Significance
Resistance-nodulation division family efflux pumps, exemplified by AcrB of Escherichia coli, play an important role in multidrug resistance of Gram-negative bacteria by actively pumping out a very wide range of antimicrobial agents. Yet it is unclear with how much affinity and with how much rate this pumping process works, except for our earlier studies on cephalosporins and penicillins. Here we succeeded in determining these parameters for the earliest-discovered inhibitor Phenylalanyl-argenine β-naphthylamide and its relatives, and examined the binding of these compounds to AcrB binding sites by docking and molecular dynamics simulations. The results suggest how an inhibitor, even when it is itself pumped out, can reduce the pumping of other substrates.
Keywords: RND transporters, efflux inhibitors, Phe-Arg-β-naphthylamide, molecular dynamics simulations
Abstract
Efflux pumps of the resistance-nodulation division superfamily, such as AcrB, make a major contribution to multidrug resistance in Gram-negative bacteria. Inhibitors of such pumps would improve the efficacy of antibiotics, and ameliorate the crisis in health care caused by the prevalence of multidrug resistant Gram-negative pathogens. Phenylalanyl-arginine β-naphthylamide (PAβN), is a well-known inhibitor of AcrB and its homologs. However, its mechanism of inhibition is not clear. Because the hydrolysis of PAβN in Escherichia coli was nearly entirely dependent on an aminopeptidase, PepN, expression of PepN in periplasm allowed us to carry out a quantitative determination of PAβN efflux kinetics through the determination of its periplasmic concentrations by quantitation of the first hydrolysis product, phenylalanine, after a short period of treatment. We found that PAβN is efficiently pumped out by AcrB, with a sigmoidal kinetics. We also examined the behavior of PAβN homologs, Ala β-naphthylamide, Arg β-naphthylamide, and Phe β-naphthylamide, as substrates of AcrB and as modulators of nitrocefin efflux through AcrB. Furthermore, molecular dynamics simulations indicated that the mode of binding of these compounds to AcrB affects the modulatory activity on the efflux of other substrates. These results, and the finding that PAβN changes the nitrocefin kinetics into a sigmoidal one, suggested that PAβN inhibited the efflux of other drugs by binding to the bottom of the distal binding pocket, the so-called hydrophobic trap, and also by interfering with the binding of other drug substrates to the upper part of the binding pocket.
The emergence and spread of antibiotic-resistant pathogenic bacteria, especially those of multidrug-resistant or even pan-resistant Gram-negative bacteria, is a major problem (1). Multidrug efflux pumps, especially those of the resistance-nodulation division (RND) family, contribute strongly to this type of resistance (2). AcrB of Escherichia coli has very wide substrate specificity (3) and its structure and mechanism have been studied extensively as a prototype RND efflux pump (4, 5). AcrB enhances the intrinsic resistance of E. coli, especially to large or lipophilic antibiotics (3), and makes the cell more resistant when overproduced (6). Because of this role, inhibitors of AcrB and its relatives are important not only in basic research but also possibly in clinical medicine, as they could reduce the resistance level of pathogens. The first such inhibitor was Phe-Arg β-naphthylamide (PAβN), reported by Lomovskaya et al. in 2001 (7). These researchers also showed that a PAβN analog, Ala β-naphthylamide, is a good substrate for the AcrB homolog MexB of Pseudomonas aeruginosa, by examining its intracellular accumulation followed by enzymatic hydrolysis that generated the fluorescent naphthylamine. The researchers suggested that PAβN is also a substrate of the AcrB/MexB pump, because pump overproducers are more resistant to the intrinsic toxicity of this compound. More recently, PAβN was predicted to bind to the distal binding pocket of AcrB (8), and a possible mechanism for inhibition was also proposed (9). Nevertheless, because the binding of PAβN to the pocket does not appear to be very strong (8, 9), it seemed essential to know more about its interaction with AcrB.
When using PAβN as an AcrB pump inhibitor in intact cells of E. coli K-12, we noted that it was degraded to yield a fluorescent product, presumably β-naphthylamine. By expressing the enzyme responsible for this hydrolysis, PepN, in the periplasm, we could show that PAβN was a good substrate for AcrB. To better understand the behavior of PAβN, we also examined the efflux kinetics and modulator functions of PAβN analogs, Ala β-naphthylamide (Ala-Naph), Arg β-naphthylamide (Arg-Naph), and Phe β-naphthylamide (Phe-Naph). These studies led us to a deeper understanding of the complexity of the interaction of these aminoacyl-naphthylamides with AcrB.
Results
Our Approach.
In our previous determination of the parameters of efflux process, we measured in intact cells the hydrolysis rates of β-lactams by periplasmic β-lactamases, a process that allowed us to determine the periplasmic concentration of these compounds. This next allowed us to calculate the influx rates of β-lactams across the outer membrane, as they are proportional to the difference in concentrations of these compounds across this membrane. The difference between the influx rate and hydrolysis rate then represented the efflux rate (10). In this study, we wanted to obtain quantitative parameters for the efflux of PAβN and other aminoacyl β-naphthylamides, and for this we needed an enzyme that hydrolyzed these compounds in the periplasm.
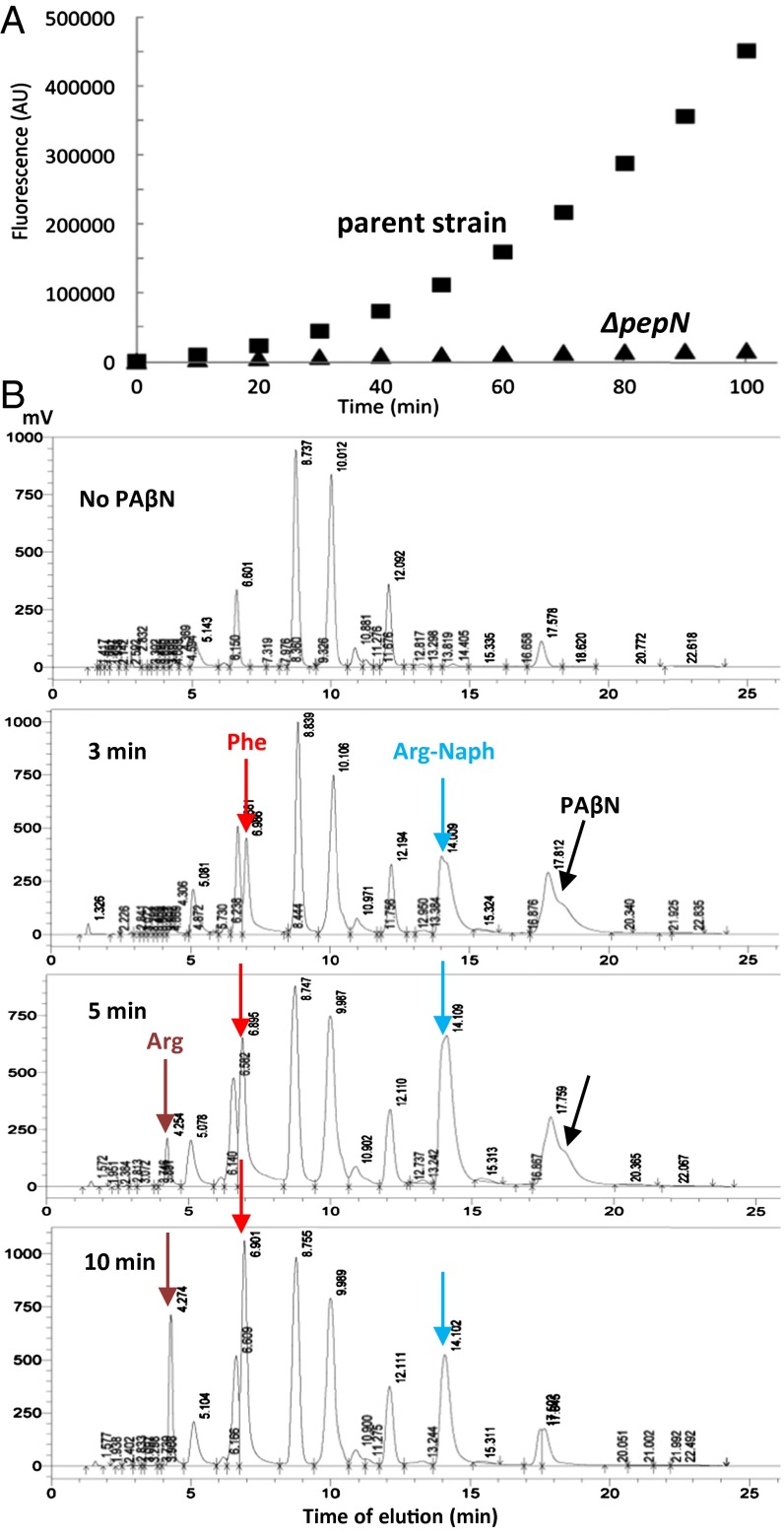
Incubation of intact E. coli cells with PAβN resulted in a time-dependent increase in fluorescence (Fig. 1A), which had an emission spectrum corresponding to that of β-naphthylamine. PAβN was thus presumably hydrolyzed by one or more of the peptidases in E. coli (11). Because aminopeptidase N (PepN) has a broad substrate range and prefers basic and hydrophobic residues at the N terminus (12), we tested the PAβN hydrolysis in a ΔpepN mutant and found it to be nearly completely negative (Fig. 1A). Because PepN is an aminopeptidase, it is expected to degrade PAβN in two steps (13), first converting it to Phe and Arg-Naph, and then the latter to Arg and β-naphthylamine. The presence of a lag in the time course of naphthylamine generation (Fig. 1A) is consistent with this idea. We could indeed confirm, by HPLC of phenylthiocarbamyl derivatives of products, that PepN first converted PAβN to Phe and Arg-Naph, and then the latter to Arg and β-naphthylamine (Fig. 1B).
Fig. 1.
Hydrolysis of PAβN by Aminopeptidase N. (A) When washed intact cells of wild-type (BW25113) and its ΔpepN::kan mutant (BW0915) of E. coli K-12 (0.2 mL at OD600 of 0.05) were incubated with 0.1 mM PAβN in 96-well microtiter plates, generation of the fluorescent product of hydrolysis, β-naphthylamine, measured with a FluoDia T70 fluorometer with excitation at 340 nm and emission at 410 nm, was nearly completely absent in the mutant. (B) Time course of generation of hydrolysis products, Phe, Arg, and Arg-Naph, by an extract of RAM121 ΔpepN::kan/pMAL-PepN detected by the HPLC of phenylthiocarbamyl derivatives.
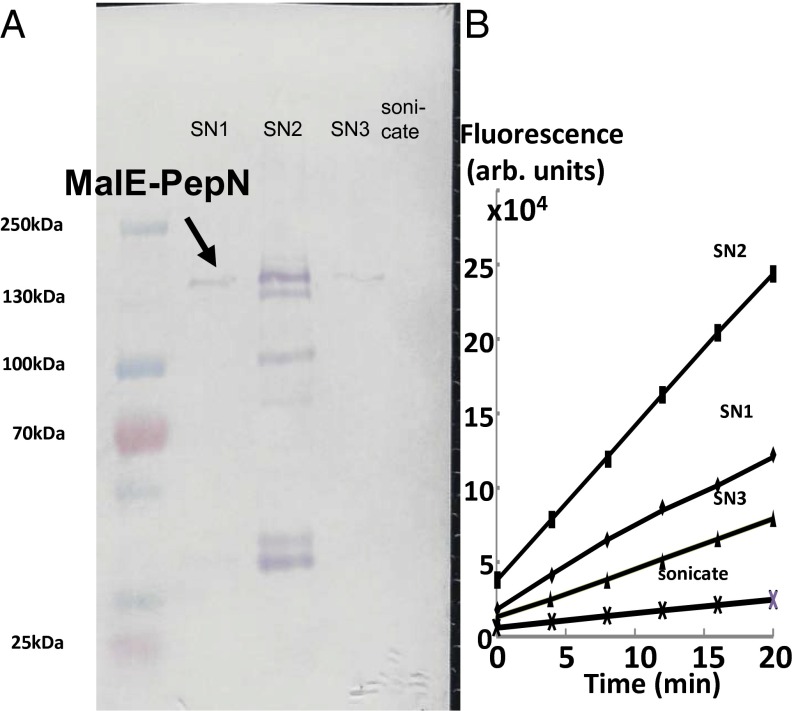
Although PepN was once claimed to be a periplasmic protein (14), it has no signal sequence (15), and indeed we could show that most of its activity was located in the cytosolic fraction (SI Appendix). Thus, to use the enzymatic hydrolysis of PAβN for exploring its efflux kinetics, it was necessary to get PepN exported into the periplasm. This was achieved by fusing the N terminus of PepN sequence to the C terminus of the periplasmic MalE protein, using the pMAL-p5X vector (Experimental Procedures). Osmotic shock experiments (Fig. 2) showed that practically all of the MalE-PepN fusion protein was located in the periplasm. Finally, to increase the precision of the assay, we used intact cells of a ΔpepN::kan/pMAL-PepN strain derived from E. coli RAM121, a mutant that produces a large channel porin (16) and allows a rapid influx of large and hydrophobic substrates, such as PAβN and other aminoacyl naphthylamides.
Fig. 2.
In the strain containing pMAL-PepN, most of the aminopeptidase activity is found in the periplasm. (A) SDS/PAGE followed by staining for MalE shows that most of the MalE-PepN is located in the osmotic shockate (SN1 through SN3) and is absent in the sonicate of the residue (i.e., the cytosol). In SN2, smaller proteins, apparently degradation products, are also seen. (B) Activity assay using 0.1 mM Arg-Naph.
PAβN Efflux Kinetics.
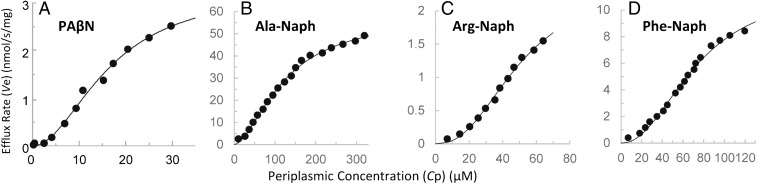
We used the initial stage in the PAβN hydrolysis by the periplasmic MalE-PepN to determine its efflux kinetics. Hydrolysis was stopped at 3 min after the addition of PAβN to intact cells. The analysis of products at this stage revealed that only Phe and Arg-Naph were present, whereas a product of the second stage of hydrolysis, Arg, was not present (Fig. 1B). Phe was quantitated by fluorescence through o-phthalaldehyde modification (17) after proteins were removed by tricholoroacetic acid precipitation. Because we know the kinetic constants of this first-stage hydrolysis by the periplasmic MalE-PepN (SI Appendix, Fig. S1), this allowed us to estimate the periplasmic concentration of PAβN, and then the efflux rate as the difference between the (calculated) influx rate across the outer membrane and the measured hydrolysis rate, as was done earlier for β-lactams (10, 18). [For calculation of influx rate, we used the permeability coefficient determined by the use of RAM121ΔpepN::kan ΔacrAB::spc/pMAL-PepN strain, following the method used for cephalosporins (10).] The results (Fig. 3A) showed that the efflux followed a sigmoidal kinetics, with the Vmax and K0.5 (the substrate concentration where the rate reaches one-half of the Vmax) of 3.0 ± 0.5 nmol/mg/s and 17.6 ± 5.0 μM, respectively, in five independent experiments. The Hill coefficient was 1.7 ± 0.4. These results show that PAβN is rapidly pumped out by AcrB, but with only a modest affinity to the pump, compared with the Vmax (about 0.03 nmol/mg/s) and KM (around 5 μM) of the strongly bound substrate, nitrocefin (10). To understand the mechanism of efflux and the inhibitor function of PAβN, we turned to the study of PAβN analogs, Ala-Naph, Arg-Naph, and Phe-Naph, both as substrates and modulators of AcrB.
Fig. 3.
(A–D) Efflux parameters of PAβN, Ala-Naph, Arg-Naph, and Phe-Naph. Curves were fitted using the Hill equation as detailed in Experimental Procedures. For this and Fig. 4, the efflux rates are shown in the unit of nmol·s·mg (dry-weight cells).
Efflux Kinetics of Aminoacyl β-Naphthylamide in Intact Cells.
Because extracts of cells harboring the pMAL-PepN plasmid showed a good hydrolytic activity against aminoacyl β-naphthylamides even without induction (Fig. 2B), this basal level activity of MalE-PepN could be used to measure the hydrolysis rate of aminoacyl β-naphthylamides in periplasm, by following the fluorescence of one of the products, β-naphthylamine.
We followed the same approach used for PAβN, above, using the KM and Vmax of MalE-PepN for aminoacyl β-naphthylamides (SI Appendix, Fig. S2), but here the assay was simpler because one of the products, β-naphthylamine, was highly fluorescent and could be used in following hydrolysis in intact cells. One difference from the β-lactam efflux assay was that, whereas with β-lactams the efflux of hydrolysis products, which are much more hydrophilic than the original ligands, could be neglected, with aminoacyl-naphthylamides the measured product, β-naphthylamine, is quite lipophilic and is expected to be a good substrate for efflux. β-Naphthylamine fluorescence, however, is not too different in organic solvents and in water at 410 nm (SI Appendix, Fig. S3), and thus its efflux is expected to have little effect on its fluorescence and, hence, the calculation of efflux rate of aminoacyl β-naphthylamides.
For each compound, four to seven independent experiments were performed, and typical results are shown in Fig. 3 B–D. There were some remarkable features on these efflux kinetics. First, for each compound, the curves were clearly sigmoidal as in the case of PAβN, showing the calculated Hill coefficients of 1.6, 2.1, and 2.3, for Ala-, Arg-, and Phe-Naph, respectively. Second, the Vmax values tended to be quite high, being 66 ± 8, 2.0 ± 0.5, and 9.5 ± 1.5 nmol/mg/s, for these three compounds, respectively. The latter two values were comparable with the Vmax of PAβN efflux above (3.0 nmol/mg/s), whereas Ala-Naph had an exceptionally high Vmax. Finally, when K0.5 values were compared, they (148 ± 21, 41 ± 15, and 62 ± 12 μM for Ala-, Arg-, Phe-Naph) were higher than those for PAβN (17.6 μM, see above), or penicillins (about 1 μM or less) (18, 19) or cephalosporins (5–26 μM, except cephalothin and cephaloridine) (10), suggesting that aminoacyl-naphthylamides probably do not become tightly bound to AcrB.
Activity of Aminoacyl β-Naphthylamides as Modulators of Nitrocefin Efflux.
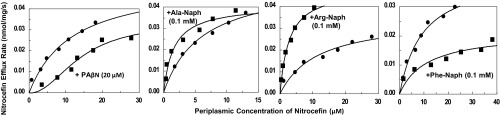
PAβN is a well-known inhibitor of AcrB-mediated drug efflux (7). In contrast, one of its homologs, Arg-Naph at 0.1 mM, was found to stimulate the efflux of nitrocefin (20). In this study we repeated this experiment and also tested if Ala-Naph or Phe-Naph also affected nitrocefin efflux. At 0.1-mM concentration, Ala-Naph stimulated nitrocefin efflux as well (Fig. 4); as with Arg-Naph, the stimulation was largely because of decrease in KM values.
Fig. 4.
Effect of 20 μM PAβN and 100 μM aminoacyl β-naphthylamides on the efflux kinetics of nitrocefin, determined as described earlier (10), using RAM121 ΔpepN strain to avoid the degradation of modulators during the assay. Unlabeled curves show nitrocefin efflux without modulators, which had an average Vmax and KM of 0.039 ± 0.008 nmol/mg/s and 8.91 ± 3.26 μM (n = 18). In comparison, in the presence of Ala-Naph, Arg-Naph, and Phe-Naph, Vmax was 0.032 ± 0.008 (n = 5), 0.046 ± 0.003 (n = 5), and 0.020 ± 0.004 (n = 4) nmol/mg/s, and KM was 2.6 ± 1.3, 3.2 ± 0.3, and 10.8 ± 4.1 μM, respectively. PAβN produced a sigmoidal kinetics with the Hill coefficient of 2.04 ± 0.10 (n = 4); Vmax was 0.038 ± 0.004 nmol/mg/s and K0.5 was 14.07 ± 0.77 μM.
In a striking contrast, Phe-Naph at 0.1 mM inhibited nitrocefin efflux (Fig. 4), apparently because of decreases in Vmax. Naphthylamine alone, or any of the component amino acids (Phe, Arg, Ala), did not produce any significant changes in nitrocefin efflux, when tested at 0.1 mM (SI Appendix, Fig. S4). PAβN, as expected, inhibits nitrocefin efflux at 20 μM (Fig. 4), and it is noteworthy that the kinetics now becomes strongly sigmoidal, with a Hill coefficient of about 2.
Molecular Dynamics Simulation of Ala-Naph, Arg-Naph, and Phe-Naph Binding to AcrB.
To understand better the behavior of aminoacyl β-naphthylamides as efflux modulators and substrates, we examined the binding of aminoacyl naphthylamides to the distal binding pocket of the binding protomer of AcrB first by docking with the program Autodock Vina (21), followed by molecular dynamics (MD) simulations, with a truncated model of AcrB used to reduce the overall computational time (8).
With Ala-Naph, docking predicted that its naphthylamine moiety will become bound to the bottom of the distal binding pocket (SI Appendix, Fig. S5A). This initial pose was not stable in the MD simulation that includes interaction with solvent, and the positively charged amino group of Ala suddenly escaped from the hydrophobic environment of the pocket, causing a reorientation of the whole ligand by about 90°. In this stable pose along the dynamics (Fig. 5B), the naphthylamine moiety interacts with the very bottom of the hydrophobic trap, particularly with residues Phe136 and Phe628, whereas the charged amino group of Ala is mainly stabilized by residues Ser135, Tyr327, and Glu673 (SI Appendix, Table S2), and by the solvent. At odds with Arg-Naph described below, Ala-Naph is not stabilized by residues lining the G-loop, a short loop containing Phe617 and anchored by two glycine residues (8), but a significant contribution comes from residues lining the access pocket, the interface between this pocket, and the distal pocket, and the external cleft (Table 1). The calculated binding free energy (without entropy correction) was −22.9 ± 3.2 kcal/mol (Table 1). This is much smaller in size than the similarly predicted binding energy of a strongly bound substrate, nitrocefin (−42.5 ± 3.9 kcal/mol), and similar to those of weakly bound substrates, such as chloramphenicol (−23.3 ± 4.6 kcal/mol) (8), although higher than that calculated for benzene (about −13 kcal/mol) (20).
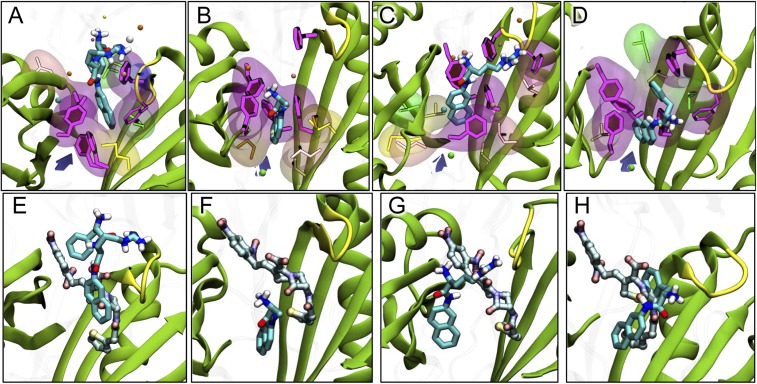
Fig. 5.
Binding of aminoacyl-naphthylamides after 320 ns of MD simulation. (A–D) Binding of PAβN (27) and Ala-, Arg-, and Phe-Naph. The ligand is shown with thick sticks colored by atom type. Side-chains of residues F136, F178, F610, F615, and F628 lining the hydrophobic trap are shown with magenta sticks and with filled rings. Side-chains of hydrophobic residues within 3.5 Å of the ligand are shown both with thinner sticks and with transparent surface. Nonpolar residues are shown with glossy beads. Subdomains PC1/PC2 and the G-loop are shown as cartoon, colored green and yellow, respectively. The entrance from the PC1/PC2 cleft is roughly indicated by a violet arrow. (E–H) Comparison of the most stable poses of PAβN and Ala-, Arg-, and Phe-Naph, with that of nitrocefin, taken from ref. 8. The modulators are shown with sticks colored as in A–D, whereas nitrocefin is shown with carbons in lighter blue.
Table 1.
Free energies of binding and surface matching coefficients
| Compound | ΔGb (kcal/mol) | Percent of ΔGb* | Surface matching | ||||||
| DP† | Hydrophobic trap | AP† | G-loop | Interface | Cleft | SMTot | SML | ||
| PAβN | −40.8 ± 5.8 | 68 | 35 | 0 | 4 | 1 | 0 | 0.71 | 0.81 |
| ALA-Naph | −22.9 ± 3.2 | 51 | 40 | 12 | 0 | 5 | 4 | 0.72 | 0.80 |
| ARG-Naph‡ | −27.2 ± 5.1 | 49 | 21 | 3 | 9 | 0 | 2 | 0.58 | 0.64 |
| PHE-Naph | −22.2 ± 3.2 | 59 | 45 | 2 | 4 | 13 | 2 | 0.76 | 0.87 |
The free energy of binding ΔGb was calculated without inclusion of entropy correction (see Experimental Procedures).
Percentage of the binding energy over that from all residues of the protein is shown. Because hydrophobic trap is a part of DP, the sum of all values sometimes exceeds 100.
DP and AP stand for distal and access (or proximal) binding pockets, respectively. Residues within the various regions are listed in SI Appendix.
These data are from the simulation whose estimate of the binding free energy was the highest.
With Arg-Naph, docking also predicted that its naphthylamine moiety will become bound to the bottom of the distal binding pocket, rich in aromatic residues [the so-called hydrophobic trap (22)] (SI Appendix, Fig. S5A). However, the introduction of water molecules in MD simulation very quickly moved Arg-Naph, with its two cationic groups in the Arg moiety, at least partially out of this hydrophobic environment, and Phe136 remained the only residue in the hydrophobic trap contributing to the binding (Fig. 5C and SI Appendix, Table S2). In all three independent MD simulations, each for 320 ns, the pseudoequilibrium positions of Arg-Naph suggested that it is a relatively weakly bound substrate. Thus, the binding free energies were −27.2 ± 5.1, −23.2 ± 2.9, and −25.0 ± 4.4 kcal/mol. The binding pose (Fig. 5C shows the result of the simulation producing the highest binding energy) also suggests only a loose interaction. The Arg moiety sticks out into the water-filled substrate channel. Indeed, the interaction with the hydrophobic trap was the smallest among the ligands studied (Table 1).
Concerning Phe-Naph, docking predicted that, similar to Ala-Naph, the ligand binds with its naphthylamine group completely in the hydrophobic trap (SI Appendix, Fig. S5A). However, because of the hydrophobic nature of the Phe extension, the orientation of the ligand in its highest-affinity pose is flipped with respect to those of Ala- and Arg-Naph, with the aminoacyl (Phe) group completely in the hydrophobic trap (SI Appendix, Fig. S5A). MD simulation resulted in the vertical turnaround of the ligand, but in the final pseudoequilibrium structure (Fig. 5D), the naphthylamine moiety interacts with the aromatic residues of the hydrophobic trap (see also Table 1 and SI Appendix, Table S2). The calculated binding energy (−22.2 ± 3.2 kcal/mol) was similar to that of Ala-Naph.
Thus, the modes of binding are clearly different among the three aminoacyl-naphthylamides examined (SI Appendix, Fig. S5B), and these differences might explain the different effects of these modulators on nitrocefin efflux. The different binding position of the naphthylamine moiety of Phe-Naph (Fig. 5D; also see SI Appendix, Fig. S6) may be the reason why this compound produces efflux inhibition, as was seen with the other inhibitors that interact with the hydrophobic trap, such as PAβN (Fig. 5A) (9) or D13-9001 (22).
Discussion
RND family efflux pumps, such as AcrB, play a major role in the emergence of multidrug resistance in Gram-negative bacteria, and inhibitors of such pumps are potentially important for human and animal health. Although a large inhibitor, D13-9001 (695 Da), was shown to bind tightly to the distal binding pocket of such pumps (22), many earlier inhibitors, such as PAβN (7) or 1-(1-naphtylmethyl)-piperazine (23), do not bind tightly (8), yet act as inhibitors. Thus, it is important to find out how PAβN, for example, functions as an inhibitor. We have examined here both PAβN and its homologs, aminoacyl naphthylamides, on their efflux parameters and their capacity to modulate the efflux of another substrate, nitrocefin.
We reported already that Arg-Naph, at 0.1 mM, stimulated nitrocefin efflux (20). When tested at the same molar concentration, Ala-Naph also stimulated nitrocefin efflux (Fig. 4), yet Phe-Naph produced a significant inhibition (Fig. 4). Ala-Naph as a substrate is pumped out extremely rapidly [as was already shown by the pioneering study by the Lomovskaya group (7)], with a high Vmax of 66 nmol/mg/s (Fig. 3), and thus it might possibly “sweep off” other substrates with it. In fact, the rather weak binding of Ala-Naph in MD simulation suggests that it might act like another stimulator, benzene, which is also a weak binder to AcrB and is apparently pumped out very rapidly (20). In addition, the simultaneous interaction with the access and distal pockets not involving the G-loop could facilitate triggering of conformational changes, such as the closure of the PC1/PC2 cleft (24) or rearrangements in the periplasmic side of the transmembrane region. The latter could induce opening of channels connecting the periplasm to the site of proton exchange (25, 26). However, the functions of Phe-Naph and Arg-Naph as modulators seem to require a somewhat different explanation. MD simulation showed that Phe-Naph binds significantly to the “hydrophobic pocket,” which binds the hydrophobic portion of D13-9001 (22), whereas Arg-Naph binding has little involvement of this pocket (Fig. 5C, Table 1, and SI Appendix, Table S2). These different modes of binding seem to offer the best explanation on why Phe-Naph and Arg-Naph act as an inhibitor and stimulator, respectively, of nitrocefin efflux (Fig. 4). Furthermore, the Phe residue is located in the groove of the pocket, and this may hinder the binding of nitrocefin (Fig. 5H). Finally the inhibitory activity of Phe-Naph is also consistent with the knowledge that Lys-Phe-β-naphthylamide is an effective inhibitor similar to PAβN (7). In contrast, in Arg-Naph, the Arg moiety is far away from the groove, and is unlikely to prevent the binding of nitrocefin to this area of the pocket (Fig. 5G). Conceivably, the effect of the double-positive charge of Arg residue on the binding of negatively charged nitrocefin may also contribute to the stimulation of its efflux (Fig. 4).
We also believe that the knowledge gained on the behavior of aminoacyl-naphthylamides helps us to understand how PAβN acts as an effective inhibitor of AcrB-mediated drug efflux. PAβN binds more strongly to the hydrophobic trap than Arg-Naph, for example (Fig. 5, Table 1, and SI Appendix, Table S2). Indeed, in the case of a recently developed more potent inhibitor, a larger fraction of the binding energy comes from the trap (9). In addition, a large portion of PAβN interacts with the substrate-binding areas of the pocket (SI Appendix, Table S2), presumably causing interference, as was also predicted (to a lesser degree) with Phe-Naph (see above). PAβN was known to be pumped out by AcrB/MexB group of pumps (7). Because PAβN binds significantly less tightly than the large inhibitor D13-9001 (9), it was not a total surprise that it acted as a good substrate and was pumped out with kinetic constants (Vmax of 3.0 nmol/mg/s and K0.5 of 17.6 μM) in the range similar to the efflux of Arg-Naph, for example (2.0 nmol/mg/s and 41 μM); the lower K0.5 value for PAβN seems to be consistent with its tighter binding to AcrB (Table 1). Because PAβN binds to the hydrophobic trap, because the simultaneous binding of other (large) substrates with PAβN would be hindered, and because the rate of PAβN efflux is not exorbitant, it cannot function as a stimulator.
We emphasize that the naphthylamides studied here represent the second group of AcrB substrates whose efflux kinetics were successfully determined, following the β-lactams (10, 18–20). An important feature of β-lactam efflux kinetics is the clearly sigmoidal nature of the rate vs. substrate concentration plots, except for nitrocefin (10, 18). The kinetics of aminoacyl-naphthylamide efflux processes studied here also showed clearly the sigmoidal kinetics (Fig. 3), with the calculated Hill coefficients of around 2. These findings are noteworthy because they indicate that sigmoidal kinetics are not limited to β-lactams and perhaps would apply to most of the substrates of AcrB, and possibly to other RND-type efflux pumps.
Sigmoidal kinetics is usually produced by positive cooperativity, and we favored the idea (10) that the trimeric nature of AcrB and its functionally rotating mechanism (27–29) are responsible. More specifically, the entry of the next ligand molecule to the proximal pocket of access or binding protomer, while a ligand is already bound to the distal pocket of binding protomer, would create a situation where two ligands are bound simultaneously to one pump complex, and this could create positive cooperativity (10). Among cephalosporins, nitrocefin showing a Michaelis–Menten kinetics has an exceptionally low KM value of around 5 μM, in comparison with other compounds showing sigmoidal kinetics, which have the K0.5 values of 20–290 μM. Also in MD simulation, nitrocefin appeared to bind to the distal binding pocket with an exceptionally strong affinity [the free energy of binding calculated with the molecular mechanics (MM)/generalized Born/solvent-accessible surface area (GBSA) approach (30) without entropy-correction was −42.5 kcal/mol], in comparison with other β-lactams [the corresponding values for cephalothin and oxacillin were −34.6 (or −28.1 in an alternative simulation run) and −23.2 kcal/mol, respectively] (8). Thus, a ligand that binds exceptionally tightly, such as nitrocefin, may not need the entry of the second ligand molecule. Aminoacyl-naphthylamides also appear to bind loosely to the binding pocket, on the basis of their high K0.5 values (41–148 μM) (see Results) of their efflux and of the low binding energy of between −22.9 and −27.2 kcal/mol found in the MD simulation; this fits with the finding of sigmoidal kinetics. The same comments apply also to the efflux of PAβN, which shows a clearly sigmoidal kinetics (Fig. 3A); its relatively high K0.5 value (17.6 μM) is consistent with its not very-tight binding to AcrB, although MD simulation suggested that it binds more tightly to the distal pocket than other aminoacyl naphthylamides (Table 1).
Finally, even nitrocefin may be pumped out with sigmoidal kinetics when its binding to the binding pocket becomes weaker, for example, by the simultaneous presence of inhibitor PAβN (Fig. 4) or by mutation within the binding site, such as F610A (31). These results further reinforce the idea that the loose binding of substrates is the main cause of the positive cooperativity and sigmoidal kinetics.
This study showed, by examining aminoacyl naphthylamides for their behaviors as modulators and substrates of AcrB, how their interaction with the AcrB transporter could be interpreted in a rational manner. We hope that knowledge of this type would be useful for the development of better inhibitors of RND pumps, which may lead to a more effective chemotherapy for multidrug-resistant Gram-negative bacteria.
Experimental Procedures
Strains.
RAM121, an E. coli K-12 strain expressing a large-channel mutant porin (16) was used to construct ΔpepN::kan derivatives. RAM121ΔpepN::kan ΔacrAB::spc was constructed by transducing ΔacrAB::spc from HN1159 (10).
Construction and Characterization of an E. coli Strain Expressing a Periplasmic Aminopeptidase.
The pepN sequence of E. coli K12 was PCR-amplified and inserted between NotI and BamHI sites of the pMAL-p5X vector (New England Biolabs). The resulting plasmid, pMAL-PepN, was then electroporated into RAM121 ΔpepN::kan, and the transformants were selected on 100-μg/mL ampicillin plates. The periplasmic location of the MalE-PepN fusion protein was ascertained by the osmotic shock procedure, as given in SI Appendix.
Time Course of PepN-Catalyzed Hydrolysis of PAβN.
A French pressure-cell extract of strain RAM121 ΔpepN::kan/pMAL-PepN was used to digest 300 µM PAβN at room temperature. Digestion was stopped at various time points, and the products were analyzed, after conversion into phenylthiocarbamyl (PTC)-derivatives, by reverse-phase HPLC using a UV detector. For details, see SI Appendix.
Efflux Assay of PAβN.
PAβN at various concentrations was added to intact cells of RAM121 ΔpepN::kan containing pMAL-PepN, and the reaction was stopped after 3 min to measure the first step in PAβN hydrolysis. The free Phe generated was measured by fluorescence after modification with o-phthalaldehyde in the presence of mercaptoethanol. The data were analyzed here and below by curve-fitting with CurveExpert (www.curveexpert.net) with a stringent tolerance (10–9) and increased possible reiterations (300). In this and other efflux assays, the efflux rates were always calculated on the basis of cell dry weight, obtained from OD600 as described by Koch (32). For details, see SI Appendix.
Efflux Assay of Aminocyl-β-Naphthylamide.
Substrates were added to intact cells of E.coli RAM121 ΔpepN::kan/pMAL-PepN, and the generation of the hydrolysis product, naphthylamine, was followed by its fluorescence, as detailed in SI Appendix.
Nitrocefin Efflux Assay.
This was carried out as described previously (10), except that RAM121 ΔpepN::kan was used to avoid the degradation of the modulators.
Computational Methods.
Docking of Ala-Naph, Phe-Naph, and Arg-Naph to the deep (or distal) pocket of the tight (or binding or B) protomer was performed using the software AutoDock Vina (21). A 20 × 20 × 20 Å grid was centered near the center of the binding pocket, and the exhaustiveness parameter was set to 64. MD simulations of Ala-Naph, Arg-Naph, and Phe-Naph binding to the distal pocket of AcrB were performed in a similar way as reported earlier (8, 9). Namely, a truncated model was used, and each system was simulated up to 320 ns (including 20 ns of equilibration phase). See SI Appendix for more details.
Supplementary Material
Acknowledgments
This study was supported in part by US Public Health Service Grant AI-009644 in Berkeley. The research of A.V.V. leading to these results was conducted as part of the Translocation consortium (www.translocation.com) and received support from the Innovative Medicines Initiatives Joint Undertaking under Grant 115525, resources of which are composed of a financial contribution from the European Union’s seventh framework program (FP7/ 2007–2013) and European Federation of Pharmaceutical Industries and Association companies’ kind contribution.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525143113/-/DCSupplemental.
References
- 1.Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27(2):128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X-Z, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28(2):337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma D, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175(19):6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol. 2011;77:1–60. doi: 10.1002/9780470920541.ch1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggerone P, Murakami S, Pos KM, Vargiu AV. RND efflux pumps: Structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13(24):3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 6.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178(1):306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lomovskaya O, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109(50):20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother. 2014;58(10):6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2009;106(14):5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller CG. Protein degradation and proteolytic modification. In: Neidhardt FC, editor. Escherichia coli and Salmonella Cellular and Molecular Biology. Vol 1. ASM Press; Washington, DC: 1996. pp. 938–954. [Google Scholar]
- 12.Chandu D, Nandi D. PepN is the major aminopeptidase in Escherichia coli: Insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology. 2003;149(Pt 12):3437–3447. doi: 10.1099/mic.0.26518-0. [DOI] [PubMed] [Google Scholar]
- 13.Addlagatta A, Gay L, Matthews BW. Structural basis for the unusual specificity of Escherichia coli aminopeptidase N. Biochemistry. 2008;47(19):5303–5311. doi: 10.1021/bi7022333. [DOI] [PubMed] [Google Scholar]
- 14.Lazdunski A, Murgier M, Lazdunski C. Evidence for an aminoendopeptidase localized near the cell surface of Escherichia coli. Regulation of synthesis by inorganic phosphate. Eur J Biochem. 1975;60(2):349–355. doi: 10.1111/j.1432-1033.1975.tb21009.x. [DOI] [PubMed] [Google Scholar]
- 15.McCaman MT, Gabe JD. Sequence of the promoter and 5′ coding region of pepN, and the amino-terminus of peptidase N from Escherichia coli K-12. Mol Gen Genet. 1986;204(1):148–152. doi: 10.1007/BF00330202. [DOI] [PubMed] [Google Scholar]
- 16.Misra R, Benson SA. Isolation and characterization of OmpC porin mutants with altered pore properties. J Bacteriol. 1988;170(2):528–533. doi: 10.1128/jb.170.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher GH, et al. A fast and sensitive method for measuring picomole levels of total free amino acids in very small amounts of biological tissues. Amino Acids. 2001;20(2):163–173. doi: 10.1007/s007260170057. [DOI] [PubMed] [Google Scholar]
- 18.Lim SP, Nikaido H. Kinetic parameters of efflux of penicillins by the multidrug efflux transporter AcrAB-TolC of Escherichia coli. Antimicrob Agents Chemother. 2010;54(5):1800–1806. doi: 10.1128/AAC.01714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima S, Nikaido H. Permeation rates of penicillins indicate that Escherichia coli porins function principally as nonspecific channels. Proc Natl Acad Sci USA. 2013;110(28):E2629–E2634. doi: 10.1073/pnas.1310333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinana AD, Vargiu AV, Nikaido H. Some ligands enhance the efflux of other ligands by the Escherichia coli multidrug pump AcrB. Biochemistry. 2013;52(46):8342–8351. doi: 10.1021/bi401303v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima R, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 23.Bohnert JA, Kern WV. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother. 2005;49(2):849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B, Weng J, Wang W. Substrate binding accelerates the conformational transitions and substrate dissociation in multidrug efflux transporter AcrB. Front Microbiol. 2015;6:302. doi: 10.3389/fmicb.2015.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eicher T, et al. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife. 2014;3:3. doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer N, Kandt C. Three ways in, one way out: Water dynamics in the trans-membrane domains of the inner membrane translocase AcrB. Proteins. 2011;79(10):2871–2885. doi: 10.1002/prot.23122. [DOI] [PubMed] [Google Scholar]
- 27.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 28.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313(5791):1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 29.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollman PA, et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 31.Soparkar K, et al. Reversal of the drug binding pocket defects of the AcrB multidrug efflux pump protein of Escherichia coli. J Bacteriol. 2015;197(20):3255–3264. doi: 10.1128/JB.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch AL. Growth measurement. In: Reddy CA, editor. Methods for General and Molecular Microbiology. 3rd Ed. ASM Press; Washington, DC: 2007. pp. 172–199. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.