Abstract
To overcome hurdles in identifying key kinases in small cell lung cancer (SCLC), we integrated a target-agnostic phenotypic screen of kinase inhibitors with target identification using activity-based protein profiling (ABPP) in which a desthiobiotin-ATP probe was used. We screened 21 SCLC cell lines with known c-MYC amplification status for alterations in viability using a chemical library of 235 small molecule kinase inhibitors. One screen hit compound was interrogated with ABPP, and through this approach we re-identified aurora kinase B as a critical kinase in MYC-amplified SCLC cells. We next extended the platform to a second compound that had activity in SCLC cell lines lacking c-MYC amplification and identified TANK-binding kinase 1 (TBK1), a kinase that affects cell viability, polo-like kinase-1 signaling, G2/M arrest, and apoptosis in SCLC cells lacking MYC amplification. These results demonstrate that phenotypic screening combined with activity-based protein profiling can identify key disease drivers, suggesting that this approach, which combines new chemical probes and disease cell screens, has the potential to identify other important targets in other cancer types.
INTRODUCTION
Identification of novel therapeutic targets in diseases such as cancer remains an important goal. Phenotypic screening of compounds can uncover dependencies, which can, for example, nominate drivers of cancer cell survival; however, this type of screening remains dependent on compounds with clear target profiles and specificity, which would ensure observations of the screen are related to their intended target (1). With compounds that affect multiple targets and/or that have poorly characterized target profiles, this method is more problematic. Target identification using RNA interference (RNAi) at face value appears optimal for target identification for these less clear compounds; however, in practice, this has been complicated by off-target effects, complicated data analysis algorithms, and in some cases an inability to validate results (2). We hypothesized that one alternative solution would be to combine phenotype-based drug screens with target identification that used activity-based protein profiling (ABPP) combined with mass spectrometry. Chemical proteomics is a powerful approach to measuring proteome-wide drug protein target spectra in an unbiased manner (3). One ABPP approach employs a desthiobiotin-ATP probe directed against the active sites of enzymes to interrogate their functional state in biological samples. The desthiobiotin-ATP probe covalently labels conserved lysine residues in or near the ATP binding pocket of kinases, which are then enriched, identified, and quantified by avidin-based purification of labeled peptides and LC-MS/MS. This approach is uniquely capable of profiling the human kinome in human disease or cell models and can identify biological targets of kinase inhibitors through competitive binding of the active sites of the kinase with an ATP probe (4–6).
In this study, we combined a kinase inhibitor library screen with the desthiobiotin-ATP probe and LC-MS/MS to identify therapeutic targets in small cell lung cancer (SCLC) (Figure 1). SCLC is poorly differentiated neuroendocrine lung malignancy, in which no significant improvements in patient outcomes have been shown with the standard treatment option of platinum-based combination chemotherapy (7).
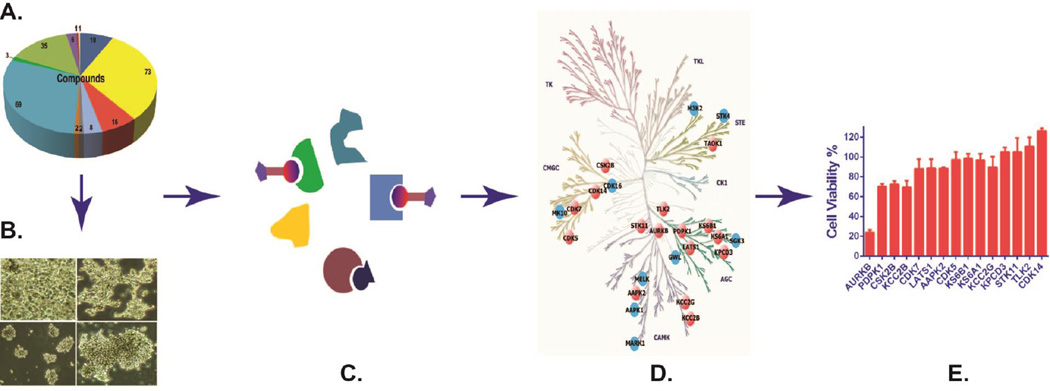
Figure 1. Workflow for therapeutic target identification in SCLC via integrated phenotypic screen with ABPP ATP probe and LC-MS/MS.
A compound library (A) was used for cell viability screen in SCLC cell lines (B), with paired active and inactive compounds based on screen results chosen for further drug profiling using desthiobiotin-ATP probe (C) tandem liquid chromatography-mass spectrometry. The identified peptides were quantified by MaxQuant, and the candidate hits (orange nodes) of active drug were determined based on those that inhibited ATP binding by >60% compared with DMSO treatment and filtered by potential hits (blue nodes) of inactive compound (D). Potential targets were validated in SCLC cells using relevant small RNA interference and drugs (E).
MATERIALS AND METHODS
Cell lines and drugs
All SCLC cell lines were provided by Dr. John V. Heymach (MD Anderson Cancer Center, Houston, TX) and Dr. Gerold Bepler (Barbara Ann Karmanos Cancer Institute, Detroit, MI). All cell lines, maintained in a central bank at Moffitt, were authenticated by STR analysis (ACTG Inc, Wheeling, IL), and all are routinely tested and shown to be negative for mycoplasma (PlasmoTest, InvivoGen, San Diego, CA). All cell lines were grown in RPMI 1640 media with 10% fetal bovine serum (GIBCO, Grand Island, NY) and maintained at 37°C in a humidified incubator with 5% CO2 atmosphere. The Roche Kinase Inhibitor Set was a gift of Hoffman-La Roche (San Diego, CA). SNS-314, danusertib, AT9283, MLN-8237, ENMD-2076, TAK-901, AZD1152, and VX-680 were purchased from Selleckchem (Houston, TX). The TANK-binding kinase 1 (TBK1) inhibitor Compound II and BX-795 were provided by Dr. Michael White (University of Texas Southwestern Medical Center, Dallas, TX).
Compound screens
We seeded 21 SCLC cell lines in 384-well plates with 1000 cells per well using a Precision™ microplate liquid handler (BioTek, Winooski, VT). All 235 tested compounds from Roche (San Diego, CA) were diluted in DMSO and added to cells at 1 µM final concentration with duplicate treatment. Cell viability assays were conducted after 72 hours of treatment according to the manufacturer’s recommendations using CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega, Madison, WI). The luminescent signal was read by the SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA). Cell viability was calculated based on the ratio to the DMSO control treatment.
ATP probe-based drug profiling
Cell lysates were prepared and labeled according to the manufacturer’s recommendations for the Pierce® Kinase Enrichment Kits and ActivX® Probes (Thermo Scientific). Briefly, cell pellets from 1×107 cells were resuspended in 600 µl of affinity purification buffer and sonicated. The lysates were cleared by centrifugation at 16,000 × g for 10 minutes at 4°C and desalted by Zeba Spin Desalting Columns. The concentration of protein was measured using the Bradford assay, and a total of 1 mg used for drug profiling per sample. MnCl2 was added to the lysate to a final concentration of 20 mM for 10 minutes before drug treatment. The final concentration of drug for the drug profiling was determined by cell viability assay in the cells. After 15-minute pre-incubation of lysate with drug and DMSO as the control, ATP probe competition reactions were performed at a final concentration of 5 µM of desthiobiotin-ATP probe for 15 minutes. All reactions were performed at the room temperature in duplicate.
MS sample preparation and LC-MS/MS analysis
The probe-labeled lysates were denatured in 5 M of urea and 5 mM of DDT at 65°C for 30 minutes and then reduced in 40 mM of iodoacetamide at room temperature for 30 minutes. The desalted proteins were digested with 20 °g of trypsin for 2 hours at 37°C, and then desthiobiotinylated peptides were captured by 50 °l slurry of high-capacity streptavidin beads for 1 hour. The beads were washed with lysis buffer, PBS buffer, and LC-MS grade water in sequence with 4 times wash cycle for each buffer, with peptides then eluted by 50% acetonitrile in 0.1% trifluoroacetic acid water. The eluted peptides were lyophilized in vacuum concentrator and resuspended in 20 °l of injection buffer containing 2% acetonitrile and 0.1% trifluoroacetic acid. We included 100 fmol of horse myoglobin tryptic peptides (25 fmol/injection) in each sample as internal standard to normalize quantification variation between MS runs. The LC-MS/MS analysis was performed as previously described (8).
Data analysis and candidate hits of active drug determination
The peptides identified by mass spectrometry were analyzed and quantified using MaxQuant version 1.2.2.5 (http://www.maxquant.org) (9). For the search parameters, desthiobiotin-ATP labeling was selected, with enzyme specificity set to tryptic with up to 4 missed cleavages, allowing 10 ppm as the main search, and up to 3 numbers of modifications per peptide. Fragment ion tolerance was set to 0.6 Da. Protein and peptide false discovery rates were set to 0.1 and 0.05, respectively, and minimum peptide length was set to 6 amino acids. MS/MS data were searched against the UniProt human database (version 2014_06) combined with common contaminants and concatenated with the reversed versions of all sequences using the Andromeda search engine integrated into MaxQuant. The protein kinases were annotated using MetaCore (http://portal.genego.com, Thomson Reuters, Philadelphia, PA), and intensity of each peptide corresponding to a protein kinase was used to calculate the inhibition based on the DMSO control treatment. The candidate binding proteins of each compound were defined as those that inhibited ATP binding by more than 60%. The final targets of the active compound E08 were obtained after filtering the binding proteins of the inactive compound.
RNAi screen
RNAi screen experiments were performed in SCLC cell lines as described previously (10). Briefly, the ON-TARGET plus Smart pool custom siRNA library was purchased from Thermo Scientific (Dharmacon, Chicago, IL). The small interfering RNA against each gene was delivered in triplicate to each well in 96-well plates using a Precision™ microplate liquid handler (BioTek, Winooski, VT). The reverse transfections were performed using RNAiMAX (Life Technologies, Grand Island, NY) according to the manufacturer’s recommendations with 5,000 cells/well were seeded in the plate. Cell viability assays were conducted 5 days after transfection using CellTiter-Glo. Viability changes were determined for each target gene after normalization on ON-TARGET plus Non-Targeting pool control siRNA.
Protein expression analysis
Western blotting was performed as previously described (10). We used the following primary antibodies: rabbit polyclonal antibody specific for PARP, phospho-TBK1/NAK (Ser172), TBK1, phospho-polo-like kinase-1 (PLK1) (Thr210), and PLK1from Cell Signaling (Beverly, MA), and β-actin antibody from Sigma (St. Louis, MO).
c-MYC amplification
We assessed MYC amplification in the 21 SCLC cell lines according to the manufacturer’s recommendations for TaqMan Copy number assay kits (Life Technologies, Grand Island, NY). Cells’ genomic DNA was extracted by the FlexiGene DNA Kit from QIAGEN (Valencia, CA). Copy number and expression were assessed using real-time PCR on an ABI Prism 7900 (Applied Biosystems) according to the manufacturer’s protocol. Copy number was calculated from triplicate reactions using Applied Biosystems CopyCaller software V2, in which the cycle threshold of the cMYC gene was normalized against the cycle threshold of an RNaseP reference assay. c-MYC amplification was defined by a copy number of more than 10.
Flow cytometry
For cell cycle assays, cells were washed in PBS and fixed by overnight drop-wise addition of ice-cold 70% ethanol at 4°C. The cells were incubated with 30 µg/mL RNaseA (Sigma-Aldrich) for 30 minutes in a 37°C water bath and then resuspended in BD Perm/Wash™ buffer (BD Biosciences, San Jose, CA) containing 50 mg/mL propidium iodide for 20 minutes at room temperature. For caspase 3 activity assays, 2×105 cells were treated with 500 nM Compound II, or 500 nM BX-795 for 48 hours, and then resuspended in 0.5 ml of Cytofix/cytoperm solution for 20 minutes on ice. Pelleted cells were washed twice with Perm/Wash buffer and incubated with PE-conjugated monoclonal active caspase-3 antibody (BD Pharmingen, San Diego, CA) for 30 minutes at room temperature. Cells were analyzed by flow cytometry (FACS 420, Becton Dickinson).
Statistical analysis
The Wilcoxon rank-sum test was applied to determine which group is more sensitive to depletion of AURKB by siRNA or AURKB inhibitors between MYC amplification (MYC+) and MYC lacking amplification (MYC-) groups. Results were considered statistically different at P < 0.05.
RESULTS
To identify survival kinases in SCLC, we screened 21 SCLC cell lines with known c-MYC amplification status for alterations in viability using a chemical library of 235 small molecule kinase inhibitors enriched as inhibitors against CDK2, CDK1, protein kinase C (PKC), GSK3, and p38 MAP kinases (Supplemental Figure S1A and S1B). MYC amplification is a known molecular alteration and frequent event in SCLC and is related to a short survival time in SCLC patients (11). Few compounds strongly affect cell viability across SCLC cell lines (Figure 2 and Supplemental Table S1); however, in our analysis, we noted two compounds with stronger effects on cell viabilty. One belongs to a series of oxindole-type compounds that have heteroatom-substituted alkynyl moieties at their C-4 position and are predominantly enriched for CDK2 as primary target (12). Of the 19 of oxindole-type compounds, only E08 demonstrated more than a 50% inhibitory effect on cell viability in 10 of 21 screened cell lines (Supplemental Figure S2A). The other group is a series of substituted bisindolylmaleimides, originally designed to compete with ATP and inhibit the PKC kinases (13). Of this group, the K14 compound inhibited cell viability across some SCLC cells. The cell-based screen implied that the active compounds E08 and K14 may target other critical kinases that drive cell survival in subgroups of SCLC beyond the cognate targets (CDK2 or PKC kinases) of these compounds, since other compounds affecting these primary targets had little effect on SCLC viability.
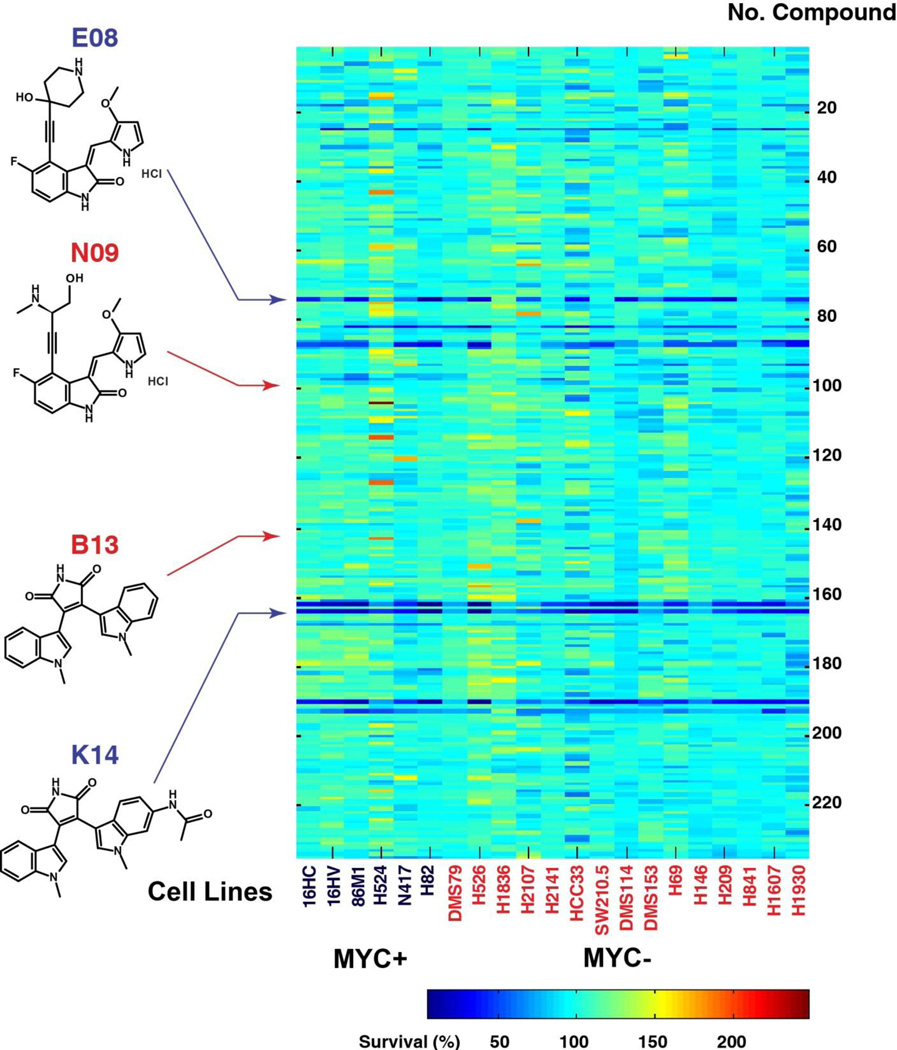
Figure 2. Compound screen across SCLC cell lines.
We investigated 235 compounds for the cell viability screen in the 21 SCLC cell lines; 1,000 cells/well were seeded in 384-well plate and treated with individual compounds at 1 µM concentration for 3 days. Cell viability was performed by CellTiter Glo. A heat map of the average percent cell survival from duplicates compared with DMSO control was plotted by the “imagesc” function in MatLab. Blue indicates less cell survival; red indicates more cell survival. Bottom right (color key legend) shows range of average values. Left panel shows the chemical structures of the active and inactive compound pairs (E08 versus N09 and K14 versus B13).
To identify these crucial kinase targets and nominate their role in survival signaling, we performed ABPP using a desthiobiotin-ATP probe in conjunction with LC-MS/MS to identify the expressed kinome in these cells and identify kinases bound to the active compounds. We chose the H82 cell line for experiments because it is the most sensitive to E08 across the 21 SCLC cell lines (Supplemental Figure S2A). To determine the target profiles of E08, which can then be further dissected for exact kinases affecting cell viability, we pre-incubated E08 with H82 proteomes to compete with the desthiobiotin-ATP probe for ATP binding sites. We reasoned that we could further filter those kinases that mediate the effects of E08 by adding experiments that pre-incubate a compound with a structure similar to E08 that had no effect on SCLC viability. We hypothesized that using a pair of inactive and active compounds with a similar chemical backbone would identify a common set of drug targets that are not critical to SCLC viability, since the two compounds have different phenotypic effects (Supplemental Figure S2B). Thus, we compared the target binding profiles of the active compound (E08) with the inactive compound (N09), which has the same chemical oxindole backbone, and explored their kinase targets using the desthiobiotin-ATP probe.
Desthiobiotin-modified peptides were analyzed by LC-MS/MS and then quantified using MaxQuant. We identified a total of 433 peptides corresponding to 188 protein kinases from two independent experiments. We found 28 peptides that corresponded to 24 protein kinases in which ATP binding was reduced (competed) >60% by active compound E08 compared with DMSO control (Supplemental Table S2). It is known that 75% of the targeted proteins contain either an ATP binding site or ATP active site in their kinase domain (14). We filtered proteins (shown as the blue nodes in Figure 3A) that are also bound to the N09 compound, reasoning that protein kinases that bind to inactive compound N09 would not be considered as the SCLC survival drivers. Through this approach, we identified 15 kinases targeted by E08 (shown as orange nodes in Figure 3A; also see Supplemental Figure S2C and Supplemental Table S2). To determine the kinase inhibition that resulted in the observed changes in cell viability caused by E08, we transfected H82 cells with RNAi corresponding to each of the 15 kinases. Aurora kinase B (AURKB) knockdown reduced cell viability (76.1% inhibition) in H82 cells compared with control siRNA (Figure 3B). We extended the 15-gene RNAi analysis to 4 other SCLC cell lines, two with MYC amplification (H524 and N417) and two without (H526 and DMS79), which further demonstrated stronger effects of AURKB knockdown compared with other kinases (Supplemental Figure S2D). MYC-amplified cell lines were significantly more sensitive to AURKB depletion compared with cell lines lacking MYC amplification (P<0.05) (Figure 3C and Supplemental Figure S2E), and the MYC-amplified lines were more sensitive to small molecule AURKB inhibitors than cells lacking the MYC amplification groups (P<0.001) (Figure 3D). Our finding suggests that AURKB is a survival kinase in SCLC with MYC amplification. Previous genomic and pharmacological vulnerability screen approaches also revealed that AURKB is a potential target for MYC-amplified SCLC (15, 16), indicating our ABPP approach combined with drug screening could be a novel way to identify targetable kinases in cancer.
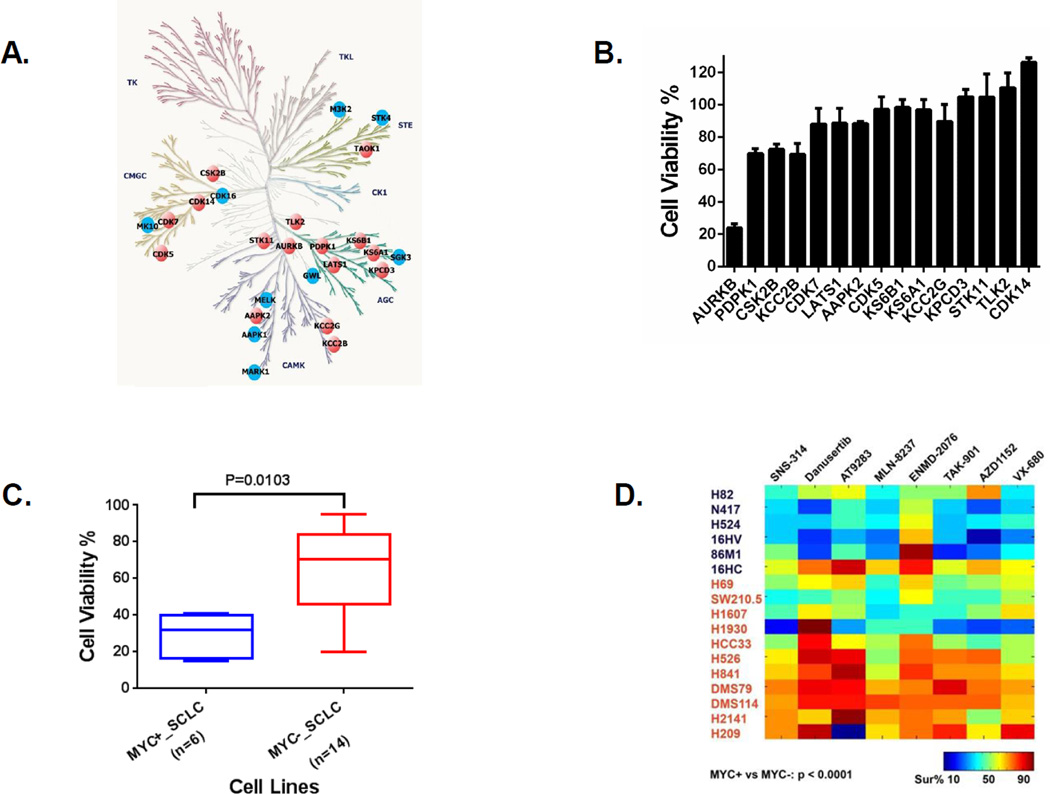
Figure 3. Aurora kinase B (AURKB) identified by combining drug screen with ABPP ATP probe in H82 cells and drive cell survival in MYC-amplified SCLC cells.
A: Binding proteins of E08 identified by ABPP ATP probe in H82 cells. Drug profiling of compounds E08 and N09 was performed in H82 cells. The compound’s candidate binding proteins (shown on the human kinome tree) were defined as those that inhibited binding of the ATP peptide by >60% compared with DMSO treatment. The orange nodes represent the final candidate targets of active compound E08, and the blue nodes represent the targets of both active and inactive compounds. B: AURKB knockdown by siRNA dramatically inhibited cell viability in H82 cells. H82 cells were transfected with pooled siRNA targeting 15 indicated genes as described in Materials and Methods. Cell viability was performed by CellTiter Glo 5 days after transfection. Cell viability changes were determined for each target gene after normalization on ON-TARGET plus Non-Targeting pool control siRNA and plotted by GraphPad Prism 6. C: MYC-amplified SCLC are more sensitive to depletion of AURKB by siRNA. SCLC cell lines with MYC amplification (n=6) and no MYC amplification (n=14) were transfected with pooled siRNA targeting AURKB. Cell viability was performed by CellTiter Glo 5 days after transfection. The box plot between MYC amplified (MYC+_SCLC) and lacking MYC amplification (MYC−_SCLC) SCLC cell lines based on cell viability changes was constructed by GraphPad Prism 6 (P value based on the Wilcoxon rank sum test). D: MYC amplified SCLC are more sensitive to AURKB inhibitors. SCLC cell lines with MYC amplification (n=6) and lacking MYC amplification (n=11) were treated with 0.5 µM of indicated AURKB inhibitors. Cell viability was performed by CellTiter Glo 3 days after treatment. A heat map of average percent cell survival from duplicates compared with DMSO control was plotted by the “imagesc” function in MatLab. Bottom right (color key legend) shows range of average values. P value shown at left bottom was based on the Wilcoxon rank sum test between MYC amplification (MYC+) and MYC lacking amplification (MYC−) groups.
Encouraged by the success of this approach in AURKB identification, we next applied the same strategy to uncover kinases affecting SCLC viability in cell lines lacking MYC amplification. We performed ABPP profiling on compound K14, which was found to have inhibitory effects on SCLC viability in cell lines lacking MYC amplification. We compared the target profile to a similar yet inactive compound B13 (both bisindolylmaleimide derivatives were originally designed to compete with ATP and inhibit the PKC kinases). We identified 7 of 15 SCLC cell lines lacking MYC amplification as being sensitive to K14 compound (>50% inhibition) (Supplemental Figure S3A). SW210.5 cells were used to perform drug profiling for the K14 and B13 pair of compounds. We identified 19 peptides corresponding to 18 protein kinases where ATP binding was competed by active compound K14 >60% compared with DMSO control (Supplemental Table S3). We identified 9 to be unique to the K14 compound after removing the kinases that were bound to the inactive compound B13 (Supplemental Figure S3B). The top peptide “TGDIFAIK*VFNNISFIRPVDVQMR,” which contained a desthiobiotin labeling site and was located at the kinase domain of TBK1, strongly binds to the active compound K14 (ATP binding inhibition of 83.1%); however, this peptide did not bind to the inactive compound B13 (ATP binding inhibition of 0%). This caught our attention, since TBK1 has been implicated as a survival kinase in some cancer cells and our previous study found TBK1 affected polo-like kinases (PLK1) (1, 17, 18). Because Aurora kinases are also critical to the success of both DNA damage and spindle assembly mitotic checkpoints, we hypothesized that TBK1 in these non-MYC amplified lines could similarly be affecting this process.
We next determined whether TBK1 inhibition is responsible for the observed changes in cell viability caused by K14 in SW210.5 cells. We examined whether RNAi-mediated depletion of TBK1 affects cell viability in SCLC cell lines lacking MYC amplification. We found that the H1607, SW210.5, H841, and DMS114 cell lines are partially sensitive to TBK1 depletion (Figure 4A), compared with A549 cells, as reported in a previous study (19). We next determined if depletion of TBK1 had measurable effects on the viability of MYC-amplified SCLC cell lines in which AURKB plays an important role in cell survival. We used RNAi-mediated knockdown of TBK1 in 5 Myc-amplified SCLC cell lines, using A549 and the TBK1-sensitive line SW210.5 as positive controls. None of these cell lines displayed any reduction in cell viability following TBK1 knockdown (Supplemental Figure S3C).
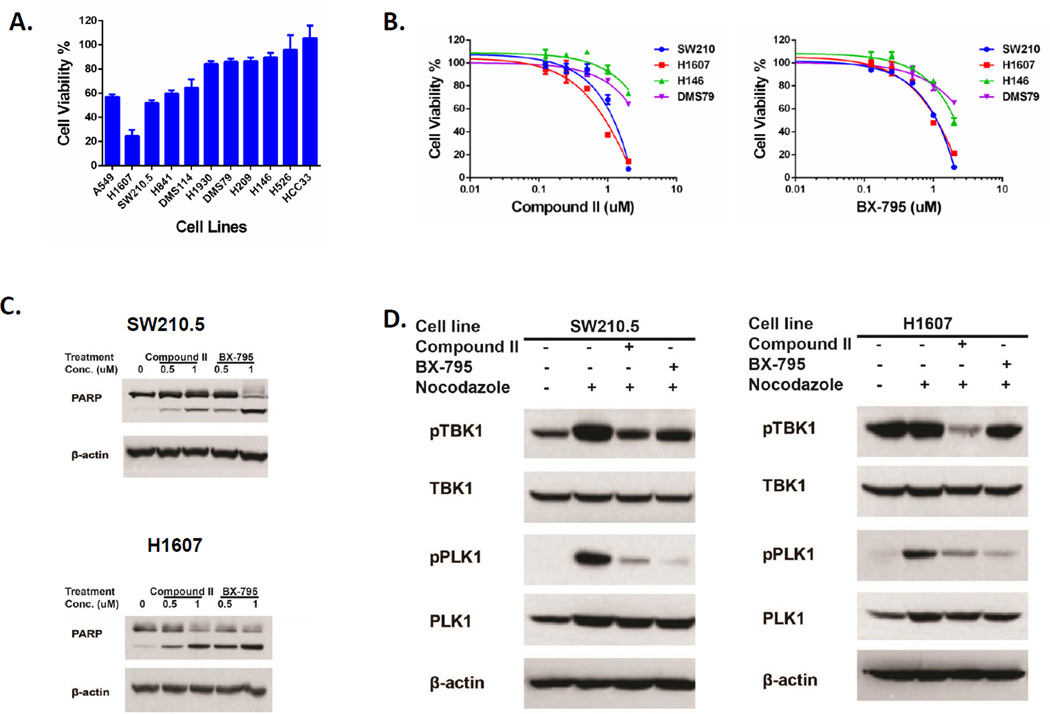
Figure 4. TBK1 identified by ABPP ATP probe in SW210.5 cells plays an important role in cell survival in a subset of SCLC cells lacking MYC amplification.
A: Depletion of TBK1 by siRNA affected cell growth in SCLC cells. Ten SCLC cell lines with no MYC amplification were transfected with pooled siRNA targeting TANK-binding kinase 1 (TBK1) and lung cancer cell line A549 as the positive control. Cell viability was performed by CellTiter Glo 5 days after transfection. Cell viability changes were constructed by GraphPad Prism 6 after normalizing with ON-TARGET plus Non-Targeting pool control siRNA. B: TBK1 inhibitors affected cell viability in SW210.5 and H1607 cells. Four SCLC cell lines with no MYC amplification were treated with TBK1 inhibitors Compound II and BX-795 at the titration concentration. Cell viability was performed by CellTiter Glo 4 days after treatment. The growth curve was constructed by GraphPad Prism 6. C. TBK1 inhibitors induced cell apoptosis in SW210.5 and H1607 cells. SW210.5 and H1607 cells were treated with indicated concentrations of TBK1 inhibitors Compound II and BX-795 for 48 hours, and PARP cleavage was evaluated by Western blotting. Equal protein loading was confirmed by β-actin evaluation. D: TBK1 mediated activation of the mitotic kinase PLK1. SW210.5 cells were treated with indicated TBK1 inhibitors for 1 hour and then exposed to 50 ng/mL of nocodazole for 18 hours. Western blot detected the signaling change using indicated antibodies, and β-actin evaluation was confirmed the equal protein loading.
We performed experiments to determine if TBK1 expression and levels of phosphorylated TBK1 relate to sensitivity of SCLC cells to TBK1 RNAi. We found no relationship between TBK1 protein expression or levels of phosphorylated TBK1 and sensitivity of cells (Supplemental Figure S3D). To confirm that SCLC tumor tissues express TBK1 protein, we identified 5 snap-frozen SCLC tissues and performed Western blotting to determine expression of total and phosphorylation of TBK1. Reassuringly, we found all 5 tumors have expression of TBK1, with 2 of 5 SCLC patient tumors having higher expression of phosphorylated TBK1 (Supplemental Figure S3D). We searched genomic profiles of CCLE and ATCC and were unable to find any different genomic features, such as p53 and Rb mutations, or MYC amplification between relative TBK1-sensitive and -resistant SCLC cell lines. We have however found one feature that appears to point toward mitotic checkpoint sensitivity as a differentiator of cell line sensitivity. Three of four TBK1-sensitive lines (SW210.5, H841, and DMS114) have increased phosphorylation of TBK1 when undergoing mitotic blockade with nocodazole, whereas three of four TBK1-resistant lines (H146, H526, and H209) do not show this effect (Supplemental Figure 3E). These results suggest an important role for TBK1 in SCLC mitotic signaling.
Next, we used two tool compound TBK1 kinase inhibitors, Compound II and BX-795, to determine effects in these same cells (17). Similar results were obtained when TBK1 was inhibited by small molecule TBK1 inhibitors Compound II and BX-795. Both H1607 and SW210.5 cells are more sensitive to these two TBK1 inhibitors than H146 and DMS79 cells (Figure 4B, Supplemental Figure S3F). These results mirror effects of TBK1 RNAi studies shown in Figure 4A. H1607 and SW210.5 cells were more sensitive to the depletion of TBK1 than H146 and DMS79 cells based on levels of TBK1 after knockdown (Supplemental Figure S3G). We examined the entire panel of cell lines in Figure 4A for IC50 using these two compounds. Overall, the results correlate between effect of TBK1 knockdown by RNAi and drug inhibition in the tested SCLC cell lines with only one exception (H526 cells). Therefore, cells that are more sensitive to TBK1 RNAi are likewise the more sensitive cells to the two TBK1 kinase inhibitor tool compounds. We confirmed that short-term exposure to these two TBK1 inhibitors did not directly affect Aurora kinase activation and phosphorylation (Supplemental Figure S3H). Both TBK1 inhibitors led to cell apoptosis with a concentration-dependent increase in PARP cleavage (Figure 4C) and caspase-3 activation (Supplemental Figure S3I) in SW210.5 and H1607 cells. Both TBK1 inhibitors induced cell cycle arrest at G2/M phase in SW210.5 cells, consistent with the hypothesis that TBK1 is important in this phase of the cell cycle through an effect on PLK1 (Supplemental Figure S3J). Consistent with this, TBK1 inhibition by Compound II and BX-795 decreased PLK1-activated phosphorylation in SW210.5 and H1607 cells (Figure 4D). Collectively, our results with RNAi and small molecule TBK1 kinase inhibitor tool compounds show that TBK1 inhibition results in reduced cell viability, reduced phosphorylation of PLK1, increased G2/M arrest in cells, and increased apoptosis in subgroups of SCLC cell lines.
DISCUSSION
Our study describes two important advances. First, we demonstrate the potential for a phenotype-based drug screen integrated with ABPP to uncover disease drivers. Second, we uncovered a new role for TBK1 in affecting cell viability, G2/M arrest, PLK1 phosphorylation, and apoptosis in subgroups of SCLC cell lines using this approach. We focused our efforts on ABPP probes that mimic ATP and can be used to enrich ATP binding proteins such as protein kinases. Protein kinases play a critical role in the development and progression of cancer by regulating key pathways that drive the hallmarks of cancer (20). Our approach complements other platforms to identify essential kinases in cancer, including synthetic lethal kinome RNAi screens (21–24), large-scale pharmacological vulnerability screens (16, 25, 26), and integrated genetic and bioinformatics strategies (27). Our approach described here has several advantages. First, we did not need to rely on high-quality drug binding data to initiate the screen. This has the potential to allow increased depth in chemical libraries, as interesting compounds can be subsequently studied with ABPP to narrow down targets. The desthiobiotin-ATP probe could be coupled with results from metabolic inhibitor screens, since this probe can identify ATP binding proteins such as metabolic kinases. Second, ABPP with high-quality chemical probes can allow for rapid identification of targets versus other approaches that require chemical modification of compounds for affinity pulldown experiments. Third, the use of an inactive compound during the workflow can be used to narrow the list of candidate targets likely to be mediating the drug effects and simplify and speed up any further validation. In light of the success of this approach, we envision broadening its application to identify targets in cancer and other diseases through combining phenotype-based compound screen disease cells along with ABPP probes that engage different target families, including hydrolases, deacetylases, or other target families of interest where one can develop ABPP probe technology. Improvements in mass spectrometry can further optimize the potential. One potential future area could be to combine screens of deubiquitinase inhibitors with ABPP probes that monitor protein ubiquitination.
There are a number of hurdles to address and limits to overcome before this approach can be more widely adopted. First, we cannot ensure the entire kinome is being covered by the desthiobiotin-ATP probe used in our studies, which could be limited by the low abundance of certain kinases or the sequences not having a solvent accessible lysine near the active site. Nonetheless, we identified 196 protein kinases in H82 cells and 179 protein kinases in SW210.5 cells, which compares favorably with other reports that employ ABPP (28, 29). However, using these probes results in a certain bias toward serine/threonine kinases over tyrosine kinases compared to approaches with immobilized kinase inhibitors. We did not include desthiobiotin-ADP probe in our analysis, and this may affect coverage of the kinome and competition results (14). Second, we made several assumptions and decisions about concentration of compounds, incubation times in our competition assays, and 60% inhibition as the criteria for candidate hits selection; thus these can leave room for error. Time dependence in replacement of the reversible inhibitor by the ATP probe can affect the inhibition profile. In addition, the ATP probes may preferentially label particular proteins resulting in further changes to the inhibition profiles. Third, LC-MS/MS-based identification and quantification of peptides can suffer from reproducibility concerns; even with the enrichment of kinases peptides in ABPP, their population and copy number are low compared to other intracellular ATP-utilizing enzymes that are captured in the same process. The intensity of certain peptides could be too weak to be detected regularly, particularly as covalent modification with ATP-desthiobiotin may negatively impact peptide identification by MS. Migrating from LC-MS/MS to targeted mass spectrometry (multiple reaction monitoring) can make peptide analyses (i.e. fewer missing values) and quantification more precise (4, 5, 30). To this end, we developed an ABPP-LC-multiple reaction monitoring approach that can enable rapid and reproducible detection as well as quantified measurements of peptides corresponding to other panels of enzymes in cells and tissues that could be used for additional target validation following screening (30). Fourth, while desthiobiotin-ATP probes are designed to engage the ATP binding site of proteins, we have observed recovery of peptides from outside the ATP binding pocket (e.g. near autophosphorylation sites). Further studies are needed to determine the relevance of these findings and the implications for the competition assays we performed.
Despite these limits and assumption, we re-identified aurora kinase through our approach, giving credence to this strategy to uncover new targets. We found that TBK1 may play a pivotal role in cell viability in subsets of SCLC, a disease lacking effective molecular therapeutics, as TBK1 may be activated in mitotic cells and associated with PLK1 activation. We have yet to identify any molecular determinants that define which SCLC subgroups are especially vulnerable to TBK1 inhibition. We found no relationship between total or phosphorylated TBK1 expression levels. However, studies using nocodazole to block cells in mitosis revealed increased TBK1 phosphorylation, suggesting that complex interplays in mitotic signaling may underlie the sensitivity that we observed. To further support this point, a recent pharmacogenomics study identified PLK1 as a potential molecular target in SCLC (27). Further studies are necessary to dissect the likely complex interplay between Aurora kinases, TBK1, and PLK1 in SCLC and other cancers. Quantitative phosphoproteomics could be used to study this interplay (31), with integrating chemical and phosphoproteomics also as investigative options. Further studies are needed in animal tumor models to fully realize the potential of TBK1 inhibitors and models mimicking cisplatin resistance. In addition, more studies are needed to understand which tumor cells are dependent on TBK1 for proliferation and survival.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Michael White (University of Texas Southwestern Medical Center, Dallas, TX) for providing TBK1 inhibitors (Compound II and BX-795), Hoffman La-Roche and Paul Gillespie for providing Roche Kinase Inhibitor Set, Cell Signaling Technology (Danvers, MA) for allowing reproduction of the kinome map, Dr. Matthew A. Smith (Moffitt Cancer Center) for assistance with Adobe illustrator application, and Rasa Hamilton (Moffitt Cancer Center) for editorial assistance with the manuscript.
FINANCIAL INFORMATION: The work was supported under NIH grant 1R21 CA169979-01A1 (E.B. Haura). Our work was supported in part by the Proteomics Core and the Flow Cytometry Core at the H. Lee Moffitt Cancer Center & Research Institute (Tampa, FL), an NCI-designated Comprehensive Cancer Center, supported under NIH grant P30-CA76292.
Abbreviations
- ABPP
Activity-based protein profiling
- AURKB
Aurora kinase B
- PKC
Protein kinase C
- PLK1
Polo-like kinase-1
- RNAi
RNA interference
- SCLC
Small cell lung cancer
- TBK1
TANK-binding kinase 1
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST: The authors declare no competing financial interests.
REFERENCES
- 1.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG., Jr Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 4.Xiao Y, Guo L, Wang Y. A targeted quantitative proteomics strategy for global kinome profiling of cancer cells and tissues. Mol Cell Proteom. 2014;13:1065–1075. doi: 10.1074/mcp.M113.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Worboys JD, Sinclair J, Yuan Y, Jorgensen C. Systematic evaluation of quantotypic peptides for targeted analysis of the human kinome. Nat Methods. 2014;11:1041–1044. doi: 10.1038/nmeth.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Bennett K, Stukalov A, Fang B, Zhang G, Yoshida T, et al. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Mol Syst Biol. 2013;9:705. doi: 10.1038/msb.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alves Rde C, Meurer RT, Roehe AV. MYC amplification is associated with poor survival in small cell lung cancer: a chromogenic in situ hybridization study. J Cancer Res Clin Oncol. 2014;140:2021–2025. doi: 10.1007/s00432-014-1769-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luk KC, Simcox ME, Schutt A, Rowan K, Thompson T, Chen Y, et al. A new series of potent oxindole inhibitors of CDK2. Bioorg Med Chem Lett. 2004;14:913–917. doi: 10.1016/j.bmcl.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Davis PD, Hill CH, Lawton G, Nixon JS, Wilkinson SE, Hurst SA, et al. Inhibitors of protein kinase C. 1. 2,3-Bisarylmaleimides. J Med Chem. 1992;35:177–184. doi: 10.1021/jm00079a024. [DOI] [PubMed] [Google Scholar]
- 14.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 15.Hook KE, Garza SJ, Lira ME, Ching KA, Lee NV, Cao J, et al. An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF-03814735. Mol Cancer Ther. 2012;11:710–719. doi: 10.1158/1535-7163.MCT-11-0184. [DOI] [PubMed] [Google Scholar]
- 16.Sos ML, Dietlein F, Peifer M, Schottle J, Balke-Want H, Muller C, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A. 2012;109:17034–17039. doi: 10.1073/pnas.1207310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41:458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng T, Liu JC, Chung PE, Uehling D, Aman A, Joseph B, et al. shRNA kinome screen identifies TBK1 as a therapeutic target for HER2+ breast cancer. Cancer Res. 2014;74:2119–2130. doi: 10.1158/0008-5472.CAN-13-2138. [DOI] [PubMed] [Google Scholar]
- 19.Kim JY, Welsh EA, Oguz U, Fang B, Bai Y, Kinose F, et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci U S A. 2013;110:12414–12419. doi: 10.1073/pnas.1220674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Chen Z, Malysa A, Li X, Oliveira P, Zhang Y, et al. A kinome screen identifies checkpoint kinase 1 (CHK1) as a sensitizer for RRM1-dependent gemcitabine efficacy. PLoS One. 2013;8:e58091. doi: 10.1371/journal.pone.0058091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murrow LM, Garimella SV, Jones TL, Caplen NJ, Lipkowitz S. Identification of WEE1 as a potential molecular target in cancer cells by RNAi screening of the human tyrosine kinome. Breast Cancer Res Treat. 2010;122:347–357. doi: 10.1007/s10549-009-0571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhola NE, Jansen VM, Bafna S, Giltnane JM, Balko JM, Estrada MV, et al. Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer. Cancer Res. 2015;75:405–414. doi: 10.1158/0008-5472.CAN-14-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wildey G, Chen Y, Lent I, Stetson L, Pink J, Barnholtz-Sloan JS, et al. Pharmacogenomic approach to identify drug sensitivity in small-cell lung cancer. PloS One. 2014;9:e106784. doi: 10.1371/journal.pone.0106784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemeer S, Zorgiebel C, Ruprecht B, Kohl K, Kuster B. Comparing immobilized kinase inhibitors and covalent ATP probes for proteomic profiling of kinase expression and drug selectivity. J Proteome Res. 2013;12:1723–1731. doi: 10.1021/pr301073j. [DOI] [PubMed] [Google Scholar]
- 29.Kunz RC, McAllister FE, Rush J, Gygi SP. A high-throughput, multiplexed kinase assay using a benchtop orbitrap mass spectrometer to investigate the effect of kinase inhibitors on kinase signaling pathways. Anal Chem. 2012;84:6233–6239. doi: 10.1021/ac301116z. [DOI] [PubMed] [Google Scholar]
- 30.Fang B, Hoffman MA, Mirza AS, Mishall KM, Li J, Peterman SM, et al. Evaluating kinase ATP uptake and tyrosine phosphorylation using multiplexed quantification of chemically labeled and post-translationally modified peptides. Methods. 2015;81:41–49. doi: 10.1016/j.ymeth.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.