Abstract
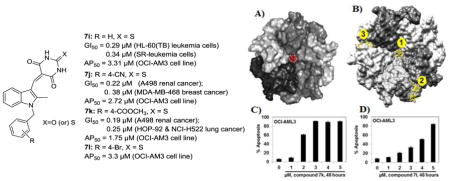
In the present study, we have designed and synthesized a series of 1-benzyl-2-methyl-3-indolyl methylene barbituric acid analogues (7a–7h) and 1-benzyl-2-methyl-3-indolylmethylene thiobarbituric acid analogues (7i–7l) as nucleophosmin 1 (NPM1) inhibitors and have evaluated them for their anti-cancer activity against a panel of 60 different human cancer cell lines. Among these analogues 7i, 7j, and 7k demonstrated potent growth inhibitory effects in various cancer cell types with GI50 values < 2 μM. Compound 7k exhibited growth inhibitory effects on a sub-panel of six leukemia cell lines with GI50 values in the range 0.22–0.35 μM. Analogue 7i also exhibited GI50 values < 0.35 μM against three of the leukemia cell lines in the sub-panel. Analogues 7i, 7j, 7k and 7l were also evaluated against the mutant NPM1 expressing OCI-AML3 cell line and compounds 7k and 7l were found to cause dose-dependent apoptosis (AP50 =1.75 μM and 3.3 μM respectively). Compound 7k also exhibited potent growth inhibition against a wide variety of solid tumor cell lines: i.e. A498 renal cancer (GI 50 = 0.19 μM), HOP-92 and NCI-H522 lung cancer (GI50 = 0.25 μM), COLO 205 and HCT-116 colon cancer (GI50 = 0.20 and 0.26 μM, respectively), CNS cancer SF-539 (GI50 = 0.22 μM), melanoma MDA-MB-435 (GI50 = 0.22 μM), and breast cancer HS 578T (GI50 = 0.22 μM) cell lines. Molecular docking studies suggest that compounds 7k and 7l exert their anti-leukemic activity by binding to a pocket in the central channel of the NPM1 pentameric structure. These results indicate that the small molecule inhibitors 7i, 7j, 7k, and 7l could be potentially developed into anti-NPM1 drugs for the treatment of a variety of hematologic malignancies and solid tumors. 2009 Elsevier Ltd. All rights reserved.
Keywords: 1-Benzyl-2-methyl-3-indolyl barbituric acid, Anticancer activity, Nucleophosmin 1, Anti-leukemic activity, Molecular docking studies, NPM1 oligomerization
Graphical Abstract
1. Introduction
Nucleophosmin 1 (NPM1) is a nucleolar phosphoprotein that shuttles between the nucleus and cytoplasm. This protein is encoded by the NPM1 gene located on chromosome 5q35 and has 12 exons.1 NPM1 plays an important role in multiple diverse biologic processes, including ribosome biogenesis, maintenance of genomic stability, centrosome duplication, regulation of cell cycle, DNA repair, histone chaperoning, inhibition of apoptosis, and regulation of the ARF-p53 tumor suppressor pathway.2–6 The NPM1 protein contains three distinct structural domains, the N-terminal oligomerization domain, the C-terminal nucleic acid binding domain, and the histone binding middle domain.7 Under native conditions, NPM1 exists as dimeric and oligomeric forms in cells.7,8 The NPM1 N-terminal oligomerization domain is believed to be critical for its molecular chaperone function and its interaction with other proteins.9
NPM1 gene mutations are most common in acute myeloid leukemia (AML), which is found in 46% to 64% of cytogenetically normal AML (CN-AML) patients and 9% to 18% of cytogenetically abnormal AML patients.10 Approximately 75–80% of AML cases will have NPM1 gene mutations in C-terminus exon 12.11 This mutation causes loss of nucleolar signal and creation of an additional nuclear export signal resulting in the aberrant cytoplasmic translocation of mutant NPM1 protein.1 NPM1 is overexpressed in several solid tumors but no mutations have been reported so far. The over-expression of NPM1 in solid tumors could promote tumor growth by inactivation of the tumor suppressor p53/ARF pathway through inhibition of apoptotic cell death, which directly implicates pathogenesis of cancer in humans.12
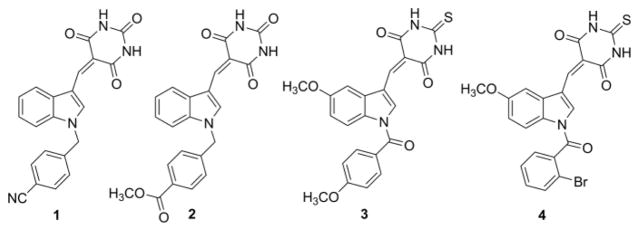
Recently, our research group has reported on the radio-sensitization of non-small cell lung cancer cells by targeting NPM1 utilizing a novel N-benzylindole barbituric acid scaffold (Fig. 1; 1–2).13 This led to the discovery of YTR-107 (1), a potent NPM1 inhibitor and radio-sensitizing agent.13,14 We have also reported on a number of structurally related N-benzoylindole thiobarbituric acids (Fig. 1; 3–4) that possess both anti-cancer and anti-inflammatory properties.3 Herein we now report on the synthesis and evaluation of a series of novel 1-benzyl-2-methyl-3-indolylmethylene barbituric acid analogues as NPM1 inhibitors that function as anticancer and anti-leukemic agents.
Fig. 1.
Chemical structures of potent radio-sensitization and anticancer agents 1–4.
Emerging data from our laboratory15 and others16 suggest that NPM1 may be a critical factor for homologous recombination (HR)-mediated repair of DNA double strand breaks (DSBs). Loss of HR imposes a radio-sensitization phenotype on cells subjected to fractionated irradiation. In response to formation of DNA DSBs, phosphorylated NPM1 (pNPM1) binds to ubiquitinated chromatin in a RNF8/RNF168-dependent manner, causing RAD51-mediated HR, and irradiation-induced foci (IRIF) that promote repair of DNA DSBs. Hence, we hypothesized that targeting pNPM1 results in radio-sensitization of cancer cells. In this respect, we have observed that the extractability of pNPM1 is reduced when cancer cells are irradiated, and 1-benzyl-2-methyl-3-indolylmethylene barbituric acid analogues such as YTR-107 reverse the effects of radiation on the extractability of pNPM1. Thus, compounds that enhance the extractability of pNPM1 upon radiation treatment have been shown to possess radio-sensitization properties.15 In contrast to cancer cells, the extractability of pNPM1 was not changed with drug plus radiation treatment in normal lung fibroblasts, suggesting that the event is specific for cancer cells.15
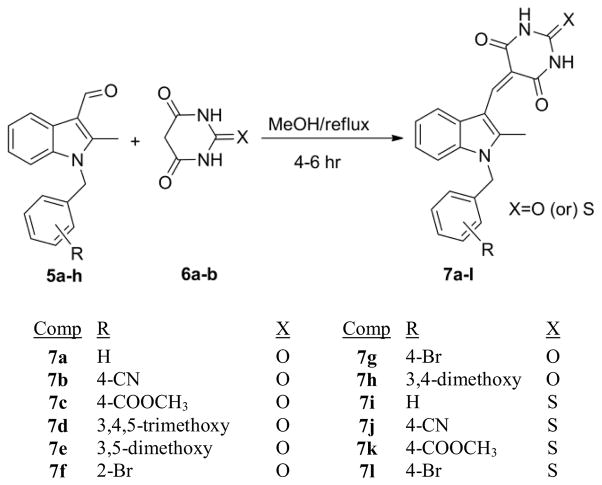
In the present work we report on the synthesis and anti-cancer activity of a series of novel 2-methyl-N-benzyl barbiturate analogues (Scheme 1); i.e. 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-oxo-dihydropyrimidine-4,6(1H,5H)-triones (7a–7h) and 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-thi-oxodihydropyrimidine-4,6(1H,5H)-diones (7i–7k). All the above compounds were initially evaluated at a single dose (10 μM) against a panel of 60 human cancer cell lines, Compounds that exhibited potent growth inhibition (7i, 7j and 7k) were further selected for five dose response in the same cell panel. The three lead compounds (7i, 7j and 7k) together with the new indole N-4-bromobenzyl derivative 7l, a sulphur isostere of compound 7g, have also been evaluated as anti-leukemic agents for their growth inhibition effects against AML cell lines, and have recently been shown to possess radio-sensitization properties.15
Scheme 1.
Synthesis of 1-benzyl-2-methyl-3-indolylmethylene barbiturates (7a–7l).
2. Results and discussion
2.1. Chemistry
For the synthesis of compounds 7a–7l, the critical 2-methyl-N-benzylindole-3-carboxaldehyde precursors (5a–5h) were synthesized using known literature procedures.17 Knoevenagel condensation of the appropriate 2-methyl-N-benzylindole-3-carboxaldehyde with compounds containing active methylene groups, i.e., barbituric acid (6a) or 2-thiobarbituric acid (6b), under reflux in methanol for 4–6 hr afforded the desired 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-oxo-dihydro- pyrimidine-4,6(1H,5H)-triones 7a–7h, or the 2-methyl-5-((1-benzyl-1H-indol-3-yl)methylene)-2-thioxo-dihydropyrimidine-4,6(1H, 5H)-diones (7i–7l) (Scheme 1) in yields ranging from 83–92% and with purities >99 % by elemental analysis. All the synthesized compounds were characterized by 1H, 13C NMR spectrometric analysis and CHN combustion analysis.
3. Biological Evaluation
3.1. In vitro growth inhibition in a panel of human cancer cell lines
Single dose evaluation of compounds 7a–7k was carried out at a concentration of 10 μM, against a panel of 60 human tumor cell lines, according to the procedure described by Rubinstein et al.18 The human tumor cell line panel included leukemia, non-small cell lung, colon, CNS, melanoma, ovarian, renal, prostate, and breast cancer cell lines. The single dose results are expressed as the percent growth inhibition of drug-treated cells following 48 hr of incubation with test compound. From these initial screening studies, compounds 7i, 7j and 7k were selected as leads for more comprehensive studies designed to determine GI50 values, which represent the molar drug concentration required for 50% cell growth inhibition. The compounds were dissolved in dimethyl sulfoxide (DMSO)/H2O and evaluated using five different concentrations at 10-fold dilutions (10−4 M, 10−5 M, 10−6 M, 10−7 M and 10−8 M) following 48 hr of incubation. Analogues containing a thiobarbituric acid group (7i–7k) exhibited higher inhibitory potency over barbituric acid containing molecules (7a–7h). Analogues 7i, 7j and 7k exhibited average GI50 values in the low micromolar level in subsequent five dose screening against all 60 human cancer cell lines in the panel (Table 1).
Table 1.
Anticancer activity (GI50/μM)a for 1-benzyl-2-methyl-3-indolylmethylene thiobarbiturate compounds (7i, 7j, and 7k)
| Panel/cell line | 7i | 7j | 7k |
|---|---|---|---|
|
| |||
| GI50 | GI50 | GI50 | |
| Leukemia | |||
| CCRF-CEM | 0.59 | 0.84 | 0.34 |
| HL-60(TB) | 0.29 | 1.08 | 0.22 |
| K-562 | 1.26 | 1.77 | 0.27 |
| MOLT-4 | 1.71 | 1.82 | 0.35 |
| RPMI-8226 | 0.35 | 0.57 | 0.31 |
| SR | 0.34 | 0.56 | 0.32 |
| Lung Cancer | |||
| A549/ATCC | 1.85 | 2.48 | 1.36 |
| HOP-62 | 1.09 | 1.96 | 0.39 |
| HOP-92 | 0.40 | 1.28 | 0.25 |
| NCI-H226 | 1.88 | 1.79 | 0.41 |
| NCI-H322M | 1.63 | 1.93 | 0.53 |
| NCI-H460 | 0.98 | 2.01 | 0.33 |
| NCI-H522 | 1.85 | 1.75 | 0.25 |
| Colon Cancer | |||
| COLO 205 | 1.33 | 1.73 | 0.20 |
| HCC-2998 | 1.18 | 1.79 | 0.40 |
| HCT-116 | 0.89 | 1.29 | 0.26 |
| HCT-15 | 1.30 | 1.74 | 0.33 |
| HT29 | 1.37 | 1.76 | 0.31 |
| KM12 | 0.95 | 1.74 | 0.31 |
| SW-620 | 1.24 | 1.36 | 0.40 |
| CNS Cancer | |||
| SF-268 | 1.26 | 1.60 | 0.42 |
| SF-295 | 0.93 | 1.88 | 0.35 |
| SF-539 | 1.14 | 1.29 | 0.22 |
| SNB-19 | 1.31 | 1.77 | 1.05 |
| SNB-75 | 1.51 | 2.04 | 0.46 |
| U251 | 1.16 | 1.61 | 0.32 |
| Melanoma | |||
| LOX IMVI | 0.53 | 0.90 | 0.32 |
| MALME-3M | 2.31 | 2.50 | 1.48 |
| M14 | 1.40 | 1.61 | 0.33 |
| MDA-MB-435 | 1.15 | 1.49 | 0.22 |
| SK-MEL-2 | 2.08 | 2.17 | 1.12 |
| SK-MEL-28 | 1.66 | 2.08 | 1.05 |
| SK-MEL-5 | 1.37 | 1.84 | 1.13 |
| UACC-257 | 1.41 | 1.67 | 0.80 |
| Ovarian Cancer | |||
| IGROV1 | 0.97 | 1.61 | 0.51 |
| OVCAR-3 | 1.18 | 1.58 | 0.30 |
| OVCAR-4 | 1.24 | 1.59 | 0.46 |
| OVCAR-5 | 1.75 | 1.84 | 0.42 |
| OVCAR-8 | 1.13 | 1.32 | 0.32 |
| NCI/ADR-RES | 1.89 | 3.83 | 0.35 |
| SK-OV-3 | 1.89 | 2.98 | 0.45 |
| Renal Cancer | |||
| 786-0 | 1.58 | 1.76 | 0.32 |
| A498 | 1.56 | 0.22 | 0.19 |
| ACHN | 1.37 | 1.70 | 0.51 |
| CAKI-1 | 1.57 | 2.21 | 0.50 |
| RXF 393 | 1.23 | 1.53 | 0.30 |
| SN12C | 1.15 | 1.40 | 0.41 |
| TK-10 | 1.47 | 1.37 | 0.31 |
| UO-31 | 0.88 | 1.46 | 0.41 |
| Prostate Cancer | |||
| DU-145 | 1.31 | 1.71 | 0.34 |
| Breast Cancer | |||
| MCF7 | 0.70 | 1.28 | 0.40 |
| MDA-MB-231/ATCC | 1.34 | 1.64 | 0.38 |
| HS 578T | 0.51 | 1.22 | 0.22 |
| BT-549 | 1.60 | 1.87 | 0.31 |
| T-47D | 1.90 | 2.54 | 1.86 |
| MDA-MB-468 | 0.51 | 0.38 | 0.34 |
GI50: 50% Growth inhibition, concentration of drug resulting in a 50% reduction in net cell growth as compared to cell numbers on day 0.
Compound 7i, exhibited GI50 values ranging from 0.29 μM to 2.31 μM in all the cancer cell lines screened (Table 1). The average GI50 value of this compound for all the cancer cell lines in the panel was 1.25 μM. Compound 7i showed good growth inhibition properties in all but two (K-562 and MOLT-4) of the cell lines in the leukemia panel, with GI50 values in the range 0.29–0.59 μM. Compound 7i also showed potent growth inhibition against HL-60(TB) leukemia cells (GI50 = 0.29 μM).
Compound 7j exhibited GI50 values ranging from 0.22 μM to 3.83 μM in all 60 cancer cell lines screened (Table 1). The average GI50 value of this compound against all the cancer cell lines in the panel was 1.65 μM. 7j showed good growth inhibition properties in CCRF-CEM (GI50 = 0.84 μM), RPMI-8226 (GI50 = 0.57 μM), and SR (GI50 = 0.56 μM) leukemia cell lines. Compound 7j also showed potent growth inhibition against A498 renal cancer (GI50 = 0.22 μM) and MDA-MB-468 breast cancer (GI50 = 0.38 μM) cell lines. Compound 7j caused growth inhibition of 82% of the all cancer cell lines screened at concentration < 2 μM.
Compound 7k exhibited GI50 values ranging from 0.19 μM to 1.86 μM in all the cancer cell lines screened. The average GI50 value for this compound against all the human cancer cell lines in the panel was 0.47 μM. 7k showed potent growth inhibition properties against all the leukemia cell lines with GI50 values ranging from 0.22 μM to 0.35 μM. Compound 7k also showed potent growth inhibition against A498 renal cancer (GI50 = 0.19 μM) and COLO 205 colon cancer (GI50 = 0.20 μM) cell lines (Table 1).
3.2. Anti-leukemic activity against NPM1-expressing OCI-AML3 cells
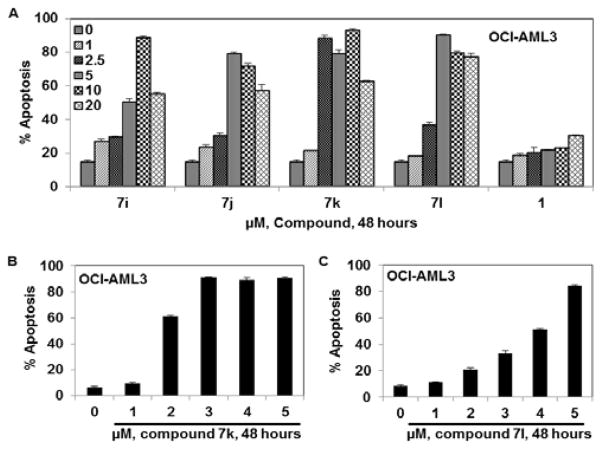
Since the NPM1 gene is mutated in a significant number of hematological malignancies, we tested 2-thioxodihydropyrimidine-4,6(1H,5H)-dione analogues 7i, 7j, and 7k, and the new analogue 7l, against an appropriate mutant NPM1-expressing OCI-AML3 cell line (Fig. 2). All four analogues showed significant apoptosis with AP50 values ranging from 1.75 μM to 3.3 μM (Table 2). Interestingly, no significant activity was observed in these studies for the structurally related radio-sensitizing molecule, compound 1 (YTR-107) (AP50 > 20 μM) against OCI-AML3 cells, indicating the importance of the 2-thioxodihydropyrimidine-4,6(1H,5H)dione moiety in the cytotoxic effects exhibited by compounds 7i, 7j, 7k and 7l. In this respect, it is important to point out that the corresponding oxygen isosteres of the latter four compounds, i.e. respectively, analogues 7a, 7b, 7c and 7g, were not found to be cytotoxic to cancer cells. Thus, a point atom change in these molecules from O to S resulted in the discovery of four 2-thioxodihydro pyrimidine-4,6(1H,5H)-dione analogues that are both potent radiosensitizers15 and anti-cancer agents.
Fig. 2.
1-Benzyl-2-methyl-3-indolylmethylene thiobarbiturate compounds induce apoptosis in mutant NPM1 expressing leukemia cells: A. OCI-AML3 cells were treated with the indicated concentrations of 1-benzyl-2-methyl-3-indolylmethylene thiobarbiturate compounds (7i, 7j, 7k, and 7l) and YTR-107 (1) for 48 hr. B. OCI-AML3 cells were treated with the indicated concentrations of compound 7k for 48 hr. C. OCI-AML3 cells were treated with the indicated concentrations of compound 7l for 48 hr. After the indicated incubation time period, cells were stained with annexin V and TO-PRO 3 and the percentages of apoptotic cells were measured by flow cytometry. Columns represent the mean of 3 independent experiments; bars represent the SEM.
Table 2.
Anti-AML activity (AP50/μM)* of compounds 7i–7l and YTR-107 (1) on the mutant NPM1-expressing OCI-AML3 cell line.
| Comp | 7i | 7j | 7k | 7l | 1 |
|---|---|---|---|---|---|
| Cell line | AP50 | AP50 | AP50 | AP50 | AP50 |
| OCI-AML3 | 3.31 | 2.72 | 1.75 | 3.3 | >20 |
AP50 represents the concentration of a drug that is required for 50% of the cells to undergo apoptosis in vitro.
3.3. Molecular docking studies
Virtual docking of small molecules to determine their binding modes to biological target molecules is a technique that is routinely employed in conjunction with validation of the potency of those molecules through bioassays. In regard to the present study, the crystal structure of the potentially drugable target, human NPM1, is available.19
Molecular docking studies were performed with the most potent compounds (7i–7l) against the target, NPM1, as well as with compound 1, a potent radio-sensitizer, but an ineffective anti-cancer agent (Fig. 2A). All docking studies were performed using SwissDock,20 which employs the EADock DSS algorithm to generate binding modes, estimate CHARMM energies,21 account for solvent effects using the FACTS implicit solvation model22 and rank binding modes with the most favorable energies. Co-ordinates for all the compounds were generated using MarvinSketch (ChemAxon), while coordinates for the crystal structure of the N-terminal domain of human NPM1 (PDB 2P1B) were obtained from the PDB database. All structure coordinates were prepared for docking and visualization using UCSF-Chimera molecular viewer. Top-scoring docked poses were considered based on the Full-Fitness scores and free energy (ΔG) values calculated by the algorithm.
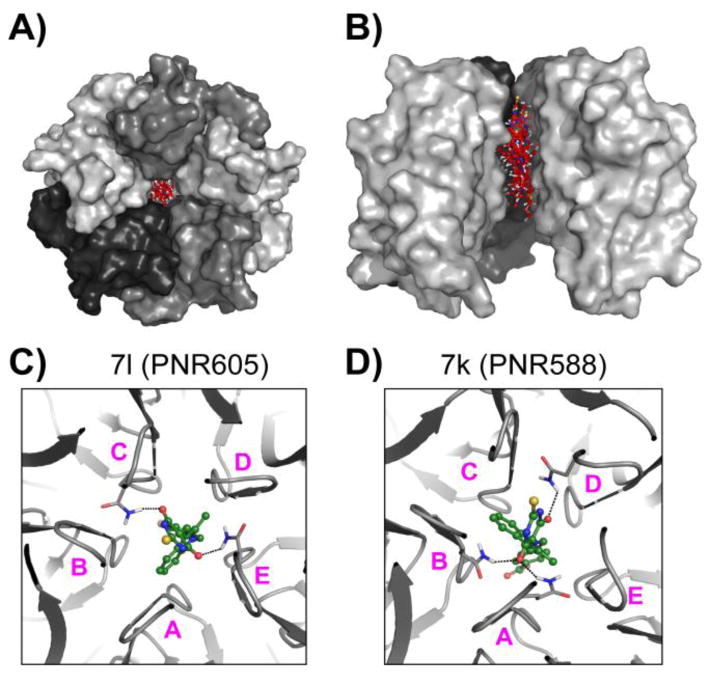
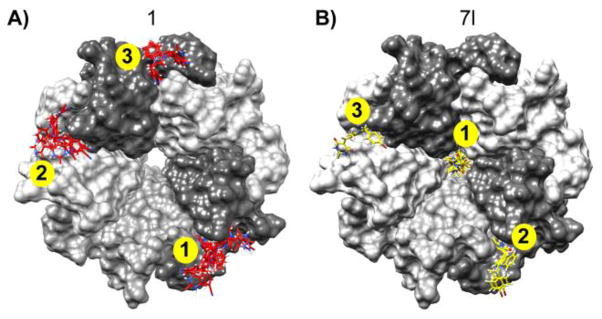
Results from our docking studies revealed that compounds 7i–7l, all of which share the presence of a 2-methyl substituted indole ring and a 2-thioxodihydro pyrimidine-4,6(1H,5H)-dione moiety, bound to the central channel formed by the NPM1 pentamer (Fig. 3). For these compounds, between four (7i) to six (7k) of the top ten highest scoring docked poses bound to the pentameric NPM1 channel (Fig. 3). As shown in Fig. 3 (panels C and D), the compounds are stabilized through H-bonds with residues from different subunits of the NPM1 pentamer in the central channel. In contrast, similar docking studies performed with compound 1 did not identify any pose (in the top ten highest scoring) localized to this central channel of NPM1. In the case of compound 1, all the top scoring docked poses were located at three inter-subunit interfaces of the outer surface of the NPM1 pentamer (Fig. 4). As shown in Fig. 4, compound 7l shares two of the top three favored binding pockets with those observed for 1 (pockets 2 and 3 for 7l correspond to pockets 1 and 2 for compound 1). We hypothesize that these common binding sites for 1 and 7l may be relevant to the radio-sensitizing effects of these two compounds. Thus, compound 7l may be able to affect NPM1 pentamer formation and function more effectively than compound 1, due to the presence of the additional favored binding pocket in the central channel of the pentamer structure. This might also explain the structural basis for the differential potency of 7l and 1 in the screening assay against OCI-AML3 leukemia cell lines (Fig. 2), and suggests that the central channel of the pentamer structure may be a new target for the development of novel anti-cancer agents.
Fig. 3.
Top-scoring docked poses of compounds 7i–7l bind to the central channel of the NPM1 pentamer. As shown in panels A and B, 5 of the top 10 hits (based on docking score) of compound 7k, bound to the central channel formed by the NPM1 pentamer (grey-shaded molecular surface). The 5 docking poses of 7k are shown as red ball-and-sticks with blue, red, white and yellow colors for N-, O-, H- and S- atoms respectively. Panel A shows the top-view of the pentamer, while panel B is a side-view, with atoms of one subunit removed for clarity. Panels C and D respectively show compounds 7l and 7k, (green ball-and-sticks), forming H-bonds (dashed lines) with the residue Q84 (grey sticks) with different subunits of the NPM1 pentamer. The five subunits are labeled A–E.
Fig. 4.
Comparison of docking results of compounds 1 and 7l bound to the NPM1 pentamer. The NPM1 pentameric subunits are depicted as grey-shaded molecular surfaces in both panels A and B. The top hits for 1 (red) and 7l (yellow) are shown as sticks, docked to their binding pockets on NPM1. The top three ranked sites are indicated in yellow circles for each molecule.
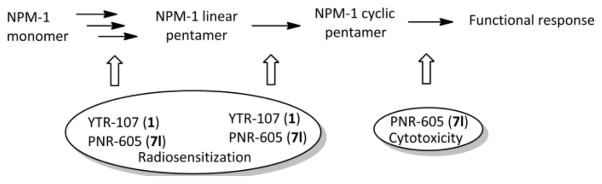
Interestingly, when docking was performed with a monomer of NPM1, almost all of the common predicted poses for 1 and 7l dock to NPM1 at a binding site that is located in the center of the interface-forming surface with the neighboring subunits of the NPM1 pentamer (data not shown), suggesting that both these molecules may be able to disrupt formation of functional pentamers of NPM1. It can therefore be proposed that binding of 1 and 7l to the same sites on NPM1 monomers may be relevant to the radio-sensitization effects of these two molecules by preventing formation of the NPM1 pentamer, while the additional binding of 7l and its analogues to the central channel of the pentameric core may be more relevant to the anti-leukemic activity of these compounds (Fig. 5), since these compounds might be capable of blocking both the oligomerization of NPM-1 and the functional response of mature (cyclic pentamer) NPM1.
Fig. 5.
Proposed mechanism and functional outcome of inhibition of NPM1 by the 1-benzyl-2-methyl-3-indolylmethylene barbituric acid analogue YTR-107 (1) and 1-benzyl-2-methyl-3-indolylmethylene thio barbituric acid analogue PNR-605 (7l).
4. Pharmacological evaluation
4.1. NCI-60 cell line anti-cancer screening assay
The methodology for the anti-cancer screening assay was carried out as per the reported literature procedure,23,24 which is also available at http://dtp.nci.nih.gov/branches/btb/ivclsp.html http://dtp.nci.nih.gov/branches/btb/ivclsp.html.
4.2. Anti-leukemic activity assay
OCI-AML3 cells were obtained and cultured as previously described.25,26 The OCI-AML2 cells (known to express WT p53 and WT NPM1) were kindly provided by Mark Minden (Ontario Cancer Institute/Princess Margaret Hospital, Ontario, Canada) and cultured in α-MEM with 20% heat inactivated FBS. Cells were passaged 2–3 times per week. Logarithmically growing cells were used for all experiments. OCI-AML3 cells have been previously demonstrated to express NPM1c+.27 Cells were treated with the indicated concentrations of 2-methyl-N-benzylindolyl barbiturate compounds (7i, 7j, 7k, and 7l) and YTR-107 (1) for 48 hr. After the indicated incubation time period, cells were stained with annexin V and TO-PRO 3 (BD Biosciences Pharmingen) as previously described,25, 26, 28 and the percentages of apoptotic cells measured by flow cytometry.
5. Conclusions
Novel 1-benzyl-2-methyl-3-indolylmethylene barbituric acid analogues (7a–7h) and 1-benzyl-2-methyl-3-indolyl- methylene thiobarbituric acid analogues (7i–7l) have been synthesized and evaluated against a panel of 60 human cancer cell lines. The 1-benzyl-2-methyl-3-indolylmethylene thiobarbituric acid analogues 7i, 7j, 7k showed potent growth inhibition properties with GI 50 values generally in the range of < 2 μM against most of the human cancer cell lines examined in the 60 cell panel. The most active compounds exhibited potent growth inhibition against human leukemia cell lines, and also showed significant apoptotic activity against the NPM1 mutant expressing leukemia cell line, OCI-AML3.
Compound 7k exhibited growth inhibitory effects on a sub-panel of six leukemia cell lines with GI50 values in the range 0.22–0.35 μM. Analogue 7i also exhibited GI50 values < 0.35 μM against three of the leukemia cell lines in the sub-panel. Analogues 7i, 7j, 7k and 7l were also evaluated against the mutant NPM1 expressing OCI-AML3 cell line and compounds 7k and 7l were found to cause dose-dependent apoptosis (AP50 = 1.75 μM and 3.3 μM respectively). Molecular docking studies suggest that compounds 7k and 7l exert their anti-leukemic activity by binding to a pocket in the central channel of the NPM1 pentameric structure. Interestingly, in the panel of human cancer cells 7k also exhibited potent growth inhibition against a wide variety of solid tumor cell lines: i.e. A498 renal cancer (GI50 = 0.19 μM), HOP-92 and NCI-H522 lung cancer (GI50 = 0.25 μM), COLO 205 and HCT-116 colon cancer (GI50 = 0.20 and 0.26 μM, respectively), CNS cancer SF-539 (GI50 = 0.22 μM), melanoma MDA-MB-435 (GI50 = 0.22 μM), and breast cancer HS 578T (GI50 = 0.22 μM) cell lines.
Molecular docking studies suggest that compound 7l exerts its anti-leukemic activity by binding to the pocket in the central channel of the NPM1 cyclic pentameric structure and that this binding site may represent a new drugable target for anti-cancer drug discovery. The docking scores (ΔG values of −8.0, −8.1, −8.3 and −8.4 kcal/mol for compounds 7i, 7j, 7k and 7l respectively) for these compounds are in excellent agreement with their trend in potencies as anti-leukemic agents. Thus, compounds 7i, 7j, 7k and 7l are considered lead compounds for further development as anti-NPM1 drugs for the treatment of various hematologic and solid tumor malignancies.
6. Experimental Part
6.1. General Chemistry
Melting points were recorded on a Kofler hot-stage apparatus and are uncorrected. TLC controls were carried out on pre-coated silica gel plates (F 254 Merck). 1H and 13C NMR spectra were recorded on a Varian 400 MHz spectrometer equipped with a Linux workstation running on vNMRj software. All the spectra were phased, baseline was corrected where necessary, and solvent signals (DMSO-d6) were used as reference for both 1H and 13C spectra. Elemental analyses was carried out by combustion analysis at Atlantic Microlabs, GA, USA.
6.2. Synthesis of 1-benzyl-2-methyl-3-indolylmethylene barbituric acids or thiobarbituric acids (7a–7h or 7i–7l)
An appropriate 2-methyl-N-benzyl-indol-3-aldehyde (1 mmol) and barbituric acid or 2-thiobarbituric acid (1.1 mmol) were refluxed in methanol for 4–6 h. On completion of the reaction (monitored by TLC), the reaction mass was cooled to room temperature, filtered, and washed with methanol, to afford the pure solid product. All the melting points of isolated, dry compounds were recorded as > 300 °C.
6.2.1. 5-((1-Benzyl-2-methyl-1H-indol-3-yl)methylene)pyrim idine-2,4,6(1H,3H,5H)-trione (7a)
Yield: 89%, 1H NMR (400 MHz, DMSO-d6): δ 2.48 (s, 3H, CH3), 5.59 (s, 2H, CH2), 7.09–7.31 (m, 8H, Ar-H), 7.53 (d, 1H, J = 7.6 Hz, C4-H), 8.53 (s, 1H, CH), 10.91 (s, 1H, NH), 11.07 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 12.6, 46.7, 110.5, 110.8, 111.9, 121.7, 122.5, 122.7, 126.2, 126.4, 127.5, 128.8, 136.5, 137.3, 145.9, 150.2, 150.5, 161.5, 164.2 ppm; Anal. Calcd for C21H17N3O3: C, 70.18; H, 4.77; N, 11.77. Found: C, 69.89; H, 4.84; N, 11.67. MW: 359.1270.
6.2.2. 4-((2-Methyl-3-((2,4,6-trioxotetrahydropyrimidi- ne-5(2H)-ylidene)methyl)-1H-indol-1-yl)methyl)benzoni- trile (7b)
Yield: 85%, 1H NMR (400 MHz, DMSO-d6): δ 2.49 (s, 3H, CH3), 5.73 (s, 2H, CH2), 7.21–7.26 (m, 4H, Ar-H), 7.33 (d, 1H, J = 6.8 Hz, C7H), 7.51 (d, J = 7.2 Hz, 1H, C4H), 7.81 (d, J = 8.4 Hz, 2H, Ar-H), 8.55 (s, 1H, CH), 10.94 (s, 1H, NH), 11.11 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.0, 46.7, 110.7, 111.0, 111.6, 112.3, 119.0, 122.2, 122.9, 123.3, 126.6, 127.7, 133.2, 137.5, 142.8, 146.3, 150.2, 150.9, 161.9, 164.6 ppm; Anal. Calcd for C22H16N4O3: C, 68.74; H, 4.20; N, 14.58. Found: C, 68.48; H, 4.36; N, 14.61. MW: 384.1222.
6.2.3. 4-((2-Methyl-3-((2,4,6-trioxotetrahydropyrimidine-5(2H)-ylidene)methyl)-1H-indol-1-yl)methyl)benzoate (7c)
Yield: 83%, 1H NMR (400 MHz, DMSO-d6): δ 2.49 (s, 3H, CH3), 3.82 (s, 3H, OCH3), 5.71 (s, 2H, CH2), 7.20–7.24 (m, 4H, Ar-H), 7.33 (d, 1H, J = 7.6 Hz, C7H), 7.53 (d, J = 8.4 Hz, 1H, C4H), 7.92 (d, J = 8.0 Hz, 2H, Ar-H), 8.56 (s, 1H, CH), 10.94 (s, 1H, NH), 11.11 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 12.5, 46.4, 52.1, 110.6, 110.9, 111.8, 121.7, 122.5, 122.8, 126.1, 126.6, 128.8, 129.7, 137.2, 142.0, 145.9, 150.0, 150.5, 161.4, 164.2, 165.8 ppm; Anal. Calcd for C23H19N3O5: C, 66.18; H, 4.59; N, 10.07. Found: C, 66.04, H, 4.68; N, 10.19. MW: 417.1325.
6.2.4. 5-((2-Methyl-1-(3,4,5-trimethoxybenzyl)-1H-indol-3-yl)methylene)pyrimidine-2,4,6(1H,3H,5H)-trione (7d)
Yield: 87%, 1H NMR (400 MHz, DMSO-d6): δ 2.55 (s, 3H, CH3), 3.60 (s, 3H, OCH3), 3.66 (s, 6H, 2xOCH3), 5.51 (s, 2H, CH2), 6.46 (s, 2H, Ar-H), 7.19–7.24 (m, 2H, C5H, C6H), 7.34 (d, J = 8.0 Hz, 1H, C7H), 7.60 (d, J = 7.6 Hz, 1H, C4H), 8.57 (s, 1H, CH), 10.92 (s, 1H, NH), 11.10 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 12.9, 46.9, 55.9, 60.0, 104.0, 109.6, 110.8, 110.9, 111.9, 121.8, 122.21, 122.8, 126.3, 132.2, 136.9, 137.3, 146.1, 150.1, 150.6, 153.2, 161.5, 164.3 ppm; Anal. Calcd for C24H23N3O6: C, 64.13; H, 5.16; N, 9.35. Found: C, 64.04; H, 5.01; N, 9.39. MW: 449.1587.
6.2.5. 5-((1-(3,5-Dimethoxybenzyl)-2-methyl-1H-indol-3-yl)- methylene)pyrimidine-2,4,6(1H,3H,5H)-trione (7e)
Yield: 90%, 1H NMR (400 MHz, DMSO-d6): δ 2.50 (s, 3H, CH3), 3.67 (s, 6H, 2xOCH3), 5.51 (s, 2H, CH2), 6.23 (s, 2H, Ar-H), 6.41 (s, 1H, Ar-H), 7.19–7.23 (m, 2H, C5H, C6H), 7.33 (d, J = 6.8 Hz, 1H, C7H), 7.53 (d, J = 8.0 Hz, 1H, C4H), 8.56 (s, 1H, CH), 10.92 (s, 1H, NH), 11.09 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.6, 47.5, 56.0, 99.4, 105.5, 111.7, 112.7, 122.6, 123.2, 127.1, 138.2, 139.8, 146.9, 151.0, 151.5, 161.7, 162.4, 165.2 ppm; Anal. Calcd for C23H21N3O5: C, 65.86; H, 5.05; N, 10.02. Found: C, 65.89; H, 4.88; N, 10.16. MW: 419.1481.
6.2.6. 5-((1-(2-Bromobenzyl)-2-methyl-1H-indol-3-yl)methy- lene)pyrimidine-2,4,6(1H,3H,5H)-trione (7f)
Yield: 91%, 1H NMR (400 MHz, DMSO-d6): δ 2.49 (s, 3H, CH3), 5.61 (s, 2H, CH2), 6.38 (t, J = 8.4 Hz, 1H, Ar-H), 7.23–7.25 (m, 4H, Ar-H), 7.33–7.43 (m, 2H, Ar-H), 7.74 (t, 1H, J = 8.8 Hz, Ar-H), 8.59 (s, 1H, CH), 12.10 (s, 1H, NH), 12.21 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 12.4, 47.3, 110.5, 110.6, 113.0, 121.4, 122.1, 123.1, 123.2, 125.9, 126.6, 128.2, 129.5, 132.8, 134.7, 137.3, 146.6, 152.1, 159.2, 162.3, 177.7 ppm; Anal. Calcd for C21H16BrN3O3: C, 57.55; H, 3.68; N, 9.59; Br, 18.23. Found: C, 57.39; H, 3.81; N, 9.65; Br, 18.13. MW: 437.0375.
6.2.7. 5-((1-(4-Bromobenzyl)-2-methyl-1H-indol-3-yl)methy- lene)pyrimidine-2,4,6(1H,3H,5H)-trione (7g)
Yield: 92%, 1H NMR (400 MHz, DMSO-d6): δ 2.50 (s, 3H, CH3), 5.59 (s, 2H, CH2), 7.05 (d, J = 8.4 Hz, 2H, Ar-H), 7.17–7.24 (m, 2H, Ar-H), 7.32 (d, J = 8.4 Hz, 1H, Ar-H), 7.53 (m, 3H, Ar-H), 8.55 (s, 1H, CH), 10.93 (s, 1H, NH), 11.10 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 12.4, 45.9, 110.6, 110.7, 111.8, 120.5, 121.6, 122.4, 122.7, 126.0, 128.5, 131.6, 135.9, 137.0, 145.8, 149.9, 150.4, 161.3, 164.1 ppm; Anal. Calcd for C21H16BrN3O3: C, 57.55; H, 3.68; N, 9.59; Br, 18.23. Found: C, 57.64; H, 3.76; N, 9.63; Br, 17.96. MW: 437.0375.
6.2.8. 5-((1-(3,4-Dimethoxybenzyl)-2-methyl-1H-indol-3-yl) methylene)pyrimidine-2,4,6(1H,3H,5H)-trione (7h)
Yield: 88%, 1H NMR (400 MHz, DMSO-d6): δ 2.54 (s, 3H, CH3), 3.68 (s, 3H, OCH3), 3.70 (s, 3H, OCH3), 5.50 (s, 2H, CH2), 6.51 (d, J = 7.2 Hz, 1H), 6.85 (d, J = 8.0 Hz, 1H), 6.92 (s, 1H, C4H), 7.18–7.22 (m, 2H, Ar-H), 7.31 (d, J = 7.2 Hz, 1H, Ar-H), 7.58 (d, J = 7.6 Hz, 1H, Ar-H), 8.55 (s, 1H, CH), 10.91 (s, 1H, NH), 11.08 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.1, 46.9, 55.8, 55.9, 110.8, 111.1, 111.3, 112.3, 112.4, 118.8, 122.1, 122.8, 123.2, 126.6, 129.1, 137.7, 146.4, 148.6, 149.2, 150.7, 151.0, 161.9, 164.7 ppm; Anal. Calcd for C23H21N3O5: C, 65.86; H, 5.05; N, 10.02. Found: C, 65.96; H, 5.04; N, 10.18. MW: 419.1481.
6.2.9. 5-((1-Benzyl-2-methyl-1H-indol-3-yl)methylene)-2-thi- oxodihydropyrimidine-4,6(1H,5H)-dione (7i)
Yield: 88%, 1H NMR (400 MHz, DMSO-d6): δ 2.57 (s, 3H, CH3), 5.63 (s, 2H, CH2), 7.13–7.33 (m, 8H, Ar-H), 7.57 (d, 1H, J = 7.6 Hz, C4-H), 8.57 (s, 1H, CH), 12.06 (s, 1H, NH), 12.17 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.1, 47.3, 110.4, 111.5, 113.6, 122.6, 123.6, 123.7, 126.5, 126.9, 128.0, 129.3, 136.6, 138.0, 147.1, 153.0, 159.8, 163.0, 178.2 ppm; Anal. Calcd for C21H17N3O2S: C, 67.18; H, 4.56; N, 11.19; S, 8.54. Found: C, 67.06; H, 4.72; N, 11.06; S, 8.67. MW: 375.1041.
6.2.10. 4-((3-((4,6-Dioxo-2-thioxotetrahydropyrimidine-5(2H) -ylidene)methyl)-2-methyl-1H-indol-1-yl)methyl)benzonitrile (7j)
Yield: 86%, 1H NMR (400 MHz, DMSO-d6): δ 2.53 (s, 3H, CH3), 5.74 (s, 2H, CH2), 7.22–7.32 (m, 6H, Ar-H), 7.53 (d, J = 7.6 Hz, 1H, C7H), 7.79 (d, 1H, J = 8.4 Hz, C4H), 8.57 (s, 1H, CH), 12.07 (s, 1H, NH), 12.19 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.0, 47.0, 110.8, 111.0, 111.3, 113.5, 118.9, 122.7, 123.7, 126.5, 127.7, 133.2, 137.8, 142.4, 147.2, 152.6, 159.8, 162.9, 178.3 ppm; Anal. Calcd for C22H16N4O2S: C, 65.98; H, 4.03; N, 13.99, S, 8.01. Found: C, 65.94; H, 4.10; N, 13.97; S, 8.13. MW: 400.0994.
6.2.11. 4-((3-((4,6-Dioxo-2-thioxotetrahydropyrimidine-5(2H) -ylidene)methyl)-2-methyl-1H-indol-1-yl)methyl)benzoate (7k)
Yield: 84%, 1H NMR (400 MHz, DMSO-d6): δ 2.55 (s, 3H, CH3), 3.82 (s, 3H, OCH3), 5.73 (s, 2H, CH2), 7.24–7.32 (m, 5H, Ar-H), 7.54 (d, 1H, J = 7.2 Hz, C4H), 7.92 (d, J = 7.2 Hz, 2H, Ar-H), 8.58 (s, 1H, CH), 12.08 (s, 1H, NH), 12.19 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.5, 47.6, 53.1, 111.2, 111.8, 114.1, 123.2, 124.2, 127.6, 129.8, 130.6, 138.4, 142.6, 147.6, 153.3, 160.3, 163.4, 166.7, 178.8 ppm; Anal. Calcd for C23H19N3O4S: C, 63.73; H, 4.42; N, 9.69; S, 740. Found: C, 63.79; H, 4.44; N, 9.84; S, 7.50. MW: 433.1096.
6.2.12. 5-((1-(4-Bromobenzyl)-2-methyl-1H-indol-3-yl)methy- lene)-2-thioxodihydropyrimidine-4,6(1H,5H)-dione (7l)
Yield: 90%, 1H NMR (400 MHz, DMSO-d6): δ 2.55 (s, 3H, CH3), 5.61 (s, 2H, CH2), 7.08 (d, J = 8.4 Hz, 2H, Ar-H), 7.22–7.31 (m, 3H, Ar-H), 7.52–7.56 (m, 3H, Ar-H), 8.56 (s, 1H, CH), 12.05 (s, 1H, NH), 12.17 (s, 1H, NH) ppm; 13C NMR (100 MHz, DMSO-d6): δ 13.0, 46.7, 110.6, 111.4, 113.6, 121.1, 122.6, 123.7, 123.7, 126.5, 129.1, 132.2, 136.1, 137.9, 147.1, 152.8, 159.8, 162.9, 178.2 ppm; Anal. Calcd for C21H16BrN3O2S: C, 55.51; H, 3.55; N, 9.25; Br, 17.59. Found: C, 55.67; H, 3.65; N, 9.28; Br, 17.27. MW: 453.0147.
Supplementary Material
Acknowledgments
We are grateful to the NCI/NIH (Grant Number CA 140409 to M.L.F. and P.A.C. and CA 183895 to R.L.E.) and to the Arkansas Research Alliance (ARA) for financial support, and to the NCI Developmental Therapeutic Program (DTP) for screening data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Falini B, Nicoletti I, Martelli MF, Mecucci C. Blood. 2007;109:874–885. doi: 10.1182/blood-2006-07-012252. [DOI] [PubMed] [Google Scholar]
- 2.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. FEBS lett. 2001;506:272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 3.Bertwistle D, Sugimoto M, Sherr CJ. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Y, Maggi LB, Jr, Brady SN, Apicelli AJ, Dai MS, Lu H, Weber JD. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, Haferlach T. Blood. 2011;117:1109–1120. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]
- 6.Poletto M, Lirussi L, Wilson DM, 3rd, Tell G. Mol Cell Biol. 2014;25:1641–1652. doi: 10.1091/mbc.E13-12-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, Mandelli F, Mecucci C, Martelli MF. Haematologica. 2007;92:519–532. doi: 10.3324/haematol.11007. [DOI] [PubMed] [Google Scholar]
- 8.Herrera JE, Correia JJ, Jones AE, Olson MO. Biochemistry. 1996;35:2668–2673. doi: 10.1021/bi9523320. [DOI] [PubMed] [Google Scholar]
- 9.Mitrea DM, Grace CR, Buljan M, Yun MK, Pytel NJ, Satumba J, Nourse A, Park CG, Madan Babu M, White SW, Kriwacki RW. Pro Nat Acad Sci USA. 2014;111:4466–4471. doi: 10.1073/pnas.1321007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon Y, Seo SW, Park S, Park S, Kim SY, Ra EK, Park SS, Seong MW. Ann Lab Med. 2013;33:60–64. doi: 10.3343/alm.2013.33.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Mol Cell Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekhar KR, Benamar M, Venkateswaran A, Sasi S, Penthala NR, Crooks PA, Hann SR, Geng L, Balusu R, Abbas T, Freeman ML. Int J Rad Onc Biol Phys. 2014;89:1106–1114. doi: 10.1016/j.ijrobp.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekhar KR, Reddy YT, Reddy PN, Crooks PA, Venkateswaran A, McDonald WH, Geng L, Sasi S, Van Der Waal RP, Roti JL, Salleng KJ, Rachakonda G, Freeman ML. Clin Cancer Res. 2011;17:6490–6499. doi: 10.1158/1078-0432.CCR-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penthala NR, Crooks PA, Freeman ML, Sekhar KR. Bioorg Med Chem. 2015;23:3681–3686. doi: 10.1016/j.bmc.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koike A, Nishikawa H, Wu W, Okada Y, Venkitaraman AR, Ohta T. Cancer Res. 2010;70:6746–6756. doi: 10.1158/0008-5472.CAN-10-0382. [DOI] [PubMed] [Google Scholar]
- 17.Reddy YT, Sekhar KR, Sasi N, Reddy PN, Freeman ML, Crooks PA. Bioorg Med Chem Lett. 2010;20:600–602. doi: 10.1016/j.bmcl.2009.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinstein LV, Shoemaker RH, Paull KD, Simon RM, Tosini S, Skehan P, Scudiero DA, Monks A, Boyd MR. J Nat Cancer Inst. 1990;82:1113–1118. doi: 10.1093/jnci/82.13.1113. [DOI] [PubMed] [Google Scholar]
- 19.Lee HH, Kim HS, Kang JY, Lee BI, Ha JY, Yoon HJ, Lim SO, Jung G, Suh SW. Proteins. 2007;69:672–678. doi: 10.1002/prot.21504. [DOI] [PubMed] [Google Scholar]
- 20.Grosdidier A, Zoete V, Michielin O. Nucleic Acids Res. 2011;39:W270–277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haberthür U, Caflisch A. J Comput Chem. 2008;29:701–715. doi: 10.1002/jcc.20832. [DOI] [PubMed] [Google Scholar]
- 23.Madadi NR, Penthala NR, Janganati V, Crooks PA. Bioorg Med Chem Lett. 2014;24:601–603. doi: 10.1016/j.bmcl.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madadi NR, Crooks PA. Lett Drug Des Discov. 2015;21:521–528. [Google Scholar]
- 25.Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, Balusu R, Koul S, Atadja P, Marquez VE, Bhalla KN. Blood. 2009;114:2733–2743. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiskus W, Buckley K, Rao R, Mandawat A, Yang Y, Joshi R, Wang Y, Balusu R, Chen J, Koul S, Joshi A, Upadhyay S, Atadja P, Bhalla KN. Cancer Biol Ther. 2009;8:939–950. doi: 10.4161/cbt.8.10.8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quentmeier H, Martelli MP, Dirks WG, Bolli N, Liso A, Macleod RA, Nicoletti I, Mannucci R, Pucciarini A, Bigerna B, Martelli MF, Mecucci C, Drexler HG, Falini B. Leukemia. 2005;19:1760–1767. doi: 10.1038/sj.leu.2403899. [DOI] [PubMed] [Google Scholar]
- 28.Rao R, Nalluri S, Fiskus W, Balusu R, Joshi A, Mudunuru U, Buckley KM, Robbins K, Ustun C, Reuther GW, Bhalla KN. Mol Cancer Ther. 2010;9:2232–2242. doi: 10.1158/1535-7163.MCT-10-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.