Abstract
Paper diagnostics have successfully been employed to detect the presence of antigens or small molecules in clinical samples through immunoassays; however, the detection of many disease targets relies on the much higher sensitivity and specificity achieved via nucleic acid amplification tests (NAAT). The steps involved in NAAT have recently begun to be explored in paper matrices, and our group, among others, has reported on paper-based extraction, amplification, and detection of DNA and RNA targets. Here, we integrate these paper-based NAAT steps onto a single paperfluidic chip in a modular, foldable system that allows for fully integrated fluidic handling from sample to result. We showcase the functionality of the chip by combining nucleic acid isolation, isothermal amplification, and lateral flow detection of human papillomavirus (HPV) 16 DNA directly from crude cervical specimens in under 1 hour for rapid, early detection of cervical cancer. The chip is made entirely of paper and adhesive sheets, making it low-cost, portable, and disposable, and offering the potential for a point-of-care molecular diagnostic platform even in remote and resource-limited settings.
INTRODUCTION
Integrated molecular diagnostics to enable sample-to-answer nucleic acid amplification testing (NAAT) have traditionally required sophisticated instrumentation to provide pressure driven fluid handling, cyclic thermal control, and optical assay detection. These requirements result in expensive equipment and costly disposables unsuitable for use in resource-limited settings. Since the start of lab-on-a-chip microfluidic technologies in the 90s, their use in remote settings has been perceived as potentially one of their most powerful applications by taking advantage of their small size, portability, low volume requirement for samples, and rapid analysis without the need for an expert operator1. However, because these systems require pumps or pressure in order to drive fluid flow, they aren't yet equipment-free and rely on instrumentation and electricity. An alternative that has garnered much attention recently is “paperfluidics”, where paper is used as a substrate to construct microfluidic devices for use in rapid diagnostic tests2-6. Paper's ability to passively transport fluids through capillary action, or wicking, eliminates the need for pumps or other fluid handling equipment. In regards to molecular diagnostics, a number of studies have reported on the success of individual NAAT steps (extraction, amplification, and detection) within paper matrices7-12. Our group has previously shown paper-based nucleic acid extraction, isothermal amplification, and lateral flow detection for DNA13 and RNA14, in separate paper-based modules. To date, only one other device has been reported that combines paper-based extraction, amplification and detection steps15, which consists of a magnetic sliding strip to run each step serially. While this device does encompass a fully integrated NAAT system, it relies on a UV light source and imaging for endpoint detection, requiring equipment that may not be readily available in remote low-resource settings. Furthermore, this device has only been shown to detect E. coli DNA that was spiked into plasma, rather than extracting DNA from clinical specimens. In this study, we sought to integrate all three modules onto a single paperfluidic chip, and demonstrate detection of a target nucleic acid analyte directly from crude clinical samples.
To showcase the full sample-to-answer functionality of the chip, we chose to design a test for cervical cancer, a disease that disproportionately affects the developing world where early detection is made difficult by a lack of screening methods suitable for these low-resource settings, and for which a rapid, low-cost, point-of-care (POC) molecular test of this nature is greatly needed. Each year, over half a million new cases of cervical cancer and over a quarter of a million deaths caused by cervical cancer occur worldwide 16,17. Despite its alarming global mortality, cervical cancer is highly preventable and easily treated through early detection and removal of precancerous lesions 16,18. Unfortunately, cervical cancer is most prevalent in areas where no effective screening programs have been established. From a total of 528,000 new cases worldwide in 2012, 445,000 cases (84%) occurred in the developing world 17, where, according to the World Health Organization (WHO), less than 5% of women have access to screening even once in their lifetime 19,20.
The traditional method to detect cervical cancer is the Pap smear, where trained pathologists analyze cervical smears under a microscope and determine if there is evidence of abnormal cells. Although cytology-based screening has dramatically reduced the incidence and mortality of cervical cancer in many developed countries, it is a qualitative process that suffers from very low sensitivity (as low as 53%) 21, and successful cytological screening programs have been difficult to implement and sustain in remote and low-income settings because they are resource-intensive and require highly trained personnel 22. Moreover, many developing regions lack the necessary infrastructure to transport Pap smears to and from a laboratory for processing and interpretation in a timely manner, thus requiring up to three patient visits to the clinic for screening, communication of results, and treatment. The delay in getting results back to patients is known to be a particularly significant barrier to screening, because large numbers of women won't return for results18. Efforts to reduce global cervical cancer incidence and mortality need to more effectively target developing regions via alternative methods that are less costly, less dependent on existing laboratory infrastructure and trained professionals, and capable of rapid, accurate diagnosis at POC.
Visual inspection with acetic acid (VIA) is a rapid and inexpensive detection method that has been promoted globally as a reasonable alternative to the Pap smear, but studies have revealed unacceptably low sensitivity and specificity, resulting in very high rates of false negative and false positive results 23,24. Moreover, like the Pap smear, VIA does not provide molecular information on the presence of the etiological agent of cervical cancer, human papillomavirus (HPV).
HPV is a common sexually transmitted infection, which in the majority of cases is transient, asymptomatic, and clinically insignificant. In some women, however, the infection becomes persistent and may lead to the development of cervical cancer. Over 99% of cervical cancer cases are caused by HPV 25, more than half of which are caused by the HPV 16 subtype26. Given the limitations of cytology, much work has been focused on molecular diagnostics for cervical cancer through HPV DNA testing. These methods typically have very high sensitivity (>96-100%) and specificity (>90-100%)21. A landmark study in rural India showed that a single round of HPV testing was associated with a significant reduction in the numbers of advanced cervical cancers and deaths from cervical cancer over time compared to cytology or VIA27. While these results validate the use of HPV DNA testing, a significant drawback is the high cost and the need for sophisticated laboratory equipment. Furthermore, current HPV DNA tests still require highly trained laboratory personnel and incur turnaround times of hours to days, depending on how far the sample has to travel to a central laboratory28,29,30.
Translating the molecular testing process to the POC can minimize these limitations by providing results faster, on the order of minutes, allowing doctors to diagnose, advise and potentially treat patients in the same visit. Asymptomatic patients positive for high-risk HPV strains like 16 could be screened more closely, thus allocating precious resources to those most at risk. A POC diagnostic device could be taken to remote settings beyond a standard clinic or laboratory, eliminating transport turnaround time. Additionally, a user-friendly, self-contained diagnostic device, with a simple visual readout similar to an at-home pregnancy test, could eliminate the need for highly trained specialists and require only minimal training of any community health worker.
Here, we present a paperfluidic chip made entirely of paper and adhesive sheets that combines nucleic acid extraction, isothermal loop-mediated amplification, and lateral flow detection via immunochromatographic strips that enable an immediate visual readout. This low-cost, portable, and disposable chip provides a simple, rapid molecular diagnostic platform for POC detection of nucleic acids. We developed a fully integrated, on-chip, sample-to-answer NAAT assay for the detection of HPV 16 DNA directly from patient cervical specimens in under an hour. This novel diagnostic platform could overcome many barriers currently faced in limited-resource settings and increase access to cervical cancer screening to those most in need.
MATERIALS AND METHODS
HPV 16 Cloned DNA Standards
HPV 16 DNA standards were generated by cloning the E7 gene for HPV 16 into the pGEM-T Easy Vector (Promega, Madison, WI). The E7 gene was PCR amplified from HPV-16 transformed cell DNA (Advanced Biotechnologies, Inc, Eldersburg, MD) with gene-specific forward and reverse cloning primers (Table S1) containing restriction endonuclease sequences SpeI and AatII, respectively, using the standard Taq Polymerase protocol (New England Biolabs, Ipswich, MA). The PCR product was purified via phenol chloroform extraction and ethanol precipitation. The cleaned PCR product was digested overnight with SpeI and AatII restriction endonucleases (New England Biolabs, Ipswich, MA). The relevant band was gel extracted and ligated to the pGEM vector and transformed into Top 10 cells from Life Technologies (Grand Island, NY). Plasmid DNA was extracted using a Mini Prep Kit (Qiagen, Valencia, CA) and sequenced (GeneWiz, Inc, Cambridge, MA) to confirm proper E7 insert. A Midi Prep Kit (Qiagen, Valencia, CA) was used to generate large scale plasmid stocks of the correctly sequenced DNA. The plasmid stocks were linearized with ZraI restriction endonuclease (New England Biolabs, Ipswich, MA). The correct size fragment was gel extracted using the QIAquick Extraction Kit (Qiagen, Valencia, CA), phenol chloroformed, and ethanol precipitated. The concentration of the purified DNA was determined by measuring the OD260 with the NanoDrop ND-2000c apparatus (Thermo Scientific, Waltham, MA). The DNA copy number was calculated and 1 mL aliquots were made and stored at −20°C.
Clinical Cervical Specimens
The cervical specimens were accrued from the BIDMC cytology laboratory, on already tested and to be discarded specimens. The IRB approval and patient consent for research use of these de-identified and discarded specimens was waived by the BIDMC Institutional Review Board. The specimens were obtained in PreservCyt® solution. Testing was done on an FDA approved plaform (Cervista; hrHPV), which evaluates 14 of the most common high-risk HPV genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) using Invader Chemistry. Any patient health identifying information was completely removed, and the specimens were labeled with a sample number and as HPV positive or negative only before they were transferred to the Klapperich Laboratory.
Samples were then transferred to 50 mL conical tubes and centrifuged for 10 min at 4000 RPM. The supernatant was removed and the cell pellet was washed with 3 mL PBS, vortexed and centrifuged for 10 min at 4000 RPM. This was repeated twice, leaving a cell pellet that was resuspended in 3 mL PBS and divided into (3) 1 mL aliquots. Each 1 mL aliquot was centrifuged for 5 min at 13000 RPM. The supernatant was removed and pellets were frozen at −80°C for long-term storage. Prior to use, pellets were resuspended in 1 mL PBS, subdivided into 200 ul aliquots, centrifuged for 10 min at 13000 RPM, and the supernatant was removed, resulting in single-use pellets for experiments. It is important to note that these centrifugation steps were required here because the patient samples we received as part of our IRB-approved study were cervical tissue specimens fixed in large volumes of ethanol-containing PreservCyt® solution, which is unsuitable for direct use in a POC device. In a real-world setting, a fresh or dried cervical swab would be placed directly into our lysis buffer, thereby eliminating these extra centrifugation steps.
For gold standard extraction experiments, DNA was extracted from a single-use pellet of each specimen using the DNeasy Blood & Tissue Kit (QIAGEN) and eluted into a final volume of 200 μL.
qPCR
To ascertain the DNA extraction yields, 5 ul of extracted DNA was amplified via quantitative PCR (qPCR). Using the Surestart Taq DNA polymerase (Agilent, Santa Clara, CA), real-time PCR was performed on an Applied Biosystems 7500 thermocyler under the following conditions: 10 min at 95°C for polymerase activation, followed by 30 cycles of 30 sec at 95°C, 15 sec at 55°C for primer annealing, and 90 sec at 60°C for amplification. The 25 μL reaction mixture contained 1X TaqMan buffer, 3.5mM MgCl2, 8% DMSO, 200uM dNTPs, 200nM primers and TaqMan probes (Table S1), 0.1X Rox Reference Dye, 0.625U Taq DNA polymerase, and 5 μL of sample or standard DNA. For clinical specimen gold standard extraction experiments, a multiplexed HPV 16 and RNaseP qPCR assay was run following the same reaction conditions where RNaseP served as a DNA control to confirm that each clinical specimen did in fact contain cells and that the Qiagen extractions were performed properly. If a clinical sample was negative for RNaseP (cycle threshold value > 30), the sample was deemed invalid and was not used for further experiments.
In each qPCR run, a cycle threshold value versus DNA concentration standard curve was generated from a dilution series of our cloned HPV 16 DNA standards. For each patient sample, the effective viral DNA concentration was quantitated via standard curve interpolations.
Isothermal Loop-Mediated Amplification Assay
An isothermal loop-mediated amplification (LAMP) assay was developed for rapid amplification and detection of the HPV 16 E7 gene using primer sequences previously designed by Luo et al.31 listed in Table S1. The assay was first optimized in tube, and then translated to a chip format. The assay takes place in situ, in a PES membrane in the sample inlet port, as previously described by our group14,32. The in-tube reaction was carried out in a final volume of 25 μl with 1 μl of the DNA sample, 1x Isothermal Amplification Buffer (New England Biolabs), 8 U large fragment Bst 2.0 DNA polymerase, 0.8 M Betaine, 1 mM dNTPs, 5 pmol each of forward and reverse outer primers (F3 and B3), 20 pmol each of forward and reverse loop primers (LF and LB), and 40 pmol each of forward and reverse inner primers (FIP and BIP). The on-chip LAMP reaction recipe was identical but was carried out in a final volume of 12.5 μL. Fresh, single-use aliquots of each reagent were used each time. The reaction was run for 30 minutes at 63°C. Forward and reverse loop primers (LF and LB) were tagged with Fluorescein isothiocyanate (FITC) and biotin, respectively, to enable immediate downstream detection of the amplified products on immunochromatographic, lateral flow detection (LFD) test strips (Ustar Biotechnologies, Hangzhou, China) consisting of streptavidin-conjugated gold nanoparticles, an anti-FITC test line, and a biotin (anti-streptavidin) flow control line.
For in-tube LAMP assay experiments, the amplified products were analyzed by 2% agarose gel electrophoresis. The specificity of the products was confirmed by restriction enzyme digestion with the PvuII restriction endonuclease (New England Biolabs) with a single cutting site within the FIP region. Following digestion at 37°C overnight, the digested products were analyzed by 2% agarose gel electrophoresis and by 10% acrylamide gel electrophoresis for higher resolution analysis.
LFD strips were imaged using an iPhone 5 camera (Apple). LFD test line and control line intensities were quantified using the Gel Analysis feature in ImageJ (National Institutes of Health) and analyzed by dividing the intensity of the test line by the intensity of the control line to obtain the percentage of control intensity for each sample. The resulting intensities were plotted as median with range. Unpaired, two-tailed Student's T-tests were used to determine the significance of each sample readout compared to the experimental negative control sample readout.
Paperfluidic Chip Fabrication
Standard letter size self-adhesive laminating sheets (Fellowes product # 5221502) served as the base material for building our paperfluidic chip, providing a hydrophobic (tape) barrier surrounding the paper components that is low-cost and optically transparent to enable our visual readout. We created blueprint drawings for the adhesive base of the chip (Figure 1b) using computer-aided design software (AutoCAD), and cut the adhesive sheets accordingly using an electronic craft cutting tool (Graphtec Craft Robo Pro S with Graphtec Studio software) using a standard blade (CB09U) and the following settings - cut force: 27, speed: 7 cm/s, acceleration: 1. The blueprint drawings include perforations in the adhesive sheets that were specifically designed to enable easily folding and ripping as needed during chip fabrication and operation, such that the integrity of the chip would remain uncompromised. The cut adhesive sheets were peeled from the protective backing and placed adhesive side-up on the benchtop as shown schematically in Figure 1c, step i. Polyethersulfone (PES) filter paper (Millipore, cat# GPWP04700) was cut into 0.375 inch diameter discs using a 3/8” craft hole punch (EK Tools, 54-10061). A single 0.375 inch diameter PES disc is manually placed directly over the 0.3 inch diameter hole in the adhesive sheet, and the top tab is folded down along the perforation over the PES (Figure 1c, step ii) to create the sample port (Figure 1c, step iii). Next, the bottom tab is folded up along the perforation to create a toehold for what will become the sample port cover film to prevent evaporation during the LAMP heat step (Figure 1c, step iv). The 0.3 inch diameter circle of tape that had been cut out of the adhesive sheet to make the sample port is peeled off the protective backing where it stayed behind and is manually placed adhesive side-down onto the adhesive sheet 2 inches down from the center of the sample port (Figure 1c, step v). This will align with the sample port when the cover film is placed during LAMP and prevent the DNA and/or PES membrane from sticking to the adhesive cover film.
Figure 1.
Paperfluidic Molecular Diagnostic Chip. a) Image of paperfluidic chip. Scale bar = 1 inch. b) Blueprint drawings and dimensions for the adhesive base of the chip. c) Schematic of chip fabrication steps: i. The cut adhesive sheets are peeled from the protective backing and placed adhesive side-up on the benchtop (the white area is adhesive sheet, the dark grey areas are holes that have been cut out of the adhesive sheet); ii. A 0.375 inch diameter PES disc is manually placed directly over the 0.3 inch diameter hole in the adhesive sheet, and the top tab is folded down along the perforation over the PES; iii. The sample port is now created (the light grey color indicates areas where the adhesive sheet has been folded over onto itself, rendering the area non-adhesive); iv. The bottom tab is folded up along the perforation to create a toehold for what will become the sample port cover film to prevent evaporation during the LAMP heat step; v. The 0.3 inch diameter circle of tape that had been cut out of the adhesive sheet to make the sample port is peeled off the protective backing where it stayed behind and is manually placed adhesive side-down onto the adhesive sheet 2 inches down from the center of the sample port. This will align with the sample port when the cover film is placed during LAMP and prevent the DNA and/or PES membrane from sticking to the adhesive cover film; vi. The absorbent pad is manually aligned and placed over the sample port extending towards the left side of the chip; vii-viii. The lower middle section of the chip is then folded over the centerline perforation over the absorbent pad to create a hydrophobic (tape) barrier between the absorbent pad and the LFD strip; ix. The LFD strip is then manually aligned with the sample port center and placed down extending over the right side of the chip; x-xi. The bottom two remaining sections of the adhesive sheet are folded up over the perforations to seal the chip from the bottom; xii. The fabrication is now complete, and the chip is then flipped over so that the PES membrane sample port is right side-up and ready for use.
Cellulose blotting paper (Whatman GB003, cat# 09-301-400) was cut using a 30W Epilog Zing laser cutter (speed=70%, power=28%, frequency=200) to make absorbent pads shaped as 2.5 inch long sectors that extend radially from 0.375 inch at the base of the sample port to an ultimate width of 0.75 inch (drawn in SolidWorks, Waltham, MA). The absorbent pad is manually aligned and placed over the sample port extending towards the left side of the chip as shown in Figure 1c, step vi. The lower middle section of the chip is then folded over the centerline perforation over the absorbent pad as shown in Figure 1c, steps vii-viii to create a hydrophobic (tape) barrier between the absorbent pad and the LFD strip. The LFD strip is then manually aligned with the sample port center and placed down extending over the right side of the chip as shown in Figure 1c, step ix. Next, the bottom two remaining sections of the adhesive sheet are folded up over the perforations to seal the chip from the bottom (Figure 1c, steps x-xi). The fabrication is now complete, and the chip is then flipped over so that the PES membrane sample port is right side-up and ready for use (Figure 1c, step xii).
Integrated On-Chip Assay
DNA Extraction and Purification
A single-step cell lysis and DNA extraction recipe was developed based on the chaotropic lysis and alcohol precipitation methods of Boom et al.33 and Linnes et al.13 A single-use pellet of each clinical cervical specimen (or 6 μL of cloned HPV16 DNA during preliminary experiments) was resuspended in a lysis buffer containing 3M guanidinium thiocyanate, 300 mM sodium chloride, 35% v/v 1-butanol (Sigma Aldrich, St. Louis, MO), and 3 μl of 15 mg/mL Glycoblue coprecipitant (Life Technologies, Grand Island, NY) in a total volume of 100 μL. This mixture was pipetted onto the sample port of the paperfluidic chip. The liquid phase wicks through the absorbent pad directly underneath the PES membrane by capillary forces, leaving the precipitated DNA-Glycoblue solid phase. A series of ethanol washes (200 μL of 70% ethanol, followed by 100 μL of 100% ethanol) were then pipetted through the sample port, removing impurities while leaving the purified DNA-glycogen precipitate on the PES membrane. The left side of the chip containing the soiled absorbent pad was then ripped along the perforation and discarded. The rest of the chip is left on the benchtop for at least 2 minutes to allow for complete drying of the ethanol prior to addition of the LAMP reaction mix.
Isothermal Amplification
A 12.5 μl LAMP reaction mix was pipetted directly onto the sample port and was fully absorbed by the PES, presumably dissociating the DNA-Glycoblue complexes. The bottom tab of the chip is then folded up along the perforation and pressed down to seal over the absorbent pad and serves as a cover film to prevent evaporation during the incubation period for LAMP. The chip is then placed face-down on a 63°C heat block or hot plate for 30 min.
Lateral Flow Detection
Following the LAMP incubation, the cover film was peeled back using the toehold tab to expose the sample port on top, and peeled under the chip to expose the sample port outlet on the bottom, thereby removing the hydrophobic (tape) barrier between the sample port and the LFD strip (see side view schematic in Figure S1). 50 μl of nuclease free water was then pipetted onto the sample port, which filtered through the PES and wicked directly onto the LFD strip for immediate detection of amplified products.
RESULTS AND DISCUSSION
Fluidic Demonstration of Chip Operation
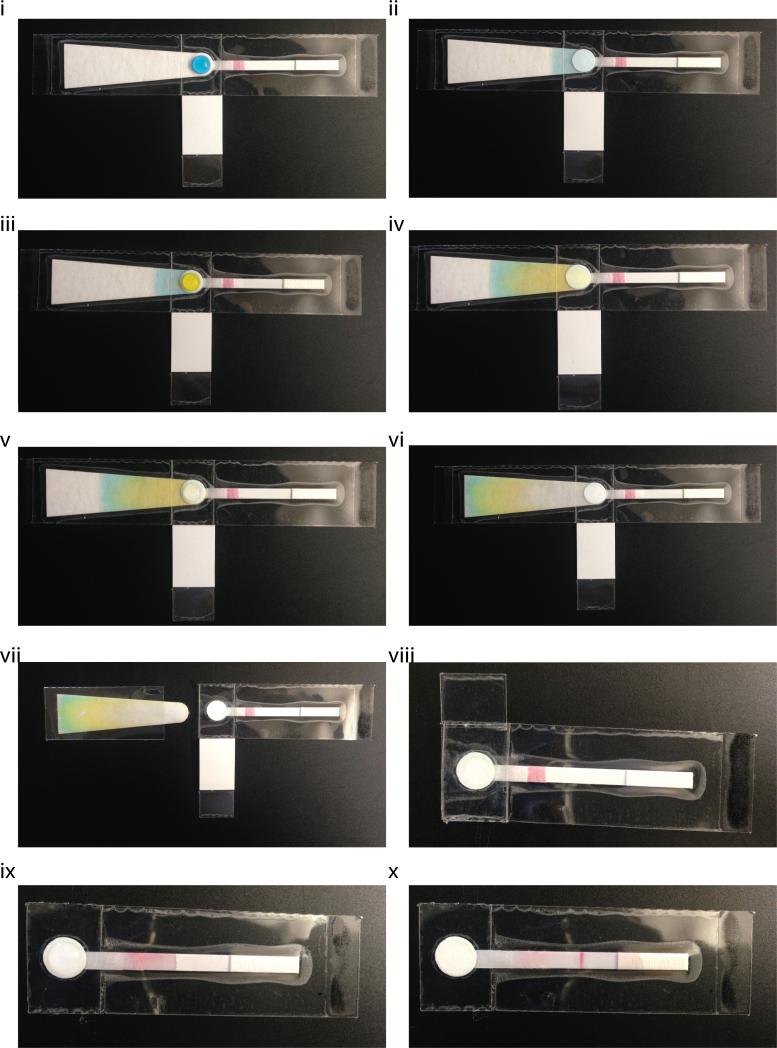
A lysed sample, demonstrated here using 100 μL of blue dye, is placed onto the sample port of the paperfluidic chip using a pipette or dropper (Figure 2i). The prevailing capillary forces generated by the absorbent pad directly underneath the sample port quickly wick the liquid waste through the PES membrane and away from the sample port (Figure 2ii). A hydrophobic (tape) barrier between the absorbent pad and the LFD strip prevents the liquid waste from wicking through to the LFD strip (see side view schematic in Figure S1). Any solid phase within the sample, most importantly the precipitated DNA, will remain on the sample port surface. Next, a first wash of 70% ethanol, demonstrated here using 200 μL of yellow dye, is filtered through the sample port (Figure 2iii-iv). The wash buffer will wick through to the absorbent pad, removing most impurities like cell debris, proteins, and salts and leaving behind the purified precipitated DNA. Because ethanol can inhibit the subsequent LAMP reaction, it is important to completely dry the sample port. To that end, a final wash of 100% ethanol, demonstrated here using 100 μL water, is filtered through the sample port (Figure 2v), leaving just the purified DNA precipitates on the PES membrane (Figure 2vi).
Figure 2.
Fluidic Demonstration of Chip Operation. i) A lysed sample, demonstrated here using 100 μL of blue dye, is placed onto the sample port of the paperfluidic chip using a pipette or dropper . ii) The prevailing capillary forces generated by the absorbent pad directly underneath the sample port quickly wick the liquid waste through the PES membrane and away from the sample port leaving the solid phase behind. iii) A first wash of 70% ethanol, demonstrated here using 200 μL of yellow dye, is filtered through the sample port. iv) The wash buffer will wick through to the absorbent pad, removing impurities and leaving behind the purified precipitated DNA. v-vi) A final wash of 100% ethanol, demonstrated here using 100 μL water, is filtered through the sample port, leaving just the purified DNA on the PES membrane. vii) The waste absorbent pad is discarded by ripping off the left side of the chip at the designated perforation. viii) The LAMP reaction mix is placed directly onto the sample port where the purified DNA remains, and the bottom tab of the chip is folded up over the designated perforation to act as a cover film for the sample port and prevent evaporation during the heat step. ix) After the heat incubation for LAMP, the cover film is peeled back using the toehold to expose the sample port on top, and peeled under the chip to expose the sample port outlet on the bottom, thereby removing the hydrophobic (tape) barrier between the sample port and the LFD strip. The PES membrane is now in direct contact with the LFD strip and the amplified products are then eluted onto the strip by adding 50 μL water to the sample port. x) The eluted products wick through the LFD strip towards the right.
The waste absorbent pad is no longer needed at this point and can be discarded by ripping off the left side of the chip at the designated perforation (Figure 2vii). Next, 12.5 μL of the LAMP reaction mix is placed directly onto the sample port where the purified DNA remains, and the bottom tab of the chip is folded up over the designated perforation to act as a cover film for the sample port and prevent evaporation during the heat step (Figure 2viii). The chip is then placed face-down onto a heat block or hot plate set to 63°C for 30 min (not shown). After the heat incubation, the cover film is peeled back using the toehold to expose the sample port on top, and peeled under the chip to expose the sample port outlet on the bottom, thereby removing the hydrophobic (tape) barrier between the sample port and the LFD strip. The PES membrane is now in direct contact with the LFD strip and the amplified products are then eluted onto the strip by adding 50 μL water to the sample port (Figure 2ix). The eluted products wick through the LFD strip towards the absorbent pad on the right. As the liquid wicks through the conjugate pad, the streptavidin-conjugated gold nanoparticles bind the biotin probes on the LB primers within the amplicons. As the liquid continues to wick over the detection zone, amplicons that also contain the FITC probe on the LF primers will aggregate at the anti-FITC test line. Any excess streptavidin-conjugated gold nanoparticles will continue to wick through the LFD strip and bind the biotin control line, which confirms that the strip functioned properly. Here, as this was just a demonstration using water, the test result is obviously negative, thus only the control line appears on the strip (Figure 2x).
HPV 16 E7 LAMP Assay
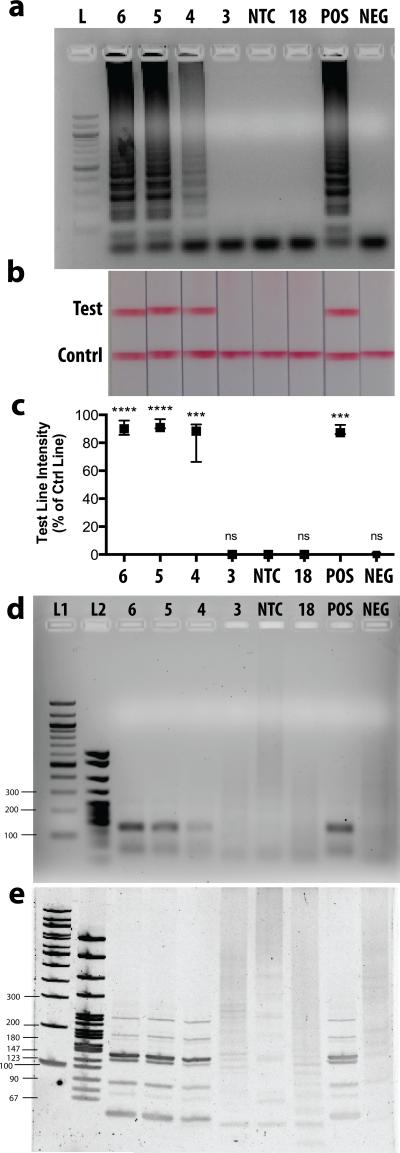
The HPV 16 E7 LAMP assay was first optimized in-tube using our cloned HPV16 DNA standards. We ran the optimized LAMP assay using serial dilutions of our DNA standards and found that our lower limit of detection was 10^4 total copies as confirmed by agarose gel electrophoresis (Figure 3a) and LFD strips (Figure 3b). The LFD strips enable immediate detection of amplified products with the naked eye. Test line intensities were quantified as a percentage of control line intensities and results from three independent experiments are plotted in Figure 3c. DNA quantities down to 10^4 total copies show a clear visible test line that is statistically different from the negative control. While 10^3 DNA copies were not amplified to detectable levels, the HPV literature has shown that a viral load below 10^4 copies is not indicative of cervical cancer progression34,35.
Figure 3.
HPV 16 E7 LAMP Assay in solution. a) 2% Agarose gel electrophoresis of LAMP products. L= 100bp DNA Ladder, 6 = 10^6 DNA copies, 5 = 10^5 DNA copies, 4 = 10^4 DNA copies, 3 = 10^3 DNA copies, NTC = no template control, 18 = 10^6 copies of HPV 18 DNA, POS = DNA extracted from an HPV16-positive patient sample, NEG = DNA extracted from an HPV16-negative patient sample. b) Representative lateral flow strips from three independent experiments show detection of LAMP products. Top line is the test line, bottom line is the flow strip control line. c) Test line intensity as percentage of control line intensity for three experiments is plotted as median with range and statistically compared to NTC control (*** p < 0.001, **** p < 0.0001, ns = not significant). d) 2% Agarose gel electrophoresis of PvuII-digested LAMP products. L1= 100bp DNA Ladder, L2 = PBR322 DNA-Msp1-digest Ladder. e) 10% Acrylamide gel electrophoresis of Pvu-II digested LAMP products.
We included a no template control (NTC), and a nonspecific DNA control (10^6 total copies of HPV 18 DNA), both of which were negative on both the gel and LFD strips, demonstrating primer specificity. Additionally, we ran our LAMP assay on Qiagen kit-extracted DNA from a patient sample that tested positive for HPV 16 and from a clinical sample that tested negative for HPV 16. It is important to note that these samples contain large amounts of human DNA and potentially other viral genomes. Our LAMP assay correctly identified these patient samples as positive and negative, respectively, thus further confirming the specificity of our LAMP assay.
One disadvantage to the LAMP method is the possibility for interaction and self-priming of the oligonucleotides during the reaction. This phenomenon has been widely reported in the literature36-39, and is usually circumvented by optimization of assay conditions and setting an assay cutoff time far before these events are likely to occur. Nonetheless, because our ultimate assay detection method is based on primer-tagged probes, it was important to ensure that a positive result on the LFD strip correlated to a LAMP product specific to our target sequence. To this end, the amplified products were digested with the PvuII restriction endonuclease and analyzed by 2% agarose gel electrophoresis (Figure 3d) and at higher resolution by 10% acrylamide gel electrophoresis (Figure 3e). The HPV 16 E7 gene sequence contains a single PvuII cutting site within the FIP region, and positive product digests were in agreement with the expected product band sizes40, while the negative product digests showed nothing on a low-resolution agarose gel (Figure 3d) and showed irregular band patterns inconsistent with expected product band sizes on a high-resolution acrylamide gel (Figure 3e).
Having confirmed the lower detection limit and specificity of our HPV16 LAMP assay in-tube, we then tested our LAMP assay in situ, directly within a paper matrix containing freshly extracted HPV16 DNA. Our group has previously reported on the successful isothermal amplification of nucleic acids within PES matrices32, as well as on the successful extraction of nucleic acids in PES followed by in situ amplification14. To test our HPV16 LAMP assay in situ, first we extracted solutions of known concentrations of HPV16 DNA mixed with our Glycoblue-containing lysis buffer through a PES membrane using an acrylic extraction setup previously described14. The extracted DNA was eluted from the PES matrices and quantified via qPCR (Figure S2a). Recovery yields were between 46% and 88% of centrifugation controls, consistent with what our group had previously reported for RNA. Next, HPV16 DNA solutions were again extracted through a PES membrane as described above, but this time instead of eluting the extracted DNA from the PES, 12.5 μL of our LAMP reaction mix was pipetted directly onto the PES membrane where it was fully absorbed. The PES disc was placed inside of a tube and incubated at 63°C for 30min. The amplified products were eluted via centrifugation from the PES membrane and analyzed via 2% agarose gel electrophoresis. As shown in Figure S2b, our LAMP assay successfully amplified as low as 10^4 total copies of HPV 16 DNA in situ.
Integrated On-Chip Assay with cloned HPV 16 DNA standards
Once we confirmed adequate extraction and amplification of HPV 16 DNA in a PES membrane, we then integrated each assay component onto our paperfluidic chip, following the step-by-step protocol outlined in Figure 2 and using solutions of known concentrations of our cloned HPV 16 DNA standards. On-chip extractions took approximately 10-15 minutes, as flow was significantly slowed after a visible blue DNA-Glycoblue film developed on the PES membrane following the initial sample filtration (images shown in Figure S3a). We also found that we needed to dispense the 100 μL sample only 50 μL at a time due to the lower surface tension of our lysis buffer containing 35% butanol, otherwise the liquid would spill over the sample port. Likewise, our 70% ethanol wash was dispensed 25 μL at a time, and the 100% ethanol wash was dispensed 10 μL at a time. Following the extraction and ripping off of the waste pad, the visible Glycoblue-containing dry precipitates were observed only on the PES membrane, and not significantly on the absorbent pad underneath, suggesting good recovery (Figure S3b). The LAMP reaction mix was then added directly to the dry sample port and immediate mixing with the Glycoblue-containing precipitates was observed. Following the 30 min heat step, the cover film tab was peeled back and the LAMP reaction liquid was visibly still present on the sample port, suggesting minimal evaporation. Following addition of 50 μL water to the sample port, elution onto the LFD strip began immediately, and test results were visible within 2 min.
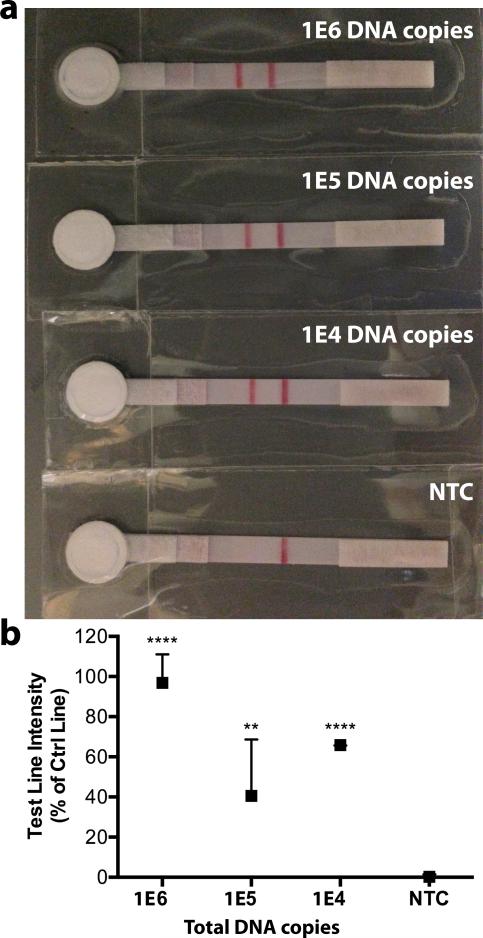
As shown in Figure 4a, our integrated on-chip assay resulted in clear, positive LFD readouts from starting material as low as 10^4 copies of HPV 16 DNA, and a clear negative readout for the negative control. The statistical analysis from three independent experiments is shown in Figure 4b.
Figure 4.
Integrated on-chip assay with cloned HPV 16 DNA standards. a) Representative lateral flow strips from three independent on-chip experiments show detection of LAMP products from 1E4 to 1E6 DNA copies, NTC = no template control. Left line is the test line, right line is the flow strip control line. b) Test line intensity as percentage of control line intensity for three experiments is plotted as median with range and statistically compared to NTC control (** p < 0.01, *** p < 0.001, **** p < 0.0001).
Integrated On-Chip Assay with Clinical Cervical Specimens
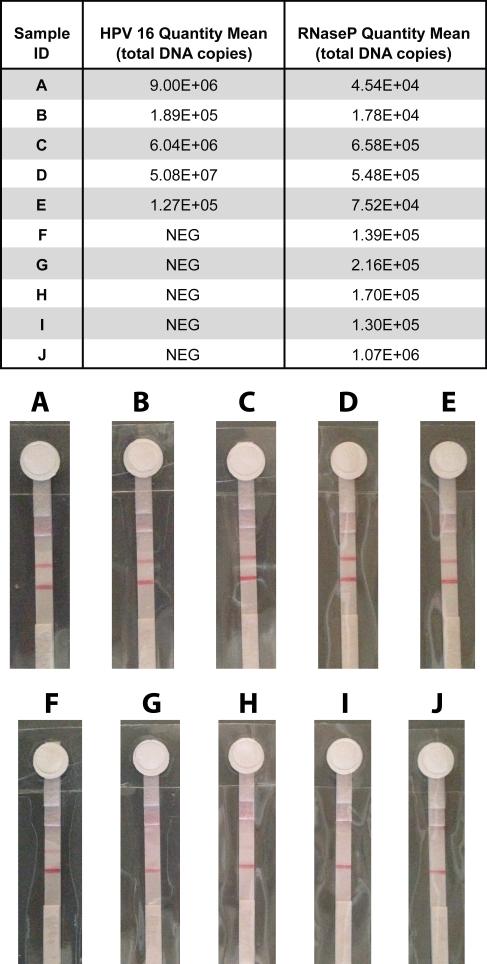
DNA from cervical tissue sample pellets was extracted via the gold standard Qiagen DNeasy Tissue Kit. Each sample extraction was analyzed by qPCR for HPV16 E7 and RNaseP DNA. RNaseP serves as a human gene internal control to ensure that the cervical swab sample contained cervical cells and that DNA was properly extracted. Any samples that tested negative for RNaseP by qPCR were considered “invalid” and were not used in further experiments. Five HPV 16 positive and five HPV 16 negative samples (Figure 5 table) were selected for on-chip testing to demonstrate proof-of-concept clinical utility of our paperfluidic chip.
Figure 5.
Integrated on-chip assay with clinical cervical specimens. Table lists gold standard assay (Qiagen-extraction quantified by qPCR) results for HPV16 and RNaseP control DNA quantities for each of 10 patient samples labeled A-J. NEG = negative result. Chip LFD strip images for each sample A-J below.
A single-use pellet from each sample A-J was resuspended in 100 μL lysis buffer, vortexed thoroughly, and pipetted onto the sample port of the chip. During preliminary experiments, significant accumulation of debris and salts from the lysed samples left a visible grainy film on the PES membrane, which greatly inhibited the subsequent LAMP reaction. This prompted an increase from 100 to 200 μL of our 70% ethanol washes, which did not entirely remove the residue in all cases, but significantly improved LAMP performance nonetheless.
All 5 positive samples resulted in clear, positive LFD results as seen in Figure 5 strips A-E. Of the 5 negative samples, 3 resulted in a negative LFD result (strips G, I, J), but 2 exhibited faint test lines (F, H) resulting in a false positive result. As mentioned above, interaction and self-priming of two or more of the six primers involved in LAMP is a widely recognized issue that may be the cause of our false positive results. Considering our LFD strip detection method, moving forward we suspect a sequence-specific probe (not primer-tagged), as reported by Curtis et al. 41, would be better suited for this type of assay.
It is also possible that cell debris and salts in the cervical samples left behind on the sample port are affecting our primer melting temperatures in ways we are unable to account for. While further primer and/or probe optimization may be required, herein we have demonstrated that a fully integrated, sample-to-answer, molecular diagnostic assay on a low-cost, disposable paperfluidic chip platform is possible.
Furthermore, it is known that with HPV testing, clinical false-positive results (positive screening tests without underlying precancerous lesions) are common42, largely due to transient levels of HPV infection in any given population that may never develop into cervical intraepithelial neoplasias or cancer. Consequently, American Cancer Society guidelines do not recommend screening by HPV testing alone for most clinical settings, but as cotesting with cytology every 5 years in women over 30 43. In resource-limited settings, where cotesting is likely not an option, our paperfluidic diagnostic platform allows for primary HPV screening, where a positive result can be followed up by cytology, thereby allocating precious resources to those most at risk. A recent systematic review concluded that HPV testing alone is very promising for the primary screening of women aged 30 years and older, particularly when coupled with cytology testing for follow-up of HPV-positive results, which may reduce the number of false-positive findings that would result from HPV testing 29,44. A study in Brazil indicated that HPV testing followed by cytology triage could be a very cost-effective strategy for lower-middle income countries, results reflecting a synergistic effect between a highly sensitive initial screening test (HPV DNA) in sequence with a highly specific test (cytology)45. In extremely resource-limited countries, use of a simple, low-cost HPV DNA test like the one presented here, followed by immediate ‘screen and treat’ strategies based on low-cost methods like VIA in those who test positive, is likely to minimize the number of patient visits and make best use of limited resources46.
Future work will be required to address some of the current limitations of the device before implementation in the field can be realized. Currently, the chip requires an external source of heat for the isothermal amplification step. The work presented here was performed using a heat block but this can also be achieved through several methods that do not require electricity such as battery operated resistance heaters47 or exothermic reactions like those in simple toe warmers48. Nonetheless, incorporating an integrated heating system within the paperfluidic platform would greatly increase its portability and usability. An additional limitation to the current chip design is that it requires a significant amount of pipetting and manipulation by the user. A number of variables can be optimized to limit the number of steps, for example, increasing the surface area of the sample port, or optimizing buffers and assay conditions to reduce the volume needed.
CONCLUSION
We present the first ever fully integrated rapid molecular diagnostic made entirely of paper and tape. Our paperfluidic chip offers an equipment-free, modular NAAT platform that is low-cost, easy to manufacture, and simple to use. We demonstrated full sample-to-answer functionality of our chip by implementing an HPV 16 DNA extraction, amplification, and detection assay directly from patient cervical samples. Our on-chip HPV 16 assay addresses many of the limitations of conventional cytology by providing highly sensitive molecular level information regarding the presence of high-risk HPV 16 in cervical samples without the need for laboratory infrastructure or highly trained pathologists. The device also addresses the limitations of current molecular diagnostic techniques by allowing for rapid, point-of-care detection in less than an hour without the need for transportation to/from a laboratory. Additionally, this paperfluidic chip can serve as a molecular diagnostic platform for any disease, requiring only changing of primer sequences and the corresponding optimization of LAMP assay conditions. The device is made from low-cost paper and tape materials, making it inexpensive, portable, and disposable, allowing it to be applied in low-resource settings and reach a larger pool of patients who may otherwise not get diagnosed due to insufficient resources and personnel.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Dr. Christopher Chen, Dr. Mario Cabodi, Dr. Jacqueline Linnes, Dr. Sharon Wong, and Dr. Andy Fan for helpful discussions, and Angelina Jockovich for her assistance with AutoCAD. NMR acknowledges funding from a National Science Foundation Graduate Research Fellowship and a Diversity Administrative Supplement under NIH grant U54-EB015403-S1.
Footnotes
ELECTRONIC SUPPLEMENTARY INFORMATION (ESI) available: Additional figures containing primer sequences, a chip sideview schematic, HPV 16 DNA extractions and in situ LAMP in PES results, and on-chip extraction images are included.
REFERENCES
- 1.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 2.Yetisen AK, Akram MS, Lowe CR. Lab Chip. 2013;13:2210. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 3.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 4.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Angew. Chem. Int. Ed. Engl. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez AW, Phillips ST, Whitesides GM. Proceedings of the National Academy of Sciences. 2008;105:19606–19611. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ, Xu F. Biosensors and Bioelectronic. 2014;54:585–597. doi: 10.1016/j.bios.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 7.Jangam SR, Yamada DH, McFall SM, Kelso DM. J. Clin. Microbiol. 2009;47:2363–2368. doi: 10.1128/JCM.r00092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govindarajan AV, Ramachandran S, Vigil GD, Yager P, Böhringer KF. Lab Chip. 2011;12:174. doi: 10.1039/c1lc20622b. [DOI] [PubMed] [Google Scholar]
- 9.Ali MM, Aguirre SD, Xu Y, Filipe CDM, Pelton R, Li Y. Chem. Commun. 2009:6640. doi: 10.1039/b911559e. [DOI] [PubMed] [Google Scholar]
- 10.Rohrman BA, Richards-Kortum RR. Lab Chip. 2012;12:3082. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan W, Zhuang Bin, Zhang P, Han J, Li C-X, Liu P. Lab Chip. 2014:1–11. doi: 10.1039/c4lc00686k. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Geva E, Mauk M, Qiu X, Abrams WR, Malamud D, Curtis K, Owen SM, Bau HH. Analyst. 2011;136:2069. doi: 10.1039/c1an00007a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linnes JC, Fan A, Rodriguez NM, Lemieux B, Kong H, Klapperich CM. RSC Adv. 2014;4:42245–42251. doi: 10.1039/C4RA07911F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez NM, Linnes JC, Fan A, Ellenson CK, Pollock NR, Klapperich CM. Anal. Chem. 2015;87:7872–7879. doi: 10.1021/acs.analchem.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connelly JT, Rolland JP, Whitesides GM. Anal. Chem. 2015;87(15):7595–7601. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- 16.Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 17.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Int. J. Cancer. 2014;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 18.Goldie SJ, Gaffikin L, Goldhaber-Fiebert JD, Gordillo-Tobar A, Levin C, Mahé C, Wright TC. Alliance for Cervical Cancer Prevention Cost Working Group. New Engl J Med. 2005;353:2158–2168. doi: 10.1056/NEJMsa044278. [DOI] [PubMed] [Google Scholar]
- 19.Denny L, Quinn M, Sankaranarayanan R. Vaccine. 2006;24:S71–S77. doi: 10.1016/j.vaccine.2006.05.121. [DOI] [PubMed] [Google Scholar]
- 20.Denny L. Reproductive Health Matters. 2008;16:18–31. doi: 10.1016/S0968-8080(08)32397-0. [DOI] [PubMed] [Google Scholar]
- 21.Ying H, Jing F, Fanghui Z, Youlin Q, Yali H. Sci. Rep. 2014:4. doi: 10.1038/srep04704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldie SJ. Virus research. 2002;89:301–309. doi: 10.1016/s0168-1702(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 23.Goel A, Gandhi G, Batra S, Bhambhani S, Zutshi V, Sachdeva P. International Journal of Gynecology & Obstetrics. 2005;88:25–30. doi: 10.1016/j.ijgo.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Hegde D, Shetty H, Shetty PK, Rai S. J Cancer Res Ther. 2011;7:454–458. doi: 10.4103/0973-1482.92019. [DOI] [PubMed] [Google Scholar]
- 25.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 26.Clifford G, Franceschi S, Diaz M, Muñoz N, Villa LL. Vaccine. 2006;24:S26–S34. doi: 10.1016/j.vaccine.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 27.Sankaranarayanan R, Nene BM, Shastri SS, Jayant K, Muwonge R, Budukh AM, Hingmire S, Malvi SG, Thorat R, Kothari A, Chinoy R, Kelkar R, Kane S, Desai S, Keskar VR, Rajeshwarkar R, Panse N, Dinshaw KA. New Engl J Med. 2009;360:1385–1394. doi: 10.1056/NEJMoa0808516. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn L, Denny L, Pollack A, Lorincz A, Richart RM, Wright TC. J. Natl. Cancer Inst. 2000;92:818–825. doi: 10.1093/jnci/92.10.818. [DOI] [PubMed] [Google Scholar]
- 29.Mayrand MH, Duarte-Franco E, Rodrigues I, Walter SD, Hanley J, Ferenczy A, Ratnam S, Coutlée F. New Engl J Med. 2007;357:1579–88. doi: 10.1056/NEJMoa071430. [DOI] [PubMed] [Google Scholar]
- 30.Villa LL, Denny L. International Journal of Gynecology & Obstetrics. 2006;94:S71–S80. doi: 10.1016/S0020-7292(07)60013-7. [DOI] [PubMed] [Google Scholar]
- 31.Luo L, Nie K, Yang MJ, Wang M, Li J, Zhang C, Liu HT, Ma XJ. J. Clin. Microbiol. 2011;49:3545–3550. doi: 10.1128/JCM.00930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linnes JC, Rodriguez NM, Liu L, Klapperich CM. Polyethersulfone improves the efficiency of nucleic acid amplification compared to current paper-based diagnostic materials. 2015. Submitted. [DOI] [PMC free article] [PubMed]
- 33.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. J. Clin. Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Duin M, Snijders PJF, Schrijnemakers HFJ, Voorhorst FJ, Rozendaal L, Nobbenhuis MAE, van den Brule AJC, Verheijen RHM, Helmerhorst TJ, Meijer CJLM. Int. J. Cancer. 2002;98:590–595. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 35.Swan DC, Tucker RA, Tortolero-Luna G, Mitchell MF, Wideroff L, Unger ER, Nisenbaum RA, Reeves WC, Icenogle JP. J. Clin. Microbiol. 1999;37:1030–1034. doi: 10.1128/jcm.37.4.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, Sugimoto C, Igarashi I. J. Clin. Microbiol. 2003;41:5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inacio J, Flores O, Spencer-Martins I. J. Clin. Microbiol. 2008;46:713–720. doi: 10.1128/JCM.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh H-Y, Shoemaker CA, Klesius PH. Journal of Microbiological Methods. 2005;63:36–44. doi: 10.1016/j.mimet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Teng P-H, Chen C-L, Sung P-F, Lee F-C, Ou B-R, Lee P-Y. Journal of Virological Methods. 2007;146:317–326. doi: 10.1016/j.jviromet.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curtis KA, Rudolph DL, Owen SM. J. Med. Virol. 2009;81:966–972. doi: 10.1002/jmv.21490. [DOI] [PubMed] [Google Scholar]
- 42.Rebolj M, Pribac I, Lynge E. European Journal of Cancer. 2011;47:255–261. doi: 10.1016/j.ejca.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Saslow D, Solomon D, Lawson HW, Killackey M, Kulasingam SL, Cain J, Garcia FAR, Moriarty AT, Waxman AG, Wilbur DC, Wentzensen N, Downs LS, Spitzer M, Moscicki AB, Franco EL, Stoler MH, Schiffman M, Castle PE, Myers ER. American Journal of Clinical Pathology. 2012;137:516–542. doi: 10.1309/AJCPTGD94EVRSJCG. [DOI] [PubMed] [Google Scholar]
- 44.Agency for Healthcare Research and Quality (AHRQ) AHRQ Publication No. 11-05156-EF-1. 2011.
- 45.Vanni T, Luz PM, Grinsztejn B, Veloso VG, Foss A, Mesa-Frias M, Legood R. Int. J. Cancer. 2011;131:E96–E104. doi: 10.1002/ijc.26472. [DOI] [PubMed] [Google Scholar]
- 46.Cuzick J, Arbyn M, Sankaranarayanan R, Tsu V, Ronco G, Mayrand M-H, Dillner J, Meijer CJLM. Vaccine. 2008;26:K29–K41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Herold KE, Sergeev N, Matviyenko A, Rasooly A. Methods in Molecular Biology. Vol. 504. Humana Press; Totowa, NJ: 2009. pp. 441–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang S, Do J, Mahalanabis M, Fan A, Zhao L, Jepeal L, Singh SK, Klapperich CM. PLoS ONE. 2013;8:e60059. doi: 10.1371/journal.pone.0060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.