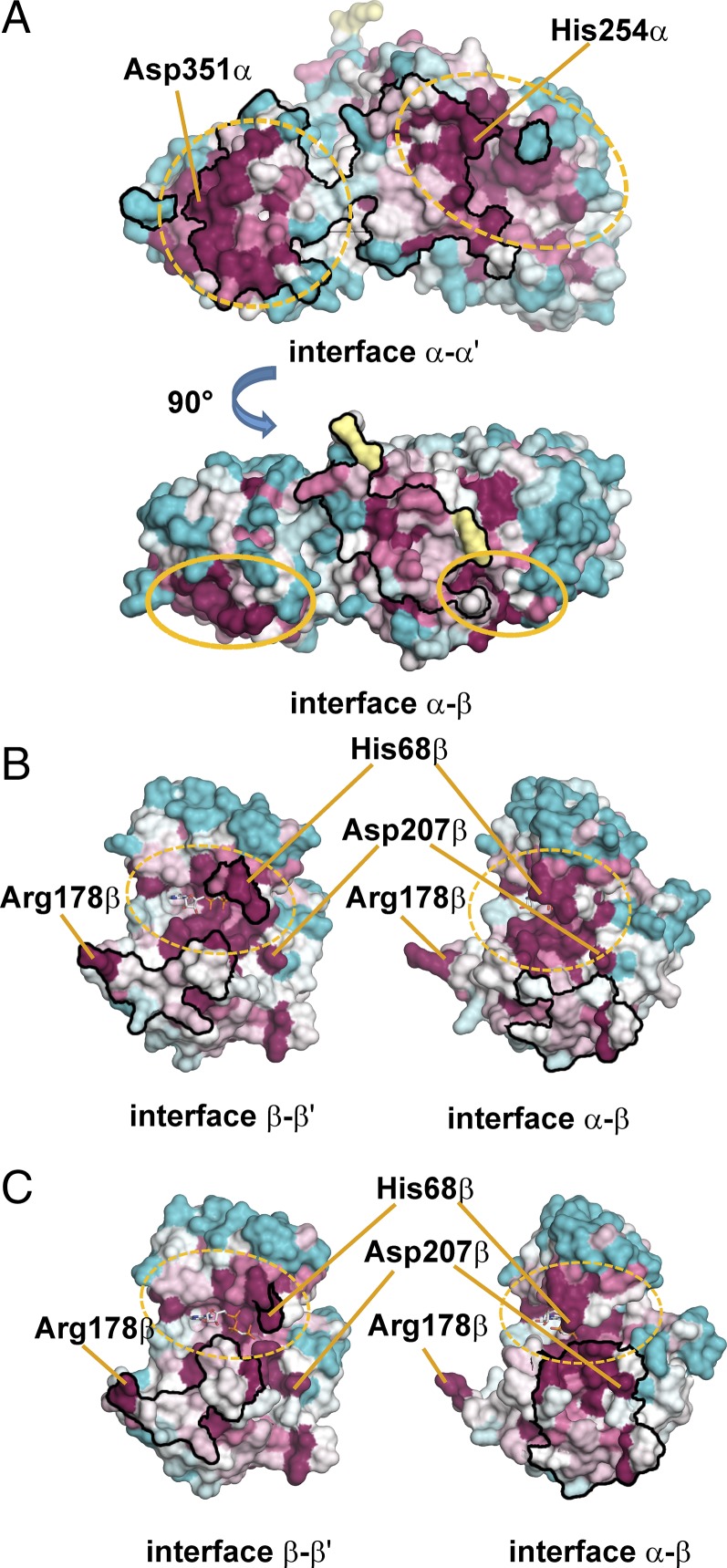
Alignment of the ckcACD1 with sequences of several members of the ACD family showed a high degree of conservation for the CoA binding subdomains (subdomains 1 and 2) and the ATP-grasp domain (subdomains 3 and 4). Larger variations are only seen for the ligase-CoA subdomain 5 (8, 20), which is observed for members of the class of two-component [composition alpha(1-2-5)/beta(3-4)], as well as for fusion-type ACDs (composition 1-2-5-3-4 or 3-4-1-2-5). To assess whether the complex structure of ckcACD1 might be archetypical for the family of ACDs, we compared the conservation of residues participating in the formation of interdomain interfaces (i.e., contacts between α-α′, α-β, and β-β′, respectively) with residues not involved in multimerization using the program ConSurf (48). The results are depicted in Fig. 6. Most of the amino acids within the interfaces are conserved, which might be a hint for common epitopes between the alpha-alpha interface (Fig. 6A, Upper) and the interface between alpha and beta subunits (Fig. 6A, Lower), as well as the heterotypic interaction to the alpha subunit or the homotypic interaction to the second beta subunit (Fig. 6 B and C). Based on these results, we propose that the ckcACD1 complex represents the prototypic assembly of at least all heterotetrameric ACDs with an α2β2 composition.
Fig. 6.
Conservation pattern for surface residues of the alpha and beta subunits of ckcACD1. Sequence alignment and calculation of the level of conservation were performed with the program ConSurf (48). Residues at the protein surface are colored according to their level of conservation (blue and light blue, less conserved; white, residues of average conservation; violet and lilac, highly conserved; yellow, insufficient data). Framed black are regions that are involved in interface formation (type of interface is indicated below the corresponding picture). (A) Interface between the two alpha subunits (derived from the crystal structure of ckcACD1-G). (Upper) Side view of the interface between both alpha subunits. Ellipses in yellow are used to depict both parts of site I (N-terminal part, broken line; C-terminal part, dashed line). (Lower) View was rotated by 90° as indicated to show the amino acids forming the interface to the beta domain. (B and C) Interface formed by the two beta subunits. (B) Interface of the beta subunit as observed in the crystal structure ckcACD1-B, with the phosphohistidine segment oriented toward active site I. (C) Interface of the beta subunit as observed in the crystal structure ckcACD1-C, with the phosphohistidine segment oriented toward active site II. (B and C, Left) Interface contact area between the two symmetry-related beta subunits. (B and C, Right) Interface contact area between one beta subunit and its corresponding alpha subunit. (Left and Right) Views are in slightly different orientation.
Conclusions
We have presented the first, to our knowledge, crystal structures of a functional nucleoside diphosphate-dependent Ac-CoA synthetase. The protein displays a so far unknown 3D arrangement of the five subdomains, which are characteristic for the underlying enzyme superfamily. The different structures represent sequential snapshots along the four-step reaction path, which starts with Ac-CoA, involves two phosphorylated His residues, and ends with a nucleoside-triphosphate. We could describe in detail the binding pockets for the Ac moiety of Ac-CoA, phosphate, and the first phosphorylated His residue. We describe the binding mode for the nucleoside diphosphate, as well as for the final product, nucleoside triphosphate. These structures provide insight into the determinants of the broad-substrate spectrum observed for ckcACD1. Most interestingly, we could present structural proof for the swinging of a protein segment containing the first phosphorylated His in close proximity to the second phosphorylated His. Our structures provide evidence for the necessity of the second His as a phosphoryl intermediate. Furthermore, we propose that the observed structure is archetypical for this group of heterotetrameric ACD enzymes with alpha(1-2-5)/beta(3-4) composition. Our study explains on the structural level the various differences described for ACD in comparison to SCS, the best-studied member of this enzyme superfamily. Overall, this study provides global information for enzymes composed of multiple subdomains and allows us to understand the transmission of activated substrates (e.g., transfer of phosphoryl group) between two distantly separated active sites within the same enzyme.
Materials and Methods
Protein Expression, Purification, and Crystallization.
Ca. K. cryptofilum ACD1 was expressed in E. coli and purified to homogeneity using heat precipitation and gel filtration. Detailed descriptions are provided in SI Appendix. Crystals of full-length ckcACD1 were grown from solutions containing 100 mM Tris⋅HCl (pH 8.3–8.7) and 14–18% (wt/vol) PEG 6000 at 278 K or 18–22% (wt/vol) PEG 6000 at 291 K, respectively. Supplementation of the solutions with 10–30 mM MgCl2 or CaCl2 resulted in better crystal growth. Crystals of ckcACD1 were indexed as orthorhombic space group P212121, with the approximate cell constants for each crystal in the range of a = 100–106 Å, b = 111–114 Å, and c = 125–127 Å and diffracted the X-ray beam up to a resolution of 2 Å (details are provided in Table 1 and SI Appendix, Table S1).
Table 1.
Data collection and refinement statistics
| ckcACD1-# | A | B | C | D | E | F | G | H | I |
| PDB entry | 4XYL | 4XYM | 4XZ3 | 4Y8V | 4YAK | 4YAJ | 4YB8 | 4YBZ | 5HBR |
| Resolution range (Å) | 83.46–1.95 (1.98–1.95) | 127.04–1.90 (1.93–1.90) | 48.84–2.40 (2.47–2.40) | 49.21–2.10 (2.14–2.10) | 83.57–2.46 (2.53–2.46) | 125.81–2.20 (2.25–2.2) | 110.61–1.90 (1.93–1.90) | 83.03–2.1 (2.14–2.1) | 126.03–1.99 (2.03–1.99) |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell a, b, c (Å) | 102.5, 112.2, 124.9 | 99.6, 114.4, 127.0 | 100.2, 111.9, 127.6 | 106.5, 111.0, 126.7 | 106.4, 111.7, 126.0 | 105.8, 111.2, 125.8 | 106.1, 110.6, 125.7 | 105.3, 109.9, 126.8 | 106.1, 110.7, 126.0 |
| Total reflections | 684,725 | 706,193 | 711,978 | 548,838 | 591,856 | 570,073 | 1,515,778 | 573,438 | 672,786 |
| Multiplicity | 6.5 (6.6) | 6.3 (2.7) | 12.5 (12.5) | 6.4 (4.4) | 10.9 (6.8) | 7.7 (3.1) | 13.0 (9.8) | 6.7 (6.9) | 6.6 (5.9) |
| Completeness (%) | 99.8 (99.7) | 97.8 (58.1) | 99.9 (98.8) | 97.6 (77.9) | 98.1 (77.3) | 97.3 (57.0) | 100.0 (99.5) | 99.5 (99.1) | 99.6 (93.1) |
| Mean I/σ(I) | 6.7 (0.6) | 8.1 (0.3) | 9.9 (0.8) | 10.1 (1.1) | 9.7 (0.6) | 6.4 (0.4) | 9.4 (0.5) | 7.5 (0.8) | 8.4 (0.5) |
| CC(1/2) | 0.992 (0.169) | 0.996 (0.103) | 0.995 (0.229) | 0.995 (0.422) | 0.996 (0.231) | 0.990 (0.105) | 0.998 (0.159) | 0.993 (0.312) | 0.998 (0.124) |
| Wilson B-factor (53) (Å2) | 25.5 | 24.9 | 40.2 | 20.4 | 45.3 | 30.8 | 37.1 | 26.2 | 37.2 |
| R-work (%) | 20.64 | 19.53 | 19.65 | 18.44 | 19.36 | 21.45 | 18.16 | 21.58 | 17.84 |
| R-free (%) | 24.64 | 23.71 | 24.44 | 22.55 | 23.82 | 24.13 | 22.05 | 25.34 | 22.22 |
Parameters for the outermost shell are shown in parentheses. CC(1/2), percentage of correlation between intensities from random half datasets. Correlation significant at the 0.1 % level (54); mean intensity over sigma, I/σ(I); R-work, R = 100 × Σhkl │ |Fobs| − |Fcalc| │ /Σ hkl |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively; R-free is equivalent to R-work but is calculated from reflections (5%) that were omitted from the refinement process (55, 56).
Structure Determination.
With a calculated Matthews coefficient of 2.4 Å3/Da (49) two alpha/beta subunits per asymmetrical unit were expected. The phase problem was solved using the molecular replacement method with models prepared from PDB ID codes 2CSU (alpha subunit) and 1WR2 (beta subunit). Positioning of two entities of the alpha subunit model yielded a clear dimeric arrangement, which was in accordance with a calculated self-rotation function indicating a twofold noncrystallographic symmetry axis. Assuming a heterotetrameric α2β2 complex, two molecular replacement solutions were also anticipated for the beta subunit. However, the molecular replacement search provided only one clear solution for the expected two molecules of the beta subunit with a reliable signal-to-noise ratio. At this stage, no reliable molecular replacement solutions could be obtained. Hence, the transformation matrix between both alpha subunits was calculated and applied on the first beta subunit to generate the symmetry-related second instance. The complete model was subjected to several rebuild cycles. Missing parts of the model became well visible in the electron density. Moreover, CoA, a component of the crystallization droplet, could clearly be identified in the difference electron density map. In subsequent steps, we prepared several crystals of ckcACD1 in complex with various cofactors. Data collection and refinement statistics for these crystals are summarized in Table 1 and SI Appendix, Table S1.
Detailed descriptions for site-directed mutagenesis, kinetic characterization, and additional structural interpretations can be found in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge access to the core facilities of the Centre for Biochemistry and Molecular Biology (BiMo)/Labor für Molekulare Biowissenschaften (LMB) of Kiel University. We thank Sebastian Krossa for helpful discussions and support during data collection. Diffraction data were collected on BL14.1, BL14.2, and BL14.3 operated by the Helmholtz-Zentrum Berlin (HZB) at the BESSYII storage ring [Berlin-Adlershof, Germany (50)], on the ID23-1 beamline at the European Synchrotron Radiation Facility (ESRF), and on P14 operated by the European Molecular Biology Laboratory (EMBL) at the Positron-Electron Tandem Ring Accelerator (PETRA) III storage ring (Deutsches Elektronen Synchrotron). We thank the beamline staff at all beamlines for providing assistance in using the beamlines. We thank the HZB for the allocation of synchrotron radiation beam time. We acknowledge financial support from the HZB. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (Grant FP7/2007-2013) for Biological Structure determination at synchrotron X-ray radiation facilities (BioStruct-X) (Grant Agreement 283570). Additionally, we are grateful for access to the High-Throughput Crystallisation (HTX) facility with a grant from P-Cube and BioStruct-X at the EMBL outstation. Beam time at P14 at the EMBL outstation was also funded by a BioStruct-X grant. This work was supported by Grant SCHO 316/10-1 from the Deutsche Forschungsgemeinschaft (to P.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4XYL, 4XYM, 4XZ3, 4Y8V, 4YAK, 4YAJ, 4YB8, 4YBZ, and 5HBR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518614113/-/DCSupplemental.
References
- 1.Schäfer T, Selig M, Schönheit P. Acetyl-CoA synthetase (ADP forming) in archaea, a novel enzyme involved in acetate formation and ATP synthesis. Arch Microbiol. 1993;159(1):72–83. [Google Scholar]
- 2.Glasemacher J, Bock AK, Schmid R, Schönheit P. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur J Biochem. 1997;244(2):561–567. doi: 10.1111/j.1432-1033.1997.00561.x. [DOI] [PubMed] [Google Scholar]
- 3.Scott JW, Poole FL, Adams MW. Characterization of ten heterotetrameric NDP-dependent acyl-CoA synthetases of the hyperthermophilic archaeon Pyrococcus furiosus. Archaea. 2014;2014:176863. doi: 10.1155/2014/176863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awano T, et al. Characterization of two members among the five ADP-forming acyl coenzyme A (Acyl-CoA) synthetases reveals the presence of a 2-(Imidazol-4-yl)acetyl-CoA synthetase in Thermococcus kodakarensis. J Bacteriol. 2014;196(1):140–147. doi: 10.1128/JB.00877-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musfeldt M, Selig M, Schönheit P. Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic Archaeon pyrococcus furiosus: Identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J Bacteriol. 1999;181(18):5885–5888. doi: 10.1128/jb.181.18.5885-5888.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bräsen C, Schönheit P. Mechanisms of acetate formation and acetate activation in halophilic archaea. Arch Microbiol. 2001;175(5):360–368. doi: 10.1007/s002030100273. [DOI] [PubMed] [Google Scholar]
- 7.Jones CP, Ingram-Smith C. Biochemical and kinetic characterization of the recombinant ADP-forming acetyl coenzyme A synthetase from the amitochondriate protozoan Entamoeba histolytica. Eukaryot Cell. 2014;13(12):1530–1537. doi: 10.1128/EC.00192-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sánchez LB, Galperin MY, Müller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (Nucleoside diphosphate-forming) J Biol Chem. 2000;275(8):5794–5803. doi: 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt M, Schönheit P. Acetate formation in the photoheterotrophic bacterium Chloroflexus aurantiacus involves an archaeal type ADP-forming acetyl-CoA synthetase isoenzyme I. FEMS Microbiol Lett. 2013;349(2):171–179. doi: 10.1111/1574-6968.12312. [DOI] [PubMed] [Google Scholar]
- 10.Bock AK, Glasemacher J, Schmidt R, Schönheit P. Purification and characterization of two extremely thermostable enzymes, phosphate acetyltransferase and acetate kinase, from the hyperthermophilic eubacterium Thermotoga maritima. J Bacteriol. 1999;181(6):1861–1867. doi: 10.1128/jb.181.6.1861-1867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sánchez LB, Morrison HG, Sogin ML, Müller M. Cloning and sequencing of an acetyl-CoA synthetase (ADP-forming) gene from the amitochondriate protist, Giardia lamblia. Gene. 1999;233(1-2):225–231. doi: 10.1016/s0378-1119(99)00134-1. [DOI] [PubMed] [Google Scholar]
- 12.Fraser ME, James MN, Bridger WA, Wolodko WT. A detailed structural description of Escherichia coli succinyl-CoA synthetase. J Mol Biol. 1999;285(4):1633–1653. doi: 10.1006/jmbi.1998.2324. [DOI] [PubMed] [Google Scholar]
- 13.Fraser ME, Joyce MA, Ryan DG, Wolodko WT. Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase. Biochemistry. 2002;41(2):537–546. doi: 10.1021/bi011518y. [DOI] [PubMed] [Google Scholar]
- 14.Joyce MA, Fraser ME, James MN, Bridger WA, Wolodko WT. ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography. Biochemistry. 2000;39(1):17–25. doi: 10.1021/bi991696f. [DOI] [PubMed] [Google Scholar]
- 15.Joyce MA, Hayakawa K, Wolodko WT, Fraser ME. Biochemical and structural characterization of the GTP-preferring succinyl-CoA synthetase from Thermus aquaticus. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 7):751–762. doi: 10.1107/S0907444912010852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser ME, Hayakawa K, Hume MS, Ryan DG, Brownie ER. Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase. J Biol Chem. 2006;281(16):11058–11065. doi: 10.1074/jbc.M511785200. [DOI] [PubMed] [Google Scholar]
- 17.Sun T, Hayakawa K, Bateman KS, Fraser ME. Identification of the citrate-binding site of human ATP-citrate lyase using X-ray crystallography. J Biol Chem. 2010;285(35):27418–27428. doi: 10.1074/jbc.M109.078667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T, Hayakawa K, Fraser ME. ADP-Mg2+ bound to the ATP-grasp domain of ATP-citrate lyase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 10):1168–1172. doi: 10.1107/S1744309111028363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolodko WT, Fraser ME, James MN, Bridger WA. The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-A resolution. J Biol Chem. 1994;269(14):10883–10890. doi: 10.2210/pdb1scu/pdb. [DOI] [PubMed] [Google Scholar]
- 20.Bräsen C, Schmidt M, Grötzinger J, Schönheit P. Reaction mechanism and structural model of ADP-forming Acetyl-CoA synthetase from the hyperthermophilic archaeon Pyrococcus furiosus: Evidence for a second active site histidine residue. J Biol Chem. 2008;283(22):15409–15418. doi: 10.1074/jbc.M710218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musfeldt M, Schönheit P. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: Heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J Bacteriol. 2002;184(3):636–644. doi: 10.1128/JB.184.3.636-644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrand JG, Spector LB. Succinyl phosphate and the succinyl coenzyme A synthetase reaction. J Biol Chem. 1969;244(10):2606–2613. [PubMed] [Google Scholar]
- 23.Srere PA. The citrate cleavage enzyme. I. Distribution and purification. J Biol Chem. 1959;234:2544–2547. [PubMed] [Google Scholar]
- 24.Mai X, Adams MW. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178(20):5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikata K, Fukui T, Atomi H, Imanaka T. A novel ADP-forming succinyl-CoA synthetase in Thermococcus kodakaraensis structurally related to the archaeal nucleoside diphosphate-forming acetyl-CoA synthetases. J Biol Chem. 2007;282(37):26963–26970. doi: 10.1074/jbc.M702694200. [DOI] [PubMed] [Google Scholar]
- 26.Elkins JG, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci USA. 2008;105(23):8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt M. 2013. ADP-abhängige Acyl-CoA Synthetasen in Archaea, Bacteria und Eukarya: Charakterisierung, Funktion und Phylogenie. PhD thesis (Kiel University, Kiel, Germany). German.
- 28.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Krissinel E. Crystal contacts as nature’s docking solutions. J Comput Chem. 2010;31(1):133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 30.Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 31.von Heijne G, Blomberg C. Early evolution of cellular electron transport: Molecular models for the ferredoxin-rubredoxin-flavodoxin region. Orig Life. 1978;9(1):27–37. doi: 10.1007/BF00929711. [DOI] [PubMed] [Google Scholar]
- 32.Fawaz MV, Topper ME, Firestine SM. The ATP-grasp enzymes. Bioorg Chem. 2011;39(5-6):185–191. doi: 10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoden JB, Firestine SM, Benkovic SJ, Holden HM. PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site. J Biol Chem. 2002;277(26):23898–23908. doi: 10.1074/jbc.M202251200. [DOI] [PubMed] [Google Scholar]
- 34.Polekhina G, Board PG, Gali RR, Rossjohn J, Parker MW. Molecular basis of glutathione synthetase deficiency and a rare gene permutation event. EMBO J. 1999;18(12):3204–3213. doi: 10.1093/emboj/18.12.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Malhi M, Deneke J, Fraser ME. Structure of GTP-specific succinyl-CoA synthetase in complex with CoA. Acta Crystallogr F Struct Biol Commun. 2015;71(Pt 8):1067–1071. doi: 10.1107/S2053230X15011188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deane CM, Allen FH, Taylor R, Blundell TL. Carbonyl-carbonyl interactions stabilize the partially allowed Ramachandran conformations of asparagine and aspartic acid. Protein Eng. 1999;12(12):1025–1028. doi: 10.1093/protein/12.12.1025. [DOI] [PubMed] [Google Scholar]
- 37.Reeves RE, Warren LG, Susskind B, Lo HS. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J Biol Chem. 1977;252(2):726–731. [PubMed] [Google Scholar]
- 38.Bräsen C, Schönheit P. Unusual ADP-forming acetyl-coenzyme A synthetases from the mesophilic halophilic euryarchaeon Haloarcula marismortui and from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Arch Microbiol. 2004;182(4):277–287. doi: 10.1007/s00203-004-0702-4. [DOI] [PubMed] [Google Scholar]
- 39.Bräsen C, Schönheit P. Regulation of acetate and acetyl-CoA converting enzymes during growth on acetate and/or glucose in the halophilic archaeon Haloarcula marismortui. FEMS Microbiol Lett. 2004;241(1):21–26. doi: 10.1016/j.femsle.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez LB, Müller M. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) from the amitochondriate protist, Giardia lamblia. FEBS Lett. 1996;378(3):240–244. doi: 10.1016/0014-5793(95)01463-2. [DOI] [PubMed] [Google Scholar]
- 41.Hidber E, Brownie ER, Hayakawa K, Fraser ME. Participation of Cys123alpha of Escherichia coli succinyl-CoA synthetase in catalysis. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 8):876–884. doi: 10.1107/S0907444907029319. [DOI] [PubMed] [Google Scholar]
- 42.Allende JE, Allende CC. Protein kinases. 4. Protein kinase CK2: An enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9(5):313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 43.Niefind K, Pütter M, Guerra B, Issinger OG, Schomburg D. GTP plus water mimic ATP in the active site of protein kinase CK2. Nat Struct Biol. 1999;6(12):1100–1103. doi: 10.1038/70033. [DOI] [PubMed] [Google Scholar]
- 44.Issinger OG. Casein kinases: Pleiotropic mediators of cellular regulation. Pharmacol Ther. 1993;59(1):1–30. doi: 10.1016/0163-7258(93)90039-g. [DOI] [PubMed] [Google Scholar]
- 45.Lambeth DO, Tews KN, Adkins S, Frohlich D, Milavetz BI. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J Biol Chem. 2004;279(35):36621–36624. doi: 10.1074/jbc.M406884200. [DOI] [PubMed] [Google Scholar]
- 46.Majumdar R, Guest JR, Bridger WA. Functional consequences of substitution of the active site (phospho)histidine residue of Escherichia coli succinyl-CoA synthetase. Biochim Biophys Acta. 1991;1076(1):86–90. doi: 10.1016/0167-4838(91)90223-m. [DOI] [PubMed] [Google Scholar]
- 47.Bailey DL, Fraser ME, Bridger WA, James MN, Wolodko WT. A dimeric form of Escherichia coli succinyl-CoA synthetase produced by site-directed mutagenesis. J Mol Biol. 1999;285(4):1655–1666. doi: 10.1006/jmbi.1998.2325. [DOI] [PubMed] [Google Scholar]
- 48.Glaser F, et al. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 49.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 50.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19(Pt 3):442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37(Database issue):D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho BK, Gruswitz F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popov AN, Bourenkov GP. Choice of data-collection parameters based on statistic modelling. Acta Crystallogr D. 2003;59(Pt 7):1145–1153. doi: 10.1107/s0907444903008163. [DOI] [PubMed] [Google Scholar]
- 54.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336(6084):1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brünger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 56.Tickle IJ, Laskowski RA, Moss DS. Rfree and the Rfree ratio. II. Calculation of the expected values and variances of cross-validation statistics in macromolecular least- squares refinement. Acta Crystallogr D. 2000;59(Pt 4):442–450. doi: 10.1107/s0907444999016868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.