Significance
A major problem in determining the crystal structures of metalloenzymes is that the reducing power of X-rays often changes the oxidation state of the metal center, thereby complicating important mechanistic conclusions on enzyme function. This reduction is especially problematic in studying Fe(IV)=O intermediates, which are powerful oxidants used by many metalloenzymes. This problem can be circumvented using the Stanford Linear Coherent Light Source (LCLS), which generates intense X-ray pulses on the femtosecond time scale and enables structure determinations with no reduction of metal centers. Here, we report the crystal structure of the Fe(IV)=O peroxidase intermediate called compound I using data obtained from the LCLS. We also present kinetic and computational results that, together with crystal structures, provide important mechanistic insights.
Keywords: compound I, peroxidase, femtosecond crystallography, XFEL
Abstract
The reaction of peroxides with peroxidases oxidizes the heme iron from Fe(III) to Fe(IV)=O and a porphyrin or aromatic side chain to a cationic radical. X-ray–generated hydrated electrons rapidly reduce Fe(IV), thereby requiring very short exposures using many crystals, and, even then, some reduction cannot be avoided. The new generation of X-ray free electron lasers capable of generating intense X-rays on the tenths of femtosecond time scale enables structure determination with no reduction or X-ray damage. Here, we report the 1.5-Å crystal structure of cytochrome c peroxidase (CCP) compound I (CmpI) using data obtained with the Stanford Linear Coherent Light Source (LCLS). This structure is consistent with previous structures. Of particular importance is the active site water structure that can mediate the proton transfer reactions required for both CmpI formation and reduction of Fe(IV)=O to Fe(III)-OH. The structures indicate that a water molecule is ideally positioned to shuttle protons between an iron-linked oxygen and the active site catalytic His. We therefore have carried out both computational and kinetic studies to probe the reduction of Fe(IV)=O. Kinetic solvent isotope experiments show that the transfer of a single proton is critical in the peroxidase rate-limiting step, which is very likely the proton-coupled reduction of Fe(IV)=O to Fe(III)-OH. We also find that the pKa of the catalytic His substantially increases in CmpI, indicating that this active site His is the source of the proton required in the reduction of Fe(IV)=O to Fe(IV)-OH.
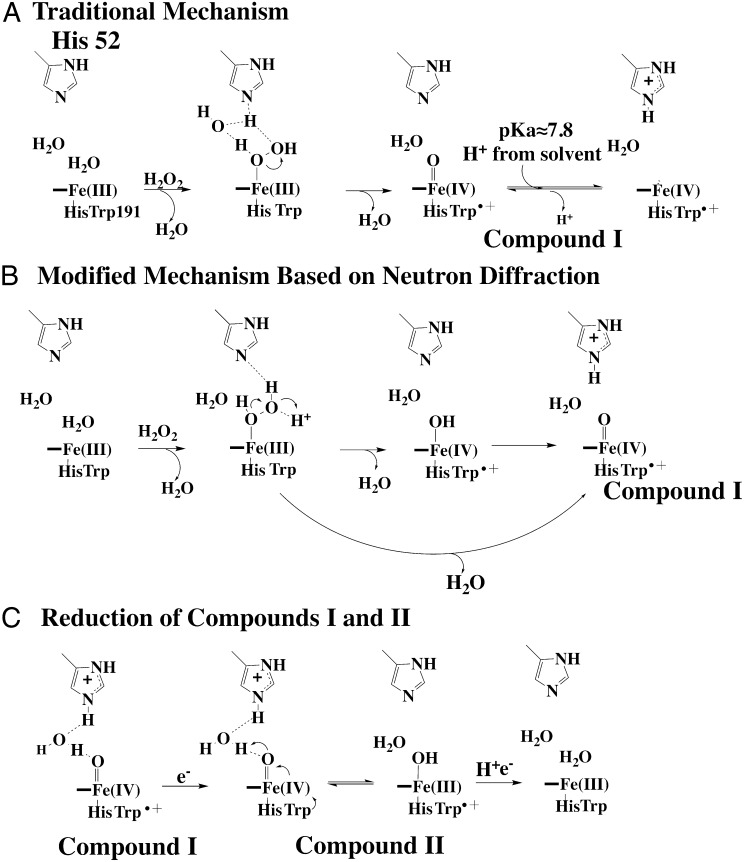
Peroxidases, and especially yeast cytochrome c peroxidase (CCP), have long played a central role in metalloenzymology owing to the relative ease of trapping and characterizing highly reactive intermediates (1). Of particular importance is compound I (CmpI; Fig. 1A), the first intermediate formed by oxidation of the heme center by H2O2. The first structure of a peroxidase (2) suggested a mechanism of CmpI formation wherein the catalytic His52 shuttles a proton from the iron-linked peroxide O atom to the distal peroxide O atom, thus promoting heterolytic fission of the O-O bond (3). This mechanism later was modified to include a water molecule (4) (Fig. 1A) that mediates the transfer of protons between the peroxide and His52. The water-modified mechanism is energetically more feasible because His52 is too far (≈3.6 Å) from the iron-linked peroxide for direct proton transfer. Crystal structures of CmpI support this water-modified mechanism. The required water bridges the Fe(IV)O oxygen atom and His52 and is perfectly positioned to serve a role in proton transfer (5, 6). However, a major problem with using crystallography to study high potential centers like Fe(IV)=O is that X-ray–generated photoelectrons can readily reduce metal centers in metalloprotein crystals. For some time, mechanistic conclusions were based on the incorrect assumption that the metal redox state remains unchanged during X-ray data collection. Quite prominent has been the discrepancy between spectroscopic studies and crystal structures of the ferryl Fe(IV)O, which is critically important not only for peroxidases but also for cytochrome P450 and NOS mechanisms (1). A majority of spectroscopic methods are most consistent with a short Fe(IV)=O double bond, whereas a number of crystal structures are consistent with an Fe(IV)-O single bond, leading to the incorrect conclusion that ferryl O atom is protonated to give Fe(IV)-OH (7). More careful low-radiation-dose composite data collection protocols, coupled with single crystal spectroscopy of CmpI (5, 6) have partially resolved this problem, and these more recent structures agree with the extensive spectroscopic data supporting Fe(IV)=O. Although peroxidase CmpI is relatively stable, it is impossible to prevent X-ray damage completely and it would be highly desirable to eliminate the X-ray–induced reduction of metal centers altogether. Until very recently, the only feasible way of obtaining such a structure was with neutron diffraction, and a 2.5-Å resolution neutron diffraction structure of CCP CmpI has recently been solved (8). Neutron diffraction offers a major advantage because it allows visualization of hydrogen atoms, thus enabling enzymatically important H-bond donor/acceptor relationships to be precisely determined. However, the experimental constraints for neutron diffraction, such as crystal size, hydrogen-deuterium exchange, and the limited facilities for neutron diffraction, have greatly limited its application. A second approach is to obtain X-ray data using the new generation of X-ray free-electron lasers (XFELs), which produce X-ray pulses on the tenths of femtosecond time scale. The extremely short, bright pulses allow diffraction to take place before significant radiation damage occurs (9–13). This method is particularly advantageous for the determination of catalytically relevant structures of metalloproteins, because diffraction is completed before atomic rearrangements occur around the metal center. Given the historical significance of CCP, we herein present a 1.5-Å structure of yeast CCP CmpI obtained from XFEL data collected at the Stanford Linear Coherent Light Source (LCLS). Although this structure, together with the neutron diffraction and other CmpI structures, is consistent with the mechanism shown in Fig. 1A, Casadei et al. (8) propose a substantially different mechanism based on the observation that the distal His52 is protonated in CmpI and that the H-bond donor/acceptor relationship does not support a water-mediated proton transfer mechanism but, rather, directs proton transfer from peroxide to His52 (Fig. 1B). Relevant to the discrepancy between the mechanisms in Fig. 1 A and B is the pKa of His52. The pKa is important, because we recently showed that the proton-coupled electron transfer reduction of CmpII (Fig. 1C) is rate-limiting in peroxidase catalysis (14) and a protonated His52 could directly participate in proton transfer to the ferryl O atom during reduction of Fe(IV)=O. Therefore, we also present computational and kinetic solvent isotope effect (KSIE) experiments that examine the pKa of His52 and address the importance of proton transfer in CmpII reduction. For the experimental work, we have used Leishmania major peroxidase (LmP) rather than yeast CCP. We and others have shown that LmP is structurally and functionally very similar to yeast CCP, including the structure of the LmP–cytochrome c (Cytc) complex, stability of CmpI, and formation of the active site Trp radical (15–18). LmP, however, offers some advantages owing to simplified kinetics because, unlike yeast CCP, LmP has only one binding site for Cytc and kcat is independent of ionic strength (14). As a result, the interpretation of kinetic results is more straightforward with LmP.
Fig. 1.
Peroxidase mechanism. (A) Traditional “water-modified” mechanism of CmpI formation. In this mechanism, peroxide first coordinates with the heme iron, followed by proton transfer to the distal peroxide O atom via an ordered water molecule and the distal His. The protonation state of the distal His depends on the His pKa. Our computational experiments indicate that the pKa substantially increases in CmpI. (B) Modified mechanism based on the observation that His52 in CCP CmpI is protonated in the neutron diffraction structure (8). Going from the initial peroxide complex to CmpI proceeds via two possible routes, one of which involves the Fe(IV)-OH intermediate. (C) Mechanism of CmpII reduction, which includes a proton-coupled electron transfer event resulting in a net transfer of a proton from the distal His to the ferryl O atom.
Results
Crystal Structures.
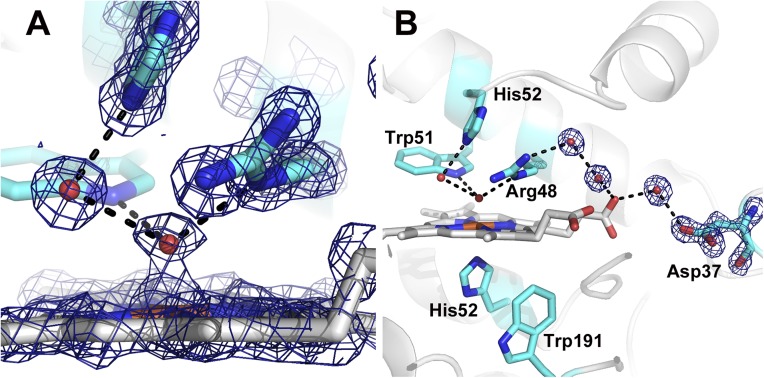
The X-ray damage-free XFEL structure of CCP CmpI is shown in Fig. 2 and is identical to our previous low-dose structure obtained by a composite data collection protocol that required several crystals, each exposed for no more than 10 s (6). The solvent structure is especially important, given that even partial reduction of CmpI alters the active site solvent architecture, possibly due to the change in charge on the heme iron and to the associated changes in the local electrostatic environment. The ordered solvent directly H-bonded to both the ferryl O atom and His52 is important to consider when discussing mechanism. Consistent with the low-dose structures, the Fe-O bond length is 1.7 Å, and thus is best described as Fe(IV)=O, whereas the Arg48 and Trp51 are in hydrogen bond contact with the ferryl oxygen. In earlier studies, we showed that the Trp191 radical is stable in the crystalline state (19), and we assume that the Trp191 radical is fully formed in our present structure. We find no changes in structure around Trp191, which is perhaps not unexpected because it has been demonstrated that the local electrostatic environment has been carefully tuned to stabilize a Trp cationic radical (1).
Fig. 2.
2Fo-Fc electron density map of the XFEL CmpI structure contoured at 2.0 σ. The dashed lines indicate H-bonding interactions, which are all less than 3.0 Å. The Fe(IV)-O bond length is 1.7 Å. Close-up view of the ferryl center is shown (A), as well as the extensive H-bonded network connecting the ferryl O atom to the surface of the enzyme (B).
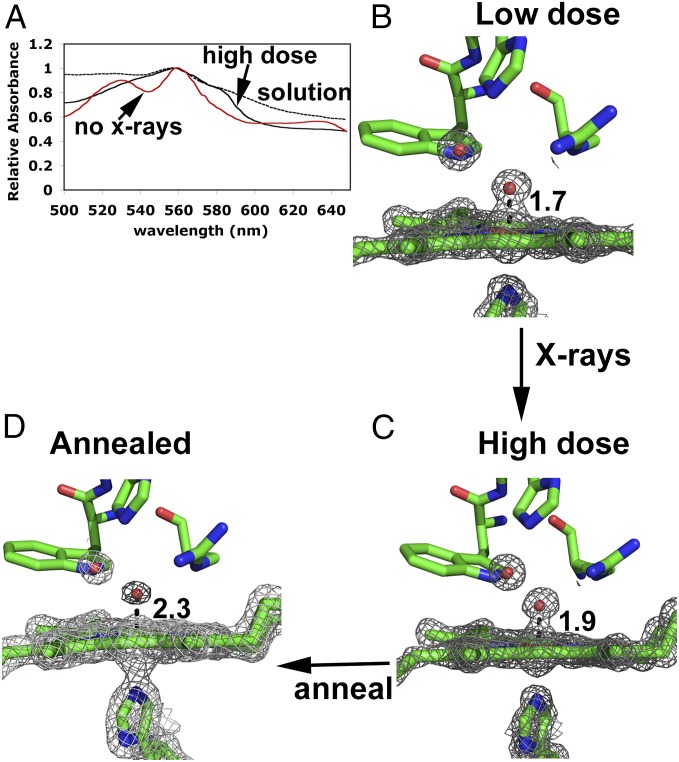
In our previous work, we found that the Fe-O bond length increases linearly with X-ray dose (6) because Fe(IV)=O is reduced by the X-ray beam. The final high-dose X-ray structure has an Fe-O bond length of 1.9 Å, but the redox state of the iron was uncertain. Fig. 3 shows the single crystal spectrum obtained from crystals before and after extensive X-ray exposure. The spectral features of the X-ray–reduced crystal are similar to the solution spectrum of dithionite–reduced CCP. Even so, a water/hydroxide only 1.9 Å from the iron would be very unusual for Fe(II) heme, whereas Fe(III)-OH would be more consistent with the known solution properties of heme proteins. Unfortunately, it is not possible to compare the solution and crystal spectra because CCP is unstable at the pH levels required to form Fe(III)-OH. When the crystal is annealed by briefly warming and then cooling back to liquid nitrogen temperatures, followed by collection of a 1.55-Å dataset, the water density moves to a distance of 2.3 Å from the iron and the electron density becomes much weaker (Fig. 3). The weaker electron density is indicative of low occupancy and/or higher thermal motion, which would be consistent with five-coordinate Fe(II) or Fe(III) or, possibly, a mixture of both. It is therefore very likely that the high-dose structure (Fig. 3C) represents a cryotrapped Fe(III)-OH or, possibly, Fe(II)-OH2. This conclusion also suggests that previous structures of CmpI or CmpII with a long Fe-O bond are Fe(III) or Fe(II) and not Fe(IV).
Fig. 3.
Spectra and structure of CmpI as a function of X-ray dose. (A) Single crystal spectra of CmpI before and after extensive (7.8 MGy) X-ray exposure and the solution spectrum of CCP reduced anaerobically with dithionite. 2Fo-Fc maps of the low-dose (B) and high-dose (C) CmpI structures taken from Meharenna et al. (6) are shown. (D) Structure (1.5 Å) of a high-dose crystal after annealing. All maps were contoured at 2.0 σ.
pKa of Catalytic Distal His.
The neutron diffraction structure of CCP CmpI shows that the distal His52 is protonated in CmpI but is unprotonated in the resting Fe(III) state. Based on these observations, Casadei et al. (8) have proposed a mechanism for CmpI formation (Fig. 1B) that differs substantially from the generally accepted mechanism (4, 20) (Fig. 1A). The main difference is that the mechanism in Fig. 1B requires a proton from some external source and Fe(IV)-OH as a possible intermediate in CmpI before movement of the proton from the ferryl hydroxide to His52, thus generating Fe(IV)=O.
There are at least two problems with this mechanism. First, the possible existence of Fe(IV)-OH is unlikely, given that extended X-ray atomic fine structure and resonance Raman studies show that the pKa of the ferryl O atom in ferryl systems with an axial His ligand is ≤4 (7, 21). Second, Casadei et al. (8) reject the water-mediated mechanism of CmpI formation (Fig. 1A) proposed by Vidossich et al. (4), based on the H-bond donor/acceptor relationships observed in the neutron diffraction structure. However, it is the H-bonding pattern in the transition state, and not CmpI, that is relevant, so the conclusion that the H-bonding pattern of the ordered water in CmpI is not consistent with water-mediated proton transfer is not sound. In addition, the peroxide iron-linked O atom is about 3.6 Å from His52, which is too long for direct H-bonding and proton transfer. As shown by the computational work of Vidossich et al. (4), a water-mediated proton transfer mechanism as depicted in Fig. 1A is energetically more feasible. This mechanism also is consistent with the CmpI X-ray crystal structures because the water molecule H-bonded to both His52 and the ferryl O atom is ideally positioned to mediate proton transfer.
One simple explanation for why His52 is observed to be protonated in the neutron diffraction structure is that the pKa of His52 increases in CmpI. This explanation is reasonable because the crystals were grown at pH 6, so one might expect a significant fraction of His52 to be protonated if the pKa were 7 or higher. As outlined in Materials and Methods, we used two computational procedures to estimate the pKa of His52 in both the resting Fe(III) state and CmpI. Both methods give a substantial increase in the pKa of His52 (Table 1) from ≈5.1–5.4 in the ferric resting state to ≈7.6–7.8 in CmpI, consistent with the neutron diffraction structure, where the crystals were grown at pH 6 (8). These results are also consistent with resonance Raman studies (22, 23), where a group, most likely the distal His, with a pKa near 8.8 (24) affects the Fe(IV)=O stretching frequency of horseradish peroxidase CmpII via changes in H-bonding strength to the ferryl O atom. The increase in pKa can be readily rationalized by the structures and the charges on the heme atoms derived from density functional calculations. In the resting Fe(III) state, there is little dielectric shielding between His52 and the heme iron. This shielding, together with the nearby Arg48, surrounds His52 with an electropositive environment that depresses the His52 pKa relative to a His free in solution. In CmpI, however, the partial negative charge on the ferryl O atom increases the basicity of His52, which is why a protonated His52 is observed in the neutron diffraction structure (8) and is not due to an unknown source of protons as depicted in Fig. 1B.
Table 1.
Calculated pKa values of His52
| Method | pKa resting | pKa CmpI |
| H++/MEAD | 5.4 | 7.4 |
| Constant pH | 5.1 | 7.8 |
KSIEs.
Despite the problematic mechanistic conclusions based on the neutron diffraction structure (8), that His52 is observed to be protonated is a critically important finding relevant to the reduction of CmpII (Fig. 1C). In the first electron transfer step to CmpI, Cytc delivers an electron to the Trp radical to give CmpII. The reduction of CmpII involves internal electron transfer from Trp to Fe(IV)=O to give the Trp radical and Fe(III)-OH (25, 26). As shown in Fig. 1C, protonation of the ferryl O atom during reduction of CmpII requires transfer of the His52 proton via ordered solvent. This step is thought to be rate-limiting for CCP at high ionic strength (25, 26), whereas for the closely related LmP, this step is limiting at all ionic strengths (14). If CmpII reduction is limiting and solvent plays an important role in proton transfer, as suggested by the CmpI structures, then one might predict a considerable KSIE.
The KSIE studies were carried out with LmP and its physiological redox partner L. major cytochrome c (LmCytc) rather than CCP. As noted in the Introduction, LmP is also a CCP and forms the same stable Trp radical (17), and the crystal structure of the LmP–LmCytc complex (16) is very similar to the CCP–Cytc complex (27). LmP, however, exhibits simpler kinetics because kcat for LmP is relatively insensitive to ionic strength (14), whereas kcat for CCP increases substantially with ionic strength (25). This increase is generally thought to be the result of a change in rate-limiting step in CCP, where product dissociation is limiting at low ionic strength and reduction of CmpII is limiting at high ionic strength (25, 26). An additional complication with CCP is the presence of a second low-affinity site for Cytc at low ionic strength (25, 28). Given that the rate-limiting step for LmP appears not to change with ionic strength and that there is no indication of a second Cytc binding site, we focused on LmP for detailed KSIE studies.
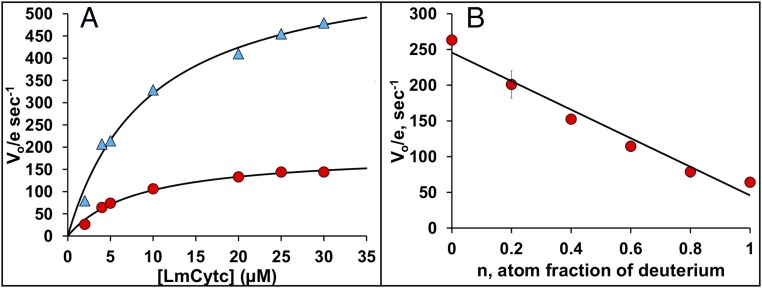
As shown in Fig. 4A, the KSIE is 3.3 with no effect on Km, clearly indicating that proton transfer is critical in the rate-limiting step. We also determined the KSIE as a function of pH and found that the KSIE increases slightly as the pH increases, to a maximum of 4.4 at pH 7.0 (Table 2), with maximum activity peaking near pH 7.0 and then decreasing at pH 8.0. These observations are consistent with previous studies (18). A plot of rate vs. fraction of D2O (proton inventory, Fig. 4B) shows a linear relationship, indicating that a single proton is involved in the rate-limiting step.
Fig. 4.
KSIEs. (A) Comparison of the LmP reaction in H2O (blue triangles) and D2O (red circles) together with kcat and Km determinations using fits of these plots to the Michaelis–Menten equation. (B) Proton inventory plot of rate vs. fraction of D2O.
Table 2.
KSIE as a function of pH/pD
| pH/pD | D2O | H2O | H2O/D2O ratio |
| Vo/e, s−1 | |||
| 6.0 | 58.9 ± 3 | 175 ± 7 | 3.0 ± 0.3 |
| 7.0 | 76.2 ± 6 | 338 ± 8 | 4.4 ± 0.4 |
| 8.0 | 66.2 ± 7 | 261 ± 10.2 | 3.9 ± 0.6 |
All measurements were done in 5 mM potassium phosphate (KH2PO4/K2HPO4) buffer titrated to the appropriate pH/pD, taking into account the following relationship between pH and pD: pD = pHobs + 0.38. Additional details are provided in Materials and Methods. Vo/e, [velocity/(enzyme)].
Conclusions
The present work, together with a wealth of data on peroxidase catalysis, supports the water-modified traditional peroxidase mechanism of CmpI formation highlighted in Fig. 1B. This conclusion includes the recent data provided by the neutron diffraction structure determined by Casadei et al. (8). One factor that was previously omitted, and that is critical to a correct interpretation of the mechanism, is that the His52 protonation observed in the CmpI neutron diffraction structure (8) is almost certainly due to an increase in the His52 pKa. This increase in the His52 pKa has important implications for CmpII reduction. Reduction of Fe(IV)=O to Fe(IV)-OH requires both electron and proton transfer, which our KSIE studies suggest is rate-limiting. As depicted in Fig. 1C, His52 is the ideal source of the required proton. Once the iron is reduced to Fe(III), the pKa returns to the resting state value of ≈5.1–5.4. Our CmpI structure is consistent with this mechanism and with previous crystallographic work based on complicated composite data protocols, which validates the earlier studies (5, 6). Our work also illustrates the utility of using the new generation of XFELs to carry out traditional crystal structure determinations with the obvious advantage of no X-ray damage or reduction of metal centers, thus providing an accessible and beneficial tool in structural biology for probing short-lived intermediates in enzyme catalysis.
Materials and Methods
Protein Expression, Purification, and Crystallization.
The N184R mutant of CmpI was produced and crystallized as previously reported (29). Briefly, 10 μL of sitting drops containing 400 μM protein, 22% (vol/vol) (4S)-2-methyl-2,4-pentanediol (MPD), and 50 mM Tris⋅phosphate (pH 6.0) were seeded and incubated at 4 °C for a few days. Freshly grown crystals were soaked in 10 mM H2O2, 35% (vol/vol) MPD, and 50 mM Tris⋅phosphate (pH 6.0). Crystals ranging between 150 μm and 1 mm in length were harvested onto Hampton-style cryoloops, flash-frozen, and stored in a Stanford Synchrotron Radiation Lightsource (SSRL) cassette.
LmP and LmCytc used in KSIE experiments were expressed and purified as previously reported (14).
Data Collection.
Data were collected at the X-ray pump probe end station at the LCLS in December 2013 and in June 2014 under a stream of liquid nitrogen. Crystals were mounted on a goniometer using the Stanford Automated Mounting System (30), and diffraction patterns were collected on a Rayonix MX325 detector as described previously (31). The helical data collection mode was used in which each crystal was translated and rotated after each exposure (31) so that multiple radiation damage-free diffraction images were obtained from each crystal. During the December 2013 beam time, a total of 96 still images were collected from nine crystals. Crystals were exposed to a 3-μm × 3-μm X-ray beam at a photon energy of 9.49 keV with a pulse length of 25 fs. Crystals were translated by 50 μm and rotated by 0.5° between exposures. During the June 2014 experiment, 275 stills were collected from 25 crystals using a 15-μm × 15-μm beam at a photon energy of 9.43 keV and a pulse length of 40 fs. Crystals were translated by 60 μm and rotated by 0.5° between exposures.
Crystallographic Data Processing.
Data were indexed and integrated separately for each experiment using nXDS (32) with profile fitting turned off, and the minimum Ewald offset correction set to 0.1. Reflections from both experiments were scaled together with XSCALE, and intensities were converted to structure factor amplitudes using XDSCONV. Two hundred fifty-three images of a total of 371 collected images were used in the final dataset.
Molecular replacement was performed using MOLREP (33) utilizing data between 18 Å and 1.5 Å. A low-radiation dose structure of CCP CmpI obtained at SSRL beamline 9-2 [Protein Data Bank (PDB) ID code 3M23] (6), from which the heme was removed, was used as a starting structure for molecular replacement. The MR structure was refined for 10 cycles at 1.5 Å using the program REFMAC (34), after which the heme was modeled in. Several more rounds of refinement were performed with all restraints on the iron removed, including the Fe-N and Fe-O restraints. Processing and refinement statistics are shown in Tables S1 and S2.
Table S1.
Data collection statistics
| Experiment | December 2013 | June 2014 |
| Source | LCLS XPP hutch | LCLS XPP hutch |
| Crystals exposed | 9 | 25 |
| Images collected | 96 | 275 |
| Images indexed | 46 | 217 |
| Wavelength, Å | 1.30591 | 1.31319 |
| Photon energy, keV | 9.436 | 9.494 |
| Pulse energy | 14.465 | 14.4150 |
| Crystal to detector distance, mm | 143.6 | 143.1 |
| Pulse length, fs | 25 | 40 |
| Electron beam energy | 14.465 | 14.415 |
XPP, X-ray pump probe.
Table S2.
Crystallographic refinement
| PDB ID code | 5EJX | 5EJT |
| Total images collected | 371 | 360 |
| Images indexed | 253 | 360 |
| Reflections used | 600,891 | 722,513 |
| Unique reflections | 65,592 | 60,803 |
| Completeness | 97.8% | 100 |
| I/σ | 3.33 | 14.5 |
| CC1/2 | 61.7% | 30.0 |
| R/Rfree | 23.4/26.1 | 15.3/17.7 or 14.4/17.3 |
| Space group | 19 | 19 |
| a, b, c; Å | 107.47, 74.99, 51.25 | 107.32, 75.19, 51.15 |
| α, β, γ; ° | 90, 90, 90 | 90, 90, 90 |
| Resolution range, Å | 18–1.5 | 39–1.55 |
CC1/2, percentage of correlation between intensities from random half data sets; I/σ, mean intensity/Σ(I) of unique reflections after merging symmetry related observations and Σ(I) = standard deviation of reflection intensity I estimated from sample statistics.
Annealed Crystal Structure.
We have shown previously that the CmpI spectrum in the crystal (Fig. 3A) is identical to the solution spectrum and that significant reduction does not occur until a dose of ≈0.1 MGy is attained (6), as calculated by RADDOSE (35). However, at a 7- to 8-MGy dose, the spectrum dramatically changes (“high-dose” spectrum in Fig. 3A) and the Fe-O distance increases to 1.9 Å (Fig. 3B). To probe the nature of the X-ray–reduced species further, data were collected at two different positions on a 0.2-mm3 × 0.2-mm3 × 0.15-mm3 CmpI crystal at 13,000 eV on BL12-2, resulting in an absorbed dose of 7.8 MGy, which ensures complete reduction. The visible spectrum of the crystal was measured on beamline BL9-1 and showed significant photoreduction. The crystal was then annealed by blocking the cryostream for 10 s. Gas bubbles were observed leaving the crystal during the warming of the crystal. The visible spectrum of the crystal was measured again, and surprisingly demonstrated some ferryl features. The crystal was exposed for an additional 1,000 s at 13,000 eV with a 200-μm X-ray beam on beamline BL9-1. The absorbed dose for the exposure was calculated to be 0.8 MGy, resulting in a cumulative absorbed dose of 8.6 MGy for the crystal. The crystal was annealed again, and the measured visible spectrum retained ferryl features. Finally, a 360° dataset with 10 s per degree was collected to a resolution of 1.55 Å. A calculated additional 2.87 MGy was absorbed by the crystal, resulting in a cumulative dose of 11.5 MGy. The dataset was processed by the SSRL’s “autoxds” script using XDS (36), POINTLESS, and AIMLESS (37), utilizing the AIMLESS CC1/2 (half data set correlation coefficient) > 30% criteria for a resolution cutoff. The previous CmpI structure (PDB ID code 3M23) was also used to initiate refinement using BUSTER (38). Data collection and refinement statistics are listed in Tables S1 and S2.
Computational Methods.
Two procedures were used for estimating the pKa of the catalytic distal His in both ferric resting state CCP (PDB ID 2CYP) and CCP CmpI (PDB ID code 3M23). In both cases, protein charges were assigned using the Amber ff99SB force field. Heme parameters, including the oxyferryl oxygen atom, were taken from density functional calculations by Harris and Loew (39). The first method used the H++ webserver (biophysics.cs.vt.edu/index.php), which is a simplified user-friendly adaption of MEAD (Macroscopic Electrostatics with Atomic Detail) (40). This approach uses a continuum dielectric model to calculate the electrostatic difference in work required to change the protonation state of a titratable group in solvent compared with the lower dielectric milieu within the protein.
The second approach is based on the methods initially developed by Warshel et al. (41) and later adapted for Amber by Mongan et al. (42). Here, the free energy required to deprotonate a titratable group in the protein (ΔGprotein) is compared with the same calculation free in solution (ΔGwater) and the ΔΔG = (ΔGprotein − ΔGwater) is used to calculate the pKa of the protein-bound group. The constant pH protocols using a Generalized Born implicit solvent model as implemented in Amber 12 were used. Provided with Amber 12 are the free energy calculations for all standard titratable amino acids free in solution, which leaves only the ΔGprotein to be calculated. This calculation is achieved by a Monte Carlo sampling of the Boltzman distribution of the two possible protonation states during a molecular dynamics run. In our case, a Monte Carlo step was performed every 100 fs over a 2-ns molecular dynamics run and the pH was set to 7.0. Because explicit solvent was not used, we found, consistent with previous results (43), that the protein can adopt unrealistic conformations. Therefore, a 1.0-kcal/mol restraint was placed on backbone atoms. The output provides the fraction of time the titratable group spends in any one of the two protonation states, which is directly related to ΔGprotein from which ΔΔG can be calculated. The pKa in the protein is readily calculated from
KSIEs.
All kinetic experiments were performed at room temperature on a Cary 300 UV/Vis spectrophotometer. LmCytc was reduced by adding a small excess of fresh sodium ascorbate and incubated on ice for 1 h. All LmCytc solutions used were ensured to contain no more than 2% LmCytc(III). Deuterated buffers for KSIE experiments were prepared in 99.9% D2O (EMD Millipore), and pD (pH in D2O) was adjusted according to the following relationship: pD = pHobs + 0.38. LmP and LmCytc stock solutions were highly concentrated using 10,000 molecular weight cutoff Amicon concentrators (Millipore) to ensure minimal protium contribution to deuterated buffers, and were equilibrated in D2O buffer before each activity measurement. All concentrations were determined using the appropriate molar extinction coefficients (ε558 of 29 mM−1⋅cm−1 for reduced LmCytc, ε408 of 113.6 mM−1⋅cm−1 for LmP, and ε240 of 0.0436 mM−1⋅cm−1 for H2O2), and LmCytc oxidation rates were calculated using a Δε558 of 19.4 mM−1⋅cm−1. The reaction was initiated by the addition of H2O2 (0.15 mM), and LmCytc(II) oxidation was monitored at 558 nm. The Km and kcat measurements were done in 25 mM potassium phosphate (KH2PO4/K2HPO4) at pH 6.8 and in the same deuterated buffer at pD 6.8. Data were fit according to the following hyperbolic term:
The pH/pD dependent assays were done in 5 mM KH2PO4/K2HPO4 buffer at pH/pD 6.0, pH/pD 7.0 and pH/pD 8.0, and in a final reaction mixture containing 0.5 nM LmP and 30 μM LmCytc.
Acknowledgments
We thank Prof. Michael T. Green for critically reviewing this manuscript and for many helpful suggestions. This work was supported by NIH Grant GM57353 (to T.L.P.). Use of the LCLS, Stanford Linear Accelerator Center (SLAC) National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by the NIH, National Institute of General Medical Sciences (including Grant P41GM103393).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5EJX and 5EJT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521664113/-/DCSupplemental.
References
- 1.Poulos TL. Heme enzyme structure and function. Chem Rev. 2014;114(7):3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulos TL, et al. The crystal structure of cytochrome c peroxidase. J Biol Chem. 1980;255(2):575–580. [PubMed] [Google Scholar]
- 3.Poulos TL, Kraut J. The stereochemistry of peroxidase catalysis. J Biol Chem. 1980;255(17):8199–8205. [PubMed] [Google Scholar]
- 4.Vidossich P, et al. On the role of water in peroxidase catalysis: A theoretical investigation of HRP compound I formation. J Phys Chem B. 2010;114(15):5161–5169. doi: 10.1021/jp911170b. [DOI] [PubMed] [Google Scholar]
- 5.Berglund GI, et al. The catalytic pathway of horseradish peroxidase at high resolution. Nature. 2002;417(6887):463–468. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 6.Meharenna YT, Doukov T, Li H, Soltis SM, Poulos TL. Crystallographic and single-crystal spectral analysis of the peroxidase ferryl intermediate. Biochemistry. 2010;49(14):2984–2986. doi: 10.1021/bi100238r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green MT. Application of Badger’s rule to heme and non-heme iron-oxygen bonds: An examination of ferryl protonation states. J Am Chem Soc. 2006;128(6):1902–1906. doi: 10.1021/ja054074s. [DOI] [PubMed] [Google Scholar]
- 8.Casadei CM, et al. Neutron cryo-crystallography captures the protonation state of ferryl heme in a peroxidase. Science. 2014;345(6193):193–197. doi: 10.1126/science.1254398. [DOI] [PubMed] [Google Scholar]
- 9.Barty A, et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat Photonics. 2012;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470(7332):73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kern J, et al. Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nat Commun. 2014;5:4371. doi: 10.1038/ncomms5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomb L, et al. Radiation damage in protein serial femtosecond crystallography using an x-ray free-electron laser Phys. Rev B Condens Matter Mater Phys. 2011;84:21411–21416. doi: 10.1103/PhysRevB.84.214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406(6797):752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 14.Chreifi G, et al. Enzymatic Mechanism of Leishmania major Peroxidase and the Critical Role of Specific Ionic Interactions. Biochemistry. 2015;54(21):3328–3336. doi: 10.1021/acs.biochem.5b00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adak S, Datta AK. Leishmania major encodes an unusual peroxidase that is a close homologue of plant ascorbate peroxidase: A novel role of the transmembrane domain. Biochem J. 2005;390(Pt 2):465–474. doi: 10.1042/BJ20050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jasion VS, Doukov T, Pineda SH, Li H, Poulos TL. Crystal structure of the Leishmania major peroxidase-cytochrome c complex. Proc Natl Acad Sci USA. 2012;109(45):18390–18394. doi: 10.1073/pnas.1213295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasion VS, Polanco JA, Meharenna YT, Li H, Poulos TL. Crystal structure of Leishmania major peroxidase and characterization of the compound i tryptophan radical. J Biol Chem. 2011;286(28):24608–24615. doi: 10.1074/jbc.M111.230524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jasion VS, Poulos TL. Leishmania major peroxidase is a cytochrome c peroxidase. Biochemistry. 2012;51(12):2453–2460. doi: 10.1021/bi300169x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonagura CA, et al. High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase. Biochemistry. 2003;42(19):5600–5608. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 20.Poulos TL, Kraut J. A hypothetical model of the cytochrome c peroxidase. cytochrome c electron transfer complex. J Biol Chem. 1980;255(21):10322–10330. [PubMed] [Google Scholar]
- 21.Behan RK, Green MT. On the status of ferryl protonation. J Inorg Biochem. 2006;100(4):448–459. doi: 10.1016/j.jinorgbio.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Mukai M, Nagano S, Tanaka M, Ishimori K. Effects of concerted hydrogen bonding of distal histidine on active site structures of horseradish peroxidase. Resonance Raman studies with Asn70 mutants. J Am Chem Soc. 1997;119(7):1758–1766. [Google Scholar]
- 23.Terner J, et al. Resonance Raman spectroscopy of oxoiron(IV) porphyrin π-cation radical and oxoiron(IV) hemes in peroxidase intermediates. J Inorg Biochem. 2006;100(4):480–501. doi: 10.1016/j.jinorgbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto S, Tatsuno Y, Kitagawa T. Resonance Raman evidence for oxygen exchange between the FeIV = O heme and bulk water during enzymic catalysis of horseradish peroxidase and its relation with the heme-linked ionization. Proc Natl Acad Sci USA. 1986;83(8):2417–2421. doi: 10.1073/pnas.83.8.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MA. A complete mechanism for steady-state oxidation of yeast cytochrome c by yeast cytochrome c peroxidase. Biochemistry. 1996;35(49):15791–15799. doi: 10.1021/bi961488c. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, et al. Design of a ruthenium-cytochrome c derivative to measure electron transfer to the radical cation and oxyferryl heme in cytochrome c peroxidase. Biochemistry. 1996;35(47):15107–15119. doi: 10.1021/bi9611117. [DOI] [PubMed] [Google Scholar]
- 27.Pelletier H, Kraut J. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science. 1992;258(5089):1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 28.Mauk MR, Ferrer JC, Mauk AG. Proton linkage in formation of the cytochrome c-cytochrome c peroxidase complex: Electrostatic properties of the high- and low-affinity cytochrome binding sites on the peroxidase. Biochemistry. 1994;33(42):12609–12614. doi: 10.1021/bi00208a011. [DOI] [PubMed] [Google Scholar]
- 29.Meharenna YT, Oertel P, Bhaskar B, Poulos TL. Engineering ascorbate peroxidase activity into cytochrome c peroxidase. Biochemistry. 2008;47(39):10324–10332. doi: 10.1021/bi8007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beam line, using compact sample cassettes and a small-scale robot. J Appl Cryst. 2002;35(6):720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen AE, et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc Natl Acad Sci USA. 2014;111(48):17122–17127. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. Processing of X-ray snapshots from crystals in random orientations. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 8):2204–2216. doi: 10.1107/S1399004714013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 34.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papagrigoriou E, et al. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23(15):2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smart OS, et al. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris DL, Loew GH. Proximal ligand effects on electronic structure and spectra of compound I of peroxidases. J Porphyr Phthalocyanines. 2001;5:334–344. [Google Scholar]
- 40.Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 41.Warshel A, Sussman F, King G. Free energy of charges in solvated proteins: Microscopic calculations using a reversible charging process. Biochemistry. 1986;25(26):8368–8372. doi: 10.1021/bi00374a006. [DOI] [PubMed] [Google Scholar]
- 42.Mongan J, Case DA, McCammon JA. Constant pH molecular dynamics in generalized Born implicit solvent. J Comput Chem. 2004;25(16):2038–2048. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 43.Swails JM, York DM, Roitberg AE. Constant pH Replica Exchange Molecular Dynamics in Explicit Solvent Using Discrete Protonation States: Implementation, Testing, and Validation. J Chem Theory Comput. 2014;10(3):1341–1352. doi: 10.1021/ct401042b. [DOI] [PMC free article] [PubMed] [Google Scholar]