Significance
Plant growth and development are mediated through a wide range of proteins, including receptor kinases and phosphatases. The receptor kinase ARABIDOPSIS CRINKLY 4 (ACR4) is part of a mechanism controlling formative cell divisions in the Arabidopsis root. However, the regulation of ACR4 signaling and how it affects cell divisions remains completely unknown. We discovered that ACR4 phosphorylates the PROTEIN PHOSPHATASE 2A-3 (PP2A-3) catalytic subunit of the PP2A phosphatase holoenzyme and that PP2A dephosphorylates ACR4. These data exposed a tightly regulated point in the associated biochemical network regulating formative cell divisions in plant roots.
Keywords: stem cells, columella, phosphorylation, kinase, phosphatase
Abstract
In plants, the generation of new cell types and tissues depends on coordinated and oriented formative cell divisions. The plasma membrane-localized receptor kinase ARABIDOPSIS CRINKLY 4 (ACR4) is part of a mechanism controlling formative cell divisions in the Arabidopsis root. Despite its important role in plant development, very little is known about the molecular mechanism with which ACR4 is affiliated and its network of interactions. Here, we used various complementary proteomic approaches to identify ACR4-interacting protein candidates that are likely regulators of formative cell divisions and that could pave the way to unraveling the molecular basis behind ACR4-mediated signaling. We identified PROTEIN PHOSPHATASE 2A-3 (PP2A-3), a catalytic subunit of PP2A holoenzymes, as a previously unidentified regulator of formative cell divisions and as one of the first described substrates of ACR4. Our in vitro data argue for the existence of a tight posttranslational regulation in the associated biochemical network through reciprocal regulation between ACR4 and PP2A-3 at the phosphorylation level.
Plants rely on coordinated formative cell division for the formation of new cell types and tissues (1). For example, in the Arabidopsis primary root tip, columella stem cells—upon formative cell division—give rise to new stem cells and daughter cells that will differentiate (2) (SI Appendix, Fig. S1A). Several plant hormones and proteins that play a role in this process have been identified, and small regulatory networks have been proposed (3–11). However, our knowledge of the mechanisms and signaling networks mediating formative cell divisions is sparse and is largely derived from transcriptional data (12).
Reversible protein phosphorylation represents a major mechanism regulating cell signaling (13), and several kinases have been shown to play a role in primary root development (5, 6, 14). For example, the evolutionarily conserved plasma membrane-localized receptor-like kinase ARABIDOPSIS CRINKLY 4 (ACR4) marks the plasma membrane in the primary root tip columella and is part of a mechanism controlling formative cell divisions in the Arabidopsis root (5, 6, 15). ACR4 possesses an extracellular ligand-binding domain, a transmembrane helix, and an intracellular domain that contains the juxtamembrane and the C-terminal subdomains, which flank the core kinase domain with serine/threonine kinase activity (Fig. 1A). The intracellular juxtamembrane domain is a likely recruitment site for interacting proteins and essential to facilitate downstream signaling (16, 17). ACR4 is expressed throughout plant development in specific cells and tissues, such as protoderm, columella, and stage I lateral roots, and ACR4 preferentially localizes at plasmodesmata (6, 18–20). In addition to its primary and lateral root phenotypes, Arabidopsis loss-of-function acr4 mutants are affected in maintaining epidermal cell identity, including disorganized cell layers in the ovule integument (18, 20). Although ACR4 was the first receptor kinase to be assigned a role in root development (5), our knowledge about its signaling pathway in the root remains limited (6, 8).
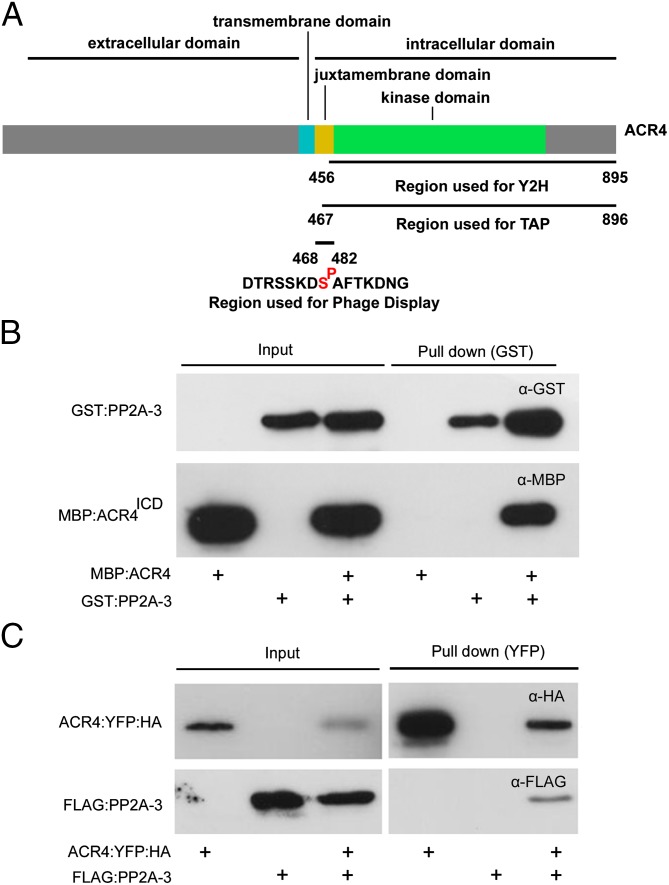
Fig. 1.
ACR4 interacts with PP2A-3. (A) Schematic representation of ACR4 with key domains and regions used for protein–protein interaction studies. (B) In vitro GST pull-down experiment using GST:PP2A-3 and MBP:ACR4ICD according to indicated combinations (+). PP2A-3 and ACR4ICD were detected by Western blotting with anti-GST and anti-MBP antibodies, respectively. (C) In planta YFP pull-down experiment using ACR4:YFP:HA and FLAG:PP2A-3 transiently coexpressed in tobacco leaves by Agrobacterium infiltration according to indicated combinations (+). PP2A-3 and ACR4 were detected by Western blotting with anti-FLAG and anti-HA antibodies, respectively.
In addition to posttranslational modifications such as phosphorylation, developmental programs and cellular functions largely rely on interactions between proteins, forming complex networks to control biological processes (21). Although membrane proteins play a crucial role in many biological processes, knowledge of the in planta membrane interactome is limited (21). Notwithstanding the recent progress with respect to global analyses of membrane protein interactions, so far ACR4 has not been represented in a membrane-linked Arabidopsis interactome (22). Therefore, the objective of this study was to use ACR4-centered, protein-focused systems biology approaches to gain insight into the ACR4-signaling cascade and to identify new potential regulators of formative cell division.
Results and Discussion
Mapping Putative ACR4 Interactions.
To identify regulators of formative cell division in Arabidopsis, we combined, in silico, tandem affinity purification (TAP), yeast two-hybrid (Y2H), and phage display approaches to define potential ACR4-interacting proteins. Given the technical difficulties associated with plasma membrane proteins, we focused on intracellular ACR4 domains for our in vitro and in vivo studies (Fig. 1A). We first interrogated available protein–protein interaction (PPI) databases for experimental and predicted interactions by applying the PPI tool within CORNET 2.0 (23) to ACR4 (AT3G59420). This resulted in a network with 85 nodes that mainly lacked experimental validation (Dataset S1 and SI Appendix, Fig. S2). Second, we applied a TAP approach to Arabidopsis cell suspension cultures expressing the N- or C-terminally tagged ACR4 intracellular kinase domain (Fig. 1A and SI Appendix, Fig. S3). This analysis resulted in four putative ACR4-interacting proteins, of which two occurred in at least two biological replicates and in both assays with N- and C-terminally tagged ACR4 intracellular kinase domain and were not present in any in house background list, namely 4-HYDROXY-TETRAHYDRODIPICOLINATE REDUCTASE 1 (HTPA REDUCTASE 1/DAPB1; AT2G44040) and HTPA REDUCTASE 2/DAPB2 (AT3G59890) (Dataset S1). Based on TAP data alone, two other candidates, PROTEIN PHOSPHATASE 2A-3 (PP2A-3; AT2G42500) and PP2A-4 (AT3G58500), could not be conclusively identified as bona fide ACR4-interacting proteins as they were detected in only one technical repeat. Third, we applied a conventional high-throughput Y2H assay to screen for potential interactions with the ACR4 intracellular domain (Fig. 1A). This revealed four potential ACR4-interacting proteins with high confidence (Dataset S1). Finally, we screened a synthetic 15-mer peptide encompassing the Ser475 phosphorylation site within the intracellular juxtamembrane domain of ACR4 (Fig. 1A) against a 21-amino acid phage-peptide library (24, 25). When the resulting consensus-binding motifs for the phosphorylated peptide were queried against the Arabidopsis protein database, over 4,000 potential ACR4-interacting proteins were identified (Dataset S1). Taken together, our complementary approaches identified several putative ACR4-interacting proteins, but when we searched for overlap between the different approaches this was limited to absent (Dataset S1). This could mean that different approaches yielded different subsets of putative ACR4-interacting proteins or that we picked a large number of likely false-positives.
In Silico Quality Assessment of Putative ACR4 Interactions.
To increase the confidence in the potential ACR4-interacting proteins listed in Dataset S1 and to select candidates for functional analyses, we performed in depth in silico quality assessment. Correlated gene expression is an indicator of cofunctionality of genes in common pathways and processes (26), and interacting proteins are often significantly coexpressed. First, we used CORNET 2.0 (23) to globally explore coexpression of ACR4 and genes encoding potential ACR4-interacting proteins, which showed some coexpression (Pearson correlation coefficient > 0.55) between ACR4 and some CORNET (2/85) and phage display hits (44/4402) (Dataset S1). Second, to further support potential PPIs in the root tip and during lateral root initiation, we used available cell- and tissue-specific transcript profiling datasets (5, 27). Visualization of root tip in silico expression patterns for TAP (3/4) and Y2H candidates (2/4) through the BAR Arabidopsis eFP Browser (28) revealed distinct expression patterns that, at least partially, overlapped with the ACR4 expression domain (SI Appendix, Fig. S2). In addition, some of the CORNET (8/85) and PHAGE DISPLAY candidates (360/4402) were—similar to ACR4—also transcriptionally differentially regulated in a transcriptome study of pericycle cells undergoing lateral root initiation (5) (Dataset S1). Based on the above observations, we generated a priority list for CORNET, TAP, Y2H, and PHAGE DISPLAY hits, narrowing down the number of candidates from 4,495 to 525 (Dataset S1). Next, to globally assess interactions between ACR4 and potential interacting proteins, we used the PPI tool within CORNET 2.0 (23). Indeed, several of the CORNET, Y2H, TAP, and prioritized PHAGE DISPLAY hits are connected with ACR4 and with each other in predicted and experimentally validated protein–protein interaction networks focusing on pairwise interactions (Dataset S1). To gain insight into the molecular functions represented in this protein–protein interaction network, we determined that several statistically overrepresented Gene Ontology categories with respect to biological process (5%) and molecular function (5%) were related to phosphorylation (Dataset S1), which is in agreement with the fact that ACR4 is a receptor kinase. Taken together, through our in silico assessment we increased the confidence in a subset of potential ACR4-interacting proteins (Dataset S1). However, our dataset is not necessarily comprehensive, as, for example, WUSCHEL RELATED HOMEOBOX 5 (WOX5), CRINKLY4-RELATED (CCR) proteins, and CLAVATA1 (CLV1), which were shown to interact with ACR4 (6, 29), were not retained.
ACR4 Interacts with PP2A-3.
Taking the results of Dataset S1 into account allowed us to impose additional criteria to select candidates for functional analyses. Among the candidates selected by at least two approaches and with a high score in the priority list (Dataset S1), we retrieved sequences that match PP2A-3 and/or PP2A-4, which are isoforms of catalytic PP2A C subunits and that form a subclade in the family of five Arabidopsis PP2A C subunits (30). In general, the PP2A heterotrimeric holoenzyme, which is a major, highly conserved eukaryotic serine/threonine phosphatase, consists of a catalytic C subunit, a type A scaffolding/regulatory subunit, and a type B regulatory subunit (31). In Arabidopsis, PP2A phosphatases have been implicated in various hormone-regulated, cellular, and developmental processes, including spatial control of cell division and columella organization, and in innate immunity, but little is known about their dynamic and highly regulated function (32–35). In the context of our focus on formative cell division, we selected PP2A-3 (and PP2A-4) for subsequent in-depth functional characterization. First, an overlay assay indicated that PP2A-3 can specifically interact with both the naive ACR4 intracellular domain (ACR4ICD) (endogenously phosphorylated at a limited number of residues in Escherichia coli through an unknown mechanism) and fully in vitro autophosphorylated ACR4ICD (24) (SI Appendix, Fig. S3 and Supplemental Notes). Second, gel-filtration analyses further confirmed the interaction between the ACR4 intracellular domain (ACR4ICD) and PP2A-3 (SI Appendix, Fig. S3 and Supplemental Notes). Subsequently, the purified recombinant MBP:ACR4ICD was effectively pulled down with GST:PP2A-3 in vitro (Fig. 1B). Moreover, in Nicotiana benthamiana transient expression assays, ACR4:YFP:HA was able to coimmunoprecipitate FLAG:PP2A-3 in planta (Fig. 1C). Taken together, these data strongly indicate that ACR4 and PP2A-3 interact with each other.
ACR4 and PP2A-3 Are Coexpressed.
As mentioned above, Arabidopsis eFP Browser data suggested that PP2A-3 is weakly expressed in columella stem cells and in the root apical meristem (SI Appendix, Fig. S2). To confirm this, we analyzed seedlings expressing a pPP2A-3::n3xGFP fusion and assessed expression in the root tip. In five independent transformants we indeed observed pPP2A-3::n3xGFP expression in the root tip, but PP2A-3 was more broadly expressed than ACR4 (5) (SI Appendix, Fig. S1). In addition, we observed PP2A-3 expression during early lateral root initiation (SI Appendix, Fig. S1), which also overlapped with ACR4 expression at this stage (5) (SI Appendix, Fig. S1). Taken together, these results show that ACR4 and PP2A-3 are expressed in overlapping domains, further supporting that they can physically interact.
PP2A-3 Is Involved in Columella Stem Cell Differentiation.
To test genetically if PP2A-3 plays a role in ACR4-mediated stem cell regulation, we analyzed primary root length and columella stem cell differentiation in a previously characterized pp2a-3 mutant (34). The primary root of pp2a-3 is slightly longer than wild type (SI Appendix, Fig. S1). With respect to columella stem cell differentiation, we observed less differentiation in pp2a-3 compared with wild type, which is similar to acr4 (Fig. 2 A and B). However, because such a columella phenotype can also be explained by altered auxin distribution, levels, or response (11), the similarity between the pp2a-3 and acr4 mutants does not provide conclusive support for a genetic and/or physical interaction. This is especially relevant because PP2A-3 has been shown to play a role in auxin transport (36). We therefore tested to what extent acr4 and pp2a-3 are affected in their sensitivity to N-1-naphthylphthalamic acid (NPA) with respect to primary root length and columella differentiation. In these assays, acr4 and pp2a-3 appeared equally sensitive to NPA treatment as Col-0 (SI Appendix, Fig. S4), suggesting that there is no apparent auxin transport-mediated effect in this case. The genetic interaction between ACR4 and PP2A-3 was further supported by the acr4 pp2a-3 double mutant, where we could not record an additive effect, arguing that both are active in the same pathway (Fig. 2 A and B). Although we cannot rule out cell-specific changes, we have excluded that the similarities in phenotype are due to a broad differential regulation of ACR4 or PP2A-3 expression in pp2a-3 and acr4 mutants, respectively (SI Appendix, Fig. S5). Taken together, our observations indicate a role for PP2A-3 in cellular patterning during primary root development that overlaps with ACR4 function and further suggest that this is possibly independent of an affected auxin transport capacity.
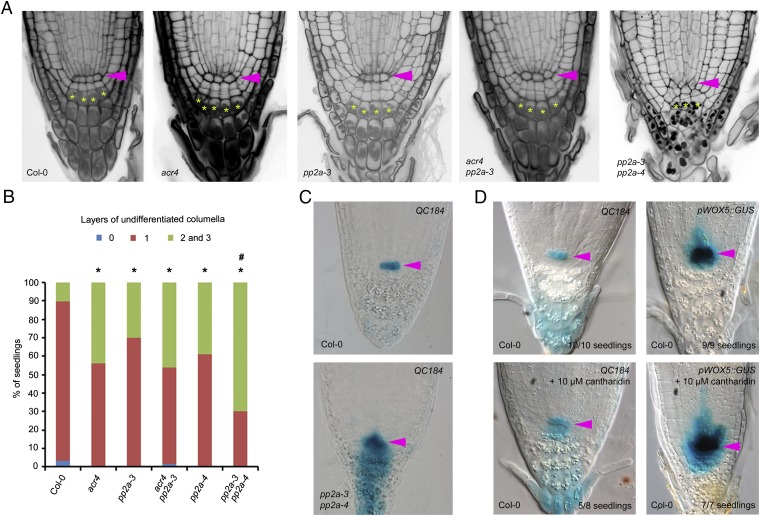
Fig. 2.
PP2A-3 mediates columella stem cell divisions. (A and B) Representative images (A) and quantification (B) of columella stem cell daughter cell differentiation (20 ≤ n ≥ 96) and irregular cellular pattern in pp2a-3 pp2a-4 (A, Right). Statistical significance (Z Test Calculator for 2 Population Proportions, P < 0.05) compared with Col-0 (*) or acr4 (#) is indicated. (C and D) Expression of QC184 and pWOX5::GUS in 5-d-old pp2a-3 pp2a-4 seedlings (C) or in 5-d-old seedlings grown on 10 µM cantharidin (D). Representative pictures with number of seedlings with similar expression pattern indicated. Pink arrowhead, quiescent center. Yellow asterisk, first columella cell layer with starch granules.
PP2A-3 and PP2A-4 Redundantly Affect Primary Root Growth.
Given that PP2A-4 is closely related to PP2A-3, we explored possible redundancy with respect to primary root growth. Indeed, a double pp2a-3 pp2a-4 mutant displayed a short primary root, further reduced columella differentiation, and severely disrupted cell organization in the root tip compared with wild type (Fig. 2 A and B and SI Appendix, Fig. S1) (34). However, this double-mutant phenotype appeared to be less severe than the one obtained by Ballesteros and coworkers (36), which is possibly due to the use of different T-DNA lines. Short root phenotypes associated with disrupted cell division and/or cellular patterning in the root tip are often associated with a loss of quiescent center cell identity (37, 38). To assess if the quiescent center was absent in the disrupted root tip of pp2a-3 pp2a-4, we analyzed the expression of the quiescent center marker QC184 (7). Surprisingly, notwithstanding the dramatic impact on the regular cellular pattern in the root tip, QC184 expression was not abolished and even appeared to expand into the columella (Fig. 2C). The latter might suggest that the stemness gradient in pp2a-3 pp2a-4 is perturbed. Furthermore, we applied cantharidin—an inhibitor of PP2A and PP2A-related phosphatases (SI Appendix, Supplemental Notes)—to the QC184 and pWOX5::GUS markers (7), demonstrating that quiescent center identity is not lost when interfering with PP2A activity (Fig. 2D and SI Appendix, Fig. S6). Interestingly, WOX5 expression was shown to be similarly affected in the clavata3/embryo surrounding region 40 (cle40) mutant, and ACR4 was identified as a target of CLE40 signaling (6, 8), further corroborating the potential connection between PP2A and ACR4. Next, evaluating sensitivity to cantharidin with respect to primary root growth revealed that acr4 is equally sensitive to cantharidin treatment as pp2a-3 and that both are not significantly more sensitive than the control (SI Appendix, Fig. S7). We furthermore established that cantharidin does not negatively affect ACR4 expression levels and observed a similar (minor) up-regulation as in pp2a-3 pp2a-4 (SI Appendix, Fig. S5). Taken together, these results further suggest that ACR4 and PP2A-3 act in the same pathway.
ACR4 Phosphorylates PP2A-3.
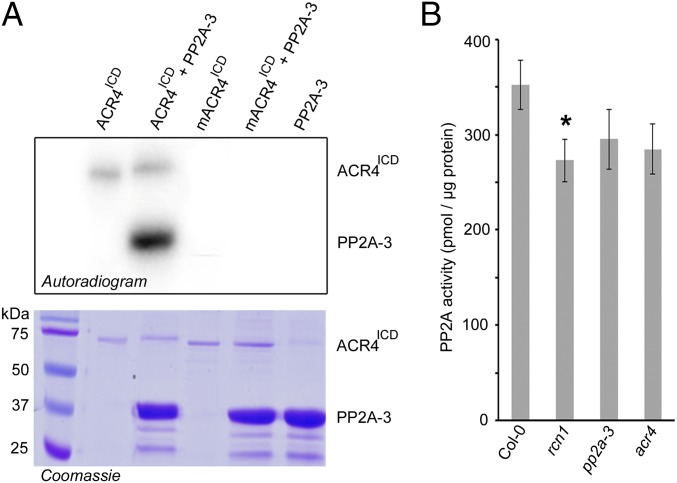
PP2A activity and function in eukaryotic cells is regulated via posttranslational modification of PP2A subunits (39). For example, PP2A complex assembly depends on the phosphorylation status of the catalytic subunit, and phosphorylation of protein phosphatases has been shown to inactivate the enzyme (39, 40), but this has not been demonstrated in plants. To evaluate if ACR4 affects the phosphorylation status of PP2A-3, we compared the phospho-proteomes of Col-0 and acr4 seedlings. However, although we could detect a peptide (NH3-GAGYTFGQDISEQFNHTNNLK-COOH) for PP2A-3 (or PP2A-4) in all samples, we did not observe any (differential) phosphorylation. Because PP2A holoenzymes act in various pathways (35, 36, 41, 42) and because ACR4 acts in only a few cells, it is likely that subtle differences mediated by ACR4 could be masked. Therefore, we explored if PP2A-3 could be phosphorylated by ACR4 in vitro. Indeed, in vitro kinase assays demonstrated that purified recombinant autophosphorylated SUMO:ACR4ICD, but not a mutant inactive version of ACR4ICD with K540A and D641A amino acid exchanges (mACR4ICD), could phosphorylate purified recombinant PP2A-3 (PP2A-3:6xHIS) (Fig. 3A). Subsequently, we identified the ACR4-dependent PP2A-3 phosphosites from the in vitro kinase assay using high-resolution mass spectrometry analyses. This revealed a total of nine—so far unknown—phosphorylated residues of which five are at Ser, three are at Thr, and one is at Tyr (Dataset S1 and SI Appendix, Fig. S8). Mapping these sites on a 3D homology model of Arabidopsis PP2A-3, based on the structure of the catalytic chain within the trimeric human PP2A enzyme, showed that these residues were predominantly solvent-exposed even in the trimer structure and, therefore, quite likely accessible for phosphorylation by ACR4 (Dataset S1 and SI Appendix, Fig. S8). Taken together, our results pinpoint PP2A-3 as a substrate for ACR4 kinase activity. Phosphorylation of the tail of the PP2A catalytic subunit plays an important role in regulating the assembly—and thus activity—of PP2A holoenzymes (39). We therefore explored PP2A activity in cellular extracts prepared from acr4 seedlings using a PP2A phosphatase assay system (35, 43, 44). In our hands, this revealed a decrease in PP2A activity of about 19% in acr4 compared with wild type, which was similar to that associated with the PP2A regulatory subunit mutant roots curl in npa 1 (rcn1) (22%) and the catalytic subunit mutant pp2a-3 (16%) (Fig. 3B). Although the results did not achieve statistical significance (P value for acr4 = 0.083), likely because the ACR4 impact on PP2A activity is diluted, they are reproducible and suggestive of ACR4 being required for some of the cellular PP2A activity.
Fig. 3.
PP2A-3 is phosphorylated by ACR4 kinase. (A) Autoradiogram for coincubated E. coli-expressed PP2A-3 and ACR4 kinase (ACR4ICD) or mutant inactive kinase mACR4ICD as indicated (Upper). The lanes in A are, from left to right, molecular weight standards, 1 µg of autophosphorylated ACR4ICD, 1 µg of autophosphorylated ACR4ICD incubated with 10 µg PP2A-3, 1 µg of mutant inactive kinase mACR4ICD, 1 µg of mutant inactive kinase mACR4ICD incubated with 10 µg PP2A-3, and 10 µg PP2A-3. Corresponding Coomassie blue-stained gel (Lower) was used as loading control. (B) Bar diagram for PP2A activity detected in whole 12-d-old seedling protein extracts as average of three biological repeats (with three technical repeats each) ± SE. Statistical significance (Student’s t test) compared with Col-0 is indicated: *P value < 0.05.
PP2A-3 Dephosphorylates ACR4.
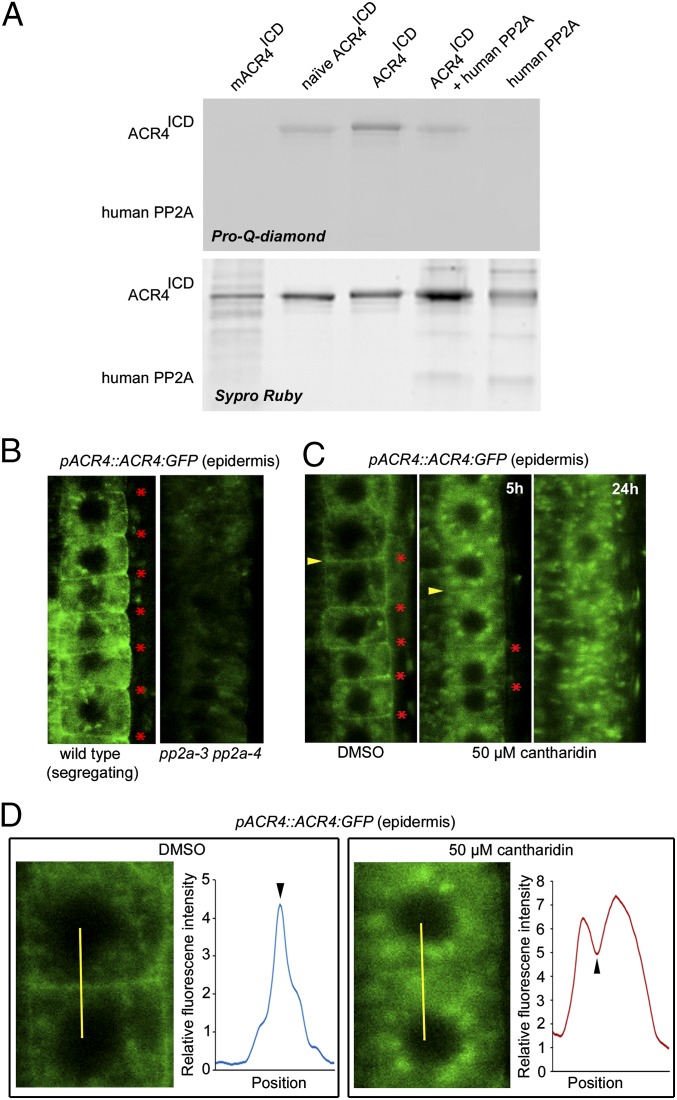
With respect to receptor kinases, PP2A has been shown to modulate the phosphostatus of BRI1 and the coreceptor BAK1 (35, 45, 46), so we also evaluated if PP2A-3 is capable of dephosphorylating ACR4. First, using E. coli-expressed PP2A-3 in a phosphatase assay did not yield a convincing difference with respect to dephosphorylation of ACR4ICD (SI Appendix, Fig. S8 and Supplemental Notes). Therefore, to determine whether ACR4 could be a substrate of PP2A, we used—in accordance with Wu and colleagues (45)—a purified, active human PP2A to dephosphorylate the ACR4ICD that had been phosphorylated in vitro. This demonstrated PP2A-mediated dephosphorylation of the phosphorylated ACR4ICD (Fig. 4A) and pinpoints ACR4 as a substrate for PP2A phosphatase activity. To assess the biological importance of altering the PP2A-mediated phosphorylation status of ACR4, we analyzed ACR4:GFP in the root of the pp2a-3 pp2a-4 double mutant. This revealed a weak GFP signal in pp2a-3 pp2a-4, with reduced membrane localization, compared with the control (Fig. 4B), which is likely not due to a change in ACR4:GFP expression levels (SI Appendix, Fig. S5). To explore this further, we investigated ACR4:GFP in the presence of the PP2A inhibitor cantharidin. This revealed a reduced membrane association of ACR4:GFP upon inhibiting PP2A activity within 5 h, whereas the membrane localization of the routinely used membrane marker FORMIN HOMOLOG 6 (FH6):GFP (47) was largely unaffected (Fig. 4 C and D and SI Appendix, Fig. S9). Overall, these results suggest that membrane localization of ACR4 is dependent on PP2A-3 (and potentially PP2A-4).
Fig. 4.
PP2A-3 dephosphorylates ACR4. (A) Pro-Q-diamond stained gel showing the phosphorylation status of ACR4ICD coincubated without or with PP2A-3 as indicated (Upper). The same gel stained with Sypro Ruby (Lower) as loading control. (B–D) Localization of ACR4:GFP in pp2a-3 pp2a-4 background (B) and following treatment with DMSO or 50 µM cantharidin for indicated hours (C and D). Red asterisk, plasma membrane with ACR4 localization; arrowhead, plasma membrane analyzed in D. (D) Detail of indicated membrane in C (yellow arrowhead) and quantification of GFP signal across the yellow line as smooth average graph. Black arrowhead, position of plasma membrane.
Conclusions
Interactions between membrane-associated proteins and soluble proteins are essential for signal transduction and for regulating plant growth and development. Here, we used various approaches to generate a prioritized list of potential ACR4-interacting proteins that are possibly involved in formative cell division, cell-to-cell communication, and root development. Taking all our interaction data together and because there is limited-to-no overlap between the different approaches, it seems that to study protein–protein interactions the use of multiple approaches is preferred as each technique seems to expose a distinct subset of potential interactors and can increase confidence in some potential interactors that would otherwise be discarded.
Starting from the prioritized, potential ACR4-interacting candidates, we identified PP2A-3 as one of the first described ACR4 substrates and showed that PP2A-3 plays an important role in the control of columella stem cell divisions and/or differentiation. Previously, it was shown that PP2A complexes associate with membranes in growing seedlings and that PP2A may interact with plasma membrane components (33, 41). Similar to the PP2A effect on BRI1 where dephosphorylated BRI1 is internalized (45, 46) and on BAK1 (35), we showed that PP2A can dephosphorylate ACR4. In this context, we also showed that PP2A activity affects the membrane localization of ACR4. The resemblance of the acr4 and pp2a-3 columella stem cell phenotype, together with the cell biological data, suggests that PP2A acts as a positive regulator of ACR4 function and that it is the dephosphorylated form of the ACR4 protein that is localized to the plasma membrane and is functional. On the basis of the available data, we propose a tentative model whereby, on the one hand, ACR4 phosphorylates the PP2A-3 catalytic subunit of the PP2A holoenzyme, possible facilitating complex assembly, and on the other hand, PP2A dephosphorylates ACR4, regulating its membrane localization and possibly activity (SI Appendix, Fig. S10). The balance between these two likely affects formative cell divisions and cell differentiation in the root, and as such ACR4 and/or PP2A appear to control their own activity. In the future, it will be important to characterize the importance of the individual ACR4 and PP2A-3 phosphosites and to evaluate these in the context of ACR4 localization and/or activity and PP2A activity and/or complex assembly, respectively. Here, it should be taken into account that, for example, in brassinosteroid signaling, PP2A type B subunits usually recruit substrates and regulate their activity (42, 48) and that membrane localization of PP2A C subunits is regulated by methylation and in turn impacts target dephosphorylation (45).
Materials and Methods
Detailed materials and methods are described in SI Appendix, Materials and Methods.
Columella Phenotyping.
For columella phenotyping, seedlings were stained with lugol and mounted in Hoyer’s solution as previously described (5).
GUS Assays.
GUS assays were performed as described previously (49).
TAP.
For the TAP approach, transformation of Arabidopsis cell suspension cultures was carried out as previously described (26, 27, 50, 51). Tandem affinity purification of protein complexes was done using the GStag (52) followed by protein precipitation and separation, according to a previously described protocol (51).
Transient Transformation.
N. benthamiana transient transformation was performed as previously described. Protein extraction and coimmunoprecipitation were performed as previously described (53) with modifications (extraction buffer at pH 9.5).
pACR4::ACR4:GFP Analyses and Quantification.
To analyze membrane localization of ACR4:GFP, we processed images in ImageJ using the Plot Profile option across a selected line. Subsequently, the plot data were processed according to a moving average calculation of five values to smoothen the graph. Multiple cells (n > 4) in multiple seedlings (n > 2) were measured and showed similar results.
Supplementary Material
Acknowledgments
We thank Jelle Van Leene, Remco Sprangers, Debbie Rombaut, and Dominique Eeckhout for experimental assistance and useful suggestions; Hyun-Sook Pai for the FLAG:PP2A-3 construct; Dr. Yasufumi Yamamoto for constructing the random 21-amino-acid phage-peptide library; and R. Reid Townsend and the Proteomics Core Laboratory at Washington University for mass spectrometry analysis. This work was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship (BB_BB/H022457/1) and Marie Curie European Reintegration Grant PERG06-GA-2009-256354 (to I.D.S.); a BBSRC CASE Studentship cofunded by Bayer CropScience (to N.C.); and a BBSRC-funded position (BB/I001875/1 to M.I.). We thank the University of Nottingham School of Biosciences for studentship funding and acknowledge the University of Nottingham research committee. The VIB Bio Imaging Core acquired the Zeiss LSM780 through a Correlative Light Electron Microscopy (CLEM) grant from Minister Lieten. Work in the T.B. laboratory was financed by the Interuniversity Attraction Poles Programme IUAP P7/29 “MARS” from the Belgian Federal Science Policy Office and by a grant of the Research Foundation Flanders (FWO G027313N). E.S. is a Postdoctoral Fellow of the Research Foundation-Flanders (FWO13/PDO/144). K.Y. was supported by a grant from the Chinese Scholarship Council (CSC).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525122113/-/DCSupplemental.
References
- 1.De Smet I, Beeckman T. Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat Rev Mol Cell Biol. 2011;12(3):177–188. doi: 10.1038/nrm3064. [DOI] [PubMed] [Google Scholar]
- 2.Scheres B. Stem-cell niches: Nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 2007;8(5):345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- 3.Aida M, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119(1):109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Wildwater M, et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123(7):1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 5.De Smet I, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322(5901):594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- 6.Stahl Y, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr Biol. 2013;23(5):362–371. doi: 10.1016/j.cub.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446(7137):811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 8.Stahl Y, Wink RH, Ingram GC, Simon R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol. 2009;19(11):909–914. doi: 10.1016/j.cub.2009.03.060. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, et al. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010;64(5):764–774. doi: 10.1111/j.1365-313X.2010.04367.x. [DOI] [PubMed] [Google Scholar]
- 10.Azpeitia E, Weinstein N, Benítez M, Mendoza L, Alvarez-Buylla ER. Finding missing interactions of the Arabidopsis thaliana root stem cell niche gene regulatory network. Front Plant Sci. 2013;4:110. doi: 10.3389/fpls.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Z, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA. 2010;107(26):12046–12051. doi: 10.1073/pnas.1000672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kajala K, et al. Omics and modelling approaches for understanding regulation of asymmetric cell divisions in Arabidopsis and other angiosperm plants. Ann Bot (Lond) 2014;113(7):1083–1105. doi: 10.1093/aob/mcu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11(6):427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 14.Van Damme D, et al. Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell. 2011;23(11):4013–4024. doi: 10.1105/tpc.111.089565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikonorova N, Vu LD, Czyzewicz N, Gevaert K, De Smet I. A phylogenetic approach to study the origin and evolution of the CRINKLY4 family. Front Plant Sci. 2015;6:880. doi: 10.3389/fpls.2015.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439(7073):168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 17.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100012. 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gifford ML, Dean S, Ingram GC. The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development. 2003;130(18):4249–4258. doi: 10.1242/dev.00634. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka H, et al. ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 2002;43(4):419–428. doi: 10.1093/pcp/pcf052. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe M, Tanaka H, Watanabe D, Machida C, Machida Y. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 2004;39(3):298–308. doi: 10.1111/j.1365-313X.2004.02132.x. [DOI] [PubMed] [Google Scholar]
- 21.Bassel GW, et al. Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell. 2012;24(10):3859–3875. doi: 10.1105/tpc.112.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones AM, et al. Border control: A membrane-linked interactome of Arabidopsis. Science. 2014;344(6185):711–716. doi: 10.1126/science.1251358. [DOI] [PubMed] [Google Scholar]
- 23.De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inzé D. CORNET 2.0: Integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 2012;195(3):707–720. doi: 10.1111/j.1469-8137.2012.04184.x. [DOI] [PubMed] [Google Scholar]
- 24.Meyer MR, Lichti CF, Townsend RR, Rao AG. Identification of in vitro autophosphorylation sites and effects of phosphorylation on the Arabidopsis CRINKLY4 (ACR4) receptor-like kinase intracellular domain: Insights into conformation, oligomerization, and activity. Biochemistry. 2011;50(12):2170–2186. doi: 10.1021/bi101935x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer MR, Shah S, Rao AG. Insights into molecular interactions between the juxtamembrane and kinase subdomains of the Arabidopsis Crinkly-4 receptor-like kinase. Arch Biochem Biophys. 2013;535(2):101–110. doi: 10.1016/j.abb.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 28.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer MR, Shah S, Zhang J, Rohrs H, Rao AG. Evidence for intermolecular interactions between the intracellular domains of the Arabidopsis receptor-like kinase ACR4, its homologs and the Wox5 transcription factor. PLoS One. 2015;10(3):e0118861. doi: 10.1371/journal.pone.0118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007;12(4):169–176. doi: 10.1016/j.tplants.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Terol J, Bargues M, Carrasco P, Pérez-Alonso M, Paricio N. Molecular characterization and evolution of the protein phosphatase 2A B′ regulatory subunit family in plants. Plant Physiol. 2002;129(2):808–822. doi: 10.1104/pp.020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhrig RG, Labandera AM, Moorhead GB. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013;18(9):505–513. doi: 10.1016/j.tplants.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Blakeslee JJ, et al. Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 2008;146(2):539–553. doi: 10.1104/pp.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spinner L, et al. A protein phosphatase 2A complex spatially controls plant cell division. Nat Commun. 2013;4:1863. doi: 10.1038/ncomms2831. [DOI] [PubMed] [Google Scholar]
- 35.Segonzac C, et al. Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 2014;33(18):2069–2079. doi: 10.15252/embj.201488698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballesteros I, et al. Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. Plant J. 2013;73(5):862–872. doi: 10.1111/tpj.12078. [DOI] [PubMed] [Google Scholar]
- 37.Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17(3):354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda M, et al. The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development. 2004;131(9):2101–2111. doi: 10.1242/dev.01096. [DOI] [PubMed] [Google Scholar]
- 39.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: In cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33(3):113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Janssens V, Goris J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(Pt 3):417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michniewicz M, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130(6):1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 42.Tang W, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13(2):124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Hu R, Zhu Y, Shen G, Zhang H. Arabidopsis PHOSPHOTYROSYL PHOSPHATASE ACTIVATOR is essential for PROTEIN PHOSPHATASE 2A holoenzyme assembly and plays important roles in hormone signaling, salt stress response, and plant development. Plant Physiol. 2014;166(3):1519–1534. doi: 10.1104/pp.114.250563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao HB, Chu YJ, Xue HW. Phosphatidic acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol Plant. 2013;6(5):1692–1702. doi: 10.1093/mp/sst076. [DOI] [PubMed] [Google Scholar]
- 45.Wu G, et al. Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal. 2011;4(172):ra29. doi: 10.1126/scisignal.2001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Rubbo S, Irani NG, Russinova E. PP2A phosphatases: The “on-off” regulatory switches of brassinosteroid signaling. Sci Signal. 2011;4(172):pe25. doi: 10.1126/scisignal.2002046. [DOI] [PubMed] [Google Scholar]
- 47.Van Damme D, Bouget FY, Van Poucke K, Inzé D, Geelen D. Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 2004;40(3):386–398. doi: 10.1111/j.1365-313X.2004.02222.x. [DOI] [PubMed] [Google Scholar]
- 48.Wang R, et al. The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B′ subunits. Mol Plant. 2016;9(1):148–157. doi: 10.1016/j.molp.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Beeckman T, Viane R. Embedding thin plant specimens for oriented sectioning. Biotech Histochem. 2000;75(1):23–26. doi: 10.3109/10520290009047981. [DOI] [PubMed] [Google Scholar]
- 50.Van Leene J, et al. A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics. 2007;6(7):1226–1238. doi: 10.1074/mcp.M700078-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Van Leene J, Witters E, Inzé D, De Jaeger G. Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 2008;13(10):517–520. doi: 10.1016/j.tplants.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Bürckstümmer T, et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3(12):1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 53.Holton N, Nekrasov V, Ronald PC, Zipfel C. The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog. 2015;11(1):e1004602. doi: 10.1371/journal.ppat.1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.