SUMMARY
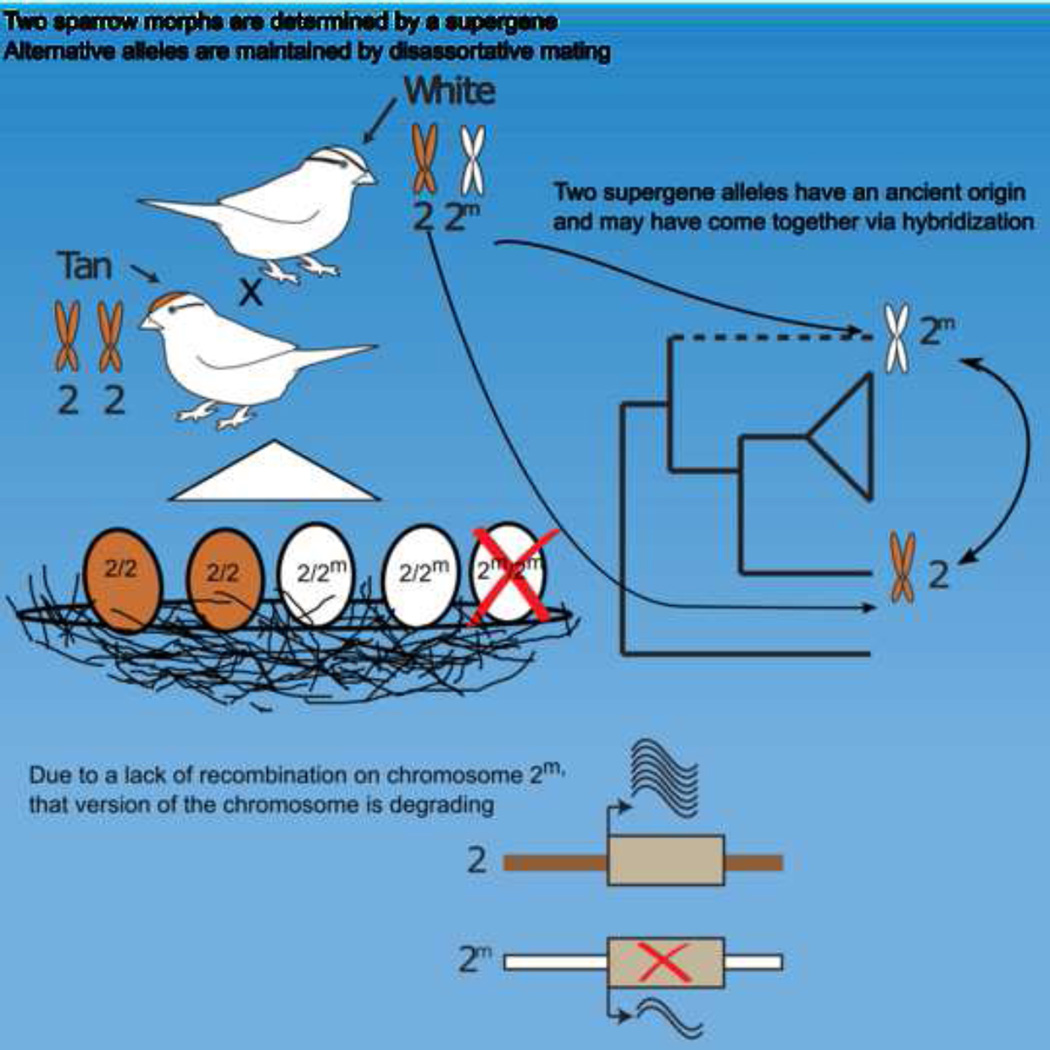
A major challenge in biology is to understand the genetic basis of adaptation. One compelling idea is that groups of tightly linked genes (i.e. ‘supergenes’ [1, 2]) facilitate adaptation in suites of traits that determine fitness. Despite their likely importance, little is known about how alternate supergene alleles arise and become differentiated, nor their ultimate fate within species. Herein we address these questions by investigating the evolutionary history of a supergene in white-throated sparrows, Zonotrichia albicollis. This species comprises two morphs, tan and white, that differ in pigmentation and components of social behavior [3–5]. Morph is determined by alternative alleles at a balanced >100Mb inversion-based supergene, providing a unique system for studying gene-behavior relationships. Using over two decades of field data we document near-perfect disassortative mating among morphs, as well as the fitness consequences of rare assortative mating. We use de novo whole genome sequencing coupled with population- and phylo-genomic data, to show that alternate supergene alleles are highly divergent at over 1,000 genes, that these alleles originated prior to the split of Z. albicollis from its sister species, and may be polymorphic in Z. albicollis due to a past hybridization event. We provide evidence that the ‘white' allele may be degrading, similar to neo-Y/Wsex chromosomes. We further show that the ‘tan’ allele has surprisingly low levels of genetic diversity, yet does not show several canonical signatures of recurrent positive selection. We discuss these results in the context of the origin, molecular evolution, and possible fate of this remarkable polymorphism.
Graphical Abstract
RESULTS AND DISCUSSION
Understanding how suites of adaptive characters remain linked despite the disruptive forces of recombination has been a major challenge in evolutionary biology. ‘Supergenes’ are linked clusters of coevolved genes that, like sex chromosomes, give rise to divergent fitness-related traits that are variable within species [1, 2]. Nonetheless, how supergenes originate, how they generate adaptive variation, and how they are maintained and evolve within a species remains unclear. To examine these basic questions, we analyzed supergene evolution in the white-throated sparrow (Zonotrichia albicollis). The two plumage morphs (Figure 1A) of the white-throated sparrow differ dramatically in reproductive behavior [3, 5], representing two extremes in reproductive tradeoffs: white males are promiscuous and invest heavily in securing additional matings at the expense of paternal care [3] whereas tan males are monogamous [3] and contribute more to parental care [6]; females exhibit similar trade-offs between investment in parental care versus mating effort [3].
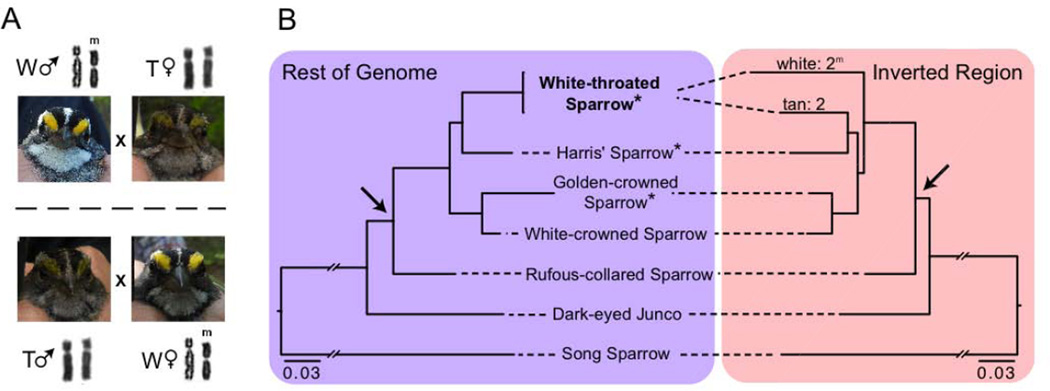
Figure 1. White-throated Sparrows comprise two morphs with an ancient origin.
(A) Tan (T) and white (W) morph sparrows (Note differences in plumage color in the head stripe and throat) differ in chromosome 2 (sometimes referred to as ZAL2) genotype and mate almost exclusively with the opposite morph, maintaining polymorphism in the species. (B) Tan (2) and white (2m) chromosomes are highly divergent within the inverted portion of chromosome, suggesting an ancient origin of the chromosomes 2 and 2m. All nodes are supported by 100% bootstrap values and the depicted topology for the inverted region is a significantly better fit to the data than trees constrained to the monophyly of 2 and 2m (p < 0.001, see Supplementary Materials). Taxa marked with an asterisk are those for which whole genomes were sequenced. The arrows highlights the ancestral node for the Zonotrichia genus. See also Table S2.
Our long-term genotypic analysis builds on previous work [6, 7], and through extensive genotyping of thousands of individuals over more than two decades, confirms that white morphs are almost always heterozygous for alternative chromosome 2 alleles (2m/2). We find that 99.7% of white morphs are heterozygous (n=1,014, Table S1), whereas tan morphs are always homozygous (2/2; 100%, n=978). Thus, chromosomes 2 and 2m segregate at ~75 and ~25% frequency, respectively, and the two plumage morphs occur in approximately equal numbers. Chromosome 2 remains polymorphic via near-obligate mating between tan and white morphs (i.e., disassortative mating) [8]. Only 17 out of 1,116 pairs (1.5%) observed in this study mated with an individual of the same morph (i.e. homotypic or assortative mating). Homotypic pairs are even less common in primary pairings (9/1106, 0.8%), but occur more commonly in secondary pairings (8/10, 80%; Table S2) following the disappearance of an opposite-morph mate and when mate-choice options are limited. As a consequence of obligate disassortative mating the species effectively has four sexes, wherein any individual can mate with only 1/4 of the individuals in the population.
Given the myriad phenotypic and behavioral differences controlled by this inversion polymorphism [3], and the unusual dynamics of the mating system, the white-throated sparrow harbors a classic example of a balanced supergene [9]. To resolve the evolutionary history of this supergene, we first sequenced and assembled the genome of a single tan male (homozygous 2/2 and Z/Z; Figure 1), resulting in a draft genome 1.03Gb long, comprising 6,018 scaffolds, 13,811 protein-coding genes, 1,104 non-coding transcripts, and 205 pseudogenes. The assembled genome has a contig N50 of 113kb and a scaffold N50 of 4.9 megabases, comparable or better than most recent short read-based bird assemblies [10].
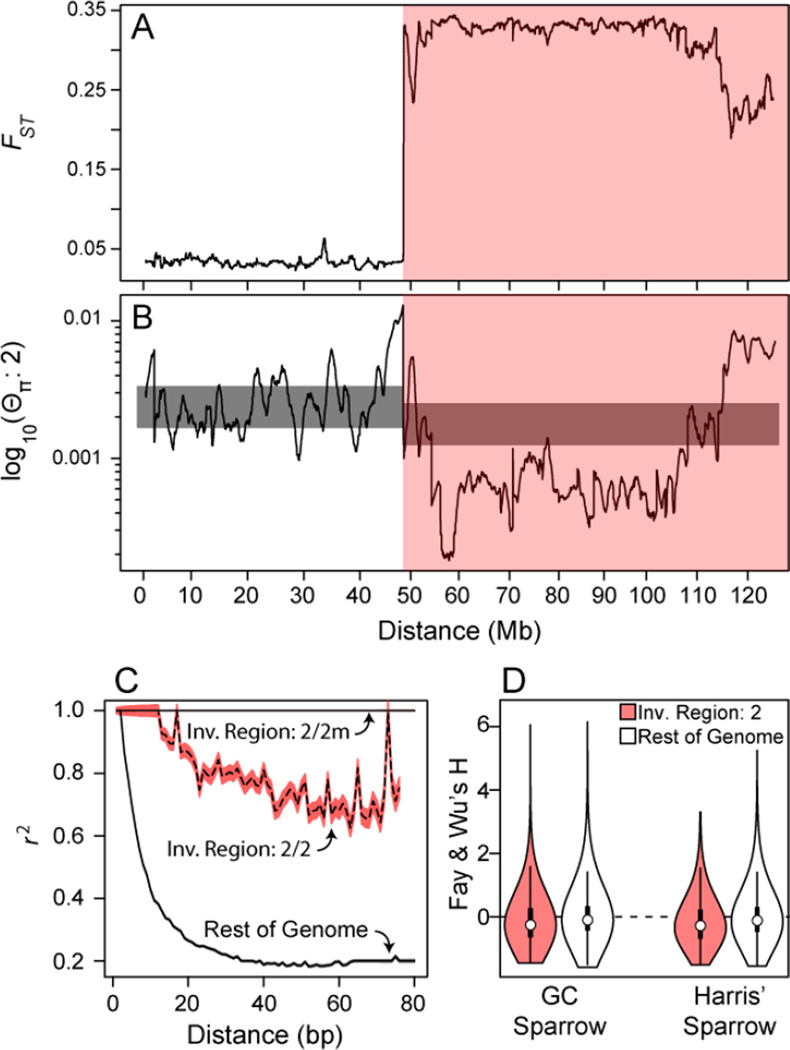
Pooled whole-genome resequencing of 24 tan and 25 white males revealed a bimodal distribution of scaffold-wide average FST between morphs estimated from 18.4 million biallelic SNPs and polymorphic indels (collectively, "variants”; Figure S1). Previous studies [11] revealed high differentiation between 2 and 2m, and we reasoned that high FST scaffolds would correspond to regions within the chromosome 2 inversion. We confirmed this by associating a subset of assembled scaffolds with FISH-mapped bacterial artificial chromosomes (BACs). Scaffolds associated with BACs known to map within the inversion had correspondingly high FST (median = 0.3; Table S3). The scaffolds that map to the region near the distal end of the inverted region on 2 have decreased average FST (~0.2, Figure 2A, Table S3) and a significantly higher rate of shared polymorphism between 2 and 2m (odds-ratio=5.73, Fisher’s Exact Test p-value < 1e-10) suggesting that limited recombination or gene conversion occurs between 2 and 2m in this region. However, the extent of genetic exchange in this region or the rest of the inverison is presently unknown. High FST (≥ 0.2) scaffolds include 1,137 genes that comprise the supergene. FST is expected to be 0.33 at variants that are fixed between 2 and 2m in comparisons between tan (2/2) and white (2/2m) birds. Approximately 78.6% of the 1.5M variants (or ~1/100 bp) within the inversion have FST of ~0.33 (Supplementary Figure 1B), demonstrating that the majority of variants in the inversion are heterozygous among all the white birds we sampled. To confirm fixed differences between 2m and 2 identified in pooled genotypes, we resequenced the genomes of three individual white morph birds sampled from a different population than the pooled sample. We find that 75% of variants within the inversion are heterozygous in all three birds; note, given limited sequencing depth of individual birds (~10X) we expect that a fraction of truly polymorphic variants to appear homozygous thus explaining the discrepancy between the rate of fixed differences (75% versus 78.6%) observed in our individual and pooled based resequencing, respectively. In addition, we estimated linkage disequilibrium (r2) within the pooled samples. As expected, r2 is ~ 1 within the inverted region among the pool of white birds (Fig. 2C).
Figure 2. Contrasting divergence, diversity and linkage disequilibrium inside (pink area) and outside the inverted region.
Sliding window estimate across FISH-mapped scaffolds of (A) FST between white and tan birds and (B) genetic diversity along the tan allele, 2. The grey bar represents the 95% confidence band (standard deviation) of neutral expectation for genetic diversity . (C) Linkage disequilibrium decays rapidly outside of the inversion, but is high, especially in 2/2m heterozygotes, within the inversion. Using golden-crowned or Harris’ sparrow as outgroups, (D) Fay and Wu’s H estimates do not show a strong signal of recent selective sweeps in the inverted region of chromosome 2. See also Figures S1, S3 and Table S3.
The inversion contains a number of well-studied genes related to the neurophysiological control of behavior, including multiple steroid hormone receptors (Table 1). ESR1 has already been implicated as contributing to behavioral differences between white-throated sparrow morphs [12], and VIP has been shown to mediate variation in aggressive behavior in other songbirds [13, 14]. Within this gene complex are also two genes with annotated roles in pigmentation (FIG4 and LYST, [15]). We note, however, that none of these genes show signatures of recent selection (see below) or unusual patterns of divergence relative to others genes within the inversion. Indeed the high levels of divergence throughout the inversion provide a challenge in resolving the relative contributions of specific genes to the suite of phenotypic differences among morphs.
Table 1.
Candidate genes for behavior and pigmentation
| Gene | Gene Name | Exon (NS/S) |
Intron | UTR (5’/3’) |
UP | DOWN |
|---|---|---|---|---|---|---|
| CGA | Glycprotein Hormones, Alpha Polypeptide | 0/1 | 34 | 0/4 | 51 | 71 |
| ESR1 | Estrogen Receptor 1 | 2/4 | 2565 | 0/14 | 53 | 54 |
| ESRRG | Estrogen Related Receptor Gamma | 1/3 | 2465 | 0/17 | 52 | 68 |
| FSHR | Follicle Stimulating Hormone Receptor | 6/11 | 1291 | 0/8 | 112 | 5 |
| HTR1B | Serotonin Receptor 1B | 0/4 | 1 | 7/3 | 46 | 59 |
| HTR1E | Serotonin Receptor 1E | 2/7 | 0 | 1/4 | 78 | 65 |
| LHCGR | Luteinizing Hormone/Choriogonadropin Receptor | 5/14 | 285 | 0/0 | 103 | 44 |
| VIP | Vasoactive Intestinal Peptide | 0/0 | 79 | 4/1 | 99 | 104 |
| FIG4 | FIG4 homologue | 3/11 | 470 | 3/ | 154 | 67 |
| LYST | Lysosomal Trafficking Regulator | 19/48 | 931 | 2/19 | 56 | 0 |
Fixed differences between chromosomes 2 and 2m in candidate genes for behavior and pigmentation (FIG4 and LYST). Polymorphisms are divided by site class including exons (nonsynonymous/synonymous substitutions), introns, UTR (5’ UTR/3’UTR), and 1 kb UP and DOWN-stream of start and stop codons, respectively.
The high level of nucleotide divergence between chromosomes 2 and 2m suggests that the divergence of these chromosomes predates divergence of white-throated sparrow and its sister species. To investigate the long-term evolutionary history of chromosomes 2 and 2m, we examined genome-wide patterns of divergence between the white-throated sparrow and its close relatives. For comparison, we analyzed closely-related taxa with previously [16, 17] or newly sequenced transcriptomes or genomes (Figure 1B). As in previous studies [11], we find that the relationship of chromosomes 2 and 2m conflicts with the species tree and suggests more recent common ancestry of chromosome 2 with other taxa than with 2m. Our genome-scale analyses and complete taxon sampling of the Zonotrichia genus revealed that outside the inversion white-throated sparrow is sister to Harris’ sparrow. This same relationship holds for the tan (2) allele within the inversion. Inclusion of rufous-collared sparrow in our analysis allowed us to resolve that the white 2m allele arose after the origin of Zonotrichia and falls sister to the rest of the clade.
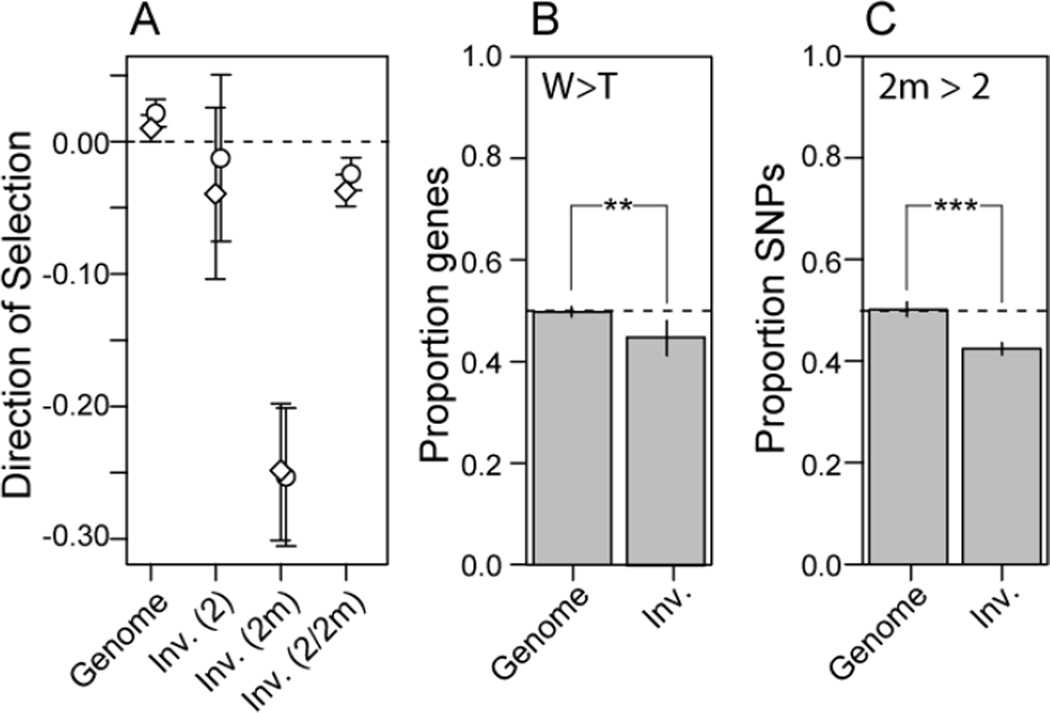
Because 2m exists almost exclusively in a non-recombining heterozygous state (Table S1), it has been hypothesized that 2m may be degrading in a manner that is similar to neo-Y/W chromosomes [18, 19]. Hyunh and colleagues [18], using a smaller set of markers, failed to find an enrichment of non-synonymous changes on 2m, and thus concluded that the chromosome was not degrading. In contrast, here we provide evidence consistent with functional degradation of 2m by using genome-wide estimates of polymorphism and divergence at non-synonymous and synonymous sites (Figure 3A). We calculated the Direction of Selection (DoS) statistic [20], which is conceptually related to the McDonald-Kreitman test, to compare the relative rates of non-synonymous and synonymous polymorphism to non-synonymous and synonymous divergence. On average, genes inside the inverted region that are linked to 2m have a significantly negative DoS (pGolden-crowned < 0.001; pHarris’ < 0.001) indicating an excess of non-synonymous polymorphism and consistent with functional degradation of 2m. Outside the inversion, DoS is slightly positive, indicating a small excess of non-synonymous fixed differences between the white-throated sparrow and either of two out-group species (pGolden-crowned = 0.059; pHarris’ < 0.001). Genes inside the inverted region that are linked to 2 do not show a significant deviation from expectation based on a randomization test (p > 0.05 for both outgroups). Intriguingly, fixed differences between 2 and 2m are only slightly more likely to be non-synonymous relative to expectations from interspecific divergence (pGolden-crowned < 0.001; pHarris’ < 0.001; see Figure 3A, ‘Inv. (2/2m)’). This latter result suggests that chromosome 2 has relatively recently become polymorphic in the white-throated sparrow: long-term heterozygosity of 2m would likely lead to fixation of putatively deleterious non-synonymous mutations giving rise to a DoS between 2 and 2m that is similar in magnitude to the DoS statistic based on polymorphisms segregating on 2m.
Figure 3. Functional degradation of 2m.
(A) Using two different outgroups (diamond = golden-crowned sparrow, circle = Harris’ sparrow), the direction of selection statistic is strongly negative on 2m, indicating an increased level of non-sysnonymous polymorphism. (B) For genes within the inversion, gene expression in higher for tan birds than heterozygous (2/2m) white-morph birds and (C) in white birds, the white (2m) allele tends to be under-expressed relative to the tan (2) allele (**: p < 0.01; ***: p < 0.001). Error bars represent 95% confidence intervals (standard error). See also Figure S2, S4, and Table S1.
In neo-sex chromosomes, functional degradation is accompanied by decreased average gene expression of Y/W linked genes [21, 22]. We contrasted gene expression levels between three tan and three white birds and also measured patterns of allele specific expression within white birds. On average, the expression level of genes within the inverted region is lower in white birds than tan birds (Figure 3B) and expression of the allele linked to 2m is lower than the allele linked to 2 within heterozygous white birds (Figure 3C). Differences in gene expression between 2- and 2m-linked genes are subtle (Figure S2) and may reflect tissue-specific effects or a relatively recent degradation of 2m. Decreased expression of 2m linked genes is not an artifact of increased general expression of 2 linked genes, as seen by a comparison of log-transformed expression levels of genes inside the inverted region versus the rest of the genome within tan birds (t = 0.0324, p = 0.97). While differences in expression levels of some genes inside the inverted region likely contributes to adaptive differentiation between morphs, our results suggest that 2m is degrading.
As in sex chromosomes, we hypothesized that pairwise genetic diversity (Θπ) on 2 and 2m would be 75 and 25% of that in the rest of the genome [23]. In contrast, Θπ on 2 is only ~30% of background diversity (0.0007 vs 0.0023; Figure 2B) and Θπ on 2m is only ~20% of background (0.00047 vs. 0.0023; Supplemental Figure S3), indicating that diversity on these chromosomes is not at the expected equilibrium under a simple neutral model. In sex chromosomes, departures from equilibrium diversity ratios have been attributed to multiple forces including differences in life-history between sexes [24] and differences in how natural selection and genetic drift operate. One component of life history that could influence patterns of diversity is differences in the variance of reproductive success between morphs. In white-throated sparrows, if tan birds have a higher variance in reproductive success the ratio of diversity between 2 and the genomic background will be >75% whereas if white birds have a higher variance in reproductive success the ratio will be <75% [24]. To assess whether the observed deviations from expected patterns of genetic diversity could be attributed to life-history variation among morphs, we calculated lifetime reproductive success among 255 white and tan adult birds. We find no significant difference in the variance of reproductive success among morphs (Levene Test, p-value = 0.84). Thus life-history variation among morphs may play a limited role in shaping patterns of genetic variation between the inverted and non-inverted regions of the genome, however we cannot reject more complicated demographic scenarios at the present time [25].
The differential action of natural selection is another factor that can influence the ratio of genetic diversity between the X/Z and autosomes and, by extension, chromosomes 2 and 2m. On the white allele, 2m, which is analogous to Y/W chromosomes, the lack of recombination makes discerning signatures of selection challenging, so we focused analyses of selection on the tan allele, 2. On 2, like X/Z chromosomes, beneficial mutations may be more likely to go to fixation (positive selection) and deleterious mutations may be more likely to be purged (purifying selection) because of functional hemizygosity in white 2/2m birds [24]. Either positive or negative selection would lead to a decrease in genetic diversity on 2 and could possibly explain the substantial reduction of diversity and elevated linkage disequilibrium observed on this chromosome (Figure 2B). Under a scenario of wide-spread positive selection along the inverted region of 2, we would expect a substantial increase in the proportion of high-frequency derived alleles which would be revealed by negative values of Fay and Wu’s H [27]. We calculated Fay and Wu’s H using either Harris’ or golden-crowned sparrow as outgroups. H is slightly more negative inside the inverted region of 2 than outside but does not show an excess of extremely negative H values (Figure 2D). Wide-spread positive selection along the inverted region of 2 may also lead to a slight excess of fixed non-synonymous differences however no such pattern is observed (Figure 3A). While positive selection has likely shaped patterns of genetic variation at certain loci the inverted region of 2, current tests fail to resolve systematic evidence for increased rates of positive selection on 2. The reduction in diversity along 2 relative to the genomic background could therefore be influenced by strengthened purifying selection arising as a consequence of functional degradation of chromosome 2m, strengthened background selection arising a consequence of reduced recombination, as well as occasional selective sweeps at specific loci.
Given the strength of disassortative mating and evidence for degradation of chromosome 2m, we hypothesized that there are fitness consequences associated with homotypic mating. White male × white female (W×W) homotypic pairs are expected to produce, on average, 25% “superwhite” 2m/2m homozygotes, potentially exposing deleterious recessive alleles that have arisen on 2m. In 27 years of study, we have found only three superwhite 2m/2m birds out of 1,989 (0.15%) genotyped birds. Based on the observed frequency of mating between white morph males and females, both via social pairing and extra pair mating, we expected 12 super whites in our sample (see Supplementary Experimental Procedures). The lower than expected number of observed superwhites (Fisher’s Exact Test p = 0.035) suggests that the 2m/2m genotype is deleterious (Table S1), that parents reduce investment in such offspring [27], or both. While the two observed female superwhites appeared normal in size, the one superwhite male we found was over 2.5 standard deviations smaller than all other age-matched chicks, and is the smallest male nestling sampled to date (Figure S4), suggesting that 2m/2m might be more deleterious in males. Occasional recombination on 2m when superwhites do survive and reproduce may also mitigate the degradation of this chromosome.
Homotypic pairings of both types (W×W and T×T) suffer additional fitness costs. For example, different pairings (W×W, T×T, T×W, W×T) suffer different rates of extra-pair paternity in their nests (F3,380 = 20.10, p < 0.0001, Table S4). Like white male × tan female (W×T) pairs, both W×W and T×T pairs experience a significant (~25%) increase in extra-pair paternity in their nests relative to tan male × white female (T×W) pairs (Tukey-Kramer post hoc tests, p < 0.05). We also find that homotypic pairs show a tendency towards producing female-biased clutches (homotypic pairs (N=12) = 37 ± 9 % males, heterotypic pairs (N=284) = 52 ± 1 % males, F1,294 = 2.66, p = 0.10; Table S4). Since many species adaptively adjust offspring sex ratios to produce a higher proportion of offspring of the smaller sex (females, in the case of the white-throated sparrow) when they are in poor condition or under stress [28], such data suggest that homotypic pairs are at a disadvantage. Lastly, although chicks raised by T×T pairs are similar in mass to disassortative pairs, chicks raised by W×W pairs are significantly smaller than those raised by T×T pairs (F1,30 = 11.57, p = 0.0019), likely a result of reduced parental care by white morph birds. This pattern is primarily driven by mass differences in male chicks, as sons from W×W pairs are significantly smaller than sons from T×T pairs (F1,12=7.45, p = 0.01), whereas the mass of daughters does not differ by pair type (F1,16 = 2.85, p = 0.11). These costs associated with homotypic pairing may contribute to the maintenance of the chromosomal polymorphism in the species by favoring disassortative pairing.
Two plausible models explain the presence of both 2 and 2m in white-throated sparrows subsequent to their ancient divergence: either these chromosomes were polymorphic in an ancestral Zonotrichia species but remain so only in white-throated sparrows, or one of the chromosomes entered white-throated sparrow via hybridization followed by adaptive introgression (Figure 1B). Previous studies favored the former scenario [11] because no potential hybridizing taxon was found [11]. As we discuss above, however, patterns of polymorphism on chromosome 2m (Figure 3A) suggest that this chromosome may have become polymorphic in the white-throated sparrow relatively recently. While the lack of a clear sister species to 2m is challenging for this interpretation, complete genus-level taxon sampling suggests that the donor species of 2m may be extinct. In support of the model that 2m is polymorphic in the white-throated sparrow via introgression, we note that hybridization between various Zonotrichia species have been observed in the wild [29] and inferred from discordance between mitochondrial and nuclear phylogenies [30]. Origination of supergene alleles in separate species provides a simple mechanism by which coadapted gene complexes can be formed, and under which recombination suppression between alleles would be expected when heterokaryotypes arise via hybridization [9].
Chromosomes 2 and 2m in white-throated sparrows are independent of sex chromosomes, but show striking parallels with patterns of sex chromosome divergence and degradation [31, 32]. Indeed, because of disassortative mating based on both chromosomes 2 and 2m and the W and Z sex chromosomes, the species operates as if there are four sexes. Sexual systems with more than two sexes are exceedingly rare among animals and theory predicts that they are unlikely to persist for long periods of time [33]. In part, this instability arises because four sex systems have a two-fold increase in some aspects reproductive effort, such as finding a mate. If such four-sex systems are truly unstable, the persistence of the inversion based plumage morph polymorphism found in the white-throated sparrow may be transient, despite the observed fitness benefits of disassortative mating.
Supplementary Material
Acknowledgments
Funding for this work was provided by the US National Institutes of Health NIGMS 1R01GM084229 (E.M.T and R.A.G.), Indiana State University (E.M.T. and R.A.G.), and East Carolina University (C.N.B). A.O.B was supported by an NIH National Research Service Award (F32 GM097837) and by the NIH grant RO1GM100366 to Dmitri Petrov and Paul Schmidt. We thank the production sequencing group of The Genome Institute at Washington University for library construction, sequencing and data curation for the white-throated sparrow genome. The University of Alaska Museum kindly provided the Z. atricapilla and Z. querula samples. Dr. Alvaro Hernandez and the high-throughput sequencing at the University of Illinois performed library preparation and genome resequencing. Marlys Houck and Oliver Ryder at the Institute for Conservation Research at the San Diego Zoo provided assistance with cell culture and karyotyping and Michael Romanov helped with BAC identification. We thank Teri Lear and Judy Lundquist from the Gluck Equine Research Center at the University of Kentucky for fluorescent in situ hybridization. Finally, we thank the 1988–2014 White-throated Sparrow field and laboratory research crews. All data has been deposited to GenBank: Whole Genome Assembly (GCA_000385455.1), Z. capensis RNA-seq PRJNA298255), pooled resequencing (SRR2012823, SRR2012824) and individual resequencing (SRS930390, SRS930391, SRS930394, Z. querula SRS1162844, Z. atricailla SRS1162859.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
EMT - project leadership, design, and manuscript preparation
AOB - project design, analysis and manuscript preparation
MLK - FISH analysis
MSB - phylogenetic analysis of transcriptome data
DJN - analysis of pooled sequencing data
AB - analysis of field data
ZAC - RNA sequencing of Z. capensis and manuscript preparation
MS - RNA sequencing of J. hyemalis and manuscript preparation
PM - genome assembly and curation
WCW - project leadership, manuscript preparation, genome sequencing
RAG - project design, field data collection
CNB- project leadership, design, analysis and manuscript preparation
References and Notes
- 1.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, Mullen SP, Kronforst MR. Doublesex is a mimicry supergene. Nature. 2014;507:229–232. doi: 10.1038/nature13112. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Wurm Y, Nipitwattanaphon M, Riba-Grognuz O, Huang Y-C, Shoemaker, Keller L. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013;494:664–668. doi: 10.1038/nature11832. [DOI] [PubMed] [Google Scholar]
- 3.Tuttle EM. Alternative reproductive strategies in the white-throated sparrow: behavioral and genetic evidence. Behav Ecol. 2003;14:425–432. [Google Scholar]
- 4.Formica VA, Tuttle EM. Examining the social landscapes of alternative reproductive strategies. J Evol Biol. 2009;22:2395–2408. doi: 10.1111/j.1420-9101.2009.01855.x. [DOI] [PubMed] [Google Scholar]
- 5.Knapton RW, Falls JB. Differences in parental contribution among pair types in the polymorphic white-throated sparrow (Zonotrichia albicollis) Can J Zool. 1983;61:1288–1292. [Google Scholar]
- 6.Thorneycroft HB. Chromosomal polymorphism in white-throated sparrow Zonotrichia albicollis (Gmelin) Science. 1966;154 doi: 10.1126/science.154.3756.1571. 1571-157&. [DOI] [PubMed] [Google Scholar]
- 7.Thorneycroft HB. Cytogenetic study of white-throated sparrow, Zonotrichia-albicollis (Gmellin) Evolution. 1975;29:611–621. doi: 10.1111/j.1558-5646.1975.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 8.Lowther JK. Polymorphism in the white-throated sparrow Zonotrichia albicollis (Gmelin) Candian Journal of Zoology. 1961;39:281–292. [Google Scholar]
- 9.Schwander T, Libbrecht R, Keller L. Supergenes and complex phenotypes. Curr Biol. 2014;24:R288–R294. doi: 10.1016/j.cub.2014.01.056. [DOI] [PubMed] [Google Scholar]
- 10.Frankl-Vilches C, Kuhl H, Werber M, Klages S, Kerick M, Bakker A, de Oliveira EHC, Reusch C, Capuano F, Vowinckel J, et al. Using the canary genome to decipher the evolution of hormone-sensitive gene regulation in seasonal singing birds. Genome Biol. 2015:16. doi: 10.1186/s13059-014-0578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas JW, Caceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearragement and suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton BM, Hudson WH, Ortlund EA, Shirk S, Thomas JW, Young ER, Zinzow-Kramer WM, Maney DL. Estrogen receptor alpha polymorphism in a species with alternative behavioral phenotypes. Proc Natl Acad Sci USA. 2014;111:1443–1448. doi: 10.1073/pnas.1317165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodson JL. Territorial aggression and dawn song are modulated by septal vasotocin and vasoactive intestinal polypeptide in male field sparrows (Spizella pusilia) Horm Behav. 1998;34:67–77. doi: 10.1006/hbeh.1998.1467. [DOI] [PubMed] [Google Scholar]
- 14.Goodson JL, Adkins-Regan E. Effect of intraseptal vasotocin and vasoactive intestinal polypeptide infusions on courtship song and aggression in the male zebra finch (Taeniopygia guttata) J Neuroendocrinol. 1999;11:19–25. doi: 10.1046/j.1365-2826.1999.00284.x. [DOI] [PubMed] [Google Scholar]
- 15.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balakrishnan CN, Mukai M, Gonser RA, Wingfield JC, London SE, Tuttle EM, Clayton DF. Brain transcriptome sequencing and assembly of three songbird model systems for the study of social behavior. PeerJ. 2014;2:e396. doi: 10.7717/peerj.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stager M, Swanson DL, Cheviron ZA. Regulatory mechanisms of metabolic flexibility in the dark-eyed junco (Junco hyemalis) J Exp Biol. 2015;218:767–777. doi: 10.1242/jeb.113472. [DOI] [PubMed] [Google Scholar]
- 18.Davis JK, Mittel B, Lowman JJ, Thomas PJ, Maney DL, Martin CL, Thomas JW. Haplotype-Based Genomic Sequencing of a Chromosomal Polymorphism in the White-Throated Sparrow (Zonotrichia albicollis) J Hered. 2011;102:380–390. doi: 10.1093/jhered/esr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh LY, Maney DL, Thomas JW. Chromosome-wide linkage disequilibrium caused by an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis) Heredity. 2011;106:537–546. doi: 10.1038/hdy.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol Biol Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- 21.Bachtrog D. Expression Profile of a Degenerating Neo-Y Chromosome in Drosophila. Curr Biol. 2006;16:1694–1699. doi: 10.1016/j.cub.2006.07.053. [DOI] [PubMed] [Google Scholar]
- 22.Hough J, Hollister JD, Wang W, Barrett SCH, Wright SI. Genetic degeneration of old and young y chromosomes in the flowering plant Rumex hastatulus. Proc Natl Acad Sci USA. 2014;111:7713–7718. doi: 10.1073/pnas.1319227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlesworth B. The effect of life-history and mode of inheritance on neutral genetic variability. Genet Res. 2001;77:153–166. doi: 10.1017/s0016672301004979. [DOI] [PubMed] [Google Scholar]
- 24.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- 25.Ramachandran S, Rosenberg NA, Feldman MW, Wakeley J. Population differentiation and migration: Coalescence times in a two-sex island model for autosomal and X-linked loci. Theor Popul Biol. 2008;74:291–301. doi: 10.1016/j.tpb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng K, Fu Y-X, Shi S, Wu C-I. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pryke SR, Griffith SC. Genetic incompatibility drives sex allocation and maternal investment in a polymorphic finch. Science. 2009;323:1605–1607. doi: 10.1126/science.1168928. [DOI] [PubMed] [Google Scholar]
- 28.Bonier F, Martin PR, Wingfield JC. Maternal corticosteroids influence primary offspring sex ration in a free-ranging passerine bird. Behav Ecol. 2007;18:1045–1050. [Google Scholar]
- 29.Payne RB. Two Apparent Hybrid Zonotrichia Sparrows. The Auk. 1979;96:595–599. [Google Scholar]
- 30.Weckstein JD, Zink RM, Blackwell-Rago RC, Nelson DA. Anomalous Variation in Mitochondrial Genomes of White-crowned (Zonotrichia leucophrys) and Golden-crowned (Z. atricapilla) Sparrows: Pseudogenes, Hybridization, or Incomplete Lineage Sorting. The Auk. 2001;118:231–236. [Google Scholar]
- 31.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho T-J, Koutseva N, Zaghlul S, Graves T, Rock S, et al. Mammalian y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grutzner F, Kaessmann H. Origins and functional evolution of y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 33.Hurst LD. Why are there only two sexes? Proceedings of the Royal Society B: Biological Sciences. 1996;263:415–422. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.