Abstract
The tumor microenvironment in the majority of cancers is known to favor polarization of tumor-associated macrophages (TAMs) to alternatively activated M2 phenotype, promoting disease progression and reducing patient survival. Effective therapy targeting this M2 macrophage population is thus a promising adjuvant to approved cancer therapies. One of the challenges in targeting M2-like TAMs is a lack of high affinity targeting ligand with good selectivity over anti-tumor M1-like TAMs. We have previously identified an M2 macrophage-targeting peptide (M2pep) that binds preferentially to murine M2 macrophages and M2-like TAMs. A fusion peptide of M2pep with pro-apoptotic peptide KLA (M2pepKLA) was further used to reduce TAM population in vivo but high concentrations and frequent dosing were required due to low binding affinity of M2pep for M2 macrophage. The goal of this study was to develop more potent TAM depletion constructs by increasing the valency of both the M2pep targeting and KLA drug domains. Divalent and tetravalent displays of M2pep ([M2pep]2-Biotin and [M2pep]4-Biotin) were synthesized and evaluated for improvement in binding avidity to the murine macrophages. High avidity and selective binding of [M2pep]2-Biotin to M2 macrophages was achieved with at least 10-fold lower concentration than required for monovalent M2pep activity. Increasing M2pep valency to four, however, resulted in a reduction in both binding activity and selectivity. Surprisingly, both divalent and tetravalent M2pep, without conjugation of any cytotoxic drug cargo, exhibited M2 macrophage-selective toxicity not observed in monovalent M2pep treatment. We next synthesized divalent M2pep with monovalent and divalent KLA ([M2pep]2-[KLA] and [M2pep]2-[KLA]2) to evaluate its enhanced potency compared to M2pepKLA. While both constructs were significantly more toxic than M2pepKLA to primary, bone marrow-derived M2 macrophage, desired selectivity was retained only with [M2pep]2-[KLA]. Finally, we evaluated all multivalent M2pep and M2pepKLA analogs using a syngeneic CT-26 tumor cell suspension. In this setting, [M2pep]4-Biotin and [M2pep]2-[KLA]2 exhibited selective toxicity to both M2-like TAMs and malignant cells but not to M1-like TAMs. Therefore, these constructs are promising anti-cancer constructs with dual-modality mechanisms: malignant cell killing and TAM-based immunomodulation.
Keywords: targeted drug delivery, M2 macrophages, multivalent display
Graphical Abstract
1. Introduction
Advances in the field of immunology and cancer biology have led to recent developments of clinically-effective cancer immunotherapies [1,2]. The complex tumor microenvironment, including the crosstalk between immune and cancer cells, is now actively investigated as a source of potential new drug targets [3]. The tumor associated macrophage (TAM) is a particularly enigmatic participant in this microenvironment. TAMs may be recruited from nearby tissue resident macrophages or from monocytes in circulation [4,5]. Due to macrophage plasticity, TAMs exist as a heterogeneous population and may be broadly categorized as classically activated M1 or alternatively activated M2 macrophages [6]. Notably, these two polarizing phenotypes also exhibit opposing functions toward cancer immunity [7]. M2-like TAMs are known to be pro-tumorigenic, promoting a local immunosuppressive environment, angiogenesis, and metastasis via secretion of TGF-β, VEGF, and MMP proteases. In contrast, M1-like TAMs bolster anti-tumor immunity by directly killing cancer cells through secretion of reactive nitric oxide and by stimulating immune response in tumor microenvironment. Clinically, high accumulation of TAMs observed in biopsy specimens has been correlated with both positive and negative disease prognosis depending on the cancer type [4,8]. This contradictory observation may be attributed to the difference in relative abundance of M2 versus M1-like TAM populations, and hence in more recent studies, analysis of more defined macrophage subpopulations and/or M1/M2 ratio is usually included [9–11]. In several types of cancers including breast, ovarian, pancreatic, and brain cancers where high density of TAMs has been correlated to poor disease prognosis, the majority of these TAMs express the M2 phenotype [9,12–14].
M2-like TAMs are therefore a potential therapeutic target, and selective depletion of this macrophage subpopulation is expected to improve patient outcome. Experimentally, bisphosphonate formulations such as clodronate liposomes and zoledronic acid have been used to eliminate macrophages; however, these approaches are not M2-specific and affect tissue-resident macrophages [15,16]. Targeted blocking of macrophage colony stimulating factor 1 receptor (CSF-1R) with anti-CSF-1R antibodies showed some promise toward M2-like TAM depletion or repolarization [17,18]. Nonetheless, systemic depletion of macrophages was observed following the treatment, and CSF-1R blocking resulted in a rise in plasma CSF-1 concentration which could mediate a rebound effect in macrophage population after therapy cessation. In addition, association between CSF-1R inhibition and increased incidence of breast cancer metastasis was also reported [19]. Another challenge for targeting M2-like TAMs arises from their recruitment to hypoxic tumor regions [20–22] which are less accessible to diffusion limited macromolecular drugs [23]. Hence, alternative M2 macrophage-targeting therapeutics are still needed that 1) readily gain access to and selectively deplete M2-like TAMs, 2) do not mediate an adverse cytokine storm effect, and 3) spare systemic macrophages to improve dose-limiting toxicity profile.
Toward this goal, we have identified a peptide targeting ligand for murine M2 macrophages using peptide phage display [24]. This peptide selectively binds both M2-like TAMs and primary, polarized M2 macrophages over other leukocytes including their M1 counterparts. Fusion of M2pep to pro-apoptotic KLA peptide (M2pepKLA) was further shown to mediate TAM depletion in vivo. However, its efficacy was limited by relatively low binding affinity of M2pep (Kd 100 µM) typical of phage display-discovered peptides. Therefore, an opportunity exists to further improve M2pepKLA by increasing receptor binding affinity of the ligand and cytotoxic potency of the cargo.
In this study, we aimed to improve our M2pep-based therapeutics for more selective and potent depletion of M2-like TAMs by 1) investigating multivalent display of M2pep to increase binding avidity to M2 macrophages and 2) investigating multivalent display of KLA as a way to enhance cargo cytotoxicity. Multivalency is used to increase binding avidity to target receptors [25–28] and also has been shown by us and others to enhance cytotoxicity of KLA [29–32]. In the first part of this study, we report the synthesis of divalent and tetravalent M2pep ([M2pep]2-Biotin and [M2pep]4-Biotin, respectively) and the binding of these constructs to primary M1 and M2 macrophages. In the second part of this study, we report synthesis and cytotoxicity evaluation of two multivalent M2pepKLA analogs; 1) [M2pep]2-[KLA] and 2) [M2pep]2-[KLA]2, having M2pep-to-KLA ratio of 2:1 and 1:1 respectively. Finally, we demonstrate cytotoxicity and selectivity of multivalent M2pep and M2pepKLA analogs in a mixed cell population derived from syngeneic CT-26 tumors.
2. Materials and methods
2.1. Materials
Fmoc-protected amino acids and 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) were purchased from AAPPTec (Louisvulle, KY) and AnaSpec (Fremont, CA). NovaPEG Rink Amide, Biotin NovaTag, and 2-chlorotrityl chloride resins were purchased from Merck Millipore (Billerica, MA). Collagenase (C0130), 3-maleimidopropionic acid, 3-(maleimido)propionic acid N-hydroxysuccinimide ester (NHS-maleimide), and tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO). 1,11-Bismaleimidotriethyleneglycol (BM(PEG)3), pacific blue anti-mouse F4/80 antibody, and LIVE/DEAD fixable far red cell stain kit were purchased from Life Technologies (Grand Island, NY). PerCP anti-mouse/human CD11b antibody and anti-mouse CD16/32 antibody (Fc receptor block) were purchased from BioLegend (San Diego, CA). FITC anti-mouse Ly-6G antibody was purchased from BD Pharmingen (San Diego, CA). Streptavidin FITC was purchased from eBioscience (San Diego, CA). Mouse macrophage colony-stimulating factor (M-CSF), interleukin-4 (IL-4), and interferon-γ (IFN-γ) were purchased from R&D Systems (Minneapolis, MN). Lipopolysaccharide (LPS) was purchased from InvivoGen (San Diego, CA). Dispase II was purchased from Roche (Indianapolis, IN). All other reagents were purchased from Fisher Scientific (Pittsburgh, PA) and were of reagent grade or better.
2.2. Multivalent peptide synthesis
2.2.1. Peptide synthesis
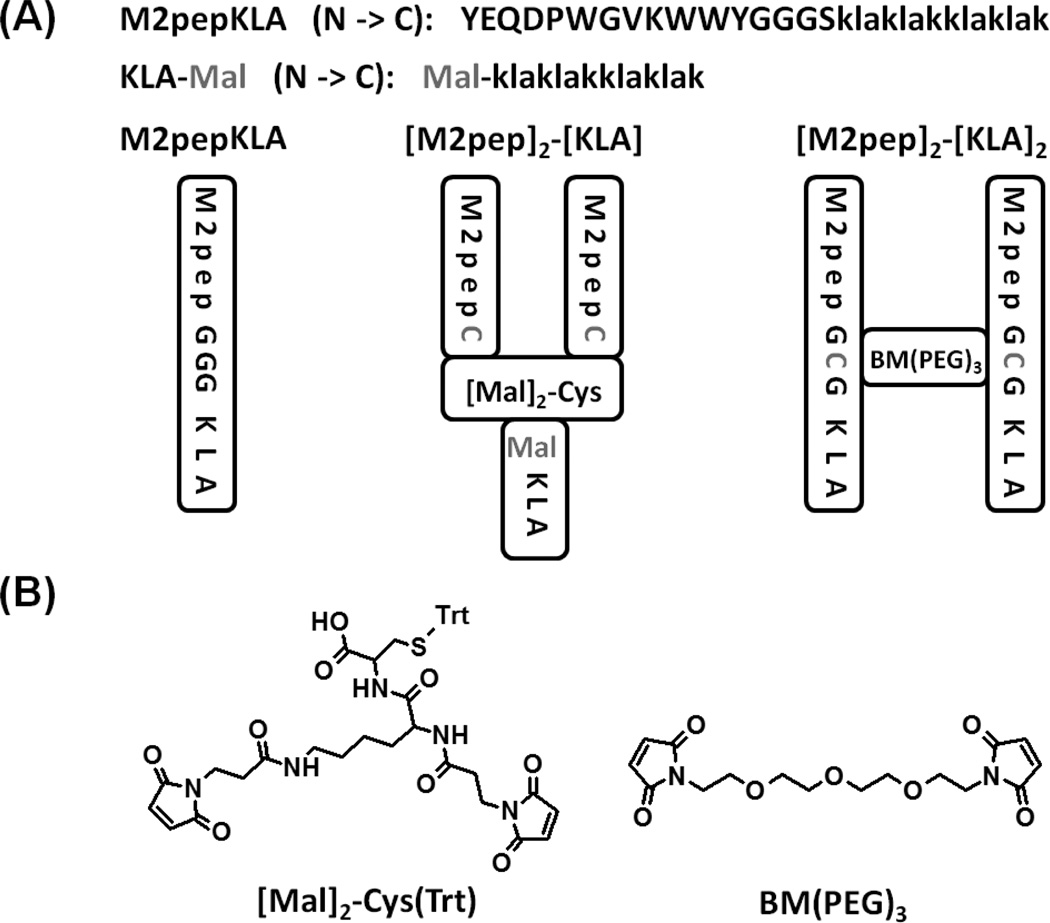
M2pepBiotin (YEQDPWGVKWWYGGGSKKKBiotin) was purchased from Elim Biopharm (Hayward, CA). M2pepKKKC (YEQDPWGVKWWYGGGSKKKC), scrambled M2pepKKKC (WEDYQWPVYKGWGGGSKKKC), M2pepKLA (YEQDPWGVKWWYGGGSklaklakklaklak), M2pepGCGKLA (YEQDPWGVKWWYGCGSklaklakklaklak), and KLA (klaklakklaklak) were synthesized on NovaPEG rink amide resin using an automated PS3 peptide synthesizer (Protein Technologies, Phoenix, AZ) following standard Fmoc solid phase peptide synthesis strategy. The peptide sequences are written from N-terminus to C-terminus, and lower case letters denote d-amino acids. When Fmoc deprotection or amino acid coupling was performed off the peptide synthesizer, the following conditions were applied: peptide resin was swelled in DCM for 15 min followed by Fmoc deprotection using 20 (v/v)% piperidine in DMF for 30 min (repeat 1 time). Amino acids were coupled by incubating peptide resin in 4 eq. of amino acid and 3.9 eq. of HATU dissolved in 0.4 M N-methylmorpholine in DMF for 3 h. Maleimide-functionalized KLA (KLA-Mal) was synthesized by on-resin coupling of KLA with 3-maleimidopropionic acid at the N terminus. Peptides were cleaved from the NovaPEG rink amide resin by incubation in TFA/triisopropylsilane (TIPS)/1,2-ethanedithiol (EDT)/1,3-dimethoxybenzene (90:2.5:2.5:5 v/v/v/v) for 2.5 h. EDT was included in the cleavage solution only for the cysteine-containing peptides. The cleaved peptides were precipitated in cold ether twice and purified to >95% purity by RP-HPLC (Agilent 1200, Santa Clara, CA) using Phenomenex Fusion-RP C18 semi-preparative column (Torrance, CA) with water (0.1% TFA) and acetonitrile (0.1% TFA) as mobile phases. Molecular weights of the purified peptides were confirmed by either MALDI-TOF MS or ESI-MS (Bruker Daltonics, Billerica, MA).
2.2.2. Synthesis of [Mal]2-Biotin and [Mal]2-Cys(Trt) divalent linkers
To synthesize the [Mal]2-Biotin divalent linker, the Fmoc protecting group on Biotin NovaTag resin was first manually deprotected and coupled with Fmoc-Lys(Fmoc)-OH following the previously-described protocol. Fmoc protecting groups were then deprotected and coupled with 3-maleimidopropionic acid (4 eq. relative to NH2 groups). The linker was cleaved in 20 (v/v)% TFA in DCM for 2 h, precipitated in cold ether twice, dried under vacuum, and used directly without purification. To synthesize the [Mal]2-Cys(Trt) divalent linker, Fmoc-Cys(Trt)-OH was coupled to 2-chlorotrityl chloride resin by incubating the resin in 4 eq. of the amino acid and 8 eq. of DIPEA in DMF for 3 h. Remaining unreacted sites were quenched by double 10 min incubations in DCM/MeOH/DIPEA (17:2:1 v/v/v). Fmoc-Lys(Fmoc)-OH and 3-maleimiopropionic acid were then coupled to the resin as noted for [Mal]2-Biotin. The linker was cleaved in acetic acid/trifluoroethanol/DCM (1:1:8 v/v/v) for 1 h, precipitated in cold ether twice, dried under vacuum, and used directly without further purification.
2.2.3. Synthesis of [Mal]4-Biotin tetravalent linker
Tetravalent linker precursor (GKGKGKGK) was synthesized on NovaPEG rink amide resin using the automated peptide synthesizer. Biotin was manually coupled to N-terminus of the peptide on resin following which the biotinylated tetravalent linker precursor (Biotin-GKGKGKGK) was cleaved in TFA/DCM (1:1 v/v) for 3 h, precipitated in cold ether twice, and purified by RP-HPLC. Biotin-GKGKGKGK was coupled with NHS-maleimide (8 eq.) and DIPEA (24 eq.) in DMF for 24 h at room temperature. As the reaction proceeded, the product ([Mal]4-Biotin) formed which became less soluble in DMF and precipitated out of the solvent. The precipitated product was washed twice with cold ether and used directly without further purification.
2.2.4. Synthesis of [M2pep]2-Biotin and [M2pep]4-Biotin
To synthesize [M2pep]2-Biotin, 0.04 eq. of TCEP (10 mg/mL in dH2O) was added to a 1 mM solution of M2pepKKKC in PBS (pH 6.5, 1 mM EDTA)/ACN (1:1 v/v). The flask was capped with a rubber septum, and the solution was purged with argon for 10 min. During purging, [Mal]2-Biotin was dissolved in methanol (10 mg/mL), and the appropriate volume (0.4 eq.) was added to the flask. Purging was continued for another 10 min after which the needle was removed, and the septum was sealed with parafilm. The reaction was left to proceed for 24 h at room temperature. The solution mixture was then evaporated under reduced pressure, resolubilized in a minimal volume of H2O/DMF (2:1 v/v). M2pepKKKC dimer formed during coupling reaction was reduced with TCEP (10 eq. relative to the starting amount of M2pepKKKC) for 30 min before the solution was desalted with a Sep-Pak C18 cartridge (Waters, Milford, MA) and purified by RP-HPLC. To synthesize [M2pep]4-Biotin, the same protocol as the synthesis of [M2pep]2-Biotin was used with some modifications as follows: 0.02 eq. of TCEP and 0.2 eq. of [Mal]4-Biotin were used in the synthesis. [Mal]4-Biotin was dissolved in DMSO to make a stock solution.
2.2.5. Synthesis of [M2pep]2-Cys(Trt) and deprotection of Trt protecting group
[M2pep]2-Cys(Trt) was synthesized in the same manner as the synthesis of [M2pep]2-Biotin. Subsequent deprotection of Trt protecting group from the peptide was done in TFA/TIPS/DMF (75:2.5:22.5 v/v/v) for 1 h and precipitated in cold ether. Successful deprotection was confirmed by ESI-MS.
2.2.6. Synthesis of [M2pep]2-[KLA] and [M2pep]2-[KLA]2
[M2pep]2-[KLA] was synthesized by conjugating KLA-Mal to [M2pep]2-Cys using the same conjugation conditions as the synthesis of [M2pep]2-Biotin except that 2 eq. of KLA-Mal was used in the synthesis. [M2pep]2-[KLA]2 was also synthesized in the same manner using 1 eq. of M2pepGCGKLA and 0.4 eq. of BM(PEG)3. BM(PEG)3 was dissolved in DMSO for use.
2.3. Generation of bone marrow-derived macrophages
All animal handling protocols were approved by the University of Washington Institutional Animal Care and Use Committee. Femurs and tibias were harvested from female c57bl6/027 mice with the age between 6–8 weeks. The bones were flushed with RPMI 1640 medium to collect bone marrow cells. Roughly 5 million cells per dish were seeded on 100 mm petri dish in RPMI 1640 medium containing 20% donor horse serum, 1% antibiotic-antimycotic (AbAm), and 20 ng/mL M-CSF. After 7 d of culture, M-CSF was replaced with either 25 ng/mL IFN-γ and 100 ng/mL LPS for M1 macrophage activation or 25 ng/mL IL-4 for M2 macrophage activation. The culture was continued for 2 d before the activated macrophages were scraped off the petri dishes for study.
2.4. Binding study
Activated macrophages were plated on a black round bottom 96-well plate at the density of 50,000 cells/well. Biotinylated multivalent M2pep analogs in 100 µL PBS containing 1% BSA were added to each well and incubated on ice for 20 min. Unbound peptides were washed off, and the bound peptides were probed by incubation with streptavidin FITC for 15 min on ice. Excess streptavidin FITC was washed off, and the macrophages were resuspended for analysis with a MACSQuant Flow Cytometer (Miltenyi Biotec, San Diego, CA). Propidium iodide (PI) was added to the samples prior to data acquisition to discriminate dead cells. Flow cytometry data were analyzed on FlowJo Analysis Software (Tree Star, Ashland, OR). Median fluorescence intensity values of FITC in the live macrophage population were normalized to the values of the control samples that were treated with only streptavidin FITC without peptide. Each condition was tested in triplicates and each experiment was also conducted three independent times.
2.5. In vitro cytotoxicity study
Activated macrophages were seeded in suspension on a black round bottom 96-well plate at the density of 50,000 cells/well. Multivalent M2pep or M2pepKLA analogs at varying concentrations in 100 µL RPMI 1640 medium were added to each well and incubated in a tissue culture (TC) incubator for 2 h at 37 °C, 5% CO2. The macrophages were washed and resuspended in PBS for flow cytometry analysis. PI was added to probe for the extent of cell death. Cell viability data were presented as percentage of live macrophages normalized to the mean percentage of the live untreated control samples. The study was conducted in three independent experiments.
2.6. Ex vivo cytotoxicity study
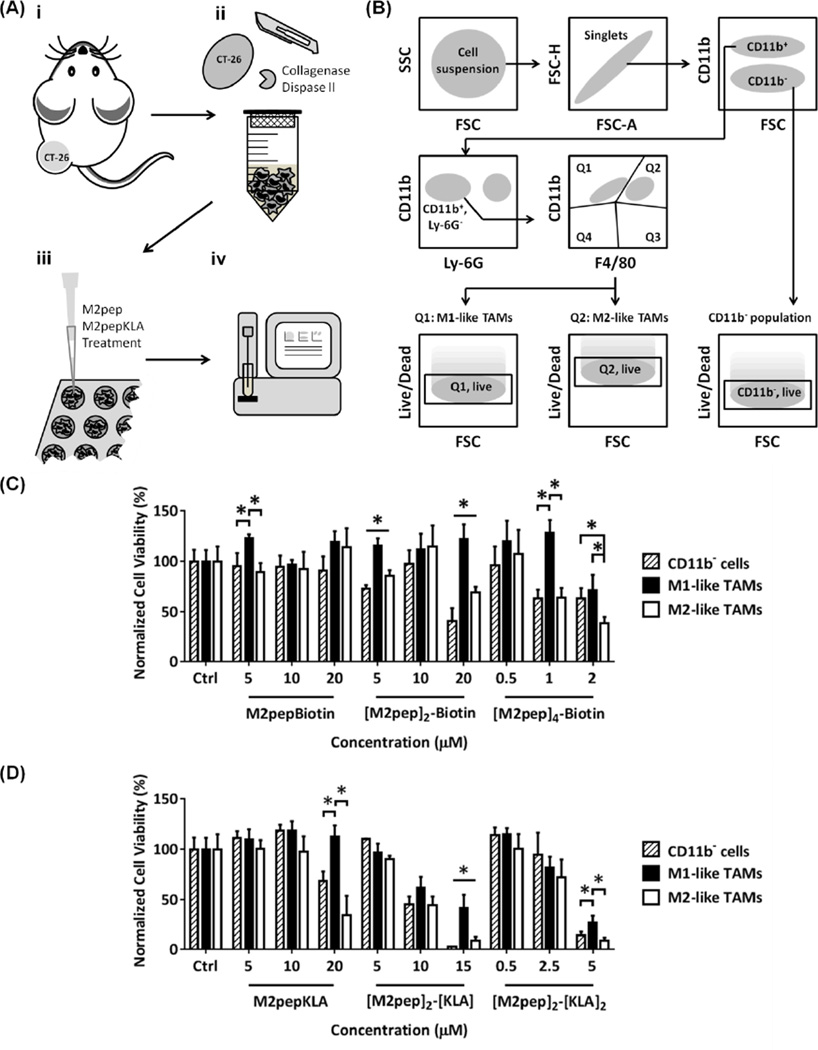
CT-26 colon carcinoma cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum and 1% AbAm. Tumors were inoculated by injection of 106 CT-26 cells into the left flanks of female BALB/c mice with the age between 6–8 weeks and left for 2 weeks to develop tumors. Each tumor was then processed into single cell suspension by mincing into small pieces, homogenizing on gentleMACS Dissociator (Miltenyi Biotec, San Diego, CA), and incubating in 5 mL RPMI 1640 medium with 100 µL collagenase (10,000 CDU/mL stock solution) and 100 µL dispase II (32 mg/mL stock solution) for 40 min in the TC incubator. The dissociated single cells were then collected through 70 µm cell strainer and plated on a black round bottom 96-well plate at the density of 200,000 cells/well. The cells were treated with multivalent M2pep and M2pepKLA analogs for 2 h in the TC incubator. To quantify cell viability of each subpopulation, the cells were first stained with LIVE/DEAD fixable far red cell stain and then with an antibody cocktail solution (anti-CD11b, anti-Ly-6G, and anti-F4/80 antibodies). After staining, the cells were fixed with 4% paraformaldehyde and analyzed with flow cytometer. The detailed list of antibodies and example flow cytometry gating schematics were included in Fig. S1.
3. Results and discussion
3.1. Multivalent targeting ligands
3.1.1. Synthesis of multivalent M2pep
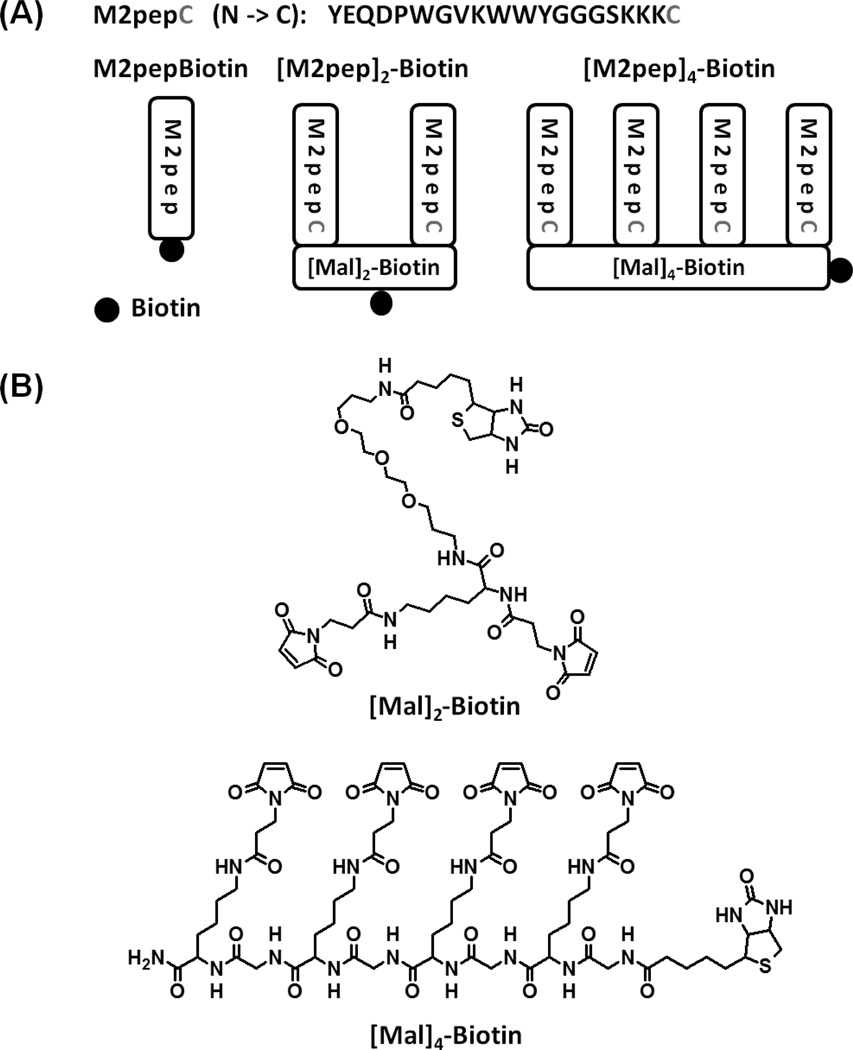
Divalent and tetravalent M2pep were synthesized with a biotin tag ([M2pep]2-Biotin and [M2pep]4-Biotin, respectively) by conjugation of M2pepKKKC to maleimide functionalized linkers, [Mal]2-Biotin and [Mal]4-Biotin, via thiol-maleimide chemistry (Scheme 1 and S1). [Mal]2-Biotin was synthesized from the Biotin NovaTag resin whereby Fmoc-Lys(Fmoc)-OH was first coupled to the resin, deprotected to expose two free amines, and then coupled with 3-maleimidopropionic acid. Cleavage of the linker from the resin and precipitation in ether afforded an amber viscous product. [Mal]4-Biotin was synthesized from GKGKGKGK scaffold with N-terminal biotin. The scaffold was first synthesized on the automated peptide synthesizer, cleaved off the resin, and purified by RP-HPLC. The purified precursor was then reacted with NHS-maleimide in solution, and a white solid product was obtained from ether precipitation of the reaction solution. In both cases, the linkers were of sufficient purity for M2pepKKKC conjugation to give [M2pep]2-Biotin and [M2pep]4-Biotin in good yields. Successful synthesis of the linkers and multivalent M2pep was confirmed by mass spectrometry (Table S1). The synthesis strategy for the multivalent linkers reported here allows for tunable display of targeting peptides at other valencies by simply extending the oligolysine peptide precursor. In addition, biotin may be replaced with other functional compounds for different applications, and spacing between each targeting peptide may also be adjusted by increasing the number of amino acids between adjacent lysines or using oligoPEG as a molecular spacer.
Scheme 1.
(A) Multivalent M2pep analogs. (B) Chemical structures of molecular linkers used in the synthesis.
3.1.2. Binding and cytotoxicity studies of multivalent M2pep with primary cultured macrophage
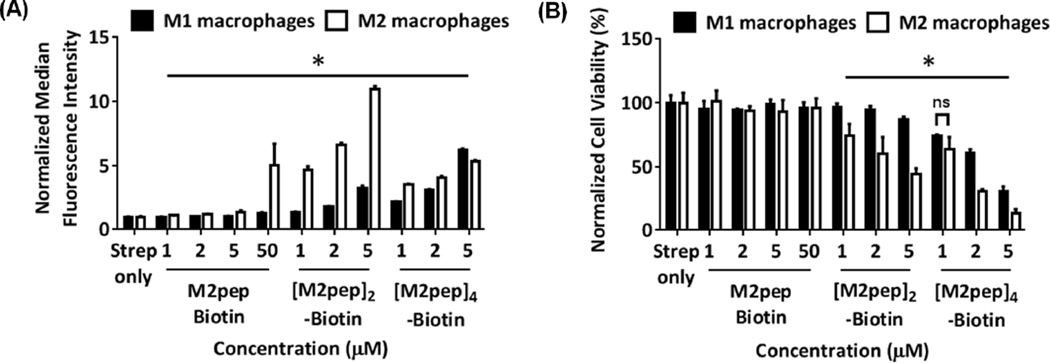
The mono-, di- and tetra-valent M2pep ligands were tested for binding selectivity to target cells using primary bone marrow-derived macrophages polarized to M1 or M2 state with IFN-γ/LPS or IL-4, respectively. Significant and selective binding of monovalent M2pep to M2 macrophages versus M1 macrophages was detected at 50 µM but not at lower concentration (≤ 5 µM) due to the relatively high KD of the peptide (~ 90 µM, reported previously [24]) (Fig. 1A). On the other hand, divalent [M2pep]2-Biotin exhibited significantly enhanced binding to M2 macrophages with good selectivity over M1 macrophages even at 1 µM, with M2 binding increasing with concentration. Unexpectedly, tetravalent [M2pep]4-Biotin bound less to cultured M2 macrophages compared to [M2pep]2-Biotin, and its selectivity over M1 macrophages was significantly compromised. Internalization of these M2pep constructs were investigated by confocal microscopy and shown to correlate with the result from the binding study (Fig. S2).
Fig. 1.
(A) In vitro binding of M2pepBiotin, [M2pep]2-Biotin, and [M2pep]4-Biotin to M1 and M2 macrophages. (B) Cell viability of macrophages following peptide incubation during binding study. Data were normalized to values from streptavidin only samples and presented as mean ± standard deviation. Stars denote statistically significant difference between M1 and M2 macrophages under the same treatment concentration. * P < 0.05.
Cytotoxicity of the multivalent peptides but not M2pepBiotin was observed during the binding study and was therefore quantified by PI staining with flow cytometry analysis (Fig 1B). Interestingly, both [M2pep]2-Biotin and [M2pep]4-Biotin were selectively toxic to M2 macrophages over M1 macrophages at low micromolar concentrations while no toxicity to either cell type was observed for M2pepBiotin even at 50 µM. Although not investigated in this study, it was possible that tetravalent M2pep display on the Biotin-GKGKGKGK base linker may not be in the optimal configuration for M2pep peptides to engage in binding to multiple M2pep receptors. Reduced binding with increasing valency beyond its optimal number was previously observed in other multivalent systems and was proposed to be attributed to steric hindrance from overcrowding of targeting ligands or over-occupation of surface receptors per individual targeting construct [33–35]. Hence, future investigation into M2pep display configuration, multivalency-associated cytotoxicity, and the effect of spacer length and flexibility may be needed to further improve M2 macrophage-binding with higher M2pep valencies. The use of flexible linkers such as oligoPEG is especially attractive as they have been shown to facilitate optimal multivalent binding over a greater span of linker lengths compared to rigid linkers which have a more stringent spacing requirement for optimal binding [36].
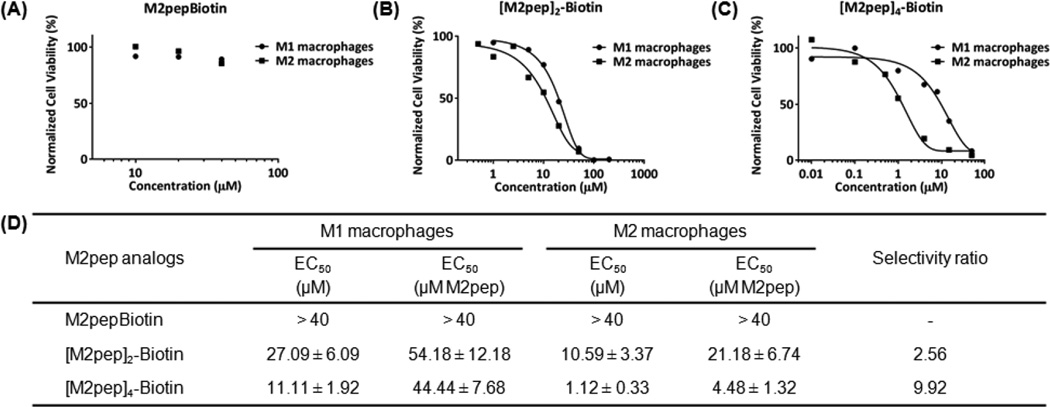
Next, we further investigated cytotoxicity of the multivalent M2pep ligands by incubating the peptides at different concentrations for 2 h at 37 °C before assessing cell viability by PI staining (Fig. 2). Values of the effective concentration which caused a 50 percent reduction in cell viability (EC50) were then calculated from the toxicity curves. Selectivity ratio in toxicity between M1 and M2 macrophages was defined as EC50 M1/EC50 M2. A relatively short incubation time was chosen because cytotoxicity was observed with a 20 min incubation of the peptides with cells on ice during the binding study. [M2pep]4-Biotin was found to be the most potent at inducing cytotoxicity to M2 macrophages (EC50 = 1.12 µM) with the highest selectivity ratio of 9.92. [M2pep]2-Biotin showed lower cytotoxicity to M2 macrophages but still retaining M2 macrophage selectivity (Selectivity ratio = 2.56). On the other hand, EC50 of M2pepBiotin was not reached even at 40 µM. We further confirmed M2pep sequence specificity on macrophage toxicity by incubating macrophages with M2pepKKKC and scrambled M2pepKKKC. Both peptides partially dimerized in solution to form dimers as confirmed by ESI-MS, but toxicity was only observed in the M2pepKKKC-treated samples (Fig. S3). In addition, we also confirmed that the cytotoxicity was not due to the linkers used (Fig. S4). The selective and potent toxicity of [M2pep]4-Biotin to M2 macrophage is surprising considering that selective binding to this population compared to M1 was not observed. M1 macrophages may simply be more resistant to this M2pep-mediated signaling pathway, or the association between tetravalent M2pep and M1 macrophages may be partially non-specific such that not all the peptide binds to the appropriate M2pep receptor to trigger the cytotoxic signal.
Fig. 2.
Representative cytotoxicity curves of (A) M2pepBiotin, (B) [M2pep]2-Biotin, and (C) [M2pep]4-Biotin on M1 and M2 macrophages. (D) Tabulated EC50 values of multivalent M2pep presented as mean ± standard deviation from 3 independent experiments.
It would be interesting to explore in future studies how a more flexible or longer linker between M2pepKKKC affects macrophage cytotoxicity. This knowledge, together with the identification of M2pep receptor, may help us elucidate the mechanism of cytotoxicity (e.g. due to crosslinking of multiple M2pep receptors on cells resulting in intracellular signaling, multivalency-induced change in conformation of individual M2pep receptors that may possess multiple M2pep-binding domains on the same receptor, or some other mechanisms). In recent years, there is a growing interest in developing drug-free macromolecular therapeutics employing the concept of receptor crosslinking-induced cell death [37]. One of the best known examples is the work by the Kopecek group which investigated into the use of coiled-coil interaction between CCE and CCK peptides to crosslink CCE-conjugated Fab’ fragments of anti-CD20 antibody with CCK-grafted polymer. Crosslinking of CD20 in this manner was shown to induce apoptosis in Raji B cells in vitro with promising preclinical results in SCID mice bearing human B-lymphoma xenografts [38]. Crosslinking of multiple M2pep receptors may thus represent a strategy to induce cell death in M2 macrophages without the need for cytotoxic drug cargos. However, a more in-depth understanding of M2pep receptor and its interaction with M2pep is required to better optimize the drug free, M2 macrophage-depleting therapeutics.
3.2. Multivalent pro-apoptotic KLA cargo
3.2.1. Synthesis of multivalent KLA constructs
Since divalent M2pep was shown here to significantly improve selective binding to M2 macrophages, we next synthesized [M2pep]2-[KLA] (Scheme 2 and S2) to evaluate how the enhanced binding would potentiate the pro-apoptotic peptide KLA toxicity to the macrophages compared to monovalent M2pepKLA. [Mal]2-Cys(Trt) was first synthesized from a 2-chlorotrityl chloride resin via manual coupling of Fmoc-Cys(Trt)-OH, Fmoc-Lys(Fmoc)-OH, and 3-maleimidopropionic acid respectively. Mild cleavage condition enabled us to obtain [Mal]2-Cys(Trt) which was conjugated to M2pepKKKC via thiol-maleimide chemistry. The Trt protecting group on the [M2pep]2-Cys(Trt) was then removed by TFA treatment exposing a free thiol for subsequent conjugation with KLA-Mal. In addition, divalent M2pep with divalent KLA ([M2pep]2-[KLA]2) was also synthesized to evaluate if increasing the number of KLA would further enhance the overall potency. M2pepGCGKLA was synthesized from the automated peptide synthesizer with cysteine replacing glycine in the spacer region of the original M2pepKLA sequence. M2pepGCGKLA was then dimerized using the BM(PEG)3 linker. Successful synthesis of both M2pepKLA analogs was confirmed by ESI-MS (Table S1).
Scheme 2.
(A) Multivalent M2pepKLA analogs. (B) Chemical structures of molecular linkers used in the synthesis.
3.2.2. Cytotoxicity study of multivalent M2pepKLA
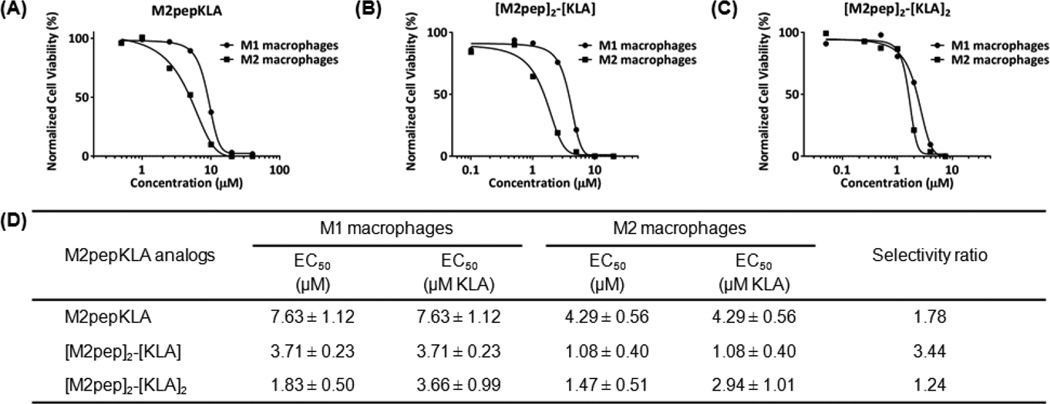
Next, the cytotoxicity of M2pepKLA, [M2pep]2-[KLA], and [M2pep]2-[KLA]2 to M1 and M2 macrophages was determined using cultured primary cells. The fusion peptide between KLA and tetravalent M2pep was not investigated in this study since [M2pep]4-Biotin did not exhibit M2 macrophage-selective binding. Toxicity curves, calculated EC50 values, and selectivity ratios are reported in Fig 3. Conjugation of KLA to M2pep imparts cytotoxicity to M2 macrophages with an EC50 value of 4.29 µM and a selectivity ratio of 1.78. Remarkably, conjugation of a single KLA to the dimerized targeting ligand ([M2pep]2-[KLA]) increases cytotoxicity by 10-fold (EC50 = 1.08 µM versus 10.58 µM for [M2pep]2-Biotin) compared to dimeric ligand alone and by 4-fold over M2pepKLA while improving selectivity (Selectivity ratio = 3.44). Thus, the increased binding avidity of the divalent M2pep ligand also increases potency when this ligand is used for internalization of KLA.
Fig. 3.
Representative cytotoxicity curves of (A) M2pepKLA, (B) [M2pep]2-[KLA], and (C) [M2pep]2-[KLA]2 on M1 and M2 macrophages. (D) Tabulated EC50 values of multivalent M2pepKLA presented as mean ± standard deviation from 3 independent experiments.
Dimerization of KLA ([M2pep]2-[KLA]2) did not further improve the overall potency compared to [M2pep]2-[KLA] but rather reduced M2 macrophage selectivity. This reduced selectivity is possibly in accordance with the improved cell penetration property of dimeric KLA recently reported by Hyun et al. [31]. Hence, this study demonstrates a significant contribution of cationic KLA on M2pep selectivity especially in the multivalent format and cautions the use of cationic antimicrobial peptides as a cytotoxic cargo in ligand-targeted therapeutics due to non-specific cell interaction. As alternatives to improve M2 macrophage-targeting selectivity, we suggest exploration of other potent drug cargos with inherently lower internalizing activity or alternatively reversibly altering charges on KLA with dimethylmaleic anhydride or benzoic imine to reduce non-specific uptake of the peptide [39–41].
In addition, we performed a cytotoxicity study with annexin V/PI staining to investigate the mechanism of macrophage death upon the treatment of multivalent M2pep and M2pepKLA constructs (Fig. S5). Interestingly, all multivalent constructs induced an increase in the annexin V-positive/PI-positive (late apoptosis/secondary necrosis) population over time whereas the annexin V-positive/PI-negative (early apoptosis) population comprised a relatively low percentage with no appreciable change over time. This implies that additional non-apoptosis mechanism may be contributing to macrophage death. It is worth noting that cationic antimicrobial peptides have been shown to induce non-apoptotic cell death by directly disrupting cell membrane [42] which could be a possible mechanism for our case.
3.3. Ex vivo evaluation of multivalent constructs with TAMs
Next, we evaluated multivalent M2pep and M2pepKLA constructs for potency and selectivity to TAMs in mixed tumor cell suspensions. Tumors were harvested from syngeneic CT-26 colon carcinoma tumors of Balb/c mice after two weeks of inoculation and processed into single cell suspension for viability studies using flow cytometry (Fig. 4A). The cell suspension included CT-26 cells, TAMs, and other tumor-associated cells (e.g. other leukocytes, endothelial cells, fibroblasts) in physiologically relevant ratios and thus represented more closely what cells the peptides would encounter in the tumor in vivo. Here, we evaluated the cytotoxic effect of multivalent M2pep and M2pepKLA analogs on 1) CD11b− population (primarily CT-26 malignant cells), 2) M1-like TAM population, and 3) M2-like TAM population following the flow cytometry gating strategy shown in Fig. 4B and Fig. S1. The cell viability data were first normalized to total cell count and then presented as percentage relative to their respective live populations in the untreated control. In general, all multivalent M2pep constructs, with or without KLA, were cytotoxic to the tumor cell suspension at high concentrations (Fig. 4C, 4D). Selective toxicity to M2-like TAMs over M1-like TAMs was observed but not over CD11b− population. Toxicity on CD11b− population was in accordance with our previously reported study that M2pep bound strongly to this cell population ex vivo although interaction and internalization in these cells in vivo was not observed [24]. Hence, we expect the cytotoxic effect on this population to be less in vivo.
Fig. 4.
(A) Tumor processing for ex vivo cytotoxicity evaluation: (i) CT-26 tumors were inoculated in Balb/c mice. (ii) The tumors were harvested and processed into single cell suspension. (iii) Cell suspension was seeded and treated with different M2pep and M2pepKLA analogs. (iv) The cells were stained and analyzed by flow cytometry. (B) Flow cytometry gating strategy for cell viability analysis of tumor sub-populations. Ex vivo cytotoxicity of multivalent M2pep (C) and M2pepKLA (D) constructs on each sub-population in CT-26 tumor-derived cell suspension. Unless labeled in pairs, stars denote statistical significance among all populations. * P < 0.05.
As observed in vitro, multimerization of M2pep resulted in cytotoxicity, with [M2pep]4-Biotin being the most potent analog. Of the KLA-containing constructs, [M2pep]2-[KLA]2 was most potent and exhibited better M2 to M1 selectivity compared with [M2pep]2-[KLA]. In tumor microenvironment where there is a complex mix of different cell populations, targeting activity of the therapeutics may be significantly altered and not fully correlated to the in vitro results. The death or stimulation of one cell population may also trigger release of cytokines that affect viability of the others making selective depletion of a desired population an even more challenging task. In addition, negatively-charged extracellular matrix components such as hyaluronan, which is abundant in the tumor microenvironment [43], may interact with the positively-charged KLA and help restore the M2 selectivity relative to M1-like TAMs observed ex vivo. However, this electrostatic interaction may impede diffusion of the drug into the deeper hypoxic region, thus limiting its in vivo efficacy [44]. Here, our ex vivo study suggested [M2pep]2-[KLA]2 and [M2pep]4-Biotin as promising candidates in term of potency for future in vivo evaluation although complementary strategies that facilitate tumor penetration may be needed. Dual killing of both cancer cells and M2-like TAMs observed ex vivo, if translated to an in vivo setting, would be desirable for cancer therapy.
Conclusions
In this work, divalent display of M2pep was shown to improve M2 macrophage-binding activity in vitro. On the other hand, tetravalent display of M2pep had reduced binding compared to divalent M2pep and was accompanied with loss in selectivity over M1 macrophages. Surprisingly, divalent and tetravalent displays of M2pep were found to exhibit M2 macrophage-selective cytotoxicity both in vitro and ex vivo. Conjugation of divalent M2pep to KLA significantly improved M2 macrophage-cytotoxicity and selectivity in vitro. In the ex vivo study, [M2pep]2-[KLA]2 and tetravalent [M2pep]4-Biotin were the most potent making them promising candidates for future in vivo study. Additionally, further improvement in multivalency-induced macrophage death by M2pep may also have a potential as a drug-free, anti-cancer construct via M2-like TAM depletion.
Supplementary Material
Acknowledgments
This work was supported by NIH 1R01CA177272. Chayanon Ngambenjawong was supported by an Anandamahidol Foundation Fellowship. Maryelise Cieslewicz was supported by a National Science Foundation Graduate Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pardoll DM. Spinning molecular immunology into successful immunotherapy. Nat. Rev. Immunol. 2002;2:227–238. doi: 10.1038/nri774. [DOI] [PubMed] [Google Scholar]
- 2.Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011;11:805–812. doi: 10.1038/nrc3153. http://dx.doi.org/10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joyce Ja. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 2005;7:513–520. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 4.Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahmar Q, Keirsse J, Laoui D, Movahedi K, Van Overmeire E, Van Ginderachter Ja. Tissue-resident versus monocyte-derived macrophages in the tumor microenvironment. Biochim. Biophys. Acta - Rev. Cancer. 2015 doi: 10.1016/j.bbcan.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 7.Lamagna C, Aurrand-Lions M, Imhof Ba. Dual role of macrophages in tumor growth and angiogenesis. J Leukoc. Biol. 2006;80:705–713. doi: 10.1189/jlb.1105656. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Liu L, Gong C, Shi H, Zeng Y, Wang X, et al. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One. 2012;7:e50946. doi: 10.1371/journal.pone.0050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colvin EK. Tumor-associated macrophages contribute to tumor progression in ovarian cancer. Front. Oncol. 2014;4:137. doi: 10.3389/fonc.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelissen R, Lievense La, Maat AP, Hendriks RW, Hoogsteden HC, Bogers AJ, et al. Ratio of Intratumoral Macrophage Phenotypes Is a Prognostic Factor in Epithelioid Malignant Pleural Mesothelioma. PLoS One. 2014;9:e106742. doi: 10.1371/journal.pone.0106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantano F, Berti P, Guida FM, Perrone G, Vincenzi B, Amato MMC, et al. The role of macrophages polarization in predicting prognosis of radically resected gastric cancer patients. J. Cell. Mol. Med. 2013;17:1415–1421. doi: 10.1111/jcmm.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurahara H, Shinchi H, Mataki Y, Maemura K, Noma H, Kubo F, et al. Significance of M2-polarized tumor-associated macrophage in pancreatic cancer. J. Surg. Res. 2011;167:211–219. doi: 10.1016/j.jss.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, Ke SQ, Huang Z, Flavahan W, Fang X, Paul J, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeisberger SM, Odermatt B, Marty C, Zehnder-Fjällman aHM, Ballmer-Hofer K, Schwendener Ra. Clodronate-liposome-mediated depletion of tumour-associated macrophages: a new and highly effective antiangiogenic therapy approach. Br. J. Cancer. 2006;95:272–281. doi: 10.1038/sj.bjc.6603240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junankar S, Shay G, Jurczyluk J, Ali N, Down J, Pocock N, et al. Real-Time Intravital Imaging Establishes Tumor-Associated Macrophages as the Extraskeletal Target of Bisphosphonate Action in Cancer. Cancer Discov. 2014;5:35–42. doi: 10.1158/2159-8290.CD-14-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ries CH, Cannarile Ma, Hoves S, Benz J, Wartha K, Runza V, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swierczak A, Cook AD, Lenzo JC, Restall CM, Doherty JP, Anderson RL, et al. The Promotion of Breast Cancer Metastasis Caused by Inhibition of CSF-1R/CSF-1 Signaling Is Blocked by Targeting the G-CSF Receptor. Cancer Immunol. Res. 2014:765–777. doi: 10.1158/2326-6066.CIR-13-0190. [DOI] [PubMed] [Google Scholar]
- 20.Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M, et al. Impeding Macrophage Entry into Hypoxic Tumor Areas by Sema3A/Nrp1 Signaling Blockade Inhibits Angiogenesis and Restores Antitumor Immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Murdoch C, Giannoudis A, Lewis CE. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood. 2004;104:2224–2234. doi: 10.1182/blood-2004-03-1109. [DOI] [PubMed] [Google Scholar]
- 22.Laoui D, Van Overmeire E, Di Conza G, Aldeni C, Keirsse J, Morias Y, et al. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res. 2014;74:24–30. doi: 10.1158/0008-5472.CAN-13-1196. [DOI] [PubMed] [Google Scholar]
- 23.Dreher MR, Liu W, Michelich CR, Dewhirst MW, Yuan F, Chilkoti A. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J. Natl. Cancer Inst. 2006;98:335–344. doi: 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- 24.Cieslewicz M, Tang J, Yu JL, Cao H, Zavaljevski M, Motoyama K, et al. Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15919–15924. doi: 10.1073/pnas.1312197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasting C, Schalley Ca, Weber M, Seitz O, Hecht S, Koksch B, et al. Multivalency as a chemical organization and action principle. Angew. Chemie - Int. Ed. 2012;51:10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 26.Locke L, Mayo M, Yoo A, Williams M, Berr S. PET imaging of tumor associated macrophages using mannose coated 64 Cu liposomes. Biomaterials. 2012;33 doi: 10.1016/j.biomaterials.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu S, Niu M, O’Mary H, Cui Z. Targeting of Tumor-Associated Macrophages Made Possible by PEG-Sheddable, Mannose-Modified Nanoparticles. Mol. Pharm. 2013;10:3525–3530. doi: 10.1021/mp400216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu M, Naguib YW, Aldayel AM, Shi Y, Hursting SD, Hersh MA, et al. Biodistribution and in Vivo Activities of Tumor-Associated Macrophage-Targeting Nanoparticles Incorporated with Doxorubicin. Mol. Pharm. 2014;11:4425–4436. doi: 10.1021/mp500565q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu DSH, Bocek MJ, Shi J, Ta A, Ngambenjawong C, Rostomily RC, et al. Multivalent display of pendant pro-apoptotic peptides increases cytotoxic activity. J. Control. Release. 2015;205:155–161. doi: 10.1016/j.jconrel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agemy L. Targeted nanoparticle enhanced proapoptotic peptide as potential therapy for glioblastoma. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17450–17455. doi: 10.1073/pnas.1114518108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyun S, Lee S, Kim S, Jang S, Yu J, Lee Y. Apoptosis Inducing, Conformationally Constrained, Dimeric Peptide Analogs of KLA with Submicromolar Cell Penetrating Abilities. Biomacromolecules. 2014;15:3746–3752. doi: 10.1021/bm501026e. [DOI] [PubMed] [Google Scholar]
- 32.Adar L, Shamay Y, Journo G, David A. Pro-apoptotic peptide-polymer conjugates to induce mitochondrial-dependent cell death. Polym. Adv. Technol. 2011;22:199–208. [Google Scholar]
- 33.Voulgaraki D, Mitnacht-Kraus R, Letarte M, Foster-Cuevas M, Brown MH, Barclay aN. Multivalent recombinant proteins for probing functions of leucocyte surface proteins such as the CD200 receptor. Immunology. 2005;115:337–346. doi: 10.1111/j.1365-2567.2005.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polizzotti BD, Kiick KL. Effects of polymer structure on the inhibition of cholera toxin by linear polypeptide-based glycopolymers. Biomacromolecules. 2006;7:483–490. doi: 10.1021/bm050672n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaffer DV, Lauffenburger Da. Optimization of cell surface binding enhances efficiency and specificity of molecular conjugate gene delivery. J. Biol. Chem. 1998;273:28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
- 36.Handl HL, Sankaranarayanan R, Josan JS, Vagner J, Mash Ea, Gillies RJ, et al. Synthesis and evaluation of bivalent NDP-alpha-MSH(7) peptide ligands for binding to the human melanocortin receptor 4 (hMC4R) Bioconjug. Chem. 2007;18:1101–1109. doi: 10.1021/bc0603642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chu T-W, Kopeček J. Drug-free macromolecular therapeutics – a new paradigm in polymeric nanomedicines. Biomater. Sci. 2015:908–922. doi: 10.1039/C4BM00442F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu K, Yang J, Liu J, Kopeček J. Coiled-coil based drug-free macromolecular therapeutics: In vivo efficacy. J. Control. Release. 2012;157:126–131. doi: 10.1016/j.jconrel.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang L, Li L, He X, Yi Q, He B, Cao J, et al. Overcoming drug-resistant lung cancer by paclitaxel loaded dual-functional liposomes with mitochondria targeting and pH-response. Biomaterials. 2015;52:126–139. doi: 10.1016/j.biomaterials.2015.02.004. doi: http://dx.doi.org/10.1016/j.biomaterials.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Cheng H, Zhu J-Y, Xu X-D, Qiu W-X, Lei Q, Han K, et al. Activable cell-penetrating peptide (ACPP) conjugated prodrug for tumor targeted drug delivery. ACS Appl. Mater. Interfaces. 2015 doi: 10.1021/acsami.5b04517. 150710083756006. [DOI] [PubMed] [Google Scholar]
- 41.Ding C, Gu J, Qu X, Yang Z. Preparation of multifunctional drug carrier for tumor-specific uptake and enhanced intracellular delivery through the conjugation of weak acid labile linker. Bioconjug. Chem. 2009;20:1163–1170. doi: 10.1021/bc800563g. [DOI] [PubMed] [Google Scholar]
- 42.Paredes-Gamero EJ, Martins MNC, Cappabianco FAM, Ide JS, Miranda A. Characterization of dual effects induced by antimicrobial peptides: Regulated cell death or membrane disruption. Biochim. Biophys. Acta - Gen. Subj. 2012;1820:1062–1072. doi: 10.1016/j.bbagen.2012.02.015. doi: http://dx.doi.org/10.1016/j.bbagen.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Itano N, Zhuo L, Kimata K. Impact of the hyaluronan-rich tumor microenvironment on cancer initiation and progression. Cancer Sci. 2008;99:1720–1725. doi: 10.1111/j.1349-7006.2008.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.