Significance
Many long noncoding RNAs (lncRNAs) play critical roles in tumor development. Here we show that an lncRNA termed UPAT [ubiquitin-like plant homeodomain and really interesting new gene finger domain-containing protein 1 (UHRF1) Protein Associated Transcript] is required for the tumorigenicity of colorectal cancer cells. UPAT interacts with and stabilizes the epigenetic factor UHRF1 by interfering with its ubiquitination and degradation. Furthermore, the UHRF1–UPAT axis up-regulates Stearoyl-CoA desaturase 1 and Sprouty 4, which are required for the survival of colon tumor cells. Our study provides evidence for an lncRNA that regulates protein ubiquitination and degradation and thereby plays a critical role in the survival and tumorigenicity of tumor cells. Our results suggest that UPAT and UHRF1 may be promising molecular targets for the therapy of colon cancer.
Keywords: long noncoding RNA, UPAT, UHRF1, ubiquitination, tumorigenicity
Abstract
Many long noncoding RNAs (lncRNAs) are reported to be dysregulated in human cancers and play critical roles in tumor development and progression. Furthermore, it has been reported that many lncRNAs regulate gene expression by recruiting chromatin remodeling complexes to specific genomic loci or by controlling transcriptional or posttranscriptional processes. Here we show that an lncRNA termed UPAT [ubiquitin-like plant homeodomain (PHD) and really interesting new gene (RING) finger domain-containing protein 1 (UHRF1) Protein Associated Transcript] is required for the survival and tumorigenicity of colorectal cancer cells. UPAT interacts with and stabilizes the epigenetic factor UHRF1 by interfering with its β-transducin repeat-containing protein (TrCP)–mediated ubiquitination. Furthermore, we demonstrate that UHRF1 up-regulates Stearoyl-CoA desaturase 1 and Sprouty 4, which are required for the survival of colon tumor cells. Our study provides evidence for an lncRNA that regulates protein ubiquitination and degradation and thereby plays a critical role in the survival and tumorigenicity of tumor cells. Our results suggest that UPAT and UHRF1 may be promising molecular targets for the therapy of colon cancer.
Among the RNA products transcribed from the mammalian genome are numerous long noncoding RNAs (lncRNAs)—that is, RNAs longer than 200 nucleotides with little or no protein-coding potential (1, 2). Many lncRNAs are expressed in a developmentally regulated and cell type-dependent manner (3, 4). Increasing evidence suggests that lncRNAs play critical roles in a diverse set of biological processes, including proliferation, differentiation, embryogenesis, neurogenesis, and stem cell pluripotency (5, 6).
It has been reported that many lncRNAs regulate gene expression by recruiting chromatin remodeling complexes to specific genomic regions (2). It has also been shown that many lncRNAs regulate transcription by modulating the activity of transcriptional regulators (1, 6–8). lncRNAs also regulate various posttranscriptional processes, including splicing, transport, translation, and degradation of mRNA. Furthermore, recent studies have shown that a number of lncRNAs play critical roles in tumor development and progression.
UHRF1 [ubiquitin-like plant homeodomain (PHD) and really interesting new gene (RING) finger domain-containing protein 1] is an epigenetic factor that consists of multiple domains (9). UHRF1 regulates transcription by regulating DNA methylation and histone modification. UHRF1 also possesses E3 ubiquitin ligase activity and ubiquitinates histones and DNA methyltransferase 1 (DNMT1), thereby regulating the chromatin structure and stability of DNMT1 (10, 11). UHRF1 plays key roles in multiple biological processes, including proliferation and development. Furthermore, UHRF1 is overexpressed in various tumors, including colon, breast, bladder, prostate, and lung cancers, and plays a critical role in the proliferation and survival of tumor cells (9).
In the present study, we attempted to identify lncRNAs critical for the tumorigenicity of colon tumor cells by performing RNA-sequencing (RNA-seq) analysis of the colon cancer cell line CCSC#P and a subclone that exhibits drastically decreased tumorigenicity, CCSC#11. We have found that an lncRNA termed UPAT (UHRF1 Protein Associated Transcript) is down-regulated in CCSC#11 and is required for the tumorigenicity of CCSC#P. We further show that UPAT interacts with UHRF1. Moreover, we show that UPAT interferes with the ubiquitination and degradation of UHRF1 and thereby plays a critical role in determining the survival and tumorigenicity of colorectal tumor cells. We also demonstrate that UHRF1 is required for the up-regulation of Stearoyl-CoA desaturase 1 (SCD1) and Sprouty 4 (SPRY4), which play critical roles in the survival of colon tumor cells.
Results
UPAT Is Required for the Tumorigenicity of Colon Tumor Cells.
We established single cell-derived subclones from the colon cancer cell line CCSC#P by limiting dilution and examined their tumorigenicity. Out of 34 clones examined, two of these, termed CCSC#11 and #22, exhibited drastically decreased tumorigenicity compared with CCSC#P when implanted s.c. into immunocompromised mice (Fig. 1A). To investigate the mechanisms underlying this decreased tumorigenicity, we performed RNA-seq analysis of CCSC#P and #11 cells (Dataset S1). We selected two lncRNAs, NR_015379 and NR_002773, down-regulated in CCSC#11 cells (Fig. S1A) and examined the effects of knocking down these genes on the survival of HCT116 and CCSC#P cells. The most significant reduction in the viability of HCT116 and CCSC#P cells was achieved by knockdown of an lncRNA, NR_002773, termed UPAT (Fig. S1 B and C), which is encoded by the pseudogene of the amine oxidase copper containing 3 (AOC3) gene (nucleotides 239–1732; 94% homology) (12–14) (Fig. S1 D–F). UPAT is also homologous to AOC2 (239–1261; homology 76%) but not to AOC1. Quantitative (q)RT-PCR analysis revealed that UPAT expression was up-regulated in highly tumorigenic colon cancer cell lines compared with weakly tumorigenic colon cancer cell lines and normal cell lines (Fig. S1G). Subcellular fractionation and qRT-PCR analysis revealed that UPAT was present in the nucleus (Fig. S1H).
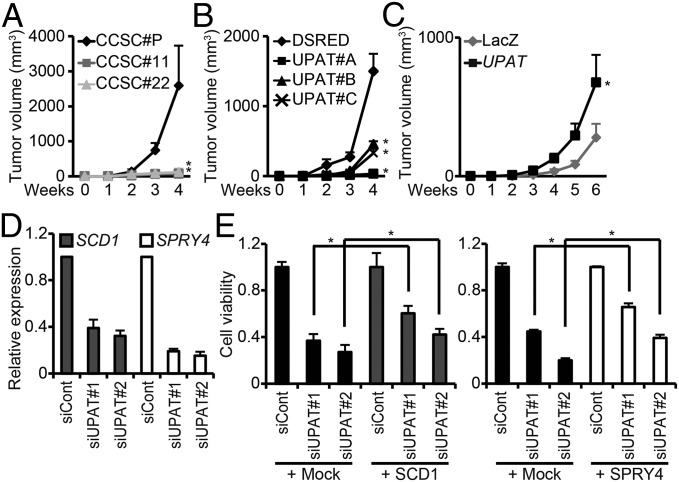
Fig. 1.
UPAT is required for the tumorigenicity of colon cancer cells. (A) CCSC#P, CCSC#11, or CCSC#22 cells were injected s.c. into nude mice and assessed for tumor growth. Results are expressed as the mean ± SEM (n = 6). *P < 0.05. (B) CCSC#P cells infected with a lentivirus expressing an shUPAT were injected into nude mice. Results are expressed as the mean ± SEM (n = 6). *P < 0.05. (C) CCSC#11 cells infected with a lentivirus expressing UPAT were injected into nude mice. Results are expressed as the mean ± SEM (n = 6). *P < 0.05. (D) qRT-PCR analysis of SCD1 and SPRY4 expression in HCT116 cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (E) Viability of HCT116 cells transfected with SCD1 (Left) or SPRY4 (Right) and/or siRNA targeting UPAT was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). *P < 0.05.
Fig. S1.
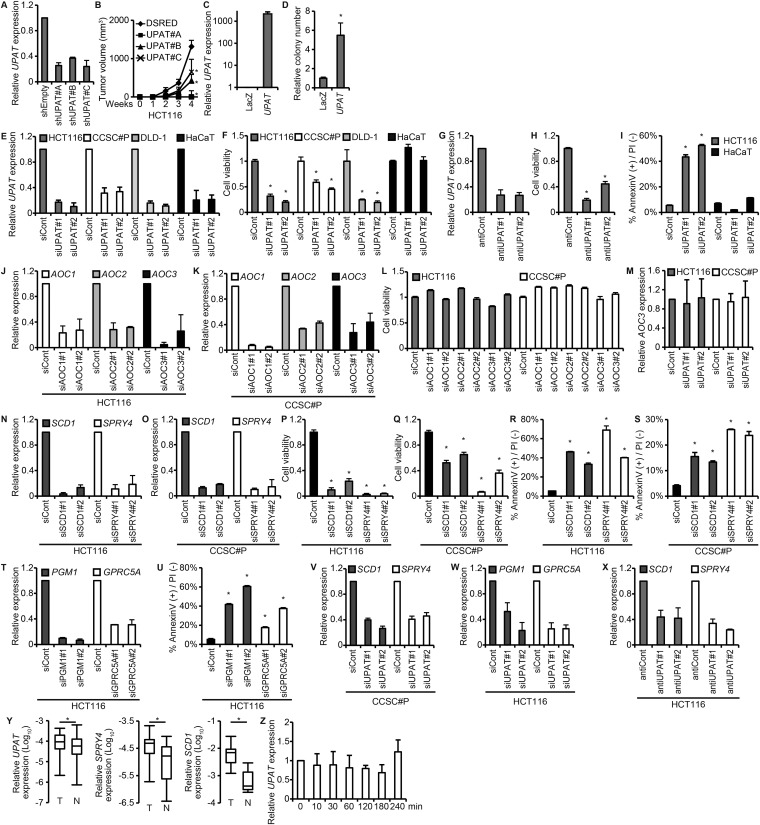
UPAT (NR_002773) is required for the survival of colon cancer cells. (A) qRT-PCR analysis of the expression of NR_015379 and UPAT (NR_002773) in CCSC#P and CCSC#11 cells. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (B) CCSC#P and HCT116 cells were transfected with siRNAs targeting the indicated genes and subjected to qRT-PCR analysis. Results are expressed as the mean ± SEM (n = 3). (C) Viability of CCSC#P and HCT116 cells transfected with siRNAs targeting NR_015379 and UPAT (NR_002773) was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). (D–F) UPAT is homologous to AOC2 and AOC3 but not to AOC1. (D and E) Alignment between UPAT and AOC2 (D) or AOC3 (E). (F) Phylogenetic trees of the AOC family of genes and UPAT. (G) Expression levels of UPAT in colon cancer and normal cell lines. UPAT expression was quantitated as the percentage relative to β-actin mRNA. (H) UPAT is localized in the nucleus. HCT116 cells were subjected to subcellular fractionation, and the amounts of UPAT in each fraction were evaluated by qRT-PCR. α-tubulin and GAPDH mRNA was used as a marker specific for the cytoplasm. Lamin A/C and U1 small nuclear RNA was used as a nuclear marker. Results are expressed as the mean ± SEM (n = 3).
To clarify the significance of UPAT in colorectal tumorigenesis, we infected HCT116 and CCSC#P cells with a lentivirus expressing an shRNA targeting UPAT (shUPAT) and examined their tumorigenicity. When transplanted into nude mice, the growth of these cells was significantly retarded compared with cells infected with a control lentivirus (Fig. 1B and Fig. S2 A and B). Moreover, we found that CCSC#11 cells infected with a lentivirus expressing UPAT exhibited increased tumorigenicity and colony formation in soft agar compared with CCSC#11 cells infected with control virus (Fig. 1C and Fig. S2 C and D). In addition, Cell Titer-Glo assays revealed that knockdown of UPAT by siRNA (siUPAT) caused a significant reduction in the growth of HCT116, CCSC#P, and DLD-1 cells but not of normal keratinocyte HaCaT cells in vitro (Fig. S2 E and F). Knockdown of UPAT by antisense oligonucleotide also resulted in the decreased growth of HCT116 cells (Fig. S2 G and H). Furthermore, Annexin V assays showed that knockdown of UPAT induced apoptosis of HCT116 but not HaCaT cells (Fig. S2 E and I). In contrast, knockdown of the AOC family of genes, AOC1∼3, did not cause a reduction in the growth of HCT116 and CCSC#P cells (Fig. S2 J–L). Moreover, knockdown of UPAT did not affect the expression of AOC3 in either HCT116 or CCSC#P cells (Fig. S2M). These results suggest that UPAT, but not the AOC family of genes, may be required for the survival and tumorigenicity of colorectal cancer cells.
Fig. S2.
UPAT is required for the tumorigenicity and the expression of RAS-, CDH1-, and hypoxia-related genes in colon cancer cells. (A) qRT-PCR analysis of UPAT expression in HCT116 cells infected with a lentivirus harboring an shUPAT. Results are expressed as the mean ± SEM (n = 3). (B) HCT116 cells infected with a lentivirus expressing an shUPAT were injected into nude mice. Results are expressed as the mean ± SEM (n = 6). *P < 0.05. (C) qRT-PCR analysis of UPAT expression in CCSC#11 cells infected with a lentivirus expressing LacZ or UPAT. Results are expressed as the mean ± SEM (n = 3). (D) CCSC#11 cells were infected with a lentivirus expressing LacZ or UPAT and subjected to colony formation assays in soft agar. The number of colonies is shown as the relative number. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (E) qRT-PCR analysis of UPAT expression in HCT116, CCSC#P, DLD-1, and HaCaT cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (F) Viability of HCT116, CCSC#P, DLD-1, and HaCaT cells transfected with siRNA targeting UPAT was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (G) qRT-PCR analysis of UPAT expression in HCT116 cells transfected with an antisense oligonucleotide targeting UPAT (antiUPAT). Results are expressed as the mean ± SEM (n = 3). (H) Viability of HCT116 cells transfected with an antisense oligonucleotide targeting UPAT (antiUPAT) was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (I) Annexin V assays were performed with HCT116 and HaCaT cells that had been transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (J–L) The AOC family of genes, AOC1∼3, are not required for the survival of colon cancer cells. (J and K) qRT-PCR analysis of AOC1, AOC2, and AOC3 expression in HCT116 (J) and CCSC#P (K) cells transfected with siRNA targeting AOC1, AOC2, or AOC3. Results are expressed as the mean ± SEM (n = 3). (L) Viability of HCT116 and CCSC#P cells transfected with siRNAs targeting AOC1, AOC2, or AOC3 was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). (M) qRT-PCR analysis of AOC3 expression in HCT116 cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (N and O) HCT116 (N) and CCDC#P (O) cells were transfected with siRNA targeting SCD1 or SPRY4 and subjected to qRT-PCR analysis. Results are expressed as the mean ± SEM (n = 3). (P and Q) Viability of HCT116 (P) and CCDC#P (Q) cells transfected with siRNA targeting SCD1 or SPRY4 was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (R and S) Annexin V assays were performed with HCT116 (R) and CCSC#P (S) cells that had been transfected with siRNA targeting SCD1 or SPRY4. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (T) HCT116 cells were transfected with siRNA targeting PGM1 or GPRC5A and subjected to qRT-PCR analysis. Results are expressed as the mean ± SEM (n = 3). (U) Annexin V assays were performed with HCT116 cells that had been transfected with siRNA targeting PGM1 or GPRC5A. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (V) qRT-PCR analysis of SCD1 and SPRY4 expression in CCSC#P cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (W) qRT-PCR analysis of PGM1 and GPRC5A expression in HCT116 cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (X) qRT-PCR analysis of SCD1 and SPRY4 expression in HCT116 cells transfected with an antisense oligonucleotide targeting UPAT (antiUPAT). Results are expressed as the mean ± SEM (n = 3). (Y) qRT-PCR analysis of UPAT, SPRY4, and SCD1 expression in human colon cancer tissues (n = 17 pairs). *P < 0.05. (Z) qRT-PCR analysis of UPAT expression in HaCaT cells that had been cultured in the presence or absence of EGF for the indicated times. Results are expressed as the mean ± SEM (n = 3).
To study the role of UPAT in colorectal cancer cells, we investigated the gene expression profiles of HCT116 cells in which UPAT expression had been suppressed by siRNA. RNA-seq analyses revealed that rat sarcoma viral oncogene homolog (RAS)-, cadherin 1 (CDH1)-, and hypoxia-related genes are regulated in UPAT knockdown cells (Dataset S2). From these genes, we selected 25 genes that were down-regulated > twofold and examined whether knockdown of any of them could cause apoptotic death in HCT116 cells. We found that siRNA knockdown of SCD1, SPRY4, phosphoglucomutase 1 (PGM1), or G protein-coupled receptor, class C, group 5, member A (GPRC5A) resulted in a marked increase in apoptotic cell death (Fig. S2 N–U). We also performed qRT-PCR analyses and confirmed that knockdown of UPAT by siRNA or antisense oligonucleotides resulted in decreased expression of these four genes (Fig. 1D and Fig. S2 V–X). Furthermore, we found that overexpression of either SCD1 or SPRY4 partially restored the growth of HCT116 cells in which UPAT had been knocked down (Fig. 1E). Moreover, we found that UPAT, SPRY4, and SCD1 mRNA expression was higher in colorectal tumors than in adjacent normal tissues (Fig. S2Y). These results suggest that UPAT-mediated up-regulation of these genes is involved in the survival of colon cancer cells. In addition, treatment of HaCaT cells with EGF did not significantly affect the expression levels of UPAT (Fig. S2Z).
UPAT Is Associated with UHRF1 in Colon Cancer Cells.
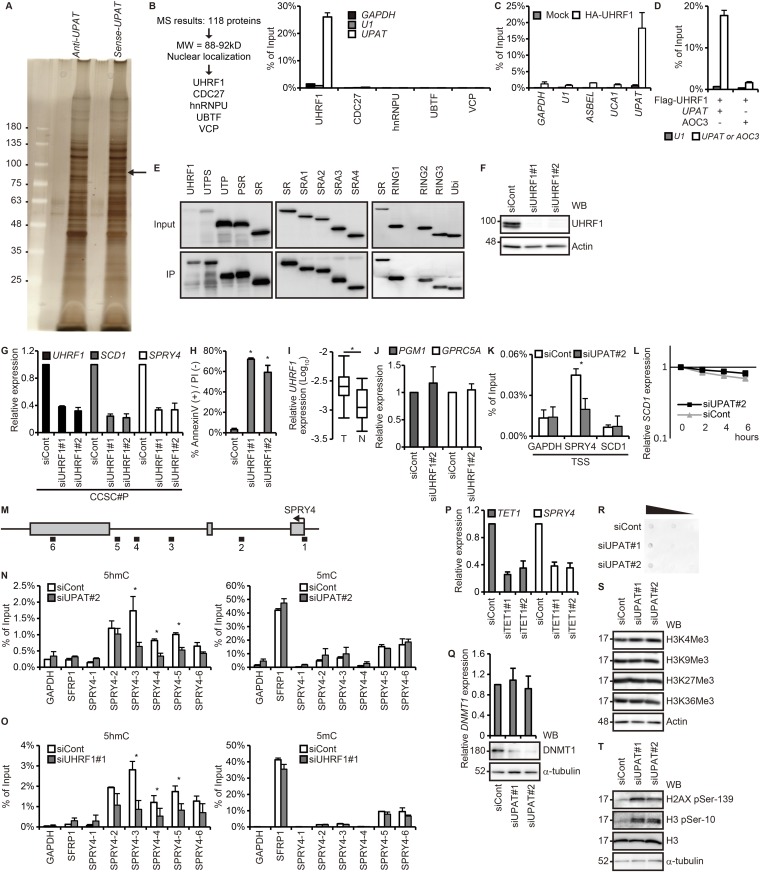
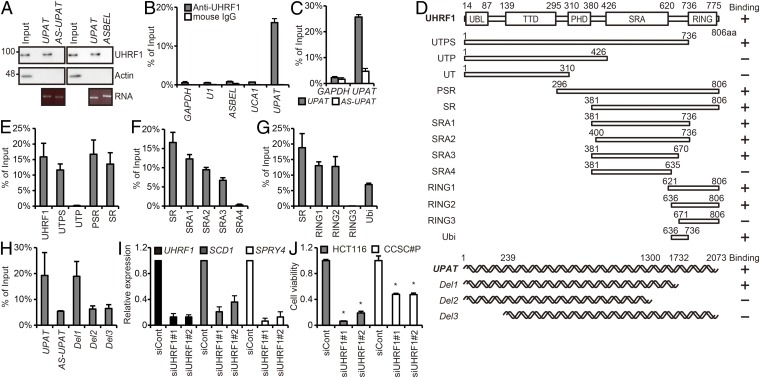
Many lncRNAs have been shown to exert their biological function by forming complexes with proteins (1, 6–8). We therefore performed RNA pull-down assays to identify proteins that potentially could associate with UPAT. Nuclear extracts from HCT116 cells were incubated with biotinylated sense or antisense UPAT RNA generated in vitro, and proteins precipitated with streptavidin beads were resolved by SDS/PAGE. A band that specifically coprecipitated with sense UPAT was excised and subjected to liquid chromatography–mass spectrometry (Fig. S3A and Dataset S3). Among the proteins identified, we selected nuclear proteins whose molecular weight were 88–92 kDa, which narrowed down our screen to five candidate proteins (Fig. S3B). We then examined whether these proteins precipitate with UPAT (RIP analysis) and from this obtained UHRF1. The association of UHRF1 with UPAT sense RNA, but not UPAT antisense or ASBEL [antisense ncRNA in the ANA/BTG3 (three) locus]RNA (15), was also confirmed by immunoblotting analysis with anti-UHRF1 antibody (Fig. 2A). To further verify this result, we performed RNA immunoprecipitation (RIP) analysis with anti-UHRF1 antibody using lysates from HCT116 cells. qRT-PCR analysis of the immunoprecipitates revealed that UHRF1 was associated with endogenous UPAT, but not with GAPDH mRNA, U1 small nuclear RNA, ASBEL, or urothelial cancer associated 1 (UCA1) (16, 17) (Fig. 2B). In addition, RIP analysis with anti-HA antibody using lysates from HCT116 cells transfected with HA-tagged UHRF1 revealed that exogenously expressed UHRF1 was also associated with UPAT but not with antisense UPAT, GAPDH mRNA, U1 small nuclear RNA, ASBEL, UCA1, or AOC3 mRNA (Fig. 2C and Fig. S3 C and D). These results suggest that UPAT is associated with UHRF1 in colon cancer cells.
Fig. S3.
UPAT is associated with UHRF1 in colon cancer cells. (A) Nuclear extracts from HCT116 cells were incubated with biotinylated sense or antisense UPAT, and proteins precipitated with streptavidine beads were resolved by SDS/PAGE followed by silver staining. The band indicated by the arrowhead was excised and subjected to liquid chromatography–mass spectrometry. (B, Left) The procedure for selection of UPAT-binding proteins. (B, Right) Lysates from HCT116 cells transfected with UPAT along with HA-tagged UHRF1, CDC27, hnRNPU, UBTF, or VCP were subjected to immunoprecipitation with anti-HA antibody followed by RT-PCR analysis to detect UPAT mRNA. GAPDH mRNA and U1 small nuclear RNA were used as negative controls. Results are expressed as the mean ± SEM (n = 3). (C) Lysates from HCT116 cells transfected with HA-UHRF1 were subjected to immunoprecipitation with anti-HA antibody followed by RT-PCR analysis to detect UPAT mRNA. GAPDH mRNA, U1 small nuclear RNA, ASBEL, and UCA1 were used as negative controls. Results are expressed as the mean ± SEM (n = 3). (D) Lysates from HCT116 cells transfected with Flag-UHRF1 along with UPAT or AOC3 were subjected to immunoprecipitation with anti-Flag antibody followed by RT-PCR analysis to detect UPAT or AOC3 mRNA. U1 small nuclear RNA was used as a negative control. Results are expressed as the mean ± SEM (n = 3). (E) Lysates from HCT116 cells that had been transfected with UPAT and the indicated HA-UHRF1 (Left) or Flag-UHRF1 (Middle and Right) deletion fragments were subjected to immunoprecipitation with anti-HA (Left) or anti-Flag (Middle and Right) antibody followed by immunoblotting analysis with anti-HA or anti-Flag antibody. (F) Lysates from HCT116 cells transfected with siRNA targeting UHRF1 were subjected to immunoblotting analysis with anti-UHRF1 or anti-Actin antibody. Actin was used as a loading control. (G) qRT-PCR analysis of UHRF1, SCD1, and SPRY4 expression in CCSC#P cells transfected with siRNA targeting UHRF1. Results are expressed as the mean ± SEM (n = 3). (H) Annexin V assays were performed with HCT116 cells that had been transfected with siRNA targeting UHRF1. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (I) qRT-PCR analysis of UHRF1 expression in human colon cancer tissues (n = 17 pairs). *P < 0.05. (J) UHRF1 is not required for the expression of PGM1 and GPRC5A. qRT-PCR analysis of PGM1 and GPRC5A expression in HCT116 cells transfected with siRNA targeting UHRF1. Results are expressed as the mean ± SEM (n = 3). (K) ChIP assays were performed with HCT116 cells transfected with control siRNA or siRNA targeting UPAT using anti-UHRF1 antibody. GAPDH-transcription start site (TSS), SPRY4-TSS, and SCD1-TSS are derived from the TSS of the GAPDH, SPRY4, and SCD1 genes, respectively. GAPDH-TSS was used as a negative control. *P < 0.05. (L) HCT116 cells transfected with control siRNA or siRNA targeting UPAT were treated with Actinomycin D for the indicated times and then subjected to qRT-PCR analysis of SCD1 expression. Results are expressed as the mean ± SEM (n = 3). (M) Schematic representation of the SPRY4 locus. (N and O) hMeDIP–qRT-PCR (Left) or MeDIP–qRT-PCR (Right) assays were performed with the HCT116 cell transfected with control siRNA or siRNA targeting UPAT (N) or UHRF1 (O) using anti-5hmC (Left) or anti-5mC (Right) antibody. SFRP1 was a positive control for 5mC. Results are expressed as the mean ± SEM (n = 2). *P < 0.05. (P) qRT-PCR analysis of TET1 and SPRY4 expression in HCT116 cells transfected with siRNA targeting TET1. Results are expressed as the mean ± SEM (n = 2). (Q, Upper) qRT-PCR analysis of DNMT1 expression in HCT116 cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (Q, Lower) Cell lysates were subjected to immunoblotting analysis with anti-DNMT1 or anti–α-tubulin antibody. α-tubulin was used as a loading control. (R) Dot blot analysis of genomic 5mC in HCT116 cells transfected with siRNA targeting UPAT. (S and T) HCT116 cells transfected with siRNA targeting UPAT were subjected to immunoblotting analysis with the indicated antibodies. Actin and α-tubulin were used as loading controls.
Fig. 2.
UPAT is associated with UHRF1 in colon cancer cells. (A) Nuclear extracts from HCT116 cells were incubated with biotinylated sense, antisense UPAT (Left), or ASBEL (Right) generated in vitro, and proteins were precipitated with streptavidin beads and subjected to immunoblotting analysis with anti-UHRF1 or anti-Actin antibody. AS-UPAT, in vitro-transcribed antisense UPAT. Actin was used as a negative control. (B) Lysates from HCT116 cells were subjected to immunoprecipitation with anti-UHRF1 antibody or anti-mouse IgG antibody followed by qRT-PCR analysis to detect UPAT mRNA. GAPDH mRNA, U1 small nuclear RNA, ASBEL, and UCA1 were used as negative controls. Results are expressed as the mean ± SEM (n = 3). (C) Lysates from HCT116 cells transfected with sense (UPAT) or antisense (AS-UPAT) UPAT and HA-UHRF1 were subjected to immunoprecipitation with anti-HA antibody followed by RT-PCR analysis to detect UPAT mRNA. AS-UPAT and GAPDH were used as negative controls. Results are expressed as the mean ± SEM (n = 3). (D) Schematic representation of the UHRF1 protein (Upper) and UPAT (Lower). Mutants used in RIP (Fig. 2 E–H), immunoprecipitation (Fig. 3E and Fig. S4H), and ubiquitination (Fig. 4A and Fig. S5B) assays are also shown. (E–G) Lysates from HCT116 cells transfected with UPAT along with wild-type, mutant HA-UHRF1 (E), or mutant Flag-UHRF1 (F and G) were subjected to immunoprecipitation with anti-HA (E) or anti-Flag (F and G) antibody followed by RT-PCR analysis to detect UPAT and U1 small nuclear RNA. Results are expressed as the mean ± SEM (n = 3). See also Fig. S3E. (H) Lysates from HCT116 cells transfected with wild-type or mutant UPAT and HA-UHRF1 were subjected to immunoprecipitation with anti-HA antibody followed by RT-PCR analysis to detect UPAT mRNA. Results are expressed as the mean ± SEM (n = 3). (I) qRT-PCR analysis of UHRF1, SCD1, and SPRY4 expression in HCT116 cells transfected with siRNA targeting UHRF1. Results are expressed as the mean ± SEM (n = 3). (J) Viability of HCT116 and CCSC#P cells transfected with siRNA targeting UHRF1 was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). *P < 0.05.
RIP assays using a series of UHRF1 deletion mutants revealed that a small region (amino acids 636–670) was required for the association of UHRF1 with UPAT (Fig. 2 D–G and Fig. S3E). Furthermore, we showed that a fragment consisting of amino acids 636–736 [termed the UBR (UPAT-binding region)] is sufficient to bind UPAT. We also attempted to delineate the region in UPAT that binds UHRF1 and found that both the 5′ and 3′ regions were required (Fig. 2 D and H).
It has been shown that UHRF1 plays a key role in the proliferation and survival of tumor cells (9). Indeed, Cell Titer-Glo assays revealed that knockdown of UHRF1 by siRNA caused a significant reduction in the growth of HCT116 and CCSC#P cells (Fig. 2 I and J and Fig. S3 F and G). Annexin V assays showed that knockdown of UHRF1 resulted in a marked increase in the apoptotic death of HCT116 cells (Fig. S3H). Moreover, we found that UHRF1 mRNA expression was higher in colorectal tumors than in adjacent normal tissues (Fig. S3I). These results raise the possibility that UHRF1 associated with UPAT is involved in the growth and survival of colon tumor cells.
UPAT and UHRF1 Epigenetically Up-Regulate SPRY4.
We next examined whether UHRF1 is involved in the up-regulation of SCD1, SPRY4, PGM1, and/or GPRC5A in HCT116 cells. We found that knockdown of UHRF1 resulted in decreased expression of the SCD1 and SPRY4 mRNAs (Fig. 2I and Fig. S3G). By contrast, knockdown of UHRF1 did not affect the expression of PGM1 or GPRC5A (Fig. S3J). Chromatin immunoprecipitation (ChIP) assays using anti-UHRF1 antibody revealed that UHRF1 was associated with the SPRY4 but not the SCD1 promoter region (Fig. S3K). Furthermore, knockdown of UPAT significantly reduced the association of UHRF1 with the SPRY4 promoter region (Fig. S3K). These results suggest that UHRF1 directly transactivates SPRY4 and that UPAT is required for this transactivation. By contrast, SCD1 may not be a direct target of UHRF1 but rather is up-regulated indirectly downstream of UHRF1 and UPAT. In addition, we found that knockdown of UPAT did not affect the stability of SCD1 mRNA (Fig. S3L).
Covalent modifications of DNA and histones can influence transcriptional activity and thereby regulate cell proliferation, survival, and tumorigenesis (18, 19). To elucidate the mechanisms underlying UHRF1-mediated transactivation of SPRY4, we performed (hydroxymethylated) methylated DNA immunoprecipitation [(h)MeDIP] analyses using anti–5-hydroxymethylcytosine (5hmC) or anti–5-methylcytosine (mC) antibody. We detected 5hmC and 5mC in the intragenic regions of the SPRY4 locus (Fig. S3 M–O). Knockdown of either UHRF1 or UPAT resulted in decreases in 5hmC levels but not in 5mC levels in the SPRY4 locus. Furthermore, knockdown of tet methylcytosine dioxygenase 1 (TET1), an enzyme that catalyzes the oxidation of 5mC to 5hmC, led to a decrease in the expression of SPRY4 (Fig. S3P). These results suggest that TET1-mediated methyl hydroxylation of the SPRY4 gene is required for the expression of SPRY4.
It has recently been reported that knockdown of UHRF1 leads to a dramatic decrease in DNMT1 (20). Indeed, qRT-PCR and immunoblotting analyses revealed that knockdown of UPAT resulted in a drastic decrease in the levels of DNMT1 protein, but not mRNA, in HCT116 cells (Fig. S3Q). Consistent with this result, dot blot analysis showed that UPAT knockdown caused a decrease in the levels of 5mC (Fig. S3R).
We also investigated whether UPAT regulates histone modification but found that knockdown of UPAT did not affect the levels of H3K4Me3, H3K9Me3, H3K27Me3, or H3K36Me3 (Fig. S3S). On the other hand, we found that knockdown of UPAT led to increases in the phosphorylation of Histone H2AX-Ser-216 and H3-Ser10, which are markers of the DNA damage response (Fig. S3T). This is consistent with a previous report showing that UHRF1 depletion caused the activation of the DNA damage response pathway (21).
UPAT Interferes with β-TrCP1– and β-TrCP2–Mediated Ubiquitination and Degradation of UHRF1.
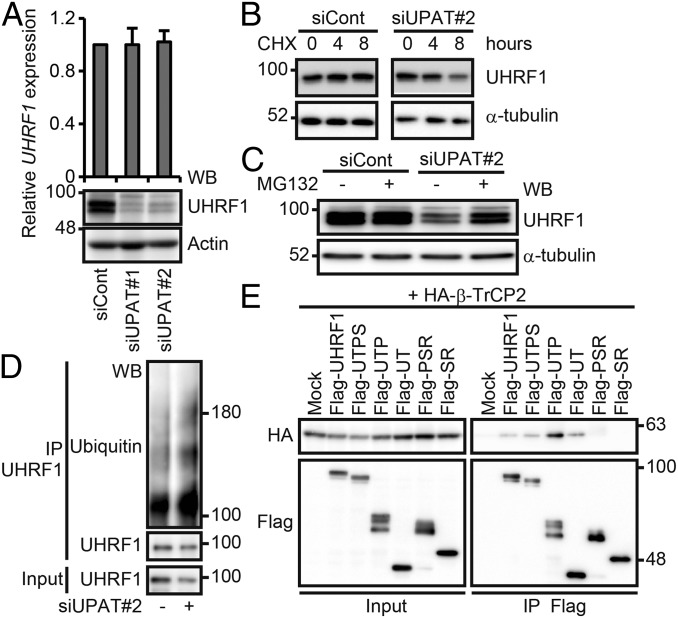
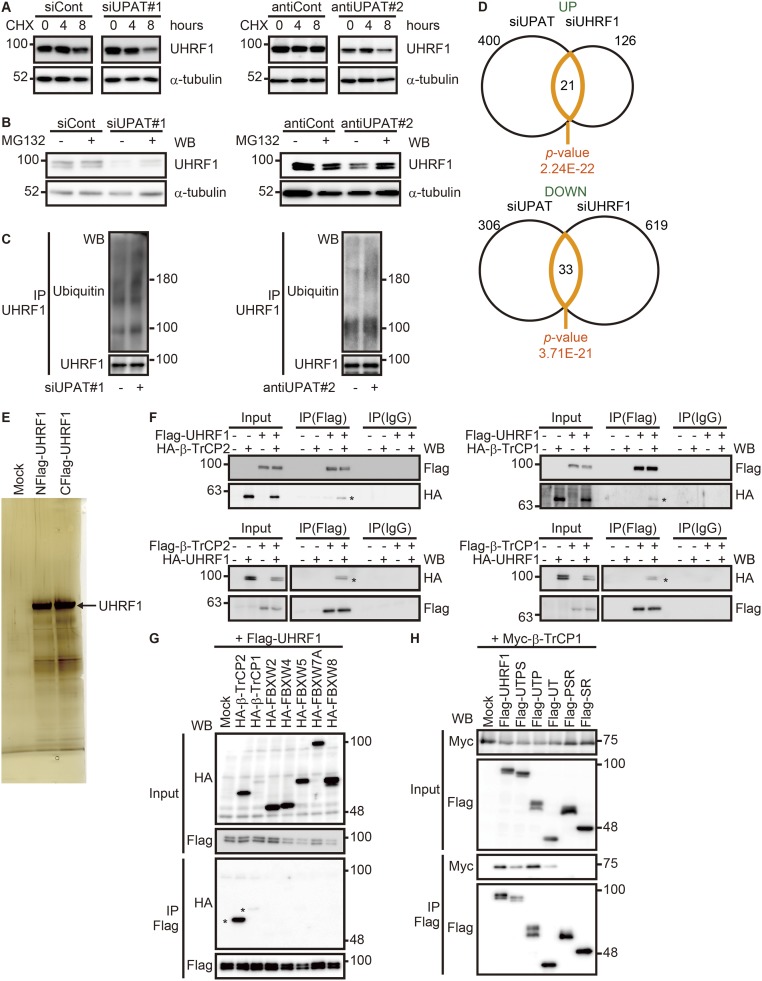
It has been reported that the stability of UHRF1 is regulated by proteasome-mediated degradation (11, 22–24). We therefore investigated whether UPAT is involved in the regulation of UHRF1 expression levels in HCT116 cells. qRT-PCR and immunoblotting analyses revealed that knockdown of UPAT by siUPAT or antisense oligonucleotides resulted in a drastic decrease in the levels of UHRF1 protein but not mRNA (Fig. 3A). We found that knockdown of UPAT reduced the stability of UHRF1 protein in HCT116 cells treated with cycloheximide (CHX) (Fig. 3B and Fig. S4A). We also found that treatment of cells with the proteasome inhibitor MG132 inhibited the decrease in the UHRF1 protein levels caused by knockdown of UPAT (Fig. 3C and Fig. S4B). Furthermore, we found that knockdown of UPAT resulted in increased ubiquitination of UHRF1 (Fig. 3D and Fig. S4C). In addition, RNA-seq analyses of HCT116 cells in which UPAT or UHRF1 expression was suppressed revealed that the expression profile of UHRF1 knockdown cells closely resembled that of UPAT knockdown cells (P values = 2.24E-22 for up-regulated genes and 3.71E-21 for down-regulated genes) (Fig. S4D and Dataset S4). Taken together, these results suggest that UPAT stabilizes UHRF1 protein by interfering with its proteasome-mediated ubiquitination and degradation.
Fig. 3.
UPAT stabilizes UHRF1 protein by interfering with its ubiquitination and degradation. (A, Upper) qRT-PCR analysis of UHRF1 expression in HCT116 cells transfected with siRNA targeting UPAT. Results are expressed as the mean ± SEM (n = 3). (A, Lower) Cell lysates were subjected to immunoblotting analysis with anti-UHRF1 or anti-Actin antibody. Actin was used as a loading control. (B) HCT116 cells transfected with siRNA targeting UPAT were treated with CHX for the indicated times and then subjected to immunoblotting analysis with anti-UHRF1 or anti–α-tubulin antibody. α-tubulin was used as a loading control. (C) HCT116 cells transfected with siRNA targeting UPAT were cultured in the presence or absence of MG132 and then subjected to immunoblotting analysis with anti-UHRF1 or anti–α-tubulin antibody. α-tubulin was used as a loading control. (D) Lysates from HCT116 cells that had been transfected with siRNA targeting UPAT and treated with MG132 were subjected to immunoprecipitation with anti-UHRF1 antibody followed by immunoblotting analysis with anti-ubiquitin or anti-UHRF1 antibody. (E) Lysates from 293FT cells transfected with HA-tagged β-TrCP2 along with empty vector (Mock) or mutants of UHRF1 were immunoprecipitated (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-HA or anti-Flag antibody.
Fig. S4.
UPAT stabilizes UHRF1 protein by interfering with its proteasome-mediated ubiquitination and degradation. (A) HCT116 cells transfected with siRNA targeting UPAT (Left, siUPAT#1) or an antisense oligonucleotide targeting UPAT (Right, antiUPAT#2) were treated with CHX for the indicated times and then subjected to immunoblotting analysis with anti-UHRF1 or anti–α-tubulin antibody. α-tubulin was used as a loading control. (B) HCT116 cells transfected with siRNA targeting UPAT (siUPAT#1) or an antisense oligonucleotide targeting UPAT (Right, antiUPAT#2) were cultured in the presence or absence of MG132 and then subjected to immunoblotting analysis with anti-UHRF1 or anti–α-tubulin antibody. α-tubulin was used as a loading control. (C) UPAT inhibits polyuiquitination of UHRF1. Lysates from HCT116 cells that had been transfected with siRNA targeting UPAT (siUPAT#1) or an antisense oligonucleotide targeting UPAT (Right, antiUPAT#2) and treated with MG132 were subjected to immunoprecipitation with anti-UHRF1 antibody followed by immunoblotting analysis with anti-ubiquitin or anti-UHRF1 antibody. (D) Venn diagram showing the overlap between genes regulated by knockdown of UPAT and those regulated by knockdown of UHRF1. The P value for the significance of the overlap is 2.24E-22 for up-regulated genes (UP) and 3.71E-21 for down-regulated genes (DOWN) as determined by hypergeometric distribution. (E) Lysates from HCT116 cells transfected with UHRF1 tagged at its NH2- or COOH terminus were subjected to immunoprecipitation with anti-Flag antibody and resolved by SDS/PAGE followed by silver staining. The arrowhead indicates UHRF1. The immunoprecipitates were subjected to liquid chromatography–mass spectrometry. (F, Upper) Lysates from HCT116 cells that had been transfected with HA-tagged β-TrCP2 (Left) or β-TrCP1 (Right) and/or Flag-tagged UHRF1 and treated with MG132 were immunoprecipitated (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-HA or anti-Flag antibody. (Lower) 293FT cells transfected with Flag-tagged β-TrCP2 (Left) or β-TrCP1 (Right) and/or HA-tagged UHRF1 were treated with MG132 and then subjected to immunoprecipitation (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-HA or anti-Flag antibody. (G) Lysates from 293FT cells transfected with Flag-tagged UHRF1 along with empty vector (Mock) or the indicated HA-tagged F-box protein expression constructs were immunoprecipitated (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-HA or anti-Flag antibody. (H) Lysates from 293FT cells transfected with Myc-tagged β-TrCP1 along with empty vector (Mock) or mutants of UHRF1 were immunoprecipitated (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-Myc or anti-Flag antibody.
To identify the ubiquitin ligase targeting UHRF1 in colorectal cancer cells, we immunoprecipitated UHRF1 from HCT116 cell lysates and analyzed the coprecipitated proteins by liquid chromatography–mass spectrometry (Fig. S4E and Dataset S3). In agreement with a recent report (24), we identified peptides corresponding to two paralogues of the F-box protein β-TrCPs, β-TrCP1/β-transducin repeat containing E3 ubiquitin protein ligase (BTRC) and β-TrCP2/F-box and WD repeat domain containing 11 (FBXW11), in addition to peptides derived from known UHRF1 binding proteins, including tripartite motif containing 28 (TRIM28) (25) and histone deacetylase 1 (HDAC1) (26) (Dataset S3). Pull-down assays using lysates from 293FT cells transfected with UHRF1 along with β-TrCP1 or β-TrCP2 confirmed that UHRF1 coprecipitated with β-TrCP1 or β-TrCP2 (Fig. S4F). Furthermore, we found that UHRF1 coprecipitated with exogenously expressed β-TrCP1 or β-TrCP2, but not with the other F-box proteins tested, FBXW2, 4, 5, 7A, or 8 (Fig. S4G). Pull-down assays using a series of UHRF1 deletion fragments revealed that a fragment (amino acids 1–282) containing the ubiquitin-like (UBL) and tandem tudor (TTD) domains (Fig. 2D), which have been reported to be histone-binding domains, was required for interaction with β-TrCP1 and β-TrCP2 (Fig. 3E and Fig. S4H).
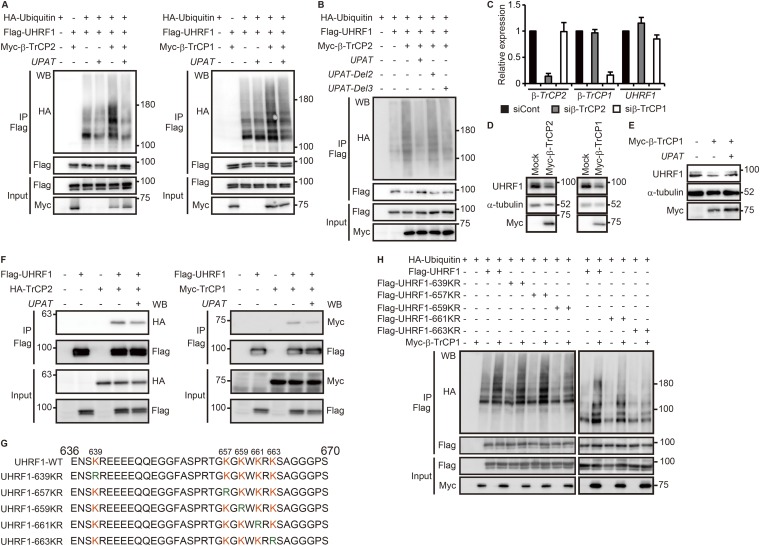
The above results suggest that β-TrCP1 and β-TrCP2 are ubiquitin ligases targeting UHRF1 for degradation. Indeed, overexpression of β-TrCP1 or β-TrCP2 resulted in the ubiquitination of UHRF1 in 293FT cells (Fig. S5A). In the absence of exogenous β-TrCP1 or β-TrCP2, ubiquitination of UHRF1 was not observed, suggesting that UHRF is not self-ubiquitinated. Furthermore, β-TrCP1– or β-TrCP2–induced ubiquitination of UHRF1 was barely detected when UPAT was overexpressed (Fig. S5A). In contrast, ubiquitination of UHRF1 was not affected by overexpression of UPAT-Del2 or -Del3 (Fig. 2D), which is unable to associate with UHRF1 (Fig. 4A and Fig. S5B). We also found that knockdown of UPAT barely induced the degradation of UHRF1 in cells transfected with an siRNA targeting β-TrCP1 or β-TrCP2 (Fig. 4B and Fig. S5C). Consistent with this, overexpression of β-TrCP1 or β-TrCP2 resulted in decreased expression of UHRF1 protein in HCT116 cells (Fig. S5D). Furthermore, overexpression of UPAT restored β-TrCP2–induced degradation of UHRF1 (Fig. 4C and Fig. S5E). On the other hand, overexpression of UPAT moderately inhibited the interaction between UHRF1 and β-TrCP1 or β-TrCP2 (Fig. S5F). Taken together, these results suggest that β-TrCP1and β-TrCP2 mediate the ubiquitination and degradation of UHRF1 and that UPAT interferes with β-TrCP1– and β-TrCP2–mediated ubiquitination of UHRF1.
Fig. S5.
UPAT inhibits β-TrCP1– or β-TrCP2–mediated polyubiqutination of UHRF1. (A and B) Lysates from 293FT cells that had been transfected with the indicated expression constructs and treated with MG132 were subjected to immunoprecipitation with anti-Flag antibody followed by immunoblotting analysis with anti-HA, anti-Flag, or anti-Myc antibody. (C) qRT-PCR analysis of β-TrCP1, β-TrCP2, and UHRF1 expression in HCT116 cells transfected with siRNA targeting β-TrCP1 or β-TrCP2. Results are expressed as the mean ± SEM (n = 3). (D) Lysates from HCT116 cells that had been transfected with Myc-tagged β-TrCP1 or Myc-tagged β-TrCP2 were subjected to immunoblotting analysis with anti-UHRF1, anti–α-tubulin, or anti-Myc antibody. α-tubulin was used as a loading control. (E) Lysates from HCT116 cells that had been transfected with Myc-tagged β-TrCP1 and/or UPAT were subjected to immunobotting analysis with anti-UHRF1, anti–α-tubulin, or anti-Myc antibody. α-tubulin was used as a loading control. (F) Overexpression of UPAT moderately inhibits the interaction between UHRF1 and β-TrCP1 or β-TrCP2. Lysates from HCT116 cells transfected with the indicated expression constructs were subjected to immunoprecipitation with anti-Flag antibody followed by immunoblotting with anti-Flag, anti-HA, or anti-Myc antibody. (G) Sequences of wild-type and mutated UHRF1 used in Fig. 4 E and F and Fig. S5H. (H) Lysates from 293FT cells transfected with the indicated expression constructs and treated with MG132 were immunoprecipitated (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-HA, anti-Flag, or anti-Myc antibody.
Fig. 4.
UPAT inhibits β-TrCP1– and β-TrCP2–mediated polyubiquitination of UHRF1. (A) Lysates from 293FT cells that had been transfected with the indicated expression constructs and treated with MG132 were subjected to immunoprecipitation with anti-Flag antibody followed by immunoblotting analysis with anti-HA, anti-Flag, or anti-Myc antibody. See also Fig. S5B. (B) Lysates from HCT116 cells transfected with siRNA targeting β-TrCP1 or β-TrCP2 and/or siUPAT were subjected to immunoblotting analysis with anti-UHRF1 or anti–α-tubulin antibody. α-tubulin was used as a loading control. (C) Lysates from HCT116 cells that had been transfected with Myc-tagged β-TrCP2 and/or UPAT were subjected to immunobotting analysis with anti-UHRF1, anti–α-tubulin, or anti-Myc antibody. α-tubulin was used as a loading control. (D) Viability of HCT116 cells transfected with UHRF1 and/or siRNA targeting UPAT was assessed by Cell Titer-Glo assays. Results are expressed as the mean ± SEM (n = 3). *P < 0.05. (E) Lysates from 293FT cells transfected with the indicated expression constructs and treated with MG132 were immunoprecipitated (IP) with anti-Flag antibody followed by immunoblotting analysis with anti-HA, anti-Flag, or anti-Myc antibody. (F) Lysates from 293FT cells that had been transfected with the indicated expression constructs were subjected to immunoblotting analysis with anti-Flag, anti–α-tubulin, or anti-Myc antibody. α-tubulin was used as a loading control. (G) UPAT binds to UHRF1 and interferes with its β-TrCP1– and β-TrCP2–mediated polyubiqutination. UHRF1 stabilized by UPAT up-regulates SCD1 and SPRY4, which are required for the survival of colon cancer cells.
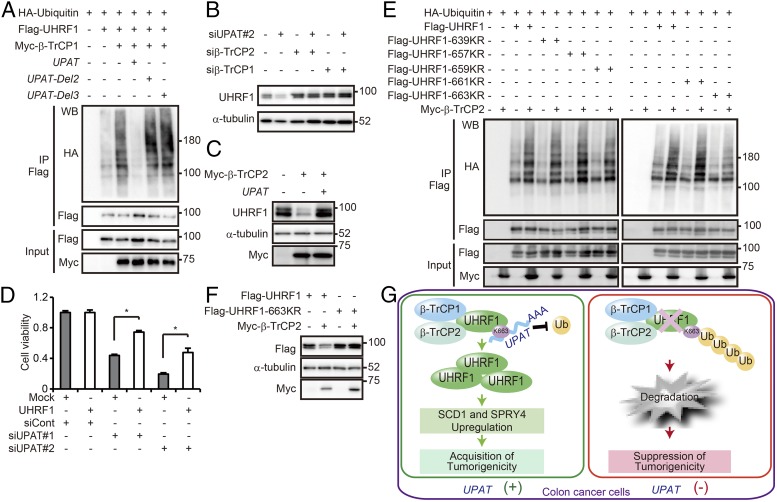
Consistent with the finding that UPAT inhibits β-TrCP1– and β-TrCP2–mediated degradation of UHRF1, we found that growth suppression of HCT116 cells by siUPAT could be partially rescued by overexpression of UHRF1 (Fig. 4D).
UHRF1 Is Ubiquitinated at Lys-663.
Because amino acids 636–670 of UHRF1 were required for its interaction with UPAT (Fig. 2 D–G), we hypothesized that UPAT may inhibit ubiquitination of this region of UHRF1. We generated mutant derivatives of UHRF1s in which Lys-639, -657, -659, -661, or -663 were replaced with Arg (UHRF1-K639R, -K657R, -K659R, -K661R, or -K663R; Fig. S5G) and examined their ubiquitination of each by transfecting these together with β-TrCP1 and β-TrCP2 and ubiquitin into 293T cells. We found that ubiquitination was normal for all fragments except UHRF1-K663R, whose ubiquitination was attenuated (Fig. 4E and Fig. S5H). Moreover, overexpression of β-TrCP2 did not induce the degradation of UHRF1-K663R (Fig. 4F). These results suggest that UHRF1 may be ubiquitinated at Lys-663 by β-TrCP1 and β-TrCP2.
Discussion
A screen for genes required for the tumorigenicity of colon tumor cells identified the lncRNA UPAT, which is encoded by a pseudogene of the AOC3 gene. We found that UPAT, but not AOC3, is required for the survival and tumorigenicity of colon tumor cells. Moreover, we found that UPAT interacts with UHRF1 and interferes with its β-TrCP1– and β-TrCP2–mediated ubiquitination and degradation. Consistent with previous reports (27–31), we confirmed that UHRF1 plays a critical role in the growth and survival of colon tumor cells. Thus, our findings suggest that UPAT-mediated stabilization of UHRF1 is critical for the proliferation and tumorigenicity of colon tumor cells.
In line with the above notion, RNA-seq analyses of HCT116 cells in which UPAT or UHRF1 expression was suppressed revealed that the expression profile of UHRF1 knockdown cells closely resembled that of UPAT knockdown cells (P values = 2.24E-22 for up-regulated genes and 3.71E-21 for down-regulated genes). However, we found many genes that are regulated by UPAT in an UHRF1-independent manner. Thus, UPAT may also have important target molecules other than UHRF1.
Our RNA-seq analyses revealed that RAS-, CDH1-, and hypoxia-related genes are down-regulated in UPAT knockdown cells. Indeed, our qRT-PCR analysis showed that siRNA knockdown of UPAT resulted in a marked decrease in SCD1, SPRY4, PGM1, and GPRC5A expression. Furthermore, we found that UPAT-mediated up-regulation of these genes is required for the survival of colon cancer cells. Consistent with our results, SCD1, the main enzyme involved in the synthesis of monounsaturated fatty acids, has been shown to be required for cancer cell proliferation, survival, transformation to cancer (32), and cancer spheroids propagation (33). The Sprouty family of proteins, key regulators of ERK signaling, has been shown to be able to function as negative or positive regulators of tumor development and/or progression in a cell type-dependent manner (34). PGM1 is known to be induced under hypoxic conditions and promotes cancer cell survival (35). In addition, it has been shown that GPRC5A is a modifier of breast cancer risk in breast cancer (BRCA)-mutation carriers and GPRC5A inactivation negatively affects BRCA1-mediated DNA repair (36). We found that UHRF1 is involved in the up-regulation of SCD1 and SPRY4 but not of PGM1 and GPRC5A. Thus, UPAT may also inhibit apoptotic cell death by mechanisms other than UHRF1 protein stabilization. We further showed that UHRF1 is associated with the SPRY4 but not the SCD1 promoter region. Thus, UHRF1 may directly transactivate SPRY4. We also found that UHRF1 increases 5hmC levels in the SPRY4 gene and thereby enhances its expression. It remains to be clarified how UHRF1 increases the level of 5hmC but not 5mC. It also remains to be investigated whether the epigenetic function of UHRF1 requires the formation of a UHRF1–UPAT complex.
The β-TrCP E3 ubiquitin ligases, β-TrCP1 and β-TrCP2, are known to play critical roles in the regulation of diverse biological processes, including cell cycle progression, differentiation, and various signal transduction pathways (37). β-TrCPs have been shown to ubiquitinate a number of important proteins, including β-catenin, cell division cycle 25 (Cdc25), RE1-silencing transcription factor (REST), mouse double minute 2 (Mdm2), and IκBβ (37). Furthermore, it has recently been reported that UHRF1 is ubiquitinated by the β-TrCP E3 ubiquitin ligases and degraded by the proteasome and that this process is accelerated in response to DNA damage (24). Consistent with these results, we found that β-TrCP1 and β-TrCP2 mediate the ubiquitination and degradation of UHRF1. Moreover, we found that UPAT interferes with the β-TrCP1– and β-TrCP2–mediated ubiquitination and degradation of UHRF1. In line with the results of Chen et al., our pull-down assays showed that β-TrCP1 and β-TrCP2 bind to amino acids 1–282 of UHRF1, which contain the UBL and TTD domains. Furthermore, we found that β-TrCP1 and β-TrCP2 ubiquitinates Lys-663 of UHRF1. Intriguingly, our RIP assays revealed that UPAT interacts with the UBR domain of UHRF1 (amino acids 636–736), which contains Lys-663. These results raise the possibility that UPAT may inhibit ubiquitination of UHRF1 by masking its ubiquitination site, Lys-663, but not by competing with β-TrCP1 and β-TrCP2 for binding to UHRF1.
It has recently been reported that the lncRNA HOX transcript antisense RNA (HOTAIR) functions as a scaffold that enhances E3-mediated ubiquitination and degradation of substrate proteins (38). It has also been shown that lincRNA-p21, originally identified as a p53-inducible lncRNA that mediates p53-induced apoptosis in mouse cells (39), is a hypoxia-responsive lncRNA that plays a critical role in the regulation of hypoxia-enhanced glycolysis by inhibiting von Hippel–Lindau (VHL)-mediated hypoxia inducible factor 1 alpha (HIF1α) ubiquitination (40). Thus, although lncRNAs are best known to regulate transcription by recruiting chromatin remodeling complexes to specific genomic regions (2), our findings, together with these earlier findings, indicate that there is a class of lncRNAs that regulate protein ubiquitination and degradation.
In summary, we have shown that the lncRNA UPAT alleviates apoptotic cell death by interfering with β-TrCP1– and β-TrCP2–mediated ubiquitination and degradation of UHRF1 (Fig. 4G). Our findings suggest that the UHRF1–UPAT axis may be a promising molecular target for colon cancer therapies.
Materials and Methods
Details are provided in SI Materials and Methods.
qRT-PCR Analysis.
Total RNA was isolated using the Total RNA Isolation kit (MACHEREY-NAGEL) and treated with DNase I (TAKARA). One microgram RNA was reverse transcribed using PrimeScript RT Master Mix (TAKARA, RR036A). qRT-PCR analysis of cDNA was performed on a LightCycler 480 (Roche Applied Science) using Syber Green PCR mastermix (Applied Biosystems). Prior to fold-change calculation, the values were normalized to the signal generated from β-actin mRNA. Primer sequences are listed in Dataset S5.
Clinical Samples.
Following written consent, resected colon cancer specimens were obtained from patients who underwent surgical treatment at the Department of Surgical Oncology, The University of Tokyo Hospital as approved by the Institutional Review Board.
SI Materials and Methods
Cell Culture.
CCSC#P cells were purchased from Celprogen (human colon cancer stem cell, cat. no. 36112–39, lot. no. 710011–05). CCSC#P and CCSC#11 cells were cultured in DMEM/F12 supplemented with 10% (vol/vol) bovine serum. HCT116 cells (ATCC) were cultured in McCoy’s 5A supplemented with 10% (vol/vol) bovine serum. DLD-1 cells (ATCC) were cultured in RPMI1640 supplemented with 10% (vol/vol) bovine serum. RKO cells (ATCC) were cultured in MEM supplemented with 10% (vol/vol) bovine serum. HAEC were cultured in endothelial basal medium-2 supplemented with 5% (vol/vol) FBS and bullet kit materials as specified by the manufacturer. HaCaT and 293FT cells (ATCC) were cultured in DMEM supplemented with 10% (vol/vol) bovine serum.
Antibodies and Regents.
Anti-FLAG (F3165) and anti-Actin (A-2066) antibodies were obtained from Sigma. Anti-UHRF1 (612264) and anti-Lamin A/C (612162) antibodies were obtained from BD Biosciences. Anti-Myc (9E10) antibody was obtained from Santa Cruz Biotechnology. Anti-DNMT1 (ab13537), anti-Histone H3K9me3 (ab8898), and anti-H3K36me3 (ab9050) antibodies were obtained from Abcam. Anti-Histone H3K4Me3 (39159) and anti–5-hydroxymethylcytidine (5hmC: 39791) antibodies were purchased from Active motif. Anti-5-methylcytidine (BI-MECY-0100) antibody was obtained from Eurogentec. Anti-Histone H3K27me3 (07-449) and anti-Histone H2AX-pSer139 (JWB301: 05–636) antibodies were obtained from Merck Millipore. Anti-histone H3 (9715) and anti-histone H3-pSer10 (6G3: 9706) antibodies were purchased from Cell Signaling. Anti–α-tubulin (CP-06) and anti-HA (16B12) antibodies were from CALBIOCHEM and COVANCE, respectively. Secondary antibodies and ECL-plus were purchased from GE Healthcare. CHX and actinomycin D were purchased from Sigma.
Lentivirus Production.
Lentiviral vector (CS-Rfa-CG) harboring an shRNA driven by the H1 promoter was transfected with the packaging vectors pCAG-HIV-gp and pCMV-VSV-G-RSV-Rev into 293FT cells using polyethylenimine ‘MAX' (PEI, Polyscience, Inc.; cat. no. 24765). All plasmids were kindly provided by H. Miyoshi, RIKEN BioResource Center, Tsukuba-shi, Ibaraki, Japan. Virus supernatants were purified by ultracentrifugation at 25,000 rpm for 90 min (SW28 rotor, Beckman). Infection efficiency was monitored by GFP expression as it is driven by the CMV promoter. The sequences of shRNAs are shown in Dataset S5.
Tumorigenesis Assay.
HCT116 or CCSC#P cells infected with a lentivirus expressing an shUPAT were injected s.c. into 6-wk-old nude mice (BALB/cAJcl-nu/nu, CLEA Japan). All animal experimental protocols were performed in accordance with the guidelines of the Animal Ethics Committee of the University of Tokyo.
Colony Formation Assays.
The base layer was prepared by mixing 1% soft agar (BD) and equivalent 2× DMEM/F12 medium and pouring 500 mL of this into six-well plates. Dispersed CCSC#11 cells were suspended in DMEM/F12 medium containing 0.33% soft agar and seeded upon the base layer at a density of 300 cells per well. Plates were maintained at 37 °C in a humidified incubator. After 10 d, the number of colonies was assessed by counting under a microscope. All experiments were conducted in triplicate.
Cell Titer-Glo Assay.
Cell viability was determined by measuring the intracellular levels of ATP using the Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega). Luminescence was measured using a Mithras LB 940 (Berthold).
RNA Interference.
Stealth siRNA duplexes targeting UHRF1, β-TrCP1, or β-TrCP2 were purchased from Invitrogen. siRNA duplexes targeting UPAT (NR_002773), NR_015379 were purchased from Dharmacon. siRNA duplexes targeting AOC1, AOC2, AOC3, SCD1, SPRY4, PGM1, or GPRC5A were purchased from Ambion. Cells were transfected with RNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Sequences of siRNAs are shown in Dataset S5. Validated Stealth negative control RNAi duplex with MED GC content #2 (Invitrogen), siGENOME NON-Targeting siRNA #3 (Dharmacon, d-001210-03), or Silencer Select negative control siRNA #1 (Ambion) was used as a control.
Antisense Oligonucleotide.
Gapmers-LNAs targeting UPAT were purchased from Exiqon. HCT116 cells were transfected with Gapmers-LNAs using Lipofectamine RNAiMAX (Invitrogen) at a final concentration of 50 nM. Gapmer-LNA sequences are shown in Dataset S5.
qRT-PCR Analysis.
Total RNA was isolated using the Total RNA Isolation kit (MACHEREY-NAGEL) and treated with DNase I (TAKARA). One microgram of RNA was reverse-transcribed using PrimeScript RT Master Mix (TAKARA, RR036A). qRT-PCR analysis of cDNA was performed on a LightCycler 480 (Roche Applied Science) using Syber Green PCR mastermix (Applied Biosystems). Before fold-change calculation, the values were normalized to the signal generated from β-actin mRNA. Primer sequences are listed in Dataset S5.
Constructs and Transfection.
UPAT, UPAT-Del1 (1–1732), UPAT-Del2 (1–1300), UPAT-Del3 (239–2073), and AOC3 were amplified by PCR using the corresponding primers and cloned into pcDNA3.1(+). For pull-down assays, UPAT was cloned into pBlueScript II SK+. UHRF1, UHRF1-UTPS (amino acids 1–736), UHRF1-UTP (1–426), UHRF1-UT (1–310), UHRF1-PSR (296–806), UHRF1-SR (381–806), UHRF1-SRA1 (381–736), UHRF1-SRA2 (400–736), UHRF1-SRA3 (381–670), UHRF1-SRA4 (381–635), UHRF1-RING1 (621–806), UHRF1-RING2 (736–806), UHRF1-RING3 (636–806), UHRF1-RING1 (671–806), UHRF1-Ubi1 (636–736), and UHRF1-Ubi2 (636–670) were amplified by PCR and cloned into pcDNA3.1(+)-Flag or pcDNA3.1(+)-HA. β-TrCP1 and β-TrCP2 were amplified by PCR and cloned into pcDNA3.1(+)-Flag, pcDNA3.1(+)-HA, or pcDNA3.1(+)-5xMyc. F-box protein expression constructs were amplified by PCR and cloned into pcDNA3.1(+)-HA. For RIP screening (Fig. S3B), CDC27, hnRNPU, UBTF, and VCP were amplified by PCR and cloned into pcDNA3.1(+)-HA. SCD1 and SPRY4 were amplified by PCR and cloned into pcDNA3.1(+)-Flag. Primer sequences are listed in Dataset S5. Plasmids were transfected into cells using polyethylenimine ‘MAX' (PEI, Polyscience, Inc.; cat. no. 24765).
Subcellular Fractionation.
Cell pellets were resuspended in one packed cell volume of Hypotonic buffer (10 mM Hepes pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.5% Nonidet P-40). After incubation on ice for 10 min, cells were disrupted by 10 passages through a 25-gauge needle. Cells were centrifuged for 10 min at 1,000 × g at 4 °C, and the supernatant containing the cytoplasmic fraction was collected by further centrifugation at 15,000 × g for 15 min. The remaining pellets were washed twice with Hypotonic buffer, resuspended in Hypertonic buffer (20 mM Hepes pH 7.5, 420 mM KCl, 1.5 mM MgCl2, 0.5% Nonidet P-40), and incubated at 4 °C for 30 min with gentle rotation. The supernatant containing the nuclear fraction was collected by centrifugation at 15,000 × g for 15 min.
RNA Pull-Down Assay.
Biotinylated UPAT or antisense-UPAT was incubated with nuclear extracts (200 μg) from HCT116 cells and then mixed with streptavidin beads, washed, and boiled in SDS buffer. The associated proteins were resolved by gel electrophoresis and visualized by silver staining or immunoblotting. Specific bands were excised and identified by a linear ion trap–orbitrap mass spectrometer (LTQ-Orbitrap Velos, Thermo Fisher Scientific) coupled with a nanoflow LC system (Dina-2A, KYA Technologies) as described previously (41). Protein identification was conducted by searching MS and MS/MS data against the RefSeq (National Center for Biotechnology Information) human protein database (32,968 protein sequences as of September 12, 2011) using Mascot version 2.4.01 (Matrix Science). Methionine oxidation, protein N-terminal acetylation, pyro-glutamination for N-terminal glutamine, and phosphorylation (Ser, Thr, and Tyr) were set as variable modifications. A maximum of two missed cleavages were allowed in our database search, and the tolerance for mass deviation was set to 3 parts per million (ppm) for peptide masses and 0.8 Da for MS/MS peaks, respectively. Protein identification was based on the criterion of having at least one MS/MS data entry with Mascot scores that exceeded the thresholds (P < 0.01).
RIP Assay.
Cells growing in six-well dishes were lysed in 0.5 mL of lysis buffer (50 mM Hepes pH 7.5, 150 mM KCl, 0.5% Nonidet P-40, 2 mM EDTA, 1 mM NaF) containing protease inhibitors and RNase Inhibitor (Promega) and centrifuged at 16,400 × g for 10 min. The supernatants were incubated with anti-UHRF1, anti-HA, anti-Flag, anti-mouse IgG, or anti-rabbit IgG antibody for 3 h at 4 °C with gentle rotation. Thirty microliters of Protein G Dynabeads (Invitrogen) were added and incubated for 3 h at 4 °C with gentle rotation. The beads were washed thrice with wash buffer (50 mM Hepes pH 7.5, 150 mM KCl, 0.05% Nonidet P-40) containing RNase Inhibitor (Promega) and then twice with PBS containing RNase Inhibitor (Promega). RNA was extracted using the Total RNA Isolation kit (MACHEREY-NAGEL), and qRT-PCR was performed as described above. Primer sequences for qRT-PCR are shown in Dataset S5.
Immunoblotting.
Cells (5 × 106) were lysed for 20 min with lysis buffer (50 mM Hepes pH 7.5, 150 mM KCl, 0.5% Nonidet P-40, 2 mM EDTA, 1 mM NaF) containing protease inhibitors. After centrifugation at 16,400 × g for 15 min at 4 °C, samples were resolved by SDS/PAGE, transferred to PVDF membranes (Immobilon-P, Millipore), and analyzed by immunoblotting using HRP-conjugated secondary antibodies. Membranes were blocked with 5% (wt/vol) skimmed milk in TBS plus Tween 20 at 4 °C overnight before probing with antibodies. Visualization was performed using the Enhanced Chemiluminescence Plus Western Blotting Detection System (GE Healthcare) and LAS-4000EPUVmini Luminescent Image Analyzer (GE Healthcare).
Immunoprecipitation.
Indicated expression plasmids were transfected into HCT116 or 293FT cells that had been treated with or without MG132. Cells were lysed in RIPA buffer [1% Nonidet-P40 (Nonidet P-40), 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 20 mM Tris·HCl, pH 7.5, 2 mM EDTA, 50 mM sodium fluoride] containing protease inhibitors and RNase Inhibitor (Promega) and centrifuged at 16,400 × g for 15 min. The supernatants were incubated with anti-FLAG M2 Magnetic Beads (SIGMA) for 2 h at 4 °C with gentle rotation. The beads were washed thrice with wash buffer (50 mM Hepes pH 7.5, 150 mM KCl, 0.05% Nonidet P-40) containing protease inhibitors and RNase Inhibitor (Promega) and then twice with PBS containing protease inhibitors and RNase Inhibitor (Promega). After washing, proteins were eluted by competition with FLAG peptide (Sigma). Immunocomplexes were analyzed by SDS/PAGE and immunoblotting with anti-Flag, anti-HA, or anti-Myc antibody.
ChIP.
ChIP assays were performed according to the manufacturer’s instructions (Upstate) with some modifications. Briefly, for each assay, cells from approximately one confluent 10-cm plate were used. When required, siRNA transfection was performed 72 h before the assays as described above. Cells washed twice with ice-cold PBS containing proteinase inhibitors were fixed with 1% formaldehyde for 10 min at 25 °C. The reaction was stopped by adding 0.5 mL of 2.5 M glycine and rotating it for 5 min. The suspension was centrifuged at 700 × g for 3 min and then resuspended in 0.3 mL nucleic lysis buffer (10 mM Tris·HCl, pH 7.5, 200 mM NaCl, 10 mM EDTA, 1% SDS) containing proteinase inhibitors. Lysates were sonicated to yield 300–1,000 bp DNA fragments. After elimination of cell debris by centrifugation, the sample was diluted with 1.8 mL of ChIP dilution buffer, and the diluted sample (0.1 mL) was used as input. Anti-UHRF1 antibody (2 μg) was added to the sample and gently mixed for 14 h at 4 °C, followed by gentle rotation with 30 μL Protein G Dynabeads (Invitrogen) for 1 h. Subsequent wash, elution, protreinase K treatment, and de–cross-linking procedures were performed according to the original protocol, except that de–cross-linking was performed for 14 h at 65 °C. Immunoprecipitated DNA was purified using the QIAquick PCR Purification Kit (QIAGEN).
Apoptosis.
Phosphatidylserine (PS) exposure at the cell surface was detected using the Annexin V-Biotin Apoptosis Detection Kit (MBL) and Streptavidin–APC conjugates (S888, Invitrogen).
Dot Blot.
Dot blot analysis was performed as described previously (42). Briefly, genomic DNA was denatured at 99 °C for 5 min and spotted onto a Hybond N+ membrane (GE Healthcare). The membrane was UV cross-linked (70,000 μJ/cm2) and then blocked with 5% (wt/vol) skimmed milk in TBS-Tween for 1 h at room temperature, followed by incubation with anti-5mC antibody (1:1,000 in TBS) at 4 °C for 14 h. Membranes were washed four times with TBS-Tween and incubated with HRP-conjugated IgG secondary antibody for 1 h at room temperature. Visualization was performed using Luminata Forte Western HRP Substrate (Millipore) and an LAS-4000EPUVmini Luminescent Image Analyzer (Fujifilm).
In Vivo Ubiquitination Assay.
293FT cells were cotransfected with HA-tagged ubiquitin, Myc-tagged β-TrCP1, Myc-tagged β-TrCP2, FLAG-tagged UHRF1, and/or UPAT. After treatment with 10 μM MG132 for 3 h, cells were lysed in Ubi Lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris·HCl, pH 8) containing 10 mM N-ethyl maleimide, protease inhibitors, and RNase Inhibitor (Promega) and were disrupted by 10 passages through a 25-gauge needle. Cells were centrifuged at 16,400 × g for 15 min at 4 °C, and the supernatants were incubated with anti-FLAG M2 Magnetic Beads (SIGMA) for 2 h at 4 °C with gentle rotation. The beads were washed thrice with Ubi Wash buffer (1% Tritone X-100, 50 mM Hepes pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.05% SDS) containing 10 mM N-ethyl maleimide, protease inhibitors, and RNase Inhibitor (Promega) and then twice with PBS containing 10 mM N-ethyl maleimide, protease inhibitors, and RNase Inhibitor (Promega). After washing, proteins were eluted by competition with FLAG peptide (Sigma). Immunocomplexes were analyzed by SDS/PAGE and immunoblotting with anti-Flag, anti-HA, or anti-Myc antibody.
Purification of UHRF1-Binding Proteins.
HCT116 cells were transfected with Flag-tagged UHRF1, lysed, and subjected to immunoprecipitation with anti-Flag antibody as described above. Eluted proteins were desalted by methanol–chloroform precipitation, digested with trypsin (Promega), and then loaded on an automated electrospray ionization (ESI)-MS/MS system, which consisted of the DiNa system (KYA Tech Corporation) equipped with a C-18 ESI capillary column (100 μm × 150 mm, NIKKYO Technos) and an LTQ Velos Orbitrap ETD instrument (ThermoFischer Scientific). For protein identification, spectra were processed using Proteome Discoverer Version 1.2 (ThermoFisher Scientific) against SEQUEST and subjected to a 5% false discovery rate (FDR) cutoff.
(h)MeDIP.
(h)MeDIP was performed as described previously (43) with minor modifications. Genomic DNA was purified from HCT116 cells using the PureLink Genomic DNA Mini kit (Life Technologies). Purified genomic DNA (5 μg/130 μL) was sheared into fragments of ∼300 base pairs in a Covaris microTube with the E2 system (Covaris, Inc.). Briefly, 1 mg of fragmented genomic DNA was denatured for 10 min at 95 °C followed by immunoprecipitation with 2.5 μg of anti-5hmC antibody (Active Motif) or anti-5mC antibody (Eurogentec) at 4 °C overnight in a 500 mL of immunoprecipitated buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl, and 0.05% Triton X-100). The mixture was incubated with 40 μL of Dynabeads Protein G at 4 °C for 2 h and washed twice with 1 mL of immunoprecipitated buffer. The beads were suspended in 50 μg of proteinase K and incubated at 55 °C for 3 h. Immunoprecipitated DNA was purified by using the QIAquick PCR Purification Kit (QIAGEN).
EGF Treatment.
HaCaT cells were plated in six-well plates. At 24 h after plating, cells were serum-starved for 16 h and then treated with EGF (20 ng/mL) for various times and washed twice with PBS. RNA was extracted using the Total RNA Isolation kit (MACHEREY-NAGEL), and qRT-PCR was performed as described above. Primer sequences for qRT-PCR are shown in Dataset S5.
Sequence Data Analysis.
For RNA-seq analyses of CCSC#P and CCSC#11 cells, 36 bp of single-end reads were sequenced on an Illumina Genome Analyzer IIx, and raw reads were mapped to the human reference genome (hg19) using TopHat 2.0.8 (tophat.cbcb.umd.edu/). Gene expression levels were calculated by Cuffdiff 2 (cufflinks.cbcb.umd.edu/) on the alignments from TopHat 2.0.8. Additional information such as gene symbol and name of mRNA was annotated according to the RefSeq database. Genes with alternative splice variants were removed. The ncRNAs with P values < 0.01 were taken as differentially expressed between CCSC#P and CCSC#11 cells. RNA-seq samples from HCT116 cells transfected with siRNA targeting UPAT or UHRF1 were sequenced using the Illumina HisEq. 2000 and analyzed similarly. Genes with fewer than one fragment per kilobase of exon per million reads mapped were removed. Genes with P values < 0.05 were considered to be differentially expressed. Functional characterization of these genes was performed using the MSigDB gene sets (software.broadinstitute.org/gsea/login.jsp). The significance of the overlap between these genes and MSigDB gene sets was calculated by hypergeometric distribution.
Statistical Analysis.
Statistical analysis was performed using the Mann–Whitney U test and the Student’s t test. A P value < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Research Program of Innovative Cell Biology by Innovative Technology (Integrated Systems Analysis of Cellular Oncogenic Signaling Networks), Grants-in-Aid for Scientific Research on Innovative Areas (Integrative Research on Cancer Microenvironment Network), the Project for Development of Innovative Research on Cancer Therapeutics, and the Takeda Science Foundation and in part by the Global Centers of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov/sra (accession no. DRA004047).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500992113/-/DCSupplemental.
References
- 1.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19(7-8):454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, et al. Noncoding RNAs in long-term memory formation. Neuroscientist. 2008;14(5):434–445. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- 5.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14(6):752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 8.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Bronner C, Krifa M, Mousli M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem Pharmacol. 2013;86(12):1643–1649. doi: 10.1016/j.bcp.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112(2):439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 12.Chassande O, Renard S, Barbry P, Lazdunski M. The human gene for diamine oxidase, an amiloride binding protein. Molecular cloning, sequencing, and characterization of the promoter. J Biol Chem. 1994;269(20):14484–14489. [PubMed] [Google Scholar]
- 13.Imamura Y, et al. Human retina-specific amine oxidase (RAO): cDNA cloning, tissue expression, and chromosomal mapping. Genomics. 1997;40(2):277–283. doi: 10.1006/geno.1996.4570. [DOI] [PubMed] [Google Scholar]
- 14.Salmi M, Jalkanen S. A 90-kilodalton endothelial cell molecule mediating lymphocyte binding in humans. Science. 1992;257(5075):1407–1409. doi: 10.1126/science.1529341. [DOI] [PubMed] [Google Scholar]
- 15.Yanagida S, et al. ASBEL, an ANA/BTG3 antisense transcript required for tumorigenicity of ovarian carcinoma. Sci Rep. 2013;3:1305. doi: 10.1038/srep01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Wang X-S, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12(16):4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 18.Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat Rev Genet. 2008;9(2):129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 20.Rothbart SB, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19(11):1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tien AL, et al. UHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J. 2011;435(1):175–185. doi: 10.1042/BJ20100840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felle M, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39(19):8355–8365. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma H, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci USA. 2012;109(13):4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, et al. DNA damage regulates UHRF1 stability via the SCF(β-TrCP) E3 ligase. Mol Cell Biol. 2013;33(6):1139–1148. doi: 10.1128/MCB.01191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44(3):361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23(46):7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 27.Li X-L, Xu J-H, Nie J-H, Fan S-J. Exogenous expression of UHRF1 promotes proliferation and metastasis of breast cancer cells. Oncol Rep. 2012;28(1):375–383. doi: 10.3892/or.2012.1792. [DOI] [PubMed] [Google Scholar]
- 28.Unoki M, et al. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer. 2009;101(1):98–105. doi: 10.1038/sj.bjc.6605123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unoki M, et al. UHRF1 is a novel diagnostic marker of lung cancer. Br J Cancer. 2010;103(2):217–222. doi: 10.1038/sj.bjc.6605717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabatino L, et al. UHRF1 coordinates peroxisome proliferator activated receptor gamma (PPARG) epigenetic silencing and mediates colorectal cancer progression. Oncogene. 2012;31(49):5061–5072. doi: 10.1038/onc.2012.3. [DOI] [PubMed] [Google Scholar]
- 31.Babbio F, et al. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene. 2012;31(46):4878–4887. doi: 10.1038/onc.2011.641. [DOI] [PubMed] [Google Scholar]
- 32.Igal RA. Stearoyl-CoA desaturase-1: A novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31(9):1509–1515. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]
- 33.Noto A, et al. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013;4:e947. doi: 10.1038/cddis.2013.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masoumi-Moghaddam S, Amini A, Morris DL. The developing story of Sprouty and cancer. Cancer Metastasis Rev. 2014;33(2-3):695–720. doi: 10.1007/s10555-014-9497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelletier J, et al. Glycogen synthesis is induced in hypoxia by the hypoxia-inducible factor and promotes cancer cell survival. Front Oncol. 2012;2:18. doi: 10.3389/fonc.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokolenko AP, et al. High prevalence of GPRC5A germline mutations in BRCA1-mutant breast cancer patients. Int J Cancer. 2014;134(10):2352–2358. doi: 10.1002/ijc.28569. [DOI] [PubMed] [Google Scholar]
- 37.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat Rev Cancer. 2008;8(6):438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon J-H, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Kozuka-Hata H, et al. Phosphoproteome of human glioblastoma initiating cells reveals novel signaling regulators encoded by the transcriptome. PLoS One. 2012;7(8):e43398. doi: 10.1371/journal.pone.0043398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 43.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.