Abstract
Oxalobacter formigenes, a member of the human colonic microbiota that plays a major role in net colonic oxalate transport and secretion, is protective against formation of calcium oxalate kidney stones. We now describe the prevalence, relative abundance and stability of O. formigenes in healthy young adults in the United States, as revealed by Human Microbiome Project (HMP) data from fecal samples from 242 healthy young adults who had 1-3 study visits. Samples underwent whole-genomic shotgun (WGS) sequencing, and/or 16S rRNA sequencing. Three datasets available from the processed sequence data were studied: WGS metagenomic analysis by alignment to reference genomes (HMSCP) or using MetaPhlAn, or QIIME analysis of the V1-3 or V3-5 16S sequences. O. formigenes was detected in fecal samples using both the WGS and 16S rRNA data. Analysis of the WGS dataset, using HMSCP, showed that 29 (31%) of 94 subjects were O. formigenes-positive while the V1-3 and V3-5 analyses were less sensitive for O. formigenes detection. When present, O. formigenes relative abundance varied over 3 log10, and was normally distributed. For 66 samples studied by all three methods, all assays agreed in 58 (88%). Of 14 subjects who were O. formigenes-positive at baseline, 13 (93%) were positive on follow-up visit, indicating the stability of colonization. O. formigenes appears to be stably present in fewer than half of healthy young USA adults and is most sensitively detected by WGS.
Keywords: kidney calculi, metagenome, microbiota, nephrolithiasis, oxalate, urolithiasis
INTRODUCTION
Oxalate is absorbed passively by the gut from dietary sources, and produced endogenously as an end-product of amino acid metabolism and excreted by the kidneys1, 2. The urinary oxalate level, reflecting overall oxalate load, is a major determinant of the risk of calcium oxalate kidney stones, the most common type3, 4. The increased prevalence of nephrolithiasis described in US adults5 and the dramatic rise in its incidence in children6 suggest that oxalate stone prevalence is rising. The major factors contributing to this trend are unknown.
Oxalobacter formigenes is a gram-negative anaerobic bacterium that depends on oxalate metabolism for energy7, and that colonizes the human intestine8. O. formigenes plays a major role in regulating net intestinal transport of oxalate9. The lack of O. formigenes in the colon is believed to increase intestinal oxalate absorption, leading to an increase in urinary oxalate excretion10. Among oxalatedegrading bacteria in humans, O. formigenes appears to be the most efficient11.
Human O. formigenes isolates are susceptible to many commonly used antibiotics12; thus, colonization status could reflect health status13, dietary habits14, or antibiotic exposure15. Because O. formigenes is difficult to culture, colonization status has been determined either indirectly using an oxalate-degradation assay or directly by polymerase chain reaction (PCR)16, 17. In healthy American adults, colonization rates varied between 38 and 62% with geographic location, age, and method of testing accounting for much of the variation12. In less developed countries, the prevalence of O. formigenes colonization is higher12. Recent studies of an uncontacted tribal group detected O. formigenes in nearly all 18. These observations suggest that O. formigenes may be disappearing as a consequence of modern lifestyles, including antibiotic use 15.
As part of the NIH-sponsored Human Microbiome Program (HMP), the microbiota and metagenome of 242 healthy young adult volunteers were surveyed19, 20. These studies provided the opportunity to address questions about the frequency of detecting O. formigenes in US subjects.
Materials and Methods
Population studied and samples interrogated
Institutional review board approval was not required since the study involved mining a publically available database consisting of microbial sequences from de-identified subjects. In our HMP data, 242 healthy subjects between the ages of 18 and 40 years old had 5300 samples taken at 15-18 body sites on up to three separate visits 20. During Phase I, the HMP investigators selected a subset of these samples for WGS and 16S rRNA sequencing analysis. The processed sequence data are available through the HMP Data Analysis and Coordination Center (DACC).
In the HMP shotgun metagenomic analysis, a subset of samples was selected for WGS sequencing and subsequent quality control tests19, 20. The data available were from an initial phase of the HMP project. Although the entire data set included 690 samples (http://hmpdacc.org/HMASM/), and the number of subjects in the HMP is 242 20, the total number of subjects included in this database was 103 with only 94 of these subjects having at least 1 stool sample.
A standardized 16S rRNA sequencing protocol was used to profile 5177 samples, and two variable regions of the 16S rRNA gene, V1-3 and V3-5, important for taxonomy assignment underwent sequence analysis 19. The samples were amplified and sequenced using the Roche-454 FLX Titanium platform 20 The V1-3 variable region of the 16S rRNA gene was sequenced for 2,971 samples (including 193 stool samples) and the V3-5 variable region was sequenced for 4,879 samples (including 328 stool samples). By including only those samples with complete records and linkage across datasets, this study focuses on a subset of 187 stool samples from 144 subjects for the V1-3 analyses, and 319 stool samples from 210 subjects for the V3-5 analyses; samples were sequenced for either both the V1-3 and V3-5 16S rRNA regions, or only one (Supplemental Figure 1).
Metagenomic and 16S rRNA sequencing data analysis
Three datasets from the DACC were available for this analysis. Two contained the results of WGS metagenomic analysis using different approaches: shotgun community profiling by alignment to reference genomes (HMSCP) and through the MetaPhlAn algorithm 21. In the first approach, WGS reads were mapped onto a reference genome database to determine the organisms present in the community. The results of the analysis included a bacterial strain abundance table for each HMP sample, available through www.hmpdacc.org/HMSCP/. The reference genome database includes organisms available in GenBank as of November 2009 and can be accessed at http://www.hmpdacc.org/HMREFG/. We used this dataset to examine prevalence of O. formigenes and to distinguish between O. formingenes Group I and Group II which are represented by strains OXCC13 and strain HOxBLS, respectively22 . In the second approach, metagenomic data were taxonomically profiled using MetaPhlAn 21, permitting estimation of relative abundances for the different taxa, with the output of the analysis at www.hmpdacc.org/HMSMCP .
The third dataset provides the output of QIIME-based analyses 23, 24 of 16S rRNA gene data. In QIIME analysis the 16S sequences are clustered into operational taxonomic units (OTUs), a representative sequence for each OTU is chosen, and then a taxonomy assigned using the RDP classifier. The complete output of QIIME analysis of the V1-3 and V3-5 16S data is available from the HMP DACC (http://www.hmpdacc.org/HMQCP/).
We constructed a MySQL database containing information from the publically available datasets above, through the HMP DACC. By restructuring the data using the MySQL database, we created a standardized pipeline for determining microbial prevalence, abundance, and diversity associations. Since particular O. formigenes 16S OTUs were identified from the HMP specimens, we sought to determine to which of the known O. formigenes Groups ( I and II) they best matched. To accomplish this, we examined the HMP sequences with reference to the O. formigenes strains in the Greengenes database, and we identified the best match from each Group.
RESULTS
Prevalence of Oxalobacter formigenes. We sought to define the prevalence of O. formigenes in the intestinal tract of the healthy young adult HMP participants. O. formigenes was detected in fecal specimens in both the WGS and 16S rRNA analyses. In the WGS dataset, using the HMSCP-analytic tools, 29(31%) of 94 subjects were positive for O. formigenes ; however, analysis of the V1-3 and V3-5 data, using QIIME, showed O. formigenes prevalence of the studied subjects only at 15% and 11%, respectively (Table 1). In comparison, the prevalence of was 71, 12, and 40%, respectively, in the same three analyses. These disparate results highlight the difficulties intrinsic to interpreting the 16S and the WGS data,
Table 1.
Prevalence of O. formigenes and E. coli in HMP subjects, based on separate analyses.
| Metagenomic WGS | V1-3 16S rRNA | V3-5 16S rRNA | ||||
|---|---|---|---|---|---|---|
| Microbe status | Subjects (n=94) |
Samples (n=139) |
Subjects (n=144) |
Samples (n=187) |
Subjects (n=210) |
Samples (n=319) |
| O. formigenes+ | ||||||
| HMSCP | 29 (31%) | 42 (30%) | 22 (15%) | 25 (13%) | 23 (11%) | 27 (9%) |
| MetaPhlAn | 27 (29%) | 36 (26%) | ||||
| E. coli+ | 67 (71%) | 86 (62%) | 17 (12%) | 18 (10%) | 84 (40%) | 99 (31%) |
Within species differences. The reference genome database used for the HMSCP WGS analysis contains strain OXCC13 that represents O. formigenes Group I and strain HOxBLS that represents Group II 25. In addition to genetic differences, the two Groups differ phenotypically in oxalate-degrading capacity, fatty acid content, oxygen tolerance and antigenic features8. In the WGS analysis, all 29 of the O. formigenes-positive subjects were colonized with sequences represented by Group I strains, and of these, 17 (59%) were simultaneously colonized with organisms with sequences represented by Group II strains. Thus, co-infection with both Group I and Group II was common, and colonization by Group II was only detected in the presence of Group I.
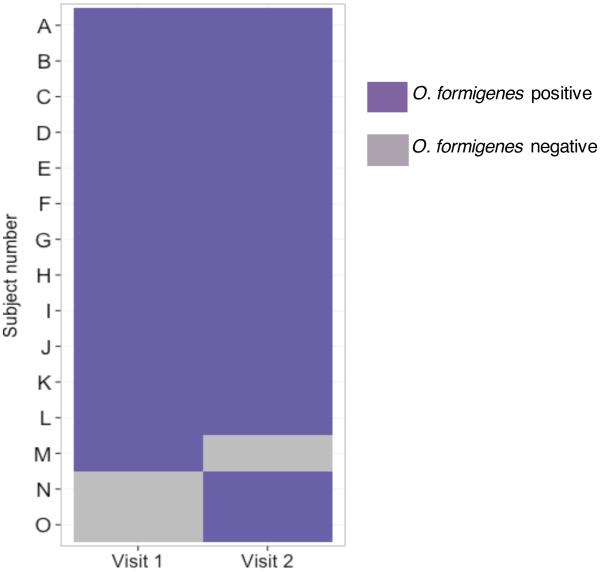
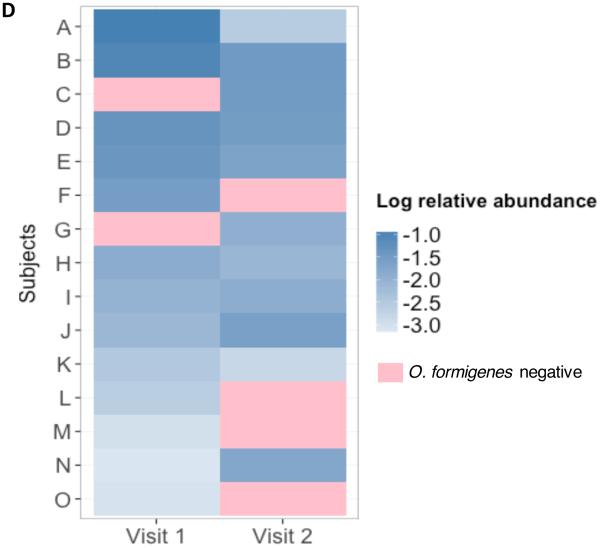
Fecal colonization stability. To examine the stability of O. formigenes colonization in the WGS-tested subjects, we studied 42 subjects who had 2 visits at which samples were obtained; the remaining 52 subjects did not complete a second visit. Of the 42 subjects, 13(33%) were positive at baseline; 12 (93%) remained positive (Figure 1). Of the 29 subjects who were negative at the first visit, 27 (93%) remained negative, but in two subjects, the presence of the organism was detected on the second visit (and for one, on the third visit). The temporal stability of O. formigenes was mirrored at the strain level, for which most (93-100%) of the positive samples were Group I, with ~40-50% simultaneously positive for Group II, depending on the visit. Only 1 (7%) of the 14 subjects who were O. formigenes-positive at the first visit carried Group II with no evidence for Group I. However, at the second visit, both Groups were detected.
Figure 1. Stability of O. formigenes over time in subjects studied twice and analyzed by shotgun community profiling.
In 42 subjects tested on two consecutive visits analyzed using the HMSCP dataset, 27 were negative on both visits and are not shown. Shown are the 15 subjects with at least one positive result.
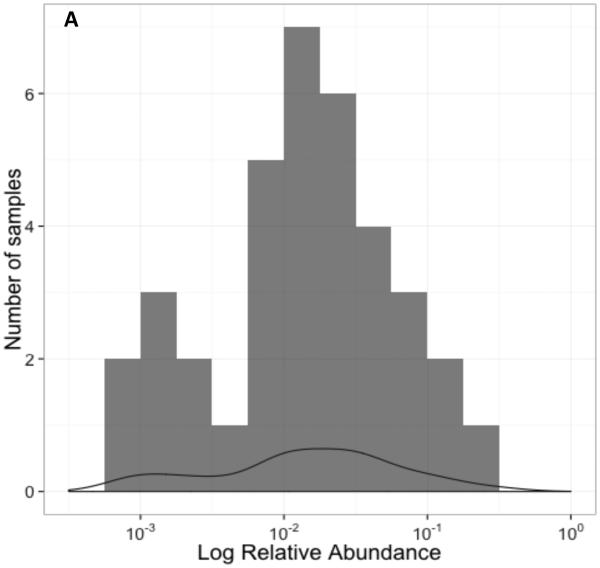
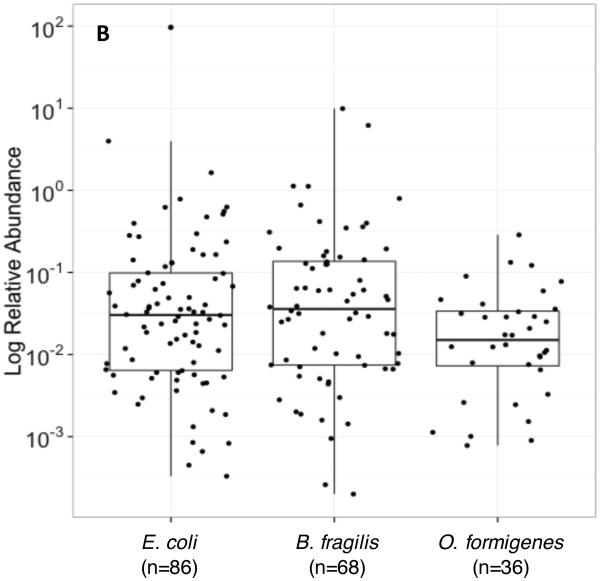
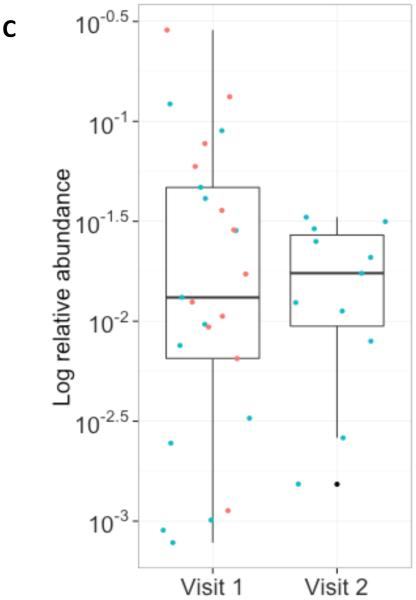
Abundance of O. formigenes: In 36 O. formigenes-positive stool samples from the WGS dataset analyzed by MetaPhlAn, the relative O. formigenes abundance was normally distributed around a median of 0.015% (Figure 2A). Abundance varied over ~3 log10 while the abundance of E. coli (median 0.03%) and Bacteroides fragilis (median 0.036%) varied over 5 log10 and 4 log10, respectively (Figure 2B). The overall mean abundance was stable on follow-up visits across the population of those positive for all three species examined. For the nine subjects studied twice, the median values for O. formigenes abundance on the first visit (0.013%) and the second visit (0.017%) were similar (Figure 2C,D).
Figure 2. Relative abundance of O. formigenes in the 36 fecal samples in the MetaPhlAn WGS dataset.
Panel A: Histogram of number of samples by log relative abundance of O. formigenes. Panel B: Log relative abundance of O. formigenes (n=36) compared to E. coli (n=86) and B. fragilis (n=68). Box indicates median and inter-quartile range, and lines indicate 95% confidence interval. Panel C: Stability of O. formigenes log relative abundance in HMP subjects examined once, (red circles; n=51) or twice (blue circles; n=43), and found to be positive on at least one sample. Panel D: O. formigenes relative abundance by visit for the 15 subjects studied twice.
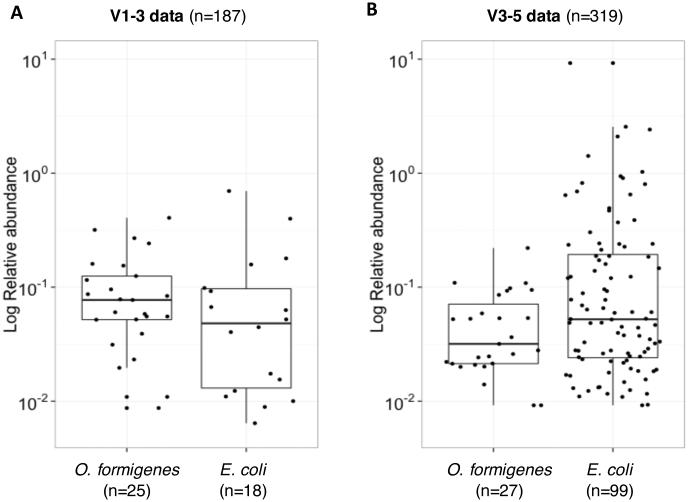
The O. formigenes and E. coli abundances also were compared in the 16S rRNA datasets. The V1-3 data showed O. formigenes abundance ranging between 0.01-0.41% (median 0.08%) and E. coli abundance ranging between 0.01-0.70%(median 0.48%) (Figure 3A). The V3-5 data showed lower O. formigenes (0.01-0.22%; median 0.03%) and slightly higher E. coli (0.01-11.67%; median 0.52%) abundance (Figure 3B). In the V1-3 and V3-5 analyses, the median abundance of O. formigenes was ~6-18-fold lower than that of E. coli, although E. coli varied over a larger range.
Figure 3. Abundance of O. formigenes and E. coli in HMP fecal samples, using QIIME 16S rRNA data.
Panel A: O. formigenes and E. coli relative abundances in 25 and 18 positive subjects, respectively, using the V1-3 dataset. Panel B: O. formigenes and E. coli relative abundance in 27 and 99 positive subjects, respectively, using the V3-5 dataset.
Relationship to other species. We applied the MetaPhlAn tool to compare O. formigenes prevalence to other gut microbiota to examine relationships between the different species in the WGS dataset. This analysis, performed on 139 stool samples from 94 subjects, detected O. formigenes in 27 subjects (29%), E. coli in 67 (71%), and B. fragilis in 49 (52%). O. formigenes presence was independent of whether or not E. coli (p=0.7) or B. fragilis (p=0.18) were present. Using the V1-3 data, O. formigenes and E. coli were detected in 15 and 12%, respectively, but using the V3-5 data, O. formigenes prevalence was 11% and E. coli was 40% (Table 1). These differences in relative proportions illustrate the limitations of microbial census-taking using primers based on the 16S variable regions 26.
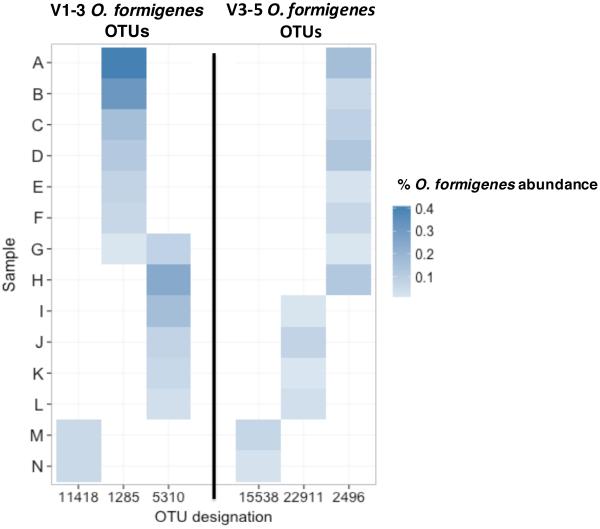
O. formigenes diversity. We also examined O. formigenes diversity in 14 samples that were positive in both the V1-3 and V3-5 datasets. We detected three O. formigenes OTUs in each of the datasets (Figure 4). Two samples had OTU 11418 in the V1-3 dataset, and in both cases, they also showed OTU 15538 in the V3-5 dataset; this suggests that the two OTUs reflect the same strains. Similarly, all but one sample that had OTU 2496 in the V3-5 dataset had OTU 1285 in V1-3. OTUs 5310 and 22911 were similarly but not identically identified in the V1-3 and V3-5 dataset samples, respectively. These data suggest the presence of two major Groups and one minor Group of O. formigenes in the positive samples.
Figure 4. O. formigenes OTU abundance for the 14 stool samples that were positive in both the V1-3 and V3-5 datasets.
The heatmap shows the percent relative abundance of the three different OTUs that each were detected in the V1-3 or V3-5 datasets, respectively.
Comparing the 14 samples that underwent both V1-3 and V3-5 16S sequencing, 12 (86%) appear concordant between both analyses and two were discordant. All but one sample, positive for OTUs 1285 and 5310, showed only a single OTU in both analyses (Figure 4). When compared to the 16S rRNA genes of known Group I and Group II O. formigenes, the sequences of OTU 1285/2496 and OTU 5310/22911 were similar to Group II and Group I strains, respectively (Supplemental Table 1). OTU 11418/15538 could represent another less prevalent O. formigenes Group, which we now call “Presumptive Group III.” Further microbiological or molecular analysis are required to identify and characterize this group.
Sensitivity of O. formigenes detection in the V1-3, V3-5, and WGS dataset analyses. We compared O. formigenes detection in 66 stool specimens for which samples were analyzed using all three HTS methods. By all three testing methods, O. formigenes was undetectable in 51 and identified in 7; thus 58 (88%) of 66 were completely concordant in all three analyses. For the non-concordant eight, their mean relative abundance (0.037%) in the WGS dataset was similar to the seven concordant positives (0.039%) (Student’s T-test, p= 0.92) All samples positive for O. formigenes in either the V1-3 or V3-5 datasets also were positive by WGS; however, O. formigenes was detected in two samples in the WGS analysis, but was negative in the V1-3 and V3-5 analyses.
O. formigenes at non-fecal sites. Sequences representative of O. formigenes also were detected in non-stool samples, ~2.2% and 1.3% of the samples subjected to V1-3 and V3-5 analyses, respectively; most were from the cutaneous sites. The positive samples in the V1-3 analysis were collected from four subjects, two of whom also had O. formigenes detected in non-stool samples by V3-5 testing (Table 2).
Table 2.
O. formigenes status by body site in V1-3 and V3-5 16S HMP data.
|
V1-3 samples
(n=2898) |
V3-5 samples
(n=4743) |
|||
|---|---|---|---|---|
|
|
||||
| Total number | O. formigenes+ | Total number | O. formigenes+ | |
|
|
||||
| Stool | 187 | 25 | 319 | 27 |
|
|
||||
| Skin | 664 | 3 | 990 | 4 |
|
|
||||
| Airways | 161 | 2 | 269 | 0 |
|
|
||||
| Oral cavity | 1622 | 1 | 2774 | 0 |
|
|
||||
| Vagina | 264 | 0 | 391 | 0 |
DISCUSSION
The HMP is of a similar scope to the Human Genome Project in that it sought to more fully characterize the micro-organisms coexisting with humans19, 20. Selection of young healthy subjects was intended to represent adults prior to the development of comorbidities. To our knowledge, study of the HMP has not yet appeared in the urology literature nor applied to the study of urolithiasis. To eventually more precisely characterize the longitudinal risk of kidney stones, studies that determine O. formigenes prevalence in unaffected, healthy adults will be essential. Developing appropriate methodologies will also be critical.
The protective role of O. formigenes against hyperoxaluria is well-established in animal models 27 and also may be applicable to humans10. The absence of O. formigenes in humans has been associated with higher urinary stone prevalence28. In the rat colon, O. formigenes induces oxalate secretion into the lumen, and decreases the oxalate gradient that drives oxalate absorption across the colonic epithelium9.
O. formigenes is detected in the gut based on the ability of anaerobic cultures to metabolize oxalate7, 16, a method that is time-consuming, limited by variation in culturability and metabolism, and non-specific, since other species may metabolize oxalate29. To improve accuracy, molecular assays have been designed to detect O. formigenes oxc which encodes Oxc (oxalyl coenzyme-A decarboxylase)30, but the heterogeneous distribution and the range in O. formigenes abundance in the human gut indicated by the present study adds to the complexity of detection strategies.
With the increasing availability of open source 16S rRNA and WGS studies of the microbiome, understanding the sensitivity, specificity, and utility of each of those methods for O. formigenes detection is needed for future investigations of its relationship to kidney stone prevalence. The HMP, with V1-3 and V3-5 16S rRNA and WGS data from the same individuals, represents a valuable resource for comparing these methods. This study serves as a model for analysis of such databases for specific bacterial species and strains to relate to pathologic conditions.
No prior studies showed that O. formigenes can be detected by WGS and HTS genomic techniques. Among the HMP methods, WGS was the most sensitive. The relatively low O. formigenes prevalence (31%) detected by WGS is consistent with prior US studies using different methodologies 15, 28. When O. formigenes is present at low abundance, it can be induced by high oxalate diet 14. Giving an oxalate load may be one way to increase assay sensitivity. With the present study design, using de-identified data, we cannot determine whether the absence of colonization is associated with higher risk of urolithiasis. We also do not have 24 hour urine data to correlate O. formigenes colonization with lower urinary oxalate excretion.
The large inter-individual variation in O. formigenes abundance could partially explain the discrepant sensitivities of the WGS and 16S rRNA methods used; variation in dietary oxalate intake also could contribute to the variation in abundance. Stable O. formigenes colonization was detected in subjects tested on multiple occasions, at both the strain and OTU levels. Our results resemble those of our prior study showing that 91.7% of patients with O. formigenes at baseline remained positive when retested one and six months later18. This relative stability is notable since neither of these studies controlled dietary oxalate content. The 16S rRNA “universal” PCR primers differ in their sensitivity for particular taxa 26; for detecting both E. coli and O. formigenes, the PCRs based on the V1-3 region were less sensitive than for V3-5.
Strain-level analysis of fecal samples has been limited, with conflicting results on the dominant O. formigenes strains in the US population. Although no prior studies have detected colonization with more than one O. formigenes strain 8, 14, our WGS analysis demonstrates that healthy adults can simultaneously harbor both Group I and Group II strains. The lack of detected colonization with Group II strains alone suggests either obligate dependence on Group I or that the methods used are more sensitive for Group I. With both sets of 16S rRNA primers, three O. formigenes OTUs were detected in the study samples, remaining stable over time. In contrast to the WGS data, Group II appears more common than Group I, and a new “Presumptive Group III” was identified. Since the relative effects of Group I and Group II strains on oxalate balance are not established, the importance of our findings to the epidemiology of stone disease remains to be determined.
That O. formigenes was present in some specimens obtained from non-fecal sites raises the possibility that it is a normal constituent of that site, or more likely represents contamination from the GI tract. Several subjects who had non-fecal positivity also had stool-positivity, favoring the hypothesis of contamination from fecal sources.
A limitation of our study is that it involves mining an existing database that was not designed to study O. formigenes. The non-uniform testing of samples/subjects adds to the limitations on the interpretability of the results. Nevertheless, using these imperfect data sets, we discovered novel information regarding O. formigenes colonization in humans, including presence of a newly identified “Presumptive Group III”.
CONCLUSIONS
In conclusion, all O. formigenes-detection methods appear insensitive, possibly due to the extensive variation in its relative abundance. Yet most (88%) of the samples examined by all three-test methods yielded completely consistent results. Although the optimal method for O. formigenes detection remains to be established, WGS was most sensitive in this study. The relatively lower rate of O. formigenes detection in relation to other non-USA populations12, provides further evidence to support a general trend of the disappearance of this organism with socioeconomic development.
Supplementary Material
Supplemental Figure 1. Venn diagrams indicating the overlap of the 335 samples included in either the V1-3 or V3-5 analyses. In total, 171 samples were examined in analyses in which both sets of primers were used. For 159 (93.0%) of these 171 samples, results from the two analyses were concordant. Of the discordant samples, 10 (83%) of 12 were V1-3-positive and V3-5-negative.
Acknowledgments
Supported in part by the National Institutes of Health (The Rare Kidney Stone Consortium (U54 DK 083908, part of the RDCRN and supported by NIDDK and NCATS), UH2 AR057506 , RO1 DK090989), by the Diane Belfer Program for Human Microbial Ecology, the C and D Fund, and the Knapp Family Foundation.
Footnotes
DISCLOSURE: No conflicts of interest.
References
- 1.Holmes RP, Assimos DG. Glyoxylate synthesis, and its modulation and influence on oxalate synthesis. J Urol. 1998;160:1617. [PubMed] [Google Scholar]
- 2.Holmes RP, Ambrosius WT, Assimos DG. Dietary oxalate loads and renal oxalate handling. J Urol. 2005;174:943. doi: 10.1097/01.ju.0000169476.85935.e2. [DOI] [PubMed] [Google Scholar]
- 3.Robertson WG, Peacock M. The cause of idiopathic calcium stone disease: hypercalciuria or hyperoxaluria? Nephron. 1980;26:105. doi: 10.1159/000181963. [DOI] [PubMed] [Google Scholar]
- 4.Siener R, Schade N, Nicolay C, et al. The efficacy of dietary intervention on urinary risk factors for stone formation in recurrent calcium oxalate stone patients. J Urol. 2005;173:1601. doi: 10.1097/01.ju.0000154626.16349.d3. [DOI] [PubMed] [Google Scholar]
- 5.Stamatelou KK, Francis ME, Jones CA, et al. Time trends in reported prevalence of kidney stones in the United States: 1976-1994. Kidney Int. 2003;63:1817. doi: 10.1046/j.1523-1755.2003.00917.x. [DOI] [PubMed] [Google Scholar]
- 6.Dwyer ME, Krambeck AE, Bergstralh EJ, et al. Temporal trends in incidence of kidney stones among children: a 25-year population based study. J Urol. 2012;188:247. doi: 10.1016/j.juro.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allison MJ, Dawson KA, Mayberry WR, et al. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 8.Duncan SH, Richardson AJ, Kaul P, et al. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol. 2002;68:3841. doi: 10.1128/AEM.68.8.3841-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatch M, Gjymishka A, Salido EC, et al. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol. 2011;300:461. doi: 10.1152/ajpgi.00434.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siener R, Bangen U, Sidhu H, et al. The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int. 2013;83:1144. doi: 10.1038/ki.2013.104. [DOI] [PubMed] [Google Scholar]
- 11.Abratt VR, Reid SJ. Oxalate-degrading bacteria of the human gut as probiotics in the management of kidney stone disease. Adv Appl Microbiol. 2010;72:63. doi: 10.1016/S0065-2164(10)72003-7. [DOI] [PubMed] [Google Scholar]
- 12.Lange JN, Wood KD, Wong H, et al. Sensitivity of human strains of Oxalobacter formigenes to commonly prescribed antibiotics. Urology. 2012;79:1286. doi: 10.1016/j.urology.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidhu H, Hoppe B, Hesse A, et al. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet. 1998;352:1026. doi: 10.1016/S0140-6736(98)03038-4. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, Knight J, Easter LH, et al. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol. 2011;186:135. doi: 10.1016/j.juro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kharlamb V, Schelker J, Francois F, et al. Oral antibiotic treatment of Helicobacter pylori leads to persistently reduced intestinal colonization rates with Oxalobacter formigenes. J Endourol. 2011;25:1781. doi: 10.1089/end.2011.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson KA, Allison MJ, Hartman PA. Isolation and some characteristics of anaerobic oxalate-degrading bacteria from the rumen. Appl Environ Microbiol. 1980;40:833. doi: 10.1128/aem.40.4.833-839.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidhu H, Holmes RP, Allison MJ, et al. Direct quantification of the enteric bacterium Oxalobacter formigenes in human fecal samples by quantitative competitive-template PCR. J Clin Microbiol. 1999;37:1503. doi: 10.1128/jcm.37.5.1503-1509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemente JC, Pehrrson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Science advances. 2015;1:1. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Human Microbiome Project, C. A framework for human microbiome research. Nature. 2012;486:215. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison MJ, MacGregor B, Sharp R, et al. Genus 1. Oxalobacter. In: Brenner D, Krieg N, Staley J, editors. Bergey's Manual of Systematic Bacteriology. Second United States of America; Springer: 2005. pp. 624–627. [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuczynski J, Stombaugh J, Walters WA, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics, Chapter. 2011;10:1. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight J, Deora R, Assimos DG, et al. The genetic composition of Oxalobacter formigenes and its relationship to colonization and calcium oxalate stone disease. Urolithiasis. 2013;41:187. doi: 10.1007/s00240-013-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales SE, Holben WE. Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl Environ Microbiol. 2009;75:2677. doi: 10.1128/AEM.02166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatch M, Cornelius J, Allison M, et al. Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int. 2006;69:691. doi: 10.1038/sj.ki.5000162. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman DW, Kelly JP, Curhan GC, et al. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. 2008;19:1197. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahin N. Oxalotrophic bacteria. Res Microbiol. 2003;154:399. doi: 10.1016/S0923-2508(03)00112-8. [DOI] [PubMed] [Google Scholar]
- 30.Sidhu H, Allison M, Peck AB. Identification and classification of Oxalobacter formigenes strains by using oligonucleotide probes and primers. J Clin Microbiol. 1997;35:350. doi: 10.1128/jcm.35.2.350-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Venn diagrams indicating the overlap of the 335 samples included in either the V1-3 or V3-5 analyses. In total, 171 samples were examined in analyses in which both sets of primers were used. For 159 (93.0%) of these 171 samples, results from the two analyses were concordant. Of the discordant samples, 10 (83%) of 12 were V1-3-positive and V3-5-negative.