Significance
Hepatocellular carcinoma (HCC) associated with hepatitis C virus (HCV) infection is the fastest-rising cause of cancer-related death in the United States. The level of intratumor HCV replication and the molecular interactions between virus and tumor remain elusive, however. Here we demonstrate that the ability of HCV to replicate in HCC is severely hampered despite unchanged miR122 expression. Surprisingly, we found that livers containing HCC harbor a more diverse viral population than that seen in cirrhotic livers without HCC. Tracking of individual variants demonstrated changes in quasispecies distribution between tumor and nontumorous areas, suggesting viral compartmentalization within the tumor. These insights into the interplay between HCV and HCC call for further investigation of whether malignant hepatocytes express or lack factors that restrict HCV entry or negatively affect viral replication.
Keywords: hepatitis C virus, hepatocellular carcinoma, HCV RNA levels, HCV quasispecies, liver
Abstract
Analysis of hepatitis C virus (HCV) replication and quasispecies distribution within the tumor of patients with HCV-associated hepatocellular carcinoma (HCC) can provide insight into the role of HCV in hepatocarcinogenesis and, conversely, the effect of HCC on the HCV lifecycle. In a comprehensive study of serum and multiple liver specimens from patients with HCC who underwent liver transplantation, we found a sharp and significant decrease in HCV RNA in the tumor compared with surrounding nontumorous tissues, but found no differences in multiple areas of control non-HCC cirrhotic livers. Diminished HCV replication was not associated with changes in miR-122 expression. HCV genetic diversity was significantly higher in livers containing HCC compared with control non-HCC cirrhotic livers. Tracking of individual variants demonstrated changes in the viral population between tumorous and nontumorous areas, the extent of which correlated with the decline in HCV RNA, suggesting HCV compartmentalization within the tumor. In contrast, compartmentalization was not observed between nontumorous areas and serum, or in controls between different areas of the cirrhotic liver or between liver and serum. Our findings indicate that HCV replication within the tumor is restricted and compartmentalized, suggesting segregation of specific viral variants in malignant hepatocytes.
Hepatitis C virus (HCV) is a hepatotropic, single-stranded RNA virus that replicates in the cytoplasm of hepatocytes and does not integrate into the host genome (1). HCV circulates in vivo as a dynamic distribution of closely related viral variants that are commonly referred to as “quasispecies” (2). Such diversity confers a remarkable advantage to the virus under host selective constraints (3–5). One of the most important features of HCV is its extraordinary ability to persist in up to 80% of infected individuals (6). Approximately 20–30% of chronic HCV carriers develop cirrhosis and its long-term sequelae, including hepatic decompensation and hepatocellular carcinoma (HCC), leading to orthotopic liver transplantation (OLT) or liver-related death (6). The incidence of HCC in the United States has more than tripled over the past 3 decades (7), an alarming trend due primarily, if not exclusively, to HCV infection (8).
Although epidemiologic evidence has linked chronic HCV infection to a significantly elevated risk of developing HCC (9), the mechanisms whereby HCV promotes hepatocarcinogenesis remain to be elucidated (10). Whether HCV elicits liver cancer indirectly, through chronic inflammation, fibrosis, and continuous liver regeneration (11), or directly, through the expression of tumor-promoting viral proteins, in a manner analogous to other oncogenic viruses, such as human papillomaviruses and Epstein–Barr virus (12, 13), remains unknown. The major challenges in defining the role of HCV in the pathogenesis of HCC include the inherent limitations of available experimental systems, the low number of infected hepatocytes, the low level of HCV antigen expression, and the limited availability and size of infected liver samples (14).
Insights into the levels of HCV replication within the tumor of patients with HCV-associated HCC may shed light on the role of HCV in hepatocarcinogenesis. This remains controversial, however, in part because of the difficulty in obtaining multiple paired liver specimens from the tumor and adjacent nontumorous tissue. Although some previous investigations have shown no differences in the presence and levels of HCV RNA between the tumor and nontumorous liver tissues (15–17), others have reported low to undetectable levels of HCV RNA within the tumor (18–20). Several reports have indicated that miR-122 is an essential cellular host factor for HCV replication (21, 22). Initial studies performed in liver cancer, regardless of the etiology, have shown a reduced expression of miR-122 (23, 24), whereas recent reports have demonstrated that its expression in HCV-associated HCC is maintained (25, 26) or even increased (27). Thus, the central questions of whether HCV actively replicates in malignant hepatocytes, its relationship with miR-122 expression, and whether viral replication is directly involved in hepatocarcinogenesis remain to be fully elucidated. A related issue is whether changes in viral replication within the tumor are associated with selection of specific viral variants; however, very limited information is available on the composition of the HCV quasispecies and the possible compartmentalization between the tumor and the surrounding nontumorous areas or serum from the same individuals with HCV-associated HCC (20, 28–30).
To investigate the role of HCV in hepatocarcinogenesis, we took advantage of a unique collection of multiple liver specimens from patients with HCV-associated HCC who underwent OLT or partial hepatectomy to simultaneously study the level of viral replication, its correlation with the intrahepatic expression of miR-122, and the composition and distribution of the viral population both within and outside the tumor.
Results
Patients.
The demographic, clinical, virologic, and histopathological features of the 12 patients with HCV-associated HCC (n = 8) or non-HCC cirrhosis (n = 4) included in this study are presented in Table S1. The mean age of the two groups was similar; all patients except one were male (92%) and all but one were infected with HCV genotype 1. The grade of tumor differentiation was G2 in four patients and G3 in four patients; in all cases, HCC was surrounded by a cirrhotic liver.
Table S1.
Baseline characteristics of the 12 patients with HCV-associated HCC or non-HCC cirrhosis
| Characteristic | HCC | Non-HCC cirrhosis |
| Patients, n | 8 | 4 |
| Age, y, mean ± SEM | 55 ± 3 | 51 ± 3 |
| Male sex, n (%) | 7 (87.5) | 4 (100) |
| Alanine aminotransferase, U/L, mean ± SEM* | 105 ± 13 | 76 ± 12 |
| Aspartate aminotransferase, U/L, mean ± SEM† | 102 ± 19 | 100 ± 15 |
| γ-Glutamyltransferase, U/L, mean ± SEM‡ | 133 ± 41 | 70 ± 18 |
| Prothombin time, INR, mean ± SEM§ | 1.2 ± 0.1 | 1.6 ± 0.1 |
| Total bilirubin, mg/dL, mean ± SEM¶ | 2.6 ± 0.7 | 1.9 ± 0.1 |
| Platelets (103/µL), mean ± SEM# | 148.5 ± 39.4 | 65.7 ± 16.0 |
| α-Fetoprotein, ng/mg, mean ± SEM‖ | 297 ± 240 | 59 ± 35 |
| Serum HCV RNA, log10 IU/mL, mean ± SEM | 5.9 ± 0.2 | 5.7 ± 0.4 |
| HCV genotype, n | ||
| 1a | 1 | 3 |
| 1b | 6 | 1 |
| 2a/2c | 1 | 0 |
| Liver pathology | ||
| Nontumorous tissue** | ||
| Activity grade, mean ± SEM | 7.4 ± 0.9 | 5.4 ± 0.5 |
| Fibrosis stage, mean ± SEM | 5.8 ± 0.2 | 6.0 ± 0.0 |
| F5, n | 2 | 0 |
| F6, n | 6 | 4 |
| Tumor grade, n†† | ||
| G2 | 4 | |
| G3 | 4 | |
| Tumor size, n | ||
| ≥2 and ≤3 cm | 4 | |
| >3 cm | 4 | |
Normal range, ≤43 U/L.
Normal range, ≤42 U/L.
Normal range, ≤38 U/L.
Normal range, 0.80–1.20 INR.
To convert serum bilirubin values to micromoles per liter, multiply by 17.1.
Normal values, ≥159 to ≤388 (103/μL).
Normal range, <10.0 ng/mL.
The degree of activity and stage of fibrosis were assessed according to the Ishak scoring system (45).
Tumors were graded using the Edmondson–Steiner grading system (46).
HCV RNA Levels in Patients with HCC and Controls with Non-HCC Cirrhosis.
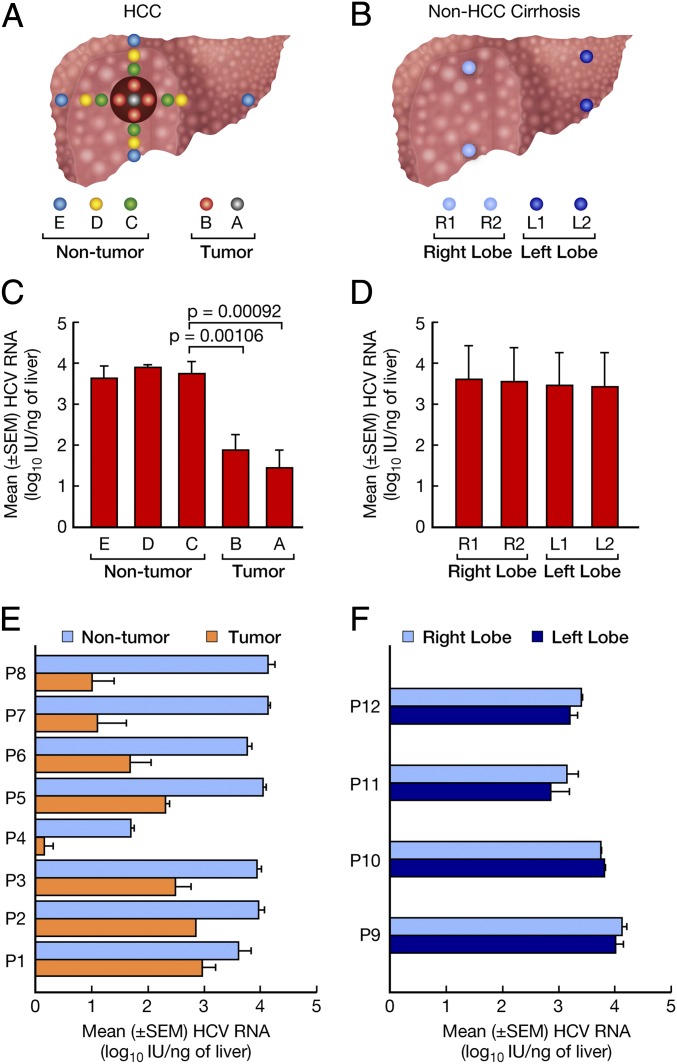
An average of 13 liver specimens from each patient with HCC and 4 liver specimens from each patient with non-HCC cirrhosis were tested for the presence and levels of intrahepatic HCV RNA by real-time PCR (Fig. 1 A and B). A sharp and significant drop in viral RNA levels was observed in all of the patients with HCC when perilesional tissue was compared with tissue inside the tumor margin (Fig. 1C), with the lowest concentration in the center of the tumor (area A). In contrast, HCV RNA levels were significantly higher and comparable among the three nontumorous areas, C, D, and E (Fig. 1C), as well as in four different regions of control livers, spanning the right and left lobes obtained from the four patients with non-HCC cirrhosis (Fig. 1D). In individual patients, the extent of the drop in mean HCV RNA levels between the surrounding nontumorous areas (C, D, and E) and the tumor (areas A and B) ranged between 0.89 and 3.15 logs. Five patients (patients 1–5) showed a <2-log drop in HCV RNA (HCC pattern 1), whereas three patients (patients 6–8) had a >2-log drop (HCC pattern 2) (Fig. 1E). In contrast, no changes in HCV RNA levels were detected between the right and left lobes of livers from the controls with non-HCC cirrhosis (Fig. 1F). The HCC and non-HCC patients had comparable levels of HCV in serum (Table S1 and Fig. S1 A and B). Thus, our study demonstrates that HCC is associated with a severe restriction of intrahepatic HCV replication.
Fig. 1.
Study design, and HCV RNA levels in multiple liver areas of patients with HCC and controls with non-HCC cirrhosis. (A and B) Study design illustrating the location of the liver specimens collected from each of eight livers containing HCC (A) and from four controls with non-HCC cirrhosis (B). In HCC, the dark-brown solid circle indicates the tumor, and the different color dots represent samples collected at various distances from the center of the tumor in the four directions (north, south, east, and west). Five biopsies were obtained from the tumor, including one from the center (area A) and four from the periphery (area B), and 12 from the surrounding nontumorous areas, including four from the perilesional tissue (area C), four taken 2–3 cm from the tumor margin (area D), and four from the edge (area E) of the liver. In non-HCC cirrhosis, four liver specimens, two from the right lobe (R1 and R2) and two from the left lobe (L1 and L2), were obtained from each liver. (C) Bars represent the mean ± SEM HCV RNA levels in different areas of the livers containing HCC from all eight patients. Data from the center of the tumor (area A) represent individual samples; data from each of the remaining liver samples (areas B–E) represent the average levels from multiple specimens obtained in the four directions for each area. (D) Bars represent the mean ± SEM HCV RNA levels in different areas of the livers from the four controls with non-HCC cirrhosis. (E) Horizontal bars indicate the mean ± SEM HCV RNA levels in tumorous and nontumorous tissues of individual patients with HCC. (F) Horizontal bars indicate the levels of HCV RNA in the left and right lobes of individual controls with non-HCC cirrhosis.
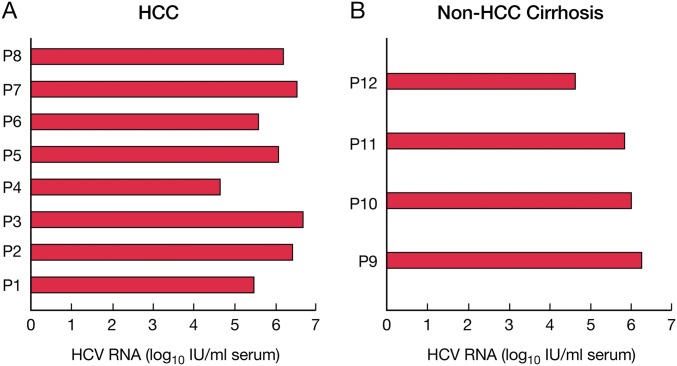
Fig. S1.
Quantification of HCV RNA in serum of individual patients with HCC (A) and controls with non-HCC cirrhosis (B).
Expression of miR-122 in Patients with HCC and Controls with Non-HCC Cirrhosis.
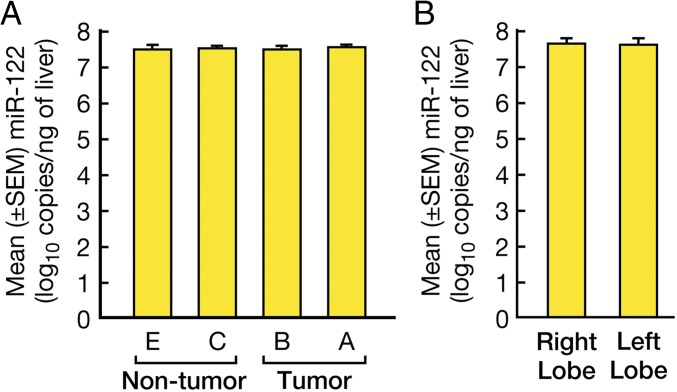
To investigate the mechanisms underlying the significant reduction in HCV replication within HCC tissues, we examined the expression of miR-122 in multiple areas of the livers containing HCC, because this miRNA has been shown to be essential for HCV replication (31). Interestingly, we found no differences in miR-122 expression among the different areas of the liver containing HCC (Fig. 2A), indicating that the significant reduction in HCV RNA within the tumor is not related to changes in miR-122 expression. Similar levels of miR-122 were also seen in different areas of the liver in controls with non-HCC cirrhosis (Fig. 2B).
Fig. 2.
Intrahepatic expression of miR-122 in HCC-containing livers. (A) Bars represent the mean ± SEM miR-122 levels in the different areas of the livers as described in Fig. 1A. (B) Intrahepatic expression of miR-122 in control livers. Bars represent the mean ± SEM miR-122 levels in different areas of the livers as described in Fig. 1B.
Genetic Diversity, Complexity, and Distribution of HCV Quasispecies in Liver Compartments and Serum of Patients with HCC and Non-HCC Cirrhosis.
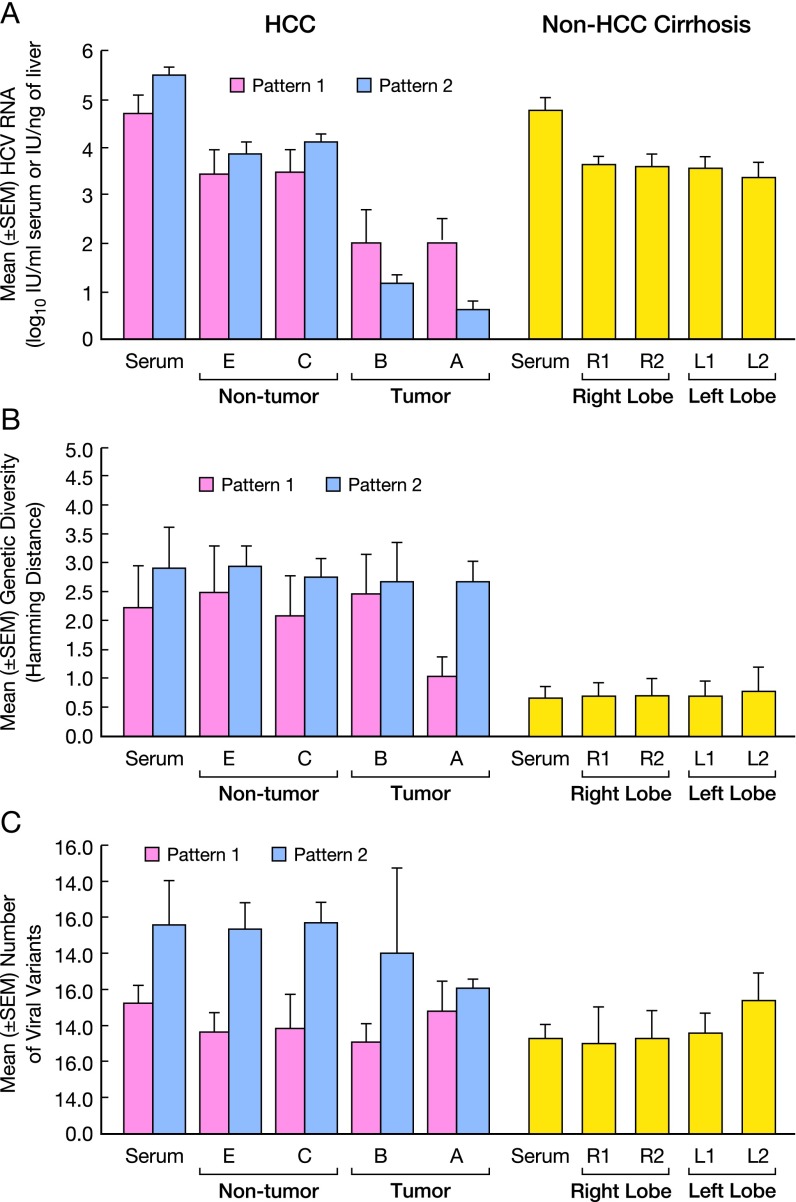
We next studied the genetic distance between the different variants (i.e., genetic diversity) and the number of viral strains (i.e., genetic complexity) of the HCV quasispecies, both within and outside the hypervariable region 1 (HVR1) of the E1/E2 region in HCC patients divided according to the extent of HCV RNA decrease (patterns 1 and 2) and in controls with non-HCC cirrhosis (Fig. S2A). We found a statistically significant decrease in HCV RNA in HCC by comparing the tumor with the surrounding nontumorous tissue (P < 0.01 for pattern 1; P < 0.000001 for pattern 2). We also found a significant difference between HCV RNA levels in HCC pattern 1 and pattern 2 within the tumor (P = 0.04), but not in the surrounding nontumorous tissues. HCV RNA levels were also significantly higher in non-HCC cirrhosis than in the tumor (P < 0.0001 for pattern 1; P < 0.000001 for pattern 2), whereas there were no differences in serum HCV RNA levels between the two groups of patients.
Fig. S2.
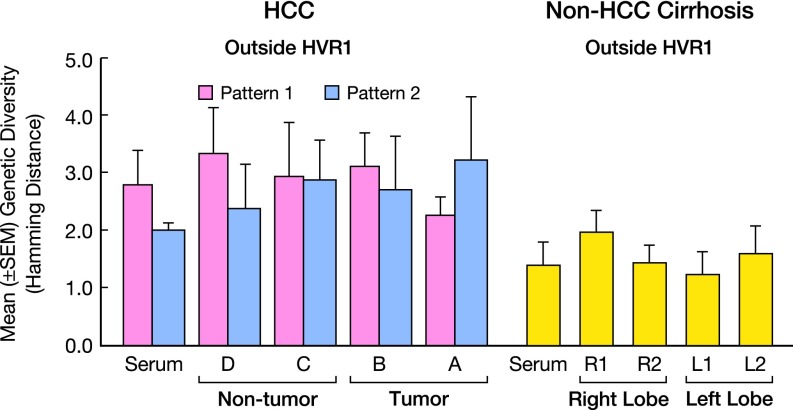
HCV RNA levels, genetic diversity, and number of viral strains in serum and multiple liver areas of patients with HCC and controls with non-HCC cirrhosis. (A) HCV RNA levels in HCC patients divided according to the drop in intrahepatic HCV RNA within the tumor in pattern 1 (<2 logs; patients 1–5) and pattern 2 (>2 logs; patients 6–8). Patient numbers are the same as in Fig. 1E. (B) Genetic diversity (distance among variants), as measured by mean Hamming distance within the HVR1 region, in serum and liver of HCC pattern 1 and 2 patients and controls. (C) Number of viral variants in serum and liver of HCC pattern 1 and 2 patients and controls. The values indicate the number of variants per 27 amino acids within the HVR1. Data represent the mean ± SEM of the results obtained from all patients within each group.
The genetic diversity, as measured by Hamming distance, was significantly higher in the livers of patients with HCC compared with those of controls with non-HCC cirrhosis both within and outside the HVR1, irrespective of the liver compartment analyzed (P < 0.0007 in all comparisons) (Figs. S2B and S3). Genetic diversity was also higher in the serum of patients with HCC, both within and outside the HVR1 (P = 0.016 and P = 0.004, respectively) (Figs. S2B and S3). Among the HCC patients, the degree of genetic diversity within HVR1 was significantly higher in the center of the tumor in patients with pattern 2 compared with those with pattern 1 (P = 0.0257) (Fig. S2B).
Fig. S3.
Genetic diversity of the HCV quasispecies within the E1/E2 region outside the HVR1 in serum and multiple liver areas of patients with HCC and controls with non-HCC cirrhosis. Genetic diversity (distance among variants) within the viral quasispecies, as measured by mean Hamming distance, in serum and liver of HCC patients and controls. HCC patients were divided according to the drop in intrahepatic HCV RNA as pattern 1 (<2 logs; patients 1–5) or pattern 2 (>2 logs; patients 6–8). Patient numbers are the same as in Fig. 1 E and F. Data represent the mean ± SEM of the results obtained from all patients within each group. The difference was significant when the livers of patients with non-HCC cirrhosis were compared both with the tumorous tissue (P = 0.002) and with the surrounding nontumorous tissue (P = 0.0035) of HCC patients.
The number of viral variants, as assessed by HVR1 sequences, was significantly higher in the liver of patients with HCC pattern 2 than in controls with non-HCC cirrhosis (P = 0.002) or in patients with HCC pattern 1 (P = 0.029), whereas no differences were detected between patients with HCC pattern 1 and these controls (Fig. S2C). A significantly higher number of variants in serum samples was also observed in patients with HCC pattern 2 compared with controls with non-HCC cirrhosis (P = 0.025).
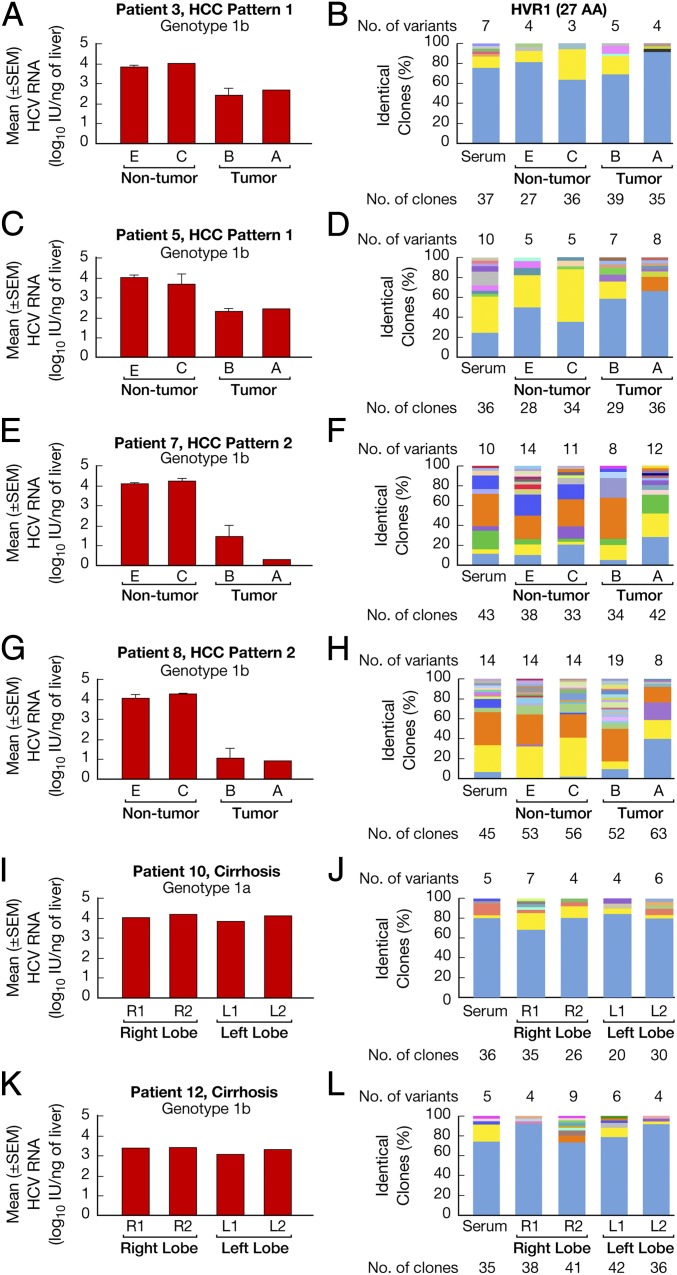
Tracking of individual viral variants in multiple liver compartments and serum demonstrated two different patterns of viral quasispecies distribution, which correlated with the drop in HCV RNA levels from the surrounding nontumorous areas to the tumor. In patients with HCC pattern 1 (patients 1–5; Fig. 1E), who had the smallest drop in HCV RNA (<2.0 log), the predominant variant within the tumor was either dominant or codominant in all liver compartments, as well as in serum (Fig. 3 B and D). Interestingly, the center of the tumor contained unique minor variants that were not detected in the periphery of the tumor, but lacked some of the variants that started to appear in the periphery of the tumor and further expanded to the surrounding nontumorous areas (Fig. 3 B and D). Patients with HCC pattern 2 (patients 6–8; Fig. 1E), who had the most dramatic drop in HCV RNA (>2.0 logs), exhibited a more complex HCV quasispecies with greater changes in the distribution of the viral variants (Fig. 3 F and H). A distinctive feature of this group was the behavior of the dominant strain found in the center of the tumor, which coexisted with other major variants and lost dominance both at the periphery of the tumor and in the surrounding nontumorous areas (Fig. 3 F and H). The virus population in the nontumorous liver compartments was similar in the patients with HCC pattern 1 and HCC pattern 2 and mirrored the quasispecies distribution in serum (Fig. 3 B, D, F, and H).
Fig. 3.
HCV quasispecies distribution in multiple liver areas of four representative patients with HCC and two representative controls with non-HCC cirrhosis. (A, C, E, G, I, and K) Bars represent the mean ± SEM HCV RNA levels in different areas of the liver from four representative HCC cases, two from pattern 1 (A and C), who exhibited a <2-log drop in HCV RNA within the tumor compared with the surrounding nontumorous areas, and two from pattern 2 (E and G), with a >2-log drop, as well as from two controls (I and K) with non-HCC cirrhosis. (B, D, F, H, J, and L) HCV quasispecies distribution in four representative HCC patients (two from pattern 1 and two from pattern 2) and in two controls with non-HCC cirrhosis. The vertical bars indicate the number and proportion of identical clones. The total number of variants identified in each liver compartment and serum is listed above the bars. The dominant viral variant found in each patient within the center of the tumor is shown in blue; other variants are shown in other colors. Within the vertical bars, each variant is identified by a different color. The same color indicates identity between viral variants detected in different liver compartments and serum within each patient but not between different patients. Patient numbers are the same as in Fig. 1 E and F.
In contrast to the changes in quasispecies distribution observed in the eight patients with HCC, analysis of multiple liver specimens and serum of the four control patients who never developed HCC showed a different pattern that was consistent across all of the liver areas analyzed, characterized by the presence of a dominant strain, representing 65–95% of the viral population, along with some minor variants (Fig. 3 J and L). The dominant strain and one or two minor variants were shared by all of the liver compartments, as well as by the viral population circulating in serum (Fig. 3 J and L).
Malignant Cell Proliferation in Patients with HCC.
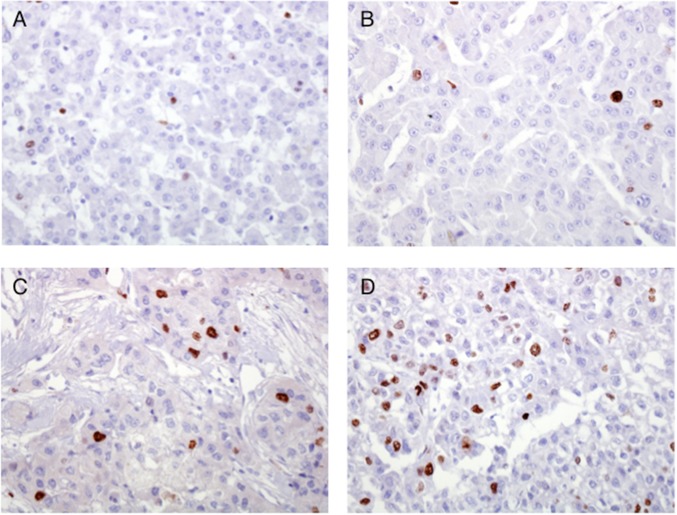
We next investigated the rate of proliferation of malignant hepatocytes in the patients with HCC pattern 1 and pattern 2. Formalin-fixed tissue obtained from the center of the tumor of four patients with HCC was available for staining for the cell proliferation marker MIB1. Two of these four patients (patients 3 and 4) exhibited pattern 1, with a <2-log drop in HCV RNA, and the other two (patients 7 and 8) exhibited pattern 2, with a >2-log drop in HCV RNA (Fig. 1E). The percentage of tumor nuclei positive for MIB1 was assessed using an ocular grid at 20×. For each patient, more than 1,000 tumor nuclei were assessed (mean, 1,640; range, 1,130–2,350). Remarkably, we found that patients with the lowest level of HCV RNA showed the highest proportion of proliferating malignant cells. The percentage of MIB1 positivity was 0.55% and 4.0% in pattern 1 patients 3 and 4, respectively (Fig. 4 A and B), compared with 12.4% and 15.8% in pattern 2 patients 7 and 8 (Fig. 4 C and D). These data suggest that the high viral diversity documented within the tumor of patients with HCC pattern 2, who had the most significant decrease in HCV RNA in the liver (>2 logs), is associated with a higher rate of cell proliferation compared with patients with HCC pattern 1.
Fig. 4.
Variation in cell proliferation rate in malignant hepatocytes of patients who exhibited different drops in HCV RNA levels within the tumor compared with the adjacent cirrhotic liver. The images illustrate the immunostaining for the proliferation marker, MIB1, in sections taken from the center of the tumors. (A and B) Pattern 1 HCC cases (patients 3 and 4) showing a <2-log drop between the HCV RNA level within the tumor compared with the surrounding nontumorous tissue. (C and D) Pattern 2 HCC cases (patients 7 and 8) with a >2-log drop in HCV RNA level in the tumor. The fraction of positive nuclei for MIB1 was higher in patients with the lowest drop in HCV RNA (C and D). Patient numbers are the same as in Fig. 1E.
HCV Compartmentalization and Selection Analysis in Patients with HCC and in Controls with Non-HCC Cirrhosis.
To determine whether the differences in HCV quasispecies distribution between the tumorous and nontumorous tissues of patients with HCC reflects compartmentalization of viral variants in different areas of the liver, we used the Mantel test (32). Compartmentalization was defined by a significant difference in virus populations (quantified as pairwise genetic distance among the sequences) between different areas of the liver or between the liver and serum. Remarkably, the viral population differed significantly between the tumor and nontumorous tissues in all eight patients with HCC (Table 1). In more than 50% of the patients (four of seven), the viral population detected in the tumor also differed from that circulating in serum (Table 1). In contrast, no significant differences were found between the viral population detected in nontumorous tissues (areas C, D, and E) and the variants circulating in serum (Table 1), indicating that the viral population circulating in serum is most likely produced in the surrounding nontumorous areas, which represents the largest area of the liver.
Table 1.
Analysis of HCV compartmentalization within the E1 and E2 envelope genes, inclusive of the HVR1, among tumor, nontumor, and serum in patients with HCC and between the right and the left lobes and between all liver compartments and serum in patients with non-HCC cirrhosis
| Disease type | P value (Mantel’s test) | ||
| Patient no. | Tumor vs. nontumor | Tumor vs. serum | Nontumor vs. serum |
| HCC | |||
| HCV RNA pattern 1 | |||
| 1 | 0.001 | 0.469 | 0.105 |
| 2 | 0.029 | 0.027 | 0.403 |
| 3 | 0.039 | 0.531 | 0.749 |
| 4 | 0.001 | 0.004 | 0.109 |
| 5 | 0.001 | 0.001 | 0.124 |
| HCV RNA pattern 2 | |||
| 6 | 0.009 | NA | NA |
| 7 | 0.021 | 0.978 | 0.836 |
| 8 | 0.009 | 0.033 | 0.239 |
| Non-HCC cirrhosis | |||
| Right vs. left lobe | Liver vs. serum | ||
| 9 | 0.411 | 0.535 | |
| 10 | 0.480 | 0.977 | |
| 11 | 0.872 | 0.825 | |
| 12 | 0.140 | 0.059 | |
The Mantel test was used to determine if sequences from a given compartment within patients with HCC (tumor, nontumorous tissues, and serum) or in controls (right lobe, left lobe, all liver compartments, and serum) were genetically closer to each other than to sequences from other compartments. Patients with HCC were divided into two groups according to the drop in HCV RNA within the tumor. Pattern 1 denotes a <2-log drop in HCV RNA; pattern 2, a >2-log drop (Fig. 1E). Patient numbers are the same as in Fig. 1 E and F. NA, not available, because the amount of serum from patient 6 was not sufficient for the analysis of the HCV quasispecies. P < 0.05 indicates that the viral population from two given compartments is significantly different, providing statistical evidence for HCV compartmentalization.
In contrast, in the controls with non-HCC cirrhosis, our analysis revealed no evidence of HCV compartmentalization between the right and left lobes, or between liver compartments and serum (Table 1). Thus, our data indicate that HCC is associated with viral compartmentalization.
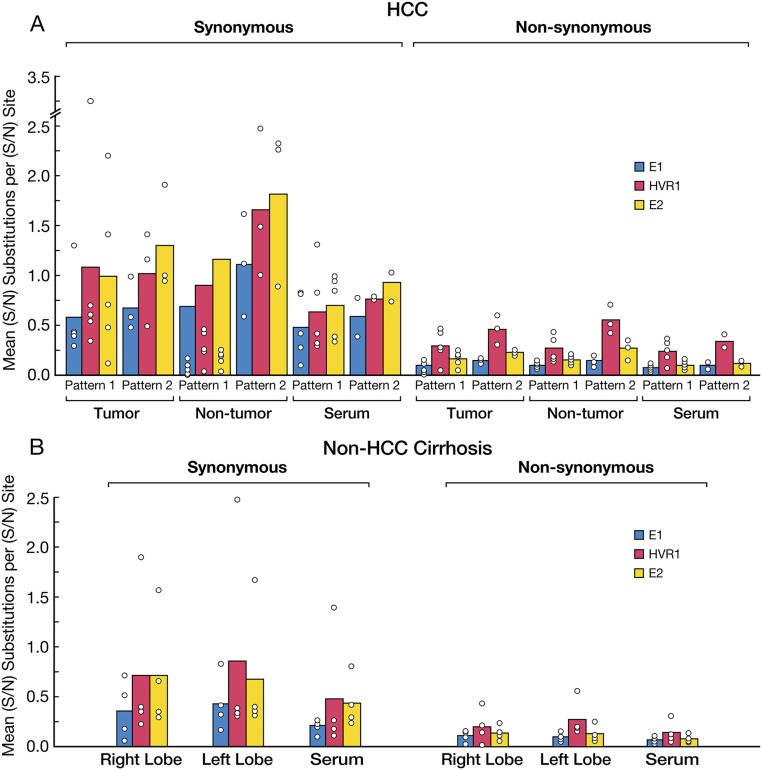
We next investigated whether the different patterns of HCV quasispecies distribution and HCV compartmentalization between the tumorous and nontumorous tissues of patients with HCC could be attributed to positive selection. We estimated the mean number of nonsynonymous and synonymous substitutions per nonsynonymous and synonymous site, respectively, across the E1/HVR1/E2 region within the tumor and the nontumorous tissues, as well as among different areas of the control non-HCC cirrhotic livers, and in the serum of both groups of patients. An excess of nonsynonymous substitutions with respect to synonymous substitutions was considered to be indicative of positive selection. Overall, the mean number of synonymous substitutions per site was greater than that of nonsynonymous substitutions per site across the E1/HVR1/E2 region, both within and outside the tumor, as well as in serum (Fig. S4A). A similar pattern was seen in cirrhotic patients (Fig. S4B). These results suggest that HCV compartmentalization and the distribution of viral variants within the tumor is not the result of a selective host immune pressure.
Fig. S4.
Synonymous and nonsynonymous substitutions in serum and multiple liver areas of patients with HCC according to the drop in HCV RNA levels (patterns 1 and 2) and controls with non-HCC cirrhosis. (A) Mean number of synonymous and nonsynonymous substitutions per site in multiple liver areas and serum of HCC patients divided into pattern 1 and pattern 2 according to the drop in HCV RNA within the tumor compared with the surrounding nontumorous areas: pattern 1, <2 logs (patients 1–5); pattern 2, >2 logs (patients 6–8). Patient numbers are the same as in Fig.1 E and F. (B) Mean number of synonymous and nonsynonymous substitutions per site in multiple liver areas and serum of patients with non-HCC cirrhosis. The bars indicate the means for all patients for each group. The results are presented separately for the E1, HVR1, and E2 regions. Circles indicate the mean values for each individual patient.
Discussion
To the best of our knowledge, this is the first study in which the levels of HCV replication and the distribution of the viral quasispecies were comprehensively investigated in serum and multiple compartments of individual livers containing HCC and compared with levels in control livers with non-HCC cirrhosis. Although the number of patients that could be included in this intensive study was limited, our patients were well characterized and devoid of confounding factors such as viral coinfections, i.v. drug use, alcohol abuse, or antiviral treatment. All were Caucasian, and all but one was infected with HCV genotype 1. Analysis of HCV RNA in up to 17 specimens from each liver containing HCC provided conclusive evidence that the tumor is associated with restricted levels of viral replication.
Previous studies performed by in situ PCR (33, 34), fluorescence microscopy (35), or laser-capture microdissection followed by RT-PCR (36, 37) indicated that the distribution of HCV RNA in the liver is focal and that the percentage of infected hepatocytes is low (never >35% of the total population). However, the focal distribution of HCV-infected cells (37) cannot explain the restricted viral replication that we detected exclusively within the tumor. Our analysis, extended to the entire liver containing HCC, clearly demonstrates that the marked decline in HCV RNA occurs exclusively within the tumor. Thus, the drop in viral RNA is highly specific to the malignant hepatocytes and cannot be related to sampling variability or to the presence of a mixture of malignant and normal hepatocytes, given that all liver specimens with a mixed-cell population were excluded from the analysis.
Consistent with these findings, and in agreement with previous studies (38–40), reduced HCV RNA levels were not detected in any of the surrounding nontumorous areas or in different regions of the right and left lobes of livers from patients with cirrhosis who never developed HCC, indicating that nontumorous liver tissue can efficiently sustain HCV replication regardless of the presence of a tumor. Thus, the decline in HCV RNA documented exclusively within the tumor would have been missed by analyzing only the levels of viremia or routine biopsy specimens of nontumor tissue.
The fact that HCV does not replicate well in malignant hepatocytes is consistent with the inability or limited efficiency of HCV to grow in hepatoma cell lines in vitro (1), suggesting that malignant hepatocytes express factors, or more likely have lost the expression of factors, that may restrict viral entry or negatively affect viral replication. Interestingly, our analysis of multiple paired liver specimens obtained from the tumor and surrounding nontumorous tissue showed no differences in miR-122 expression, indicating that the reduced levels of HCV RNA in the tumor were not the result of a reduction in miR-122 expression. The level of miR-122 in HCC compared with the surrounding nontumorous tissue has been a matter of debate (23–27). Our observation is consistent with the results of recent studies that showed no changes in miR-122 expression between tumorous and nontumorous tissue in HCV-associated HCC (25, 26).
Our access to a unique collection of liver and serum samples provided us with the opportunity to study the relationship between HCV RNA replication and viral quasispecies distribution within and outside the tumor. Surprisingly, we found that livers containing HCC harbor a more complex viral population with a significantly higher genetic diversity compared with cirrhotic livers without HCC. Although there was no difference in overall viral diversity between the tumor and surrounding nontumorous tissues, despite the significant drop in HCV RNA within the tumor, tracking of individual viral variants showed distinct changes in the composition and distribution of the viral quasispecies. Of note, the extent of the variation in quasispecies distribution appeared to correlate with the differential decline in HCV RNA between nontumorous and tumorous compartments; the greater the drop in HCV RNA within the tumor, the greater the shift in viral quasispecies distribution between the tumor and surrounding nontumorous tissues.
Our data also show that the differences in quasispecies distribution are not the result of differences in host selective pressure in malignant hepatocytes, as was suggested previously (20). The molecular mechanisms leading to high viral diversity within the tumor despite the significant drop in viral replication remain to be elucidated. We found a higher rate of proliferation of malignant hepatocytes in the center of the tumor in the patients with HCC pattern 2, who maintained a high degree of diversity despite the greatest drop in HCV RNA, suggesting that proliferation of neoplastic cells may play a role in maintaining a high degree of viral diversity within the tumor. This is an interesting observation that needs to be confirmed in a large series of patients.
Changes in malignant hepatocytes may lead to the expression, or lack of expression, of factors that contribute to maintaining a segregation of viral variants between tumor and nontumor compartments. Consistent with this hypothesis, Mantel test analysis demonstrated significant differences in HCV quasispecies distribution between the tumorous and nontumorous compartments, providing evidence of HCV compartmentalization between these two liver areas. In contrast, analysis of multiple liver areas in the control patients with non-HCC cirrhosis demonstrated no evidence of HCV compartmentalization within the liver or between the liver and serum, further indicating that the presence of HCV compartmentalization is unique to tumor-containing livers (20, 41). Interestingly, viral compartmentalization between plasma and extrahepatic sites, including leukocytes (42), PBMCs (43), and the brain (44), also has been reported in HCV-infected patients.
In summary, our study shows that patients with HCC harbor a more complex HCV population in the liver compared with patients with non-HCC cirrhosis. Moreover, our data provide conclusive evidence that HCC is associated with restricted HCV replication within the tumor despite unchanged miR122 expression levels, as well as with HCV compartmentalization. Whether and to what extent HCV-infected tumor cells harbor replication-competent virus remain to be defined, although this question is difficult to address owing to the lack of small animal models and efficient in vitro systems for growing primary HCV isolates. Our results provide insights into the role of HCV in hepatocarcinogenesis and may help to elucidate whether malignant hepatocytes express or lack factors that restrict viral entry or replication.
Materials and Methods
Study Subjects and Design.
Eight patients with HCV-associated HCC were selected for this intensive study, In these patients, serum and up to 17 liver specimens were collected from the same liver of each patient at the time of OLT (six patients) or partial hepatectomy (two patients) (SI Materials and Methods). As a control group, we studied four patients with non-HCC cirrhosis who underwent OLT for end-stage non-HCC cirrhosis associated with HCV. All patients were followed at the Liver Transplantation Center of Brotzu Hospital in Cagliari, Italy. All patients provided written informed consent, and the protocol was approved by the hospital’s Ethical Committee. The study was also approved by the US National Institutes of Health’s Office of Human Subjects Research, on the condition that all samples were deidentified.
Methods.
Details of the liver pathology, serologic, and virologic assays, including quantification of serum HCV RNA by TaqMan, HCV genotype, detection of miR-122 by real time PCR, reverse-transcriptase reaction and PCR amplification for the analysis of the HCV quasispecies, and molecular cloning and sequencing, are provided in SI Materials and Methods.
Analysis of HCV Quasispecies.
Details on the methods used to calculate the amino acid genetic diversity, HCV compartmentalization, and selection analysis are provided in SI Materials and Methods.
Statistical Analysis.
Differences in the levels of HCV RNA in serum and in different areas of the liver of patients with and without HCC were assessed using the t test for samples with unequal variances. Details are given in SI Materials and Methods.
SI Materials and Methods
Patients.
Multiple liver specimens and serum were obtained from eight well-characterized patients with HCV-associated HCC who underwent OLT (six patients) or partial hepatectomy (two patients). As a control group, we studied four patients with non-HCC cirrhosis who underwent OLT for end-stage non-HCC cirrhosis associated with HCV. All patients were negative for serologic markers of active infection with hepatitis B virus, hepatitis D virus, or HIV type 1. Demographic, clinical, virologic, and histopathological features of the 12 patients are presented in Table S1.
Study Design and Sample Collection.
To investigate the levels of HCV RNA in serum and liver of patients with HCV-associated HCC, we collected up to 17 liver biopsy specimens from the same liver of each patient with HCV-associated HCC in the four directions, termed for simplicity north (N), south (S), east (E), and west (W), relative to the center of the tumor (Fig. 1A), along with serum samples at the time of OLT. The study design included five liver specimens from the tumor, including one from the center (region A) and four from the periphery of the tumor (region B: N, S, E, W), and 12 liver specimens from the surrounding nontumorous tissue, including four specimens from the immediate perilesional area (region C: N, S, E, W), four taken 2–3 cm from the tumor margin (region D: N, S, E, W), and four taken from the edge of the liver (region E: N, S, E, W) (Fig. 1A). In some cases, owing to the location of the tumor, collection of nontumorous liver specimens at all distances and directions from the center of the tumor was not possible.
Out of a total of 115 liver specimens collected from the eight patients with HCV-associated HCC, 6 were not used because of the histopathology showing a mixed population of tumor and nontumor hepatocytes. The remaining 109, including 34 from tumors and 75 from nontumorous tissue, were analyzed, with an average of 13 liver specimens per patient. Four liver specimens were collected from each cirrhotic liver, two from the right lobe (R1 and R2) and two from the left lobe (L1 and L2) (Fig. 1B), with the exception of patient 11, for whom only three liver specimens were studied, for a total of 15 liver specimens.
To investigate the relationship between intrahepatic HCV RNA levels and expression of miR-122, for each of the eight patients with HCC, four liver specimens, including the center and periphery of the tumor, the perilesional area, and the most distant nontumorous area, were tested for miR-122 expression by real-time PCR.
Liver Pathology.
Each liver specimen was divided into two pieces. One piece was snap-frozen and stored at −80 °C for molecular studies, and the other was formalin-fixed and paraffin-embedded (FFPE) for pathological examination by two expert hepatopathologists (D.E.K. and S.G.). When FFPE sections obtained from the tumor or the perilesional area showed a mixed population of tumor and nontumor hepatocytes, the corresponding frozen liver specimens were excluded from the study. For each liver specimen, activity grade and stage of fibrosis were established according to the Ishak scoring system (45). The grade of tumor differentiation was evaluated according to the Edmondson and Steiner grading system (46), and the proportion of proliferating malignant cells was measured by staining of MIB1 (47).
Analysis of HCV Quasispecies in Serum and Liver of Patients with HCV-Associated HCC and Non-HCC Cirrhosis.
To investigate whether the composition and distribution of the HCV quasispecies varied among different compartments, we analyzed for each patient with HCC serum and four liver specimens, including two from the tumor (A and B) and two from the surrounding nontumorous tissue, comprising the perilesional area (C) and the most distant nontumorous area (E) along a single direction. As controls, we studied serum samples and four liver specimens for each of the four patients with non-HCC cirrhosis, with the exception of patient 11, in whom only three liver specimens were studied. The number of viral variants and the genetic distance among different variants (i.e., genetic diversity), in parallel with the statistical analysis of quasispecies compartmentalization, were performed by examining viral sequences from the envelope genes [envelope glycoproteins 1 (E1) and 2 (E2)], inclusive of the HVR1 (5), in 57 liver specimens. A total of 2,097 molecular clones were sequenced, each 528 nt long, with a mean of 36 per sample. A total of 57 specimens, including 38 from the eight patients with HCC (15 tumorous specimens, 16 nontumorous tissue specimens, and 7 serum samples), and 19 from the controls (including 15 liver specimens and 4 serum samples) were examined for the study of viral quasispecies.
RNA Extraction and Quantification of Liver and Serum HCV RNA by Real-Time PCR.
Total RNA was extracted from stored frozen liver specimens using TRIzol reagent (Invitrogen) according to the manufacturer’s recommendations, and RNA quality and integrity were assessed using the RNA 6000 Nano assay on an Agilent 2100 Bioanalyzer. For the quantification of serum HCV RNA, total RNA was extracted from 140 µL of serum (or plasma) using the QIAamp Viral RNA Mini Kit (Qiagen), and total RNA was eluted in 60 µL. For analysis of the HCV quasispecies in serum, total RNA was extracted from 100 µL of serum using TRIzol reagent (Invitrogen), as described previously (5). HCV RNA levels in liver tissue were quantified using TaqMan real-time PCR, as described previously (48). The primers and probe were derived from the highly conserved 5′ UTR of the HCV genome. In each reaction, 50 ng of total liver RNA was tested; results were expressed as log10 IU per nanogram, based on the World Health Organization’s 96/790 reference standard (48). A standard curve comprising a 6-log dynamic range was constructed with the OptiQuant HCV RNA nucleic acid reference panel (Acrometrix) (48). Levels of serum HCV RNA were measured by commercial assays (COBAS Amplicor HCV 2.0 and COBAS Amplicor HCV Monitor 2.0; Roche Molecular Systems).
Detection of miR-122 by Real-Time PCR.
Detection of miR-122 was performed following the method described by Lanford et al. (31) with minor modifications. In brief, high-quality microRNA was extracted from liver tissue using the miRNeasy Mini Kit (Qiagen). Then 50 ng of total miRNA was transcribed with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems; 4366596) and hsa-miR-122 Assay Kit (Applied Biosystems; 4427975), which supplies reverse-transcriptase and PCR primers and probes, following the manufacturer’s recommendations. TaqMan real-time PCR was carried out in 20-µL reactions containing TaqMan 2× Universal PCR Master Mix, AmpErase UNG (Applied Biosystems; 4324018), and the PCR primers and probe as described above. The cycling conditions were 10 min at 95 °C for enzyme activation, followed by 45 cycles of 15 s at 95 °C and 1 min at 60 °C. miR-122 quantity was estimated through comparison with an 8-log10 serial-dilution line of a synthetic miR-122 mimic standard (Dharmacon; C300591-05).
Reverse-Transcription PCR for Analysis of the HCV Quasispecies.
Total liver RNA and RNA extracted from 100 µL of serum were reverse-transcribed in 20 µL, and the resulting cDNA was amplified using one set of nested primers from the E1/E2 region, including HVR1 (4). The sensitivity, specificity, and details of our nested PCR assay have been reported previously (4). Appropriate precautions were taken to reduce the risk of contamination (4). The HCV genotype was determined by sequence analysis of part of the E1 gene, as reported previously (4).
Molecular Cloning and Sequencing.
The PCR products obtained from the E1 and E2 genes were purified using the QIAquick Gel Extraction Kit (Qiagen), cloned into the pCR2.1-TOPO vector (Life Technologies), and transformed into TOP10 competent cells (Life Technologies). Plasmid DNA was extracted with the QIAprep Spin Miniprep Kit (Qiagen) and sequenced with an Applied Biosystems 3730XL automated DNA sequencer using a modified Sanger method.
DNA Sequence Alignment and Translation to Amino Acid Sequences.
Complete sequences of the E1/E2 region, including HVR1, for all samples from each patient were aligned using ClustalW2 (49) and subsequently edited manually. These alignments were then partitioned into the HVR1 and surrounding E1/E2 regions. Amino acid sequences were inferred by automated translation of HVR1 and E1/E2 sequences.
Genetic Diversity.
Genetic diversity was assessed by analyzing 176 amino acids and calculated using the Hamming distance (50), defined as the number of amino acid differences between two sequences. The mean Hamming distance, defined the average of the values obtained for all sequence pairs derived from a single sample, was calculated separately for the HVR1 (27 amino acids) and on the entire sequence outside the HVR1 (the E1/E2 region; 149 amino acids). Data for all available samples within each liver compartment were averaged. The center of the tumor for patient P4 was not included in the analysis of genetic diversity, because PCR amplification using the set of primers derived from the E1/E2 region turned out to be negative.
HCV Compartmentalization.
Genetic distances within and between different liver regions were calculated using the Dayhoff substitution model (51). For each patient, a matrix of pairwise genetic distances between HCV found in liver regions and in serum was calculated using MEGA version 5.2.2 (52), based on the alignment of full-length amino acid sequences spanning the E1/E2 region, inclusive of the HVR1. To test for HCV compartmentalization, a parallel matrix of group membership was constructed using values of 0 for distances between sequences from the same region and 1 for sequences from different regions.
To determine whether sequences from a given compartment (tumor, nontumor, or serum) were genetically closer to each other than to sequences from other compartments, the correlation between the matrix of genetic distances and the matrix of group membership were evaluated using the Mantel test (32) with 999 permutations. This test was performed using the mantel.randtest program from the ade4 package (53) in R (R Development Core Team, www.R-project.org).
Selection Analysis.
Selection analysis was done using complete sequence alignments, 528 nt long (176 codons). These alignments were inspected and corrected manually to maintain the correct reading frame. Sequences with premature stop codons were removed. For each codon, estimates of the numbers of inferred synonymous (s) and nonsynonymous (n) substitutions were calculated, along with the numbers of sites that were estimated to be synonymous (S) and nonsyonymous (N). Sites within codons were defined as synonymous or silent when a substitution would not result in an amino acid change, and as nonsynonymous when a substitution would induce an amino acid change. These estimates were produced using joint maximum likelihood (ML) reconstructions of ancestral states under the Muse–Gaut model of codon substitution (54) and the Felsenstein 1981 model of nucleotide substitution (55). To estimate ML values, a neighbor-joining (56) tree topology was computed automatically. ML computations of dN and dS were performed using the HyPhy software package (57) as implemented in MEGA version 6.0.6 (58).
Statistical Analysis.
Levels of HCV RNA were measured in different areas of the liver containing HCC (regions A, B, C, D, and E in the four directions) of eight well-characterized patients with HCV-associated HCC. In each patient, multiple measurements of the same region were averaged, then the overall mean HCV RNA concentration in each liver region was calculated. HCV RNA concentrations of tumor and adjacent nontumor regions (A vs. C and B vs. C) were statistically compared using the t test for samples with unequal variances. As controls, HCV RNA levels were measured in serum samples and liver specimens of four patients with chronic HCV infection with non-HCC cirrhosis, using four samples for each patient, two from the right lobe (R1 and R2) and two from the left lobe (L1 and L2), with the exception of patient 11, for whom only three liver specimens were studied.
Differences in HCV genetic diversity, as well as in the number of viral variants, between patients with HCC and patients with non-HCC cirrhosis were evaluated using the t test for samples with unequal variances. Statistical comparisons between patients were always made using patients as test units. Compartmentalization of the HCV quasispecies was evaluated using the Mantel test (32).
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases, Clinical Center, and National Cancer Institute. G.D. received a grant from Fondazione Banco di Sardegna (739/2011.1045).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KR674246–KR676342).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516879113/-/DCSupplemental.
References
- 1.Lemon SM, Walker C, Alter MJ, Yi MK. Hepatitis C viruses. In: Knipe D, Howley P, editors. Fields Virology. 5th Ed Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 2.Martell M, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: Quasispecies nature of HCV genome distribution. J Virol. 1992;66(5):3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farci P. New insights into the HCV quasispecies and compartmentalization. Semin Liver Dis. 2011;31(4):356–374. doi: 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- 4.Farci P, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288(5464):339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 5.Farci P, et al. Profibrogenic chemokines and viral evolution predict rapid progression of hepatitis C to cirrhosis. Proc Natl Acad Sci USA. 2012;109(36):14562–14567. doi: 10.1073/pnas.1210592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: A perspective on long-term outcome. Semin Liver Dis. 2000;20(1):17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention Hepatocellular carcinoma— United States, 2001-2006. MMWR Morb Mortal Wkly Rep. 2010;59(17):517–520. [PubMed] [Google Scholar]
- 9.Saito I, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87(17):6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGivern DR, Lemon SM. Virus-specific mechanisms of carcinogenesis in hepatitis C virus-associated liver cancer. Oncogene. 2011;30(17):1969–1983. doi: 10.1038/onc.2010.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Read SA, Douglas MW. Virus-induced inflammation and cancer development. Cancer Lett. 2014;345(2):174–181. doi: 10.1016/j.canlet.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 12.Arvanitakis L, Yaseen N, Sharma S. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J Immunol. 1995;155(3):1047–1056. [PubMed] [Google Scholar]
- 13.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 14.Sheahan TP, Rice CM. Single cell analysis of HCV-infected patient hepatocytes: The science is no longer science fiction. Gastroenterology. 2013;145(6):1199–1202. doi: 10.1053/j.gastro.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber MA, et al. Detection of replicative hepatitis C virus sequences in hepatocellular carcinoma. Am J Pathol. 1992;141(6):1271–1277. [PMC free article] [PubMed] [Google Scholar]
- 16.Haruna Y, et al. Expression of hepatitis C virus in hepatocellular carcinoma. Cancer. 1994;73(9):2253–2258. doi: 10.1002/1097-0142(19940501)73:9<2253::aid-cncr2820730904>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, Hayashi H, Itoh Y, Asano T, Isono K. Detection of minus-strand hepatitis C virus RNA in tumor tissues of hepatocellular carcinoma. Cancer. 1994;73(1):48–52. doi: 10.1002/1097-0142(19940101)73:1<48::aid-cncr2820730110>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 18.Dash S, et al. HCV RNA levels in hepatocellular carcinomas and adjacent non-tumorous livers. J Virol Methods. 2000;90(1):15–23. doi: 10.1016/s0166-0934(00)00199-3. [DOI] [PubMed] [Google Scholar]
- 19.Horiike N, et al. Hepatitis C virus plus- and minus-strand RNA in hepatocellular carcinoma and adjoining nontumorous liver. J Med Virol. 1993;41(4):312–315. doi: 10.1002/jmv.1890410410. [DOI] [PubMed] [Google Scholar]
- 20.Sobesky R, et al. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology. 2007;46(6):1704–1712. doi: 10.1002/hep.21898. [DOI] [PubMed] [Google Scholar]
- 21.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 22.Masaki T, et al. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015;17(2):217–228. doi: 10.1016/j.chom.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gramantieri L, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 24.Kutay H, et al. Down-regulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 26.Spaniel C, et al. microRNA-122 abundance in hepatocellular carcinoma and non-tumor liver tissue from Japanese patients with persistent HCV versus HBV infection. PLoS One. 2013;8(10):e76867. doi: 10.1371/journal.pone.0076867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakatsanis A, et al. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52(4):297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 28.Rüster B, et al. Comparative sequence analysis of the core- and NS5-region of hepatitis C virus from tumor and adjacent non-tumor tissue. J Med Virol. 2001;63(2):128–134. [PubMed] [Google Scholar]
- 29.Saito S, et al. Comparison of hypervariable regions (HVR1 and HVR2) in positive- and negative-stranded hepatitis C virus RNA in cancerous and non-cancerous liver tissue, peripheral blood mononuclear cells and serum from a patient with hepatocellular carcinoma. Int J Cancer. 1996;67(2):199–203. doi: 10.1002/(SICI)1097-0215(19960717)67:2<199::AID-IJC9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 30.Young KC, et al. Variation of hepatitis C virus load, hypervariable region 1 quasispecies and CD81 hepatocyte expression in hepatocellular carcinoma and adjacent non-cancerous liver. J Med Virol. 2002;68(2):188–196. doi: 10.1002/jmv.10195. [DOI] [PubMed] [Google Scholar]
- 31.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209–220. [PubMed] [Google Scholar]
- 33.Gosálvez J, et al. Relative quantification and mapping of hepatitis C virus by in situ hybridization and digital image analysis. Hepatology. 1998;27(5):1428–1434. doi: 10.1002/hep.510270534. [DOI] [PubMed] [Google Scholar]
- 34.Lau GK, et al. Hepatic expression of hepatitis C virus RNA in chronic hepatitis C: A study by in situ reverse-transcription polymerase chain reaction. Hepatology. 1996;23(6):1318–1323. doi: 10.1002/hep.510230604. [DOI] [PubMed] [Google Scholar]
- 35.Liang Y, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137(4):1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 36.Kandathil AJ, et al. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology. 2013;145(6):1404–1413. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiffler JD, et al. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4(8):e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasui T, Shimomura H, Tsuji H, Wato M, Tsuji T. Quantitation of hepatitis C virus RNA in liver tissue as a predictive marker of the response to interferon therapy in chronic hepatitis C. Acta Med Okayama. 1994;48(3):151–157. doi: 10.18926/AMO/31121. [DOI] [PubMed] [Google Scholar]
- 39.Idrovo V, et al. Hepatitis C virus RNA quantification in right and left lobes of the liver in patients with chronic hepatitis C. J Viral Hepat. 1996;3(5):239–246. doi: 10.1111/j.1365-2893.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 40.Terrault NA, et al. Hepatitis C virus: Quantitation and distribution in liver. J Med Virol. 1997;51(3):217–224. [PubMed] [Google Scholar]
- 41.Rüster B, Zeuzem S, Roth WK. Hepatitis C virus sequences encoding truncated core proteins detected in a hepatocellular carcinoma. Biochem Biophys Res Commun. 1996;219(3):911–915. doi: 10.1006/bbrc.1996.0340. [DOI] [PubMed] [Google Scholar]
- 42.Schramm F, et al. Frequent compartmentalization of hepatitis C virus with leukocyte-related amino acids in the setting of liver transplantation. J Infect Dis. 2008;198(11):1656–1666. doi: 10.1086/592986. [DOI] [PubMed] [Google Scholar]
- 43.Zehender G, et al. Compartmentalization of hepatitis C virus quasispecies in blood mononuclear cells of patients with mixed cryoglobulinemic syndrome. J Virol. 2005;79(14):9145–9156. doi: 10.1128/JVI.79.14.9145-9156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishman SL, et al. Molecular and bioinformatic evidence of hepatitis C virus evolution in brain. J Infect Dis. 2008;197(4):597–607. doi: 10.1086/526519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishak K, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 46.Edmondson HA, Steiner PE. Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer. 1954;7(3):462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 47.Ekholm M, et al. Immunohistochemical assessment of Ki67 with antibodies SP6 and MIB1 in primary breast cancer: A comparison of prognostic value and reproducibility. Histopathology. 2014;65(2):252–260. doi: 10.1111/his.12392. [DOI] [PubMed] [Google Scholar]
- 48.Engle RE, Russell RS, Purcell RH, Bukh J. Development of a TaqMan assay for the six major genotypes of hepatitis C virus: Comparison with commercial assays. J Med Virol. 2008;80(1):72–79. doi: 10.1002/jmv.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamming WR. Coding and Information Theory. 2nd Ed Prentice-Hall; Englewood Cliffs, NJ: 1986. [Google Scholar]
- 51.Schwartz RM, Dayhoff MO. Matrices for detecting distant relationships. In: Dayhoff MO, editor. Atlas of Protein Sequence and Structure. 3rd Ed. Vol 5. National Biomedical Research Foundation; Washington, DC: 1978. pp. 353–358. [Google Scholar]
- 52.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dray S, Dufour AB. The ade4 package: Implementing the duality diagram for ecologists. J Stat Softw. 2007;22(4):1–20. [Google Scholar]
- 54.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11(5):715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- 55.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 56.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 57.Kosakovsky Pond SL, Frost SD. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 58.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]