Abstract
Rationale
In Drosophila, the Hippo signaling pathway negatively regulates organ size by suppressing cell proliferation and survival through the inhibition of Yorkie, a transcriptional co-factor. Yes-associated protein (YAP), the mammalian homolog of Yorkie, promotes cardiomyocyte growth and survival in postnatal hearts. However, the underlying mechanism responsible for the beneficial effect of YAP in cardiomyocytes remains unclear.
Objectives
We investigated whether miR-206, a microRNA known to promote hypertrophy in skeletal muscle, mediates the effect of YAP upon promotion of survival and hypertrophy in cardiomyocytes.
Methods and Results
Microarray analysis indicated that YAP increased miR-206 expression in cardiomyocytes. Increased miR-206 expression induced cardiac hypertrophy and inhibited cell death in cultured cardiomyocytes, similar to that of YAP. Down regulation of endogenous miR-206 in cardiomyocytes attenuated YAP-induced cardiac hypertrophy and survival, suggesting that miR-206 plays a critical role in mediating YAP function. Cardiac-specific overexpression of miR-206 in mice induced hypertrophy and protected the heart from ischemia/reperfusion (I/R) injury, whereas suppression of miR-206 exacerbated I/R injury and prevented pressure overload-induced cardiac hypertrophy. miR-206 negatively regulates FoxP1 expression in cardiomyocytes and overexpression of FoxP1 attenuated miR-206-induced cardiac hypertrophy and survival, suggesting that FoxP1 is a functional target of miR-206.
Conclusions
YAP increases the abundance of miR-206, which in turn plays an essential role in mediating hypertrophy and survival by silencing FoxP1 in cardiomyocytes.
Keywords: Hippo pathway, YAP, microRNA, heart, apoptosis, signal transduction, cardiac hypertrophy
INTRODUCTION
The Drosophila Hippo pathway is an evolutionarily conserved signaling cascade and a key regulator of tissue growth and organ size1, 2. Activation of the core complex (consisting of the kinases Hippo and Warts and the adapter proteins Salvador and Mats) leads to phosphorylation and inhibition of Yorkie, the transcription co-factor and terminal effector of the Hippo pathway. Conversely, Hippo pathway loss of function, or direct phosphorylation by homeodomain interacting protein kinase 2 (HIPK2), can elicit Yorkie activation3. Through association and activation of the transcription factor Scalloped, Yorkie regulates expression of cyclin E, diap1 and the microRNA (miRNA) bantam to promote proliferation and inhibit apoptosis (reviewed in1, 2). Hippo pathway components are highly conserved between flies and mammals, and mammalian homologs including Mst1/2 (Hippo), Lats1/2 (Warts), Yes-associated protein (YAP;Yorkie) and TEAD family transcription factors (Scalloped) exhibit similar, but not identical, regulation and function outputs (reviewed in4).
Elucidation of Hippo signaling and the functional implications of Hippo modulation in the heart are highly relevant and of great interest. Our previous work demonstrated that transgenic mice with cardiac-specific overexpression of Mst1 develop dilated cardiomyopathy with increased cardiomyocyte (CM) apoptosis. Interestingly, CM size in these mice was smaller than non-transgenic controls despite dilation of the heart and increased wall stress. These findings suggested that activation of Hippo promotes CM apoptosis while compensatory hypertrophy is suppressed by Mst15. We also observed that Lats2, a negative regulator of YAP, inhibits both hypertrophy and survival of CMs during pressure overload stress 6. Furthermore, disruption of endogenous cardiac YAP led to impaired compensatory hypertrophy, proliferation, and survival of CMs during chronic myocardial infarction (MI) and ischemia/reperfusion (I/R)7, 8. On the other hand, overexpression of constitutively active YAP promoted myocardial regeneration by stimulating CM proliferation 9, 10. Previous reports have demonstrated the importance of crosstalk between Hippo and IGF-Wnt/β-catenin pathways for CM proliferation during heart development 11, 12. These results suggest that the Hippo pathway plays an essential role in CM biology and that YAP is an important mediator of the actions of the Hippo pathway in the heart. However, the mechanisms underlying how Hippo/YAP modulates CM growth and survival remain largely undefined.
miRNAs regulate gene expression to affect cell growth, proliferation and survival, and are important mediators of heart development, homeostasis and disease13. miR-206 belongs to the miR-1/miR-206 family of miRNAs. miR-206 is expressed in several known tissues/cell types including the brain 14, 15, cancer cell lines 16 and brown adipocytes 17 and mediates multiple functions, including tissue growth and differentiation 14, 15, 17, 18, suppression of angiogenesis 19, as well as tumor suppression 16. miR-206 and miR-1 have been shown to form clusters with miR-133b and miR-133a, respectively, and are critical factors for skeletal and cardiac muscle development and function (reviewed in 20). In skeletal muscle, miR-206 positively regulates myogenesis 21–23, hypertrophy 24 and regeneration, and delays the progression of amyotrophic lateral sclerosis 25 and Duchenne muscular dystrophy 26. These findings suggest that miR-206 is an important modulator of striated muscle growth. Importantly, miR-206 is also present in CMs27, but the cellular function of miR-206 in these cells is not well understood.
In our current study, we explored the possibility that YAP regulates expression of miRNAs that modulate CM growth and survival. Through a microarray-based screening of miRNAs that showed altered expression in response to YAP, we found that the abundance of miR-206 was increased by YAP in CMs. Despite some functional overlap between YAP and miR-206, a link between them has not been demonstrated. Therefore, the goals of this work were (i) to investigate the role of miR-206 in mediating the pro-hypertrophic and anti-apoptotic actions of YAP in CMs and (ii) to identify the functional target of miR-206 in CMs. Our results demonstrate that miR-206 mediates hypertrophy and cell survival stimulated by YAP through targeted degradation of the tumor suppressor, Forkhead box protein P1 (FoxP1).
METHODS
An expanded Methods section is available in the online Data Supplement.
Adenoviral vectors
Adenoviruses harboring genes of interest were made using the AdMax system (Microbix). Short hairpin RNA knockdown adenovirus for FoxP1 or miR-206 was made using the adenoviral shuttle vector pDC311 (Microbix), into which the U6 RNA polymerase III promoter and the polylinker region of pSilencer 1.0U6 expression vector (Ambion) were subcloned.
Sequence of sh-FoxP1: 5′—3′ GCCCACACTGCAGAGGAAATTCAAGAGA TTTCCTCTGCAGTGTGGGC TTTTTT;
Sequence of anti-miR-206: 5′—3′ CGCGTCTCGAGCCACACACTTCCTTACATTCCAGCCCACACACTTCCTTACATTCCAA
Transgenic mice
Tg-miR-206 and Tg-206-SPONGE mice were generated on an FVB background, using the α-myosin heavy chain promoter to achieve cardiac-specific expression of around 300bp of the genomic region of miR-206 or two repeats of GFP-tagged miR-206 antisense, respectively. All protocols concerning animal use were approved by the Institutional Animal Care and Use Committee at Rutgers, New Jersey Medical School.
miRNA microarray
Total RNA was isolated from YAP- and LacZ-transduced NRCMs. Ten micrograms of RNA were sent to L.C. Sciences for a miRNA microarray 28. Samples were enriched for small RNA, labeled with Cy3 and Cy5 fluorescent dyes and hybridized to a single Atactic μParaFlo microfluidics chip that held all 334 mature rodent miRNA probes identified to date, as well as perfectly matched and mismatched probes for quality control. Each miRNA probe is represented 9X on the microarray. Among the control probes, PUC2PM-20B and PUC2MM-20B are the perfect match and single-based match detection probes, respectively, of a 20-mer RNA-positive control sequence that is spiked into the RNA samples before labeling. One may assess assay stringency from the intensity ratio of PUC2PM-20B to PUC2MM-20B, which is normally larger than 30. After signal amplification, the background was subtracted and normalized using the LOWESS (locally weighted regression) method. For a transcript to be listed as detectable, it must meet the following criteria: signal intensity higher than 3X (background SD), spot coefficient of variation<0.5 (coefficient of variation=SD/signal intensity) and signals from at least 50% of the repeating probes above detection level.
Northern blot
Total RNA, extracted using TRIzol reagent (Invitrogen), was separated on a 1% agarose gel, transferred to an uncharged nylon membrane, Hybond-NX (Amersham Biosciences), and UV cross-linked. The membrane was pre-hybridized/hybridized with MiracleHyb Hybridization solution according to the instruction manual (Stratagene). DNA oligonucleotides, anti-sense sequences of mature miRNAs, were obtained from Integrated DNA Technologies. The probes were 5′end-labeled with ET adenosine 5′-triphosphate[γ-32P] (PerkinElmer) using a T4 Polynucleotide Kinase kit (NEB) and used for hybridization (1X106cpm/ml).
Statistics
All values are expressed as mean±SEM. Statistical analyses were performed using ANOVA or t-test with a P<0.05 considered significant.
RESULTS
The abundance of miR-206 is increased by YAP and decreased by Mst1
To identify miRNAs involved in YAP-induced cardiac hypertrophy and survival, total RNA was extracted from neonatal rat CMs (NRCMs) overexpressing wild-type YAP and enriched for small RNAs. We used microarrays containing rodent miRNAs to analyze the enriched extracts and selected miRNAs that showed the greatest increase in abundance, including miR-711, miR-483, and miR-206 (Online Table I). Of note, we found no significant similarity in the nucleotide sequences between these miRNAs and the established Yorkie target bantam (Online Fig. I). Importantly, we found that overexpression of miR-206 significantly increased CM size (Online Fig. IIA), while neither miR-711 nor miR-483 overexpression significantly affected cell size (Online Fig. IIB). Additionally, a link between miR-206 expression and skeletal muscle hypertrophy has been demonstrated previously29. We therefore focused our efforts on investigating the possible role of miR-206 in mediating YAP-induced cardiac hypertrophy and survival for this study.
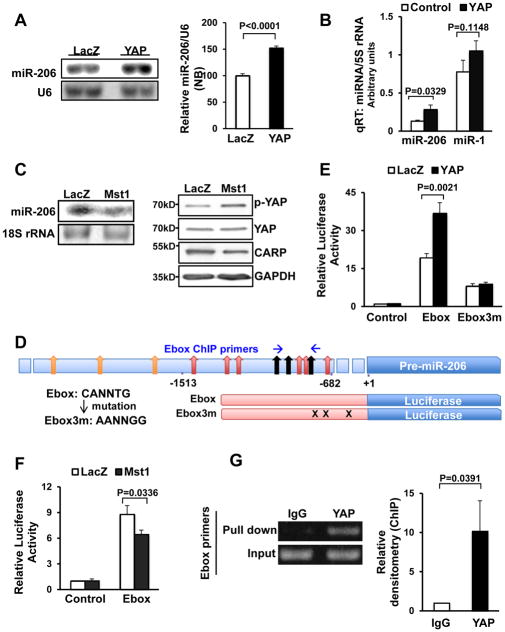
Both Northern blot and qRT-PCR confirmed our microarray result that the abundance of miR-206 was significantly increased (+1.44 fold by Northern blot, +2.13 fold by qRT-PCR) in CMs overexpressing wild-type YAP, in which the activity of YAP was confirmed by TEAD luciferase activation (Fig. 1A,B and Online Fig. IIF). Interestingly, YAP did not alter miR-206 levels in C2C12 mouse myoblasts, which have a higher abundance of miR-206 than NRCMs at baseline (Online Fig. IIC), suggesting that upregulation of miR-206 is cell type specific. Conversely, overexpression of Mst1, which induced inhibitory phosphorylation of YAP and reduced expression levels of the YAP target gene cardiac ankyrin repeat protein (CARP), decreased the abundance of miR-206 (Fig. 1C, Online Fig. IID). Additionally, co-expression of an Mst1-resistant form of YAP (Serine-127-Alanine) abolished the repressive effect of Mst1 on miR-206 abundance (Online Fig. IIE) indicating that the mammalian Hippo pathway suppresses miR-206 expression in CMs.
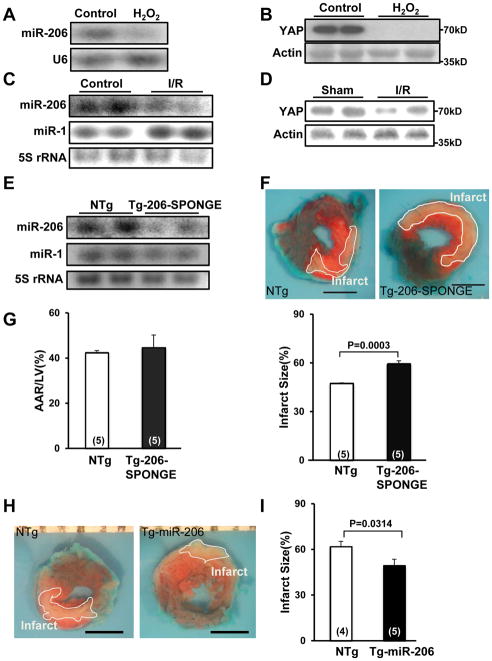
Figure 1. The abundance of miR-206 in CMs is regulated by YAP.
(A and B) Neonatal rat CMs (NRCMs) were transduced with Ad-LacZ or Ad-YAP. In A, Northern blot analyses were conducted using miR-206 and U6 probes, and the results were quantified by densitometry. In B, qRT-PCR analyses were conducted with miR-1, miR-206 and 5S rRNA primers. N=3. (C) CMs were transduced with either Ad-LacZ or Ad-Mst1 and Northern blot analyses were conducted using a miR-206 probe (left panel) or western blot was performed to determine phosphorylation status of YAP (right panel). N=3. (D) A schematic representation showing the location of E-boxes and ChIP primers in the miR-206 enhancer (upper). The wild-type and mutant E-box sequences are shown (lower left). Nine wild-type or mutated E-box sites were included in the promoters of the luciferase reporter used in (E) and (F) (lower right). (E and F) CMs were transduced with Ad-YAP, Ad-Mst1 or Ad-LacZ and then transfected with luciferase reporter constructs harboring either a control, wild-type E-box or mutated E-box sequence in the promoter. N=4 (E) and 3 (F). (G) YAP-overexpressing CMs were harvested for chromatin immunoprecipitation (ChIP) assays. ChIP assays were conducted with antibodies against YAP or a control immunoglobulin G (IgG) and specific Ebox primers. PCR using input DNA as a template served as an internal control. N=3.
Given the reported similarity between miR-1 and miR-206 30, we sequenced the qPCR products detected by the miR-206 and miR-1 qPCR primers to ensure their intended mature miRNA sequences were measured (Online Fig. IIIA). The specificity of the Taqman® MicroRNA Assay was further verified in that qRT-PCR did not detect an increase in miR-206 in CMs transduced with miR-1 adenovirus (Online Fig. IIIB).
To investigate the mechanism by which YAP increases the abundance of miR-206, we focused on the role of enhancer box (E-box) elements in modulating miR-206 transcriptional activation. We constructed luciferase reporters containing either the wild-type miR-206 enhancer with 9 E-box sites or a mutant enhancer with mutations in the 3 most conserved E-box sites (E-box-3m) (Fig. 1D). YAP significantly activated the wild-type E-box, but not E-box-3m enhancer-luciferase reporter, compared to control luciferase reporter (Fig. 1E). On the other hand, Mst1 significantly decreased the E-box luciferase activity (Fig. 1F). We also performed chromatin immunoprecipitation to test whether YAP associates with the miR-206 E-box. ChIP showed that YAP binds to the E-box located in the miR-206 promoter (Fig. 1G). This suggests that the Hippo pathway inhibits miR-206 transcription through the E-box elements. Moreover, YAP did not stimulate the miR-1 enhancer-luciferase reporter, indicating that YAP selectively up regulates miR-206 and not miR-1 expression in CMs (Online Fig. IVA).
miR-206 induces cardiac hypertrophy and protects myocytes from death
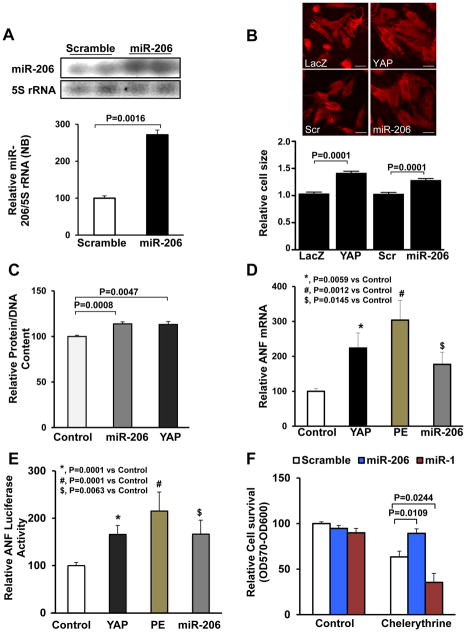
Transduction of NRCMs with adenovirus harboring miR-206 (Ad-miR-206) induced upregulation of mature miR-206, as evaluated by Northern blot (Fig. 2A). Adenoviral-mediated expression of either miR-206 or YAP for 48 hours induced CM hypertrophy, as evidenced by an increase in cell size and the total protein content normalized to DNA content (Fig. 2B,C). Similarly, miR-206 and YAP both significantly increased mRNA expression of ANF, a hypertrophic marker gene 31, as well as activation of the ANF-luciferase reporter gene in CMs. ANF-luciferase activation was comparable to that elicited by phenylephrine (PE), an α1 adrenergic agonist known to stimulate cardiac hypertrophy 32 and used here as a positive control to promote cardiac hypertrophy (Fig. 2D,E). In contrast, miR-1 expression did not increase total protein normalized to DNA content in CMs (Online Fig. IVB). These results suggest that upregulation of miR-206 is sufficient to induce CM hypertrophy in vitro while upregulation of miR-1 is not.
Figure 2. miR-206 promotes CM hypertrophy in vitro.
(A–C) Neonatal rat CMs (NRCMs) were transduced with Ad-Control (Ad-Sc or Ad-LacZ), Ad-YAP or Ad-miR-206. (A) After 48 hours, cells were harvested for Northern blot analyses, using a miR-206 probe. (B) NRCMs were fixed and stained with anti-Troponin T to visualize CMs. Cell image analysis of the relative cell surface area (cell size) was performed. Scale bar, 30 μm. (C) Evaluation of total protein content normalized to DNA content. N=4. (D) CMs were transduced with Ad-LacZ (control), Ad-YAP or Ad-miR-206, or treated with phenylephrine (PE, 100 μM) for 48 hours, and then harvested for qRT-PCR analyses of ANF mRNA. N=4. (E) CMs were co-transfected with ANF promoter luciferase reporter gene and either control, YAP or miR-206 plasmid, or treated with PE for 48 hours. Cells were harvested for ANF luciferase activity measurement. N=4. (F) Forty-eight hours after transduction of Ad-scrambled, Ad-miR-206, or Ad-miR-1, CMs were treated with 5 μM chelerythrine for 1 hour. Cells were harvested for viability assays measured by Cell Titer Blue. N=3.
To examine the effect of miR-206 upon cell death, CMs were treated with chelerythrine, a known inducer of apoptosis 33. Adenoviral-mediated expression of miR-206 in CMs inhibited chelerythrine-induced cell death as evaluated with the Cell Titer Blue viability assay, caspase-3 cleavage and TUNEL-positive nuclei (Fig. 2F, Online Fig. IVC,D). These results indicate that miR-206 protects against CM death. On the other hand, miR-1 facilitated chelerythrine-induced apoptosis (Fig. 2F, Online Fig. IVC,D), evidence of further functional distinction between miR-206 and miR-1 in CMs. Of note, neither miR-206 nor miR-1 expression affected proliferation of NRCMs (Online Fig. IVE).
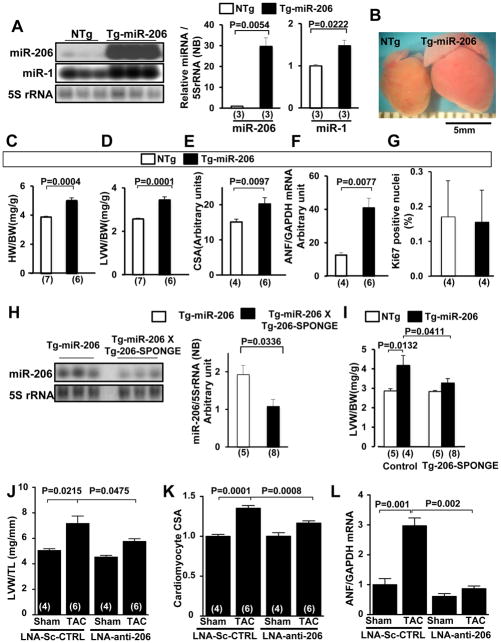
To test whether miR-206 is sufficient to induce cardiac hypertrophy in vivo, we generated 4 lines of transgenic (Tg) mice with cardiac-specific expression of the genomic region (~300bp) of miR-206, driven by the α-myosin heavy chain promoter (Tg-miR-206). Mature miR-206 was upregulated in Tg-miR-206 mouse myocardium compared to non-transgenic (NTg) controls, as shown by Northern blot analysis (Fig. 3A, Online Fig. VA, left). While we observed a modest increase in miR-1 levels by Northern blot, this was most likely due to detection of ectopic miR-206 by the miR-1 Northern probe, which, due to the nature of its design, has less selectivity compared to the qRT-PCR probes. In fact, we did not observe any increase in miR-1 expression levels using qRT-PCR analysis of Tg-miR-206 hearts compared to NTg (Online Fig. VA, right). In all lines, Tg-miR-206 mice developed baseline cardiac hypertrophy compared to NTg controls as evidenced by increased heart weight/body weight (HW/BW) and left ventricle (LV) weight/body weight (LVW/BW) ratios, LV myocyte cross-sectional area, and ANF mRNA expression (Fig. 3B–F, Online Fig. VB,C). Left ventricular function was maintained in Tg-miR-206 mice as evidenced by normal left ventricular ejection fraction (LVEF) and lung weight/body weight ratio (Online Fig. VB,D). The increase in LVW/BW can likely be attributed to individual CM hypertrophy since overexpression of miR-206 did not affect myocyte proliferation, as indicated by no change in the number of Ki67-positive nuclei or the estimated total myocyte number (Fig. 3G, Online Fig. VE,F).
Figure 3. miR-206 promotes cardiac hypertrophy in vivo.
(A–G) Characterization of the lowest expressing line of Tg-miR-206 mice at 3 months of age. (A) Tg-miR-206 (line24) and non-transgenic control (NTg) mouse hearts were harvested for Northern blot analysis with a miR-206 probe, a miR-1 probe, or a 5S rRNA probe. (B) Representative heart pictures taken in PBS under a microscope. (C, D) Postmortem measurements of heart weight/body weight (HW/BW, mg/g) and left ventricular weight/body weight (LVW/BW, mg/g). (E) Quantitative analysis of myocyte cross-sectional area (CSA) by wheat germ agglutinin (WGA) staining is shown. (F) The level of ANF mRNA was evaluated by qRT-PCR analyses. (G) Ki67-positive nuclei/total CM nuclei was evaluated with anti-Ki67 and anti-Troponin T staining. (H and I) Tg-miR-206 and Tg-miR-206/Tg-206-SPONGE bi-transgenic mouse hearts were harvested for Northern blot analyses with a miR-206 probe and a 5S rRNA probe (H) and postmortem measurement of LVW/BW (mg/g) (I). (J–L) Wild-type C57B6 male mice were treated with LNA scrambled control or LNA-anti-206 by tail vein injection 7 days prior to intervention. Groups were subjected to 1-week TAC or sham operation. (J) Left ventricular weight/tibia length (LVW/TL) was determined. (K) Heart sections were stained with WGA and CM CSA was determined. (L) mRNA was isolated from ventricles and subjected to qRT-PCR to detect ANF levels. N=4–6.
To confirm that cardiac hypertrophy is elicited specifically due to increased miR-206 expression, we crossed Tg-miR-206 mice with a separately generated transgenic mouse, Tg-206-SPONGE. These mice harbor two repeats of antisense miR-206 downstream of GFP driven by the α–MHC promoter (Tg-206-SPONGE mouse). Expression of miR-206, as well as the extent of cardiac hypertrophy, was significantly reduced in the double-Tg mice compared to Tg-miR-206 mice (Fig. 3H,I). Taken together, these results suggest that miR-206 expression induces cardiac hypertrophy in vivo. Although we generated multiple lines of transgenic mice, none of the lines exhibited low levels of miR-206 overexpression. Thus, it is possible that the cardiac phenotype observed in Tg-miR-206 may not reflect physiological function of miR-206.
In order to address this issue in part, we also employed a complimentary loss-of-function approach to test the involvement of miR-206 in cardiac hypertrophy. Wild-type C57B/6J mice were administered LNA inhibitor designed to selectively inhibit miR-206, or control scrambled LNA, by tail vein injection. Specificity of the LNA inhibitor was confirmed by qRT-PCR analysis of miRNA expression 7 days after treatment (Online Fig. VG). After 7 days, mice from both treatment groups were subjected to sham operation or transverse aortic constriction (TAC) to elicit hemodynamic overload. After 7 days of TAC, echocardiography was performed and mice were sacrificed. Importantly, we found up regulation of myocardial miR-206 expression levels after 7 days TAC, consistent with its observed pro-hypertrophic effect (Online Fig. VH). A significant increase in LVW/TL in LNA control-treated mice following TAC was observed; however, no increase was observed in LNA-anti-206 treated mice (Fig. 3J). We also noted significant differences in chamber wall thickness between the control and anti-206 TAC groups ((Supplement) Table II). Additionally, CM cross sectional area and ANF expression were significantly increased in the LNA control-treated TAC group, and these responses were attenuated in the LNA-anti-206 treated mice (Fig. 3K,L, Online Fig. VI). These data demonstrate that inhibition of miR-206 impairs pressure overload-induced hypertrophy and indicates that miR-206 is an important endogenous promoter of heart growth in response to stress.
miR-206 is an important mediator of YAP-induced cardiac hypertrophy and inhibition of cardiac cell death
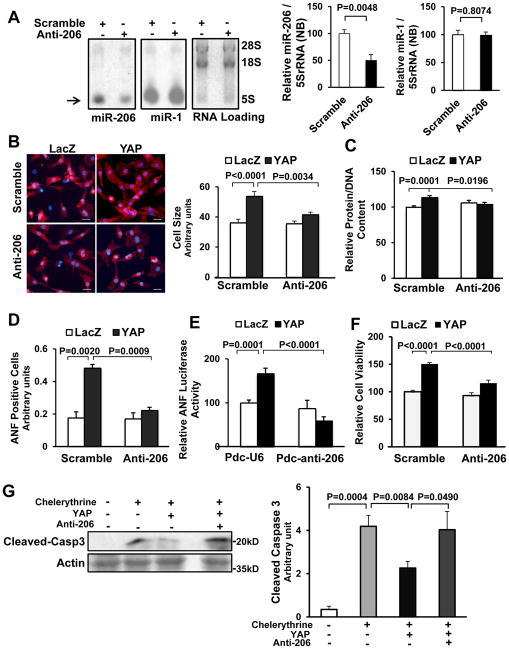
In order to further clarify the cell autonomous effect of endogenous miR-206 in mediating YAP-induced cardiac hypertrophy, adenovirus harboring anti-miR-206 (Ad-anti-miR-206), comprising two repeats of a complementary sequence to miR-206 under the control of a U6 promoter, was generated. Adenovirus harboring a scrambled sequence (Ad-scrambled) was used as a control. Transduction of CMs with Ad-anti-miR-206 significantly (−50%) reduced expression of miR-206, but not miR-1 (Fig. 4A), and attenuated YAP-induced CM hypertrophy, as evidenced by decreases in cell size, protein/DNA content, perinuclear ANF staining and activation of the ANF-luciferase reporter (Fig. 4B–E, Online Fig. VIA). We also used a miRIDIAN inhibitor to inhibit miR-206 in CMs. The miRIDIAN miR-206 inhibitor, but not control miRIDIAN inhibitor, significantly inhibited miR-206 function, as shown by an increase of luciferase reporter containing miR-206 antisense sequence in the 3′UTR, but not empty luciferase reporter (Online Fig. VIB), and attenuated YAP-induced hypertrophy of CMs, as evidenced by suppression of ANF luciferase reporter and CM size (Online Fig. VID,E). These results suggest that endogenous miR-206 is critical in mediating YAP-induced CM hypertrophy in vitro. Importantly, specific down regulation of miR-1 did not abolish YAP-induced CM hypertrophy, indicating that miR-1 does not mediate hypertrophy elicited by YAP (Online Fig. VIF,G).
Figure 4. miR-206 mediates YAP-induced hypertrophy and inhibition of cell death.
Twelve to 24 hours after transduction of either Ad-sh-scr or Ad-anti-206, CMs were transduced with Ad-LacZ or Ad-YAP for 48 hours. (A) CMs were harvested for Northern blot, using a miR-206 probe or a miR-1 probe. RNA loading was evaluated with ethidium bromide staining. The level of miR-206 and miR-1 was normalized by that of 5S-rRNA. N=5. (B) CMs were stained with anti-α-actinin antibody (red) and DAPI (blue), and relative cell surface area (cell size) was evaluated. Scale bar, 30 μm. N=3. (C) CMs were harvested for the measurement of total protein content normalized by DNA content. N=3. (D) CMs were stained with anti-ANF antibody and DAPI. ANF-positive cells were quantified. N=3. (E) CMs were transfected with ANF luciferase reporter gene and either Pdc-U6 or Pdc-anti-206 plasmids before transduction with Ad-LacZ or Ad-YAP. After 48 hours, myocytes were harvested for ANF luciferase activity measurements. N=4. (F) CMs were harvested for Cell Titer Blue assays. N=3. (G) CMs were treated with or without chelerythrine (5 μM) for 1 hour and harvested for immunoblot analyses with anti-cleaved caspase 3 (Cleaved-Casp3) antibody and the results were quantified by densitometry. N=4.
We also tested the role of miR-206 in mediating CM survival elicited by YAP. miR-206 knockdown not only inhibited YAP-induced CM viability at baseline (Fig. 4F), but it also reversed the YAP-induced protection against chelerythrine-induced cell death, as shown by caspase-3 cleavage (Fig. 4G). These results suggest that endogenous miR-206 is an important mediator of YAP-induced protection of CMs in vitro.
Endogenous miR-206 is protective during I/R injury
In order to examine the physiological function of miR-206 during stress, we first examined miR-206 expression in response to oxidative stress. H2O2 treatment reduced RNA and protein levels of miR-206 and YAP, respectively, in CMs (Fig. 5A,B, Online Fig. VIIA). Similarly, miR-206 and YAP were down regulated in mouse hearts subjected to ischemia followed by reperfusion compared to sham-operated controls (Fig. 5C,D, Online Fig. VIIB,C). In contrast, miR-1 was upregulated in response to ischemia followed by reperfusion (Online Fig. VIID). To evaluate the role of endogenous miR-206 in mediating CM survival in vivo, we used Tg-206-SPONGE mice, which have reduced miR-206, but not miR-1, expression compared to NTg controls (Fig. 5E, Online Fig. VIIE,F). We found that I/R-induced cardiac injury was exacerbated in Tg-206-SPONGE compared to NTg mice, as indicated by a greater size of myocardial infarction normalized by the area at risk (Fig. 5F,G). In contrast, the size of myocardial infarction normalized by the area at risk following I/R was significantly reduced in Tg-miR-206 compared to NTg hearts (Fig. 5H,I). To test whether increased miR-206 expression can mitigate the chronic effects of I/R injury, including decreased cardiac function, we subjected NTg and Tg-miR-206 mice to I/R and followed them with serial echocardiography. Although there was no difference in basal heart function between Tg-miR-206 and NTg mice, we found that Tg-miR-206 mice had significantly better cardiac function, as determined by percent fractional shortening (%FS), post I/R compared to NTg controls (Online Fig. VIIG). These results suggest that down regulation of endogenous miR-206 promotes myocardial I/R injury whereas restoration of miR-206 levels with exogenous miR-206 protects against I/R injury and counteracts the progression to heart failure.
Figure 5. miR-206 protects the heart against I/R injury.
(A, B) CMs were treated with vehicle or H2O2 (50 μM) for 1 hour. Representative results of Northern blot analyses with miR-206 and U6 probes (A) or immunoblot analyses with anti-YAP antibody (B) are shown. N=4. (C, D) FVB mice at 3 months of age were subjected to 45 minutes of ischemia and 24 hours of reperfusion (I/R) or sham operation and the hearts were harvested for Northern blotting with miR-206 and miR-1 probes (C) and immunoblot analyses performed with anti-YAP antibody (D). N=5. (E) Tg-206-SPONGE and NTg mouse hearts were harvested for Northern blot analyses with miR-206 and miR-1 probes. N=3. (F, G) Tg-206-SPONGE and NTg mice at 3 months of age were subjected to 45 minutes of ischemia and 24 hours of reperfusion. The hearts were subjected to Alcian blue (1%) and TTC (1%) staining. In F, representative images of Alcian Blue dye and TTC staining are shown. Boundaries of the infarct areas are indicated by white lines. Scale bar, 2 mm. In G, area at risk (AAR)/LV (%) (left) and infarct size/AAR (%) (right) are shown. (H, I) Tg-miR-206 and NTg mice at 3 months of age were subjected to 45 minutes of ischemia and 24 hours of reperfusion. In H, representative images of Alcian Blue dye and TTC staining are shown. Boundaries of the infarct areas are indicated by white lines. In I, infarct size/AAR (%) is shown.
miR-206 induces cardiac hypertrophy through inhibition of FoxP1
A search of miR-206 targets using TargetScan (http://www.targetscan.org, Release 5.2) allowed us to identify FoxP1, a tumor suppressor protein, as a potential target and mediator of the pro-hypertrophic and pro-survival effects of miR-206 in CMs. FoxP1 was ranked as 7th among 478 conserved targets of miR-206 by total context score predicted by TargetScan. Since our gain-of-function analyses suggested that miR-206 has growth promoting effects, we predicted that molecules known to inhibit cell growth responses might mediate the effect of miR-206. FoxP1 was ranked highest among the proteins known to have growth inhibitory function. In addition, FoxP1 has 4 predicted miR-206 binding sites in its 3′UTR, two of which are conserved sites (Online Fig. VIIIA) further strengthening our rationale to investigate FoxP1.
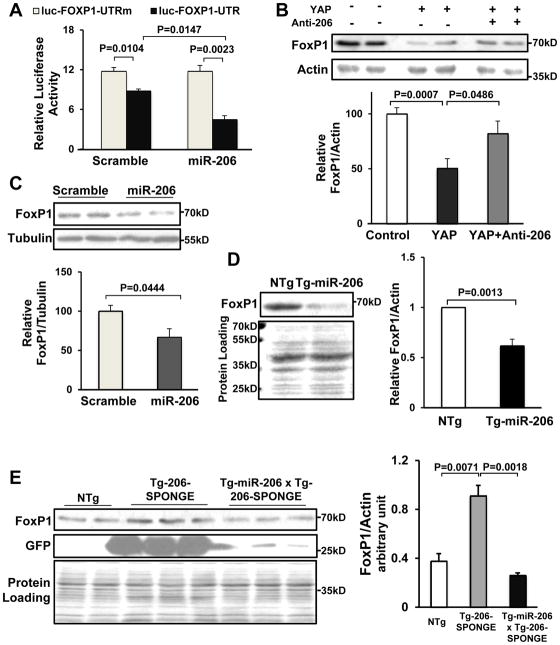
To demonstrate that FoxP1 is a target of miR-206, we first conducted luciferase assays using the FoxP1 3′ UTR. The 3′UTR of the luciferase gene was replaced with either the WT FoxP1 3′UTR or a mutated FoxP1 3′UTR in which the miR-206 binding sites were altered. Transduction of NRCMs with Ad-miR-206, but not Ad-scrambled, significantly inhibited the luciferase reporter containing the WT FoxP1 3′UTR; however, the luciferase reporter containing the mutated FoxP1 3′ UTR was not affected (Fig. 6A). Conversely, a miRIDIAN miR-206 inhibitor, but not control miRIDIAN inhibitor, significantly stimulated the luciferase reporter containing the WT FoxP1 3′UTR but did not affect the control reporter containing the mutated FoxP1 3′ UTR (Online Fig. VIC). Adenoviral-mediated expression of either YAP or miR-206 in CMs inhibited FoxP1 expression, as determined by Western blotting (Fig. 6B,C). Furthermore, YAP-induced down regulation of FoxP1 was attenuated in the presence of anti-miR-206, suggesting that YAP down regulates FoxP1 through miR-206 (Fig. 6B). Adenoviral-mediated expression of miR-1 did not affect protein levels of FoxP1 in CMs (Online Fig. VIIIB, C). FoxP1 was also down regulated in Tg-miR-206 hearts (Fig. 6D), whereas it was upregulated in Tg-206-SPONGE mice, in which endogenous miR-206 is suppressed (Fig. 6E). The upregulation of FoxP1 observed in Tg-206-SPONGE mice was normalized when Tg-206-SPONGE mice were crossed with Tg-miR-206 mice (Fig. 6E). Taken together, these results suggest that FoxP1 is a specific target of miR-206 in the heart and the CMs therein.
Figure 6. YAP-miR-206 signaling down regulates FoxP1 expression in CMs.
(A) CMs were transfected with luciferase reporter constructs harboring either wild-type (UTR) or mutated (UTRm) FoxP1 3′UTR sequences and then transduced with Ad-scrambled or Ad-miR-206. N=3. (B, C) CMs were transduced with Ad-LacZ, Ad-YAP, Ad-scrambled or Ad-miR-206 for 48 hours. Cells were harvested for immunoblot analyses with anti-FoxP1 antibody and either anti-actin or anti-α-tubulin antibodies. The results of densitometric analyses are also shown. N=4. (D, E) Tg-206-SPONGE, Tg-miR206/Tg-206-SPONGE and NTg mouse hearts were harvested for immunoblot analyses, using anti-FoxP1, anti-GFP and anti-actin antibodies. N=4.
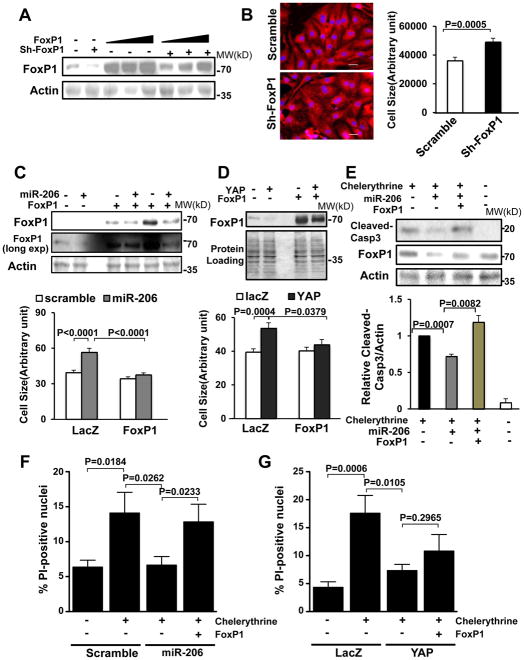
Based on our hypothesis, we predicted that FoxP1 inhibits cardiac hypertrophy. To test this, cultured CMs were transduced with an adenovirus harboring shRNA targeting FoxP1 (Ad-sh-FoxP1). We confirmed that Ad-sh-FoxP1 efficiently down regulated both endogenous and overexpressed FoxP1 in CMs (Fig. 7A, Online Fig. VIIID). Down regulation of FoxP1 significantly increased cell size at baseline (Fig. 7B) and increased cell viability after H2O2 treatment (Online Fig. VIIIG), indicating that endogenous FoxP1 negatively regulates CM hypertrophy and survival.
Figure 7. miR-206 promotes CM hypertrophy and survival through down regulation of FoxP1.
(A, B) CMs were transduced with Ad-sh-scr or Ad-sh-FoxP1 with or without Ad-FoxP1 for 72 hours. Cells were harvested for immunoblot analyses with anti-FoxP1 antibody (A) or stained with anti-α-actinin (red) antibody/DAPI (blue) for relative cell size measurements (B). N=3. (C–E) Twelve to 24 hours after transduction with Ad-LacZ, Ad-miR-206 or Ad-YAP, CMs were transduced with Ad-LacZ or Ad-FoxP1 for 48 hours. Cells were harvested for immunoblot analyses using anti-FoxP1 antibody (C, D upper), stained with anti-α-actinin antibody and DAPI for relative cell surface area (cell size) measurements (C, D lower). N=4. (E) CMs were treated with 5 μM chelerythrine for 1 hour and then harvested for immunoblot analyses with anti-cleaved caspase 3 (Cleaved-Casp3) and FoxP1 antibodies. N=4. (F) CMs were transduced with Ad-sh-scr, miR-206 and/or FoxP1 expression constructs and treated with vehicle or chelerythrine (5 μM). Cell viability was determined by propidium iodine (PI) staining. (G) Cells were transduced with Ad-LacZ, Ad-YAP and Ad-FoxP1 constructs and treated with vehicle or chelerythrine (5 μM). Cell viability was determined by PI staining. N=3.
To elucidate the contribution of FoxP1 down regulation to the overall effect of miR-206 in CMs, we generated an adenoviral vector harboring FoxP1 (Ad-FoxP1). We confirmed that FoxP1 is overexpressed in Ad-FoxP1-transduced CMs, and that miR-206- or YAP-induced down regulation of FoxP1 was partially attenuated by overexpression of FoxP1 (Fig. 7C,D and Online Fig. VIIIE,F). Both miR-206- and YAP-induced CM hypertrophy, as evaluated by cell size, was significantly attenuated when FoxP1 levels were normalized by adenoviral-mediated FoxP1 expression (Fig. 7C,D lower panels). These results indicate that YAP/miR-206 induce CM hypertrophy through down regulation of FoxP1. We then tested the involvement of FoxP1 in mediating the protective effect of YAP and miR-206. We found that when FoxP1 levels were normalized, the ability of miR-206 to inhibit chelerythrine-induced CM death was abrogated (Fig. 7E,F, Online Fig. VIIIH). Interestingly, protection conferred by YAP was partially but not completely inhibited by FoxP1 (Fig. 7G) suggesting that miR-206 promotes cell survival through down regulation of FoxP1, whereas YAP likely promotes survival through additional mechanisms7.
DISCUSSION
Considering the remarkable level of homology present between each component of the Hippo signaling pathway, it is reasonable to predict that YAP targets in mammalian cells might include miRNAs, similar to that observed in Drosophila (i.e. bantam). To identify possible miRNAs that are functional targets of YAP, we conducted microarray analysis and detected several miRNAs whose expression was upregulated by YAP in CMs. Here we demonstrate that miR-206 acts as an important mediator downstream of YAP to promote hypertrophy and survival of CMs.
Recent work has demonstrated that YAP upregulates miR-29, which in turn inhibits PTEN and activates the mTOR pathway34. In CMs, we found that YAP upregulated miR-29 1.9-fold (Online Table I) indicating that regulation of miR-29 by YAP may occur in multiple cell and tissue types. Although one study reported that miR-29c induces modest enlargement of NRCMs 35, our results show that overexpression of miR-29 did not induce hypertrophy in NRCMs (Online Fig. IIB). Previous work has shown that miR-29 is enriched in cardiac fibroblasts and inhibits cardiac fibrosis 36. Additionally, miR-29 was found to negatively regulate CM proliferation during post-natal cardiac development demonstrating that miR-29 does play a role in cardiovascular biology37. Thus, miR-29 is likely to mediate additional non-growth related functions of YAP in CMs.
The enhancer region of miR-206 does not contain typical binding sites for the reported mammalian transcriptional partners of YAP, including TEAD, RUNX, ErbB4, p73, Tbx5 or Smad (reviewed in4). Instead, we found that E-box elements are present in the miR-206 enhancer region and that three conserved E-box elements on the miR-206 enhancer are critical for basal, as well as YAP-induced, miR-206 expression in CMs (Fig. 1E). It has been shown that myogenic factors induce expression of muscle-specific miRNAs (including miR-1, miR-206 and miR-133) by binding to E-box regions in the promoter/enhancer 22, 38. However, the identity of the E-box binding transcription factor responsible for YAP-induced upregulation of miR-206 and its functional relationship to YAP remain to be demonstrated and warrant future investigation.
We have demonstrated that both YAP and miR-206 stimulate hypertrophy and survival of CMs in vitro. In vivo, exogenous expression of either YAP or miR-206 protects CMs against I/R-induced death and reduces heart injury8. Furthermore, cardiac-specific heterozygous deletion of Yap1 promotes cardiac dysfunction but inhibits cardiac hypertrophy during chronic MI, suggesting that endogenous YAP plays an important role in promoting both survival and hypertrophy during cardiac remodeling in the adult mouse heart7. Additionally, we found that overexpression of miR-206 in the mouse heart induces hypertrophy without cardiac dysfunction, and that selective inhibition of miR-206 prevents 1-week TAC-induced hypertrophy without altering cardiac function. Taken altogether, we speculate that the YAP-miR-206 pathway may mediate physiological hypertrophy, and serves a compensatory function to help protect the heart against increased demand and stress. Interestingly, although phosphatidylinositol 3-kinase has been shown to promote physiological hypertrophy 39, we previously reported that YAP-induced cardiac hypertrophy was not affected by phosphatidylinositol 3-kinase inhibition7, suggesting that the YAP-miR-206 pathway mediates physiological hypertrophy through PI3K-independent mechanisms. We have shown previously that YAP is activated in the peri-infarct area in the post-MI heart 7. Although YAP is activated by mechanical stress and agonists of G protein-coupled receptors in other cell types 40, 41, how YAP is activated in response to hypertrophic stimuli remains to be elucidated in CMs.
Expression of miR-206 is relatively low at baseline and is further down regulated in response to oxidative stress in CMs and I/R in the heart. This is consistent with our previous findings that both oxidative stress and I/R activate Mst1 and Lats2, which would in turn inhibit YAP 5, 42, 43. Since further down regulation of miR-206 exacerbated I/R injury (Fig, 5F,G), whereas exogenous miR-206 expression reduced injury, endogenous miR-206 appears to have an essential role in promoting survival of CMs during I/R. We also found that miR-206 expression was sufficient to counteract the decreased cardiac function observed in response to chronic I/R, further evidence of the cardioprotective effect of miR-206 in vivo.
We speculate that the effect of YAP upon growth and survival of CMs functions through multiple mechanisms in a context-dependent manner. YAP regulates proliferation of CMs through YAP-TEAD interaction 44. We have shown recently that YAP acts as a nuclear co-factor of FoxO1 to protect the heart during myocardial I/R 8. Furthermore, YAP has been shown to regulate global biogenesis and processing of miRNA through an interaction with p72 (DDX17) 45 and let-7-dependent down regulation of Dicer 46, respectively. Further investigation is required to elucidate the relative importance of these mechanisms in the regulation of growth and death of CMs by YAP.
Our results indicate that FoxP1 is a physiological mediator of miR-206 function in CMs. First, we found that its 3′UTR is specifically targeted by miR-206. In NRCMs, miR-206 expression down regulated FoxP1 protein and inhibited FoxP1-UTR luciferase activity. Our results suggest that miR-206 also targets FoxP1 in vivo. We observed down regulation of FoxP1 in Tg-miR-206 hearts while FoxP1 expression was increased in Tg-206-SPONGE mouse hearts (Fig. 6E). Although FoxP1 has been identified as a target of miR-1 in certain cell types 47, we clearly demonstrate that FoxP1 is specifically targeted by miR-206, and not by miR-1, in CMs (Online Fig. VIIIC).
Second, we demonstrate that FoxP1 is an endogenous negative regulator of cardiac hypertrophy and survival. Indeed, down regulation of FoxP1 was sufficient to induce hypertrophy and promote cell survival in cultured CMs (Fig. 7B, Online Fig. VIIIG). Furthermore, both the stimulation of hypertrophy and the suppression of apoptosis by miR-206 were attenuated when FoxP1 down regulation was reversed by adenoviral supplementation of FoxP1 in CMs (Fig. 7, C,E,F, Online Fig. VIIIH). Taken together, our results suggest that down regulation of FoxP1 plays an important role in mediating the effects of miR-206 in CMs.
FoxP1 systemic knockout mice die at E14.5 due to heart failure 48. These mice exhibit defects in valve formation caused by impaired apoptosis in the endocardial cushion mesenchyme, and have a thinner ventricular compact zone due to increased proliferation of the ventricular trabecular zone 48. Cardiac-specific FoxP1 knockout mice die within 24 hours of birth and have a thickened myocardium resulting from increased myocyte proliferation in a cell-autonomous fashion 49. These data suggest that FoxP1 promotes apoptosis and inhibits CM proliferation during cardiac development. FoxP1 also negatively regulates cardiac hypertrophy by interacting with and inhibiting Nfat3 transcriptional activity 50. It will be interesting to clarify the involvement of miR-206 in these cardiac events through FoxP1 targeting. In addition, FoxP1 is abundant in the nuclei of CMs in failing human hearts 51. Since Mst1, a negative regulator of YAP, is activated in the failing heart52, upregulation of FoxP1 may be mediated through down regulation of miR-206. The roles of miR-206 and FoxP1 in regulating myocardial cell death in the failing heart remain to be elucidated.
Some limitations in this study should be noted. First, the absolute level of miR-206 in the heart and the cardiomyocytes therein is low compared to skeletal muscle and the skeletal myocytes therein. Although we conducted multiple loss-of-function experiments, using anti-miRNA, SPONGE, LNA anti-miR, and miRIDIAN inhibitor, the functional importance of endogenous miR-206 requires further scrutiny, for example, by using cardiac specific miR-206 knock-out mice. Second, increasing lines of evidence suggest that organs can take up miRNA from the circulation 53. Although the level of miR-206 in the circulation is low, it can be released from skeletal muscle after heavy work load, such as marathon running 54. Thus, we cannot formally exclude the possibility that exogenous, rather than endogenous, miR-206, such as that which originates from skeletal muscle, affects the function of the heart. Again, studies using skeletal muscle specific miR-206 knock-out mice would be helpful to address this issue.
In summary, we show here that miR-206 is upregulated by YAP and mediates YAP-induced hypertrophy and CM survival. The expression of miR-206 is regulated in response to I/R and oxidative stress. miR-206 promotes CM survival and cardiac hypertrophy in vivo, which in turn contributes to the regulation of myocardial injury and organ size. We also identify FoxP1 as a novel and critical downstream target of miR-206 that mediates its effects on CM survival and hypertrophy. To our knowledge, this is the first report demonstrating a functional miRNA target of YAP in CMs that impacts heart growth, death and function.
Supplementary Material
Novelty and Significance.
What Is Known?
YAP is a downstream effector of the Hippo signaling pathway, which controls organ size by modulating cell growth, proliferation and apoptosis.
YAP is a transcription co-factor that can regulate multiple transcription factors, including TEAD, RUNX, TBX5 and FOXO.
YAP promotes cardiomyocyte survival and proliferation in the postnatal heart, and cardiac regeneration and remodeling after myocardial infarction.
What New Information Does This Article Contribute?
YAP upregulates miR-206, which, in turn, plays an important role in mediating the effect of YAP upon cardiomyocyte survival and hypertrophy.
miR-206 promotes cardiomyocyte survival during ischemia/reperfusion in the heart.
miR-206 mediates cardiomyocyte hypertrophy and survival through inhibitory targeting of FOXP1.
Increasing lines of evidence suggest that the Hippo pathway plays an important role in regulating the growth and death of cardiomyocytes by negatively regulating YAP, a transcription co-factor. The nuclear localization of YAP is negatively regulated by two upstream kinases, MST1 and LATS2, which constitute the core of the Hippo pathway. YAP that is localized in the nucleus positively regulates cardiomyocyte proliferation, hypertrophy and survival. YAP is known to exert its function through TEAD and other transcription factors. However, the role of YAP in regulating miRNAs in the heart is unknown. We show that YAP transcriptionally upregulates miR-206, which, in turn, mediates the effect of YAP upon cardiomyocyte hypertrophy and survival. Endogenous miR-206 also plays an essential role in mediating pressure overload-induced cardiac hypertrophy and cardiomyocyte survival during ischemia/reperfusion. One of the important targets of miR-206 is FOXP1. Inhibition of FOXP1 mediates the effect of miR-206 on cardiomyocyte hypertrophy and survival. Taken together, our results suggest that miR-206 is an important downstream effector of YAP that mediates both hypertrophy and cardiomyocyte survival at baseline and in response to stress. The YAP-miR-206 pathway appears to be a promising target for treatment of heart disease through modulation of myocardial growth and death mechanisms.
Acknowledgments
The authors thank Daniela Zablocki and Christopher D. Brady for critical reading of the manuscript, and Linda Reed for assistance with tail vein injections.
SOURCES OF FUNDING
This work was supported in part by U.S. Public Health Service Grants HL 67724, HL91469, HL102738, HL112330, HL127339, HL122669 and AG23039, the Fondation Leducq Transatlantic Network of Excellence and the American Heart Association (11SDG7240067).
Nonstandard Abbreviations and Acronyms
- AAR
Area at risk
- Ad-
adenovirus-
- αMHC
alpha myosin heavy chain
- ANF
Atrial natriuretic factor
- Anti-206
Anti-microRNA-206
- CARP
cardiac ankyrin repeat protein
- CM
cardiomyocytes
- FoxP1
Forkhead box protein P1
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- I/R
Ischemia/reperfusion
- Mdn-206
miRIDIAN miR-206 inhibitor
- MI
myocardial infarction
- miRNA
MicroRNA
- miR-206
MicroRNA-206
- NRCMs/CMs
Neonatal rat cardiomyocytes
- NTg
non-transgenic control mouse
- PE
Phenylephrine
- sc206
Scramble miR-206 adenovirus
- sh-sc
Scramble shRNA adenovirus
- TTC
Triphenyltetrazolium chloride
- 3′UTR
3 prime untranslated region
- YAP
Yes-associated protein
Footnotes
DISCLOSURES
None.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–71. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon CL, Zhang X, Lin JI, Manning SA, Harvey KF. Homeodomain-interacting protein kinase regulates Hippo pathway-dependent tissue growth. Curr Biol. 2012;22:1587–94. doi: 10.1016/j.cub.2012.06.075. [DOI] [PubMed] [Google Scholar]
- 4.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto S, Yang G, Zablocki D, Liu J, Hong C, Kim SJ, Soler S, Odashima M, Thaisz J, Yehia G, Molina CA, Yatani A, Vatner DE, Vatner SF, Sadoshima J. Activation of Mst1 causes dilated cardiomyopathy by stimulating apoptosis without compensatory ventricular myocyte hypertrophy. J Clin Invest. 2003;111:1463–74. doi: 10.1172/JCI17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsui Y, Nakano N, Shao D, Gao S, Luo W, Hong C, Zhai P, Holle E, Yu X, Yabuta N, Tao W, Wagner T, Nojima H, Sadoshima J. Lats2 is a negative regulator of myocyte size in the heart. Circ Res. 2008;103:1309–18. doi: 10.1161/CIRCRESAHA.108.180042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Re DP, Yang Y, Nakano N, Cho J, Zhai P, Yamamoto T, Zhang N, Yabuta N, Nojima H, Pan D, Sadoshima J. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J Biol Chem. 2013;288:3977–88. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao D, Zhai P, Del Re DP, Sciarretta S, Yabuta N, Nojima H, Lim DS, Pan D, Sadoshima J. A functional interaction between Hippo-YAP signalling and FoxO1 mediates the oxidative stress response. Nature communications. 2014;5:3315. doi: 10.1038/ncomms4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, van Gorp PR, Zhou B, Chen J, Seidman JG, Wang DZ, Pu WT. Cardiac-Specific YAP Activation Improves Cardiac Function and Survival in an Experimental Murine MI Model. Circ Res. 2014;115:354–63. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, Richardson JA, Sadek HA, Bassel-Duby R, Olson EN. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–44. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Science translational medicine. 2014;6:239ps3. doi: 10.1126/scitranslmed.3009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee ST, Chu K, Jung KH, Kim JH, Huh JY, Yoon H, Park DK, Lim JY, Kim JM, Jeon D, Ryu H, Lee SK, Kim M, Roh JK. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72:269–77. doi: 10.1002/ana.23588. [DOI] [PubMed] [Google Scholar]
- 15.Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G. Differential expression of microRNAs in mouse pain models. Mol Pain. 2011;7:17. doi: 10.1186/1744-8069-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol. 2009;218:444–9. doi: 10.1002/jcp.21621. [DOI] [PubMed] [Google Scholar]
- 18.Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, Saito K, Nakamura T, Siomi H, Ito H, Arai Y, Shinomiya K, Takeda S. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A. 2009;106:20794–9. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin CY, Lee HC, Fu CY, Ding YY, Chen JS, Lee MH, Huang WJ, Tsai HJ. miR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nature communications. 2013;4:2829. doi: 10.1038/ncomms3829. [DOI] [PubMed] [Google Scholar]
- 20.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–5. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–78. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 25.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–54. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, Pu WT. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Molecular and cellular biology. 2009;29:2193–204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westendorp B, Major JL, Nader M, Salih M, Leenen FH, Tuana BS. The E2F6 repressor activates gene expression in myocardium resulting in dilated cardiomyopathy. Faseb J. 2012;26:2569–79. doi: 10.1096/fj.11-203174. [DOI] [PubMed] [Google Scholar]
- 28.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 30.Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. The Journal of cell biology. 2010;191:347–65. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadoshima J, Jahn L, Takahashi T, Kulik TJ, Izumo S. Molecular characterization of the stretch-induced adaptation of cultured cardiac cells. An in vitro model of load-induced cardiac hypertrophy. The Journal of biological chemistry. 1992;267:10551–60. [PubMed] [Google Scholar]
- 32.Aoki H, Richmond M, Izumo S, Sadoshima J. Specific role of the extracellular signal-regulated kinase pathway in angiotensin II-induced cardiac hypertrophy in vitro. The Biochemical journal. 2000;347(Pt 1):275–84. [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto S, Seta K, Morisco C, Vatner SF, Sadoshima J. Chelerythrine Rapidly Induces Apoptosis through Generation of Reactive Oxygen Species in Cardiac Myocytes. J Mol Cell Cardiol. 2001;33:1829–48. doi: 10.1006/jmcc.2001.1446. [DOI] [PubMed] [Google Scholar]
- 34.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, Bardeesy N, Camargo FD, Guan KL. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nature cell biology. 2012;14:1322–9. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jentzsch C, Leierseder S, Loyer X, Flohrschutz I, Sassi Y, Hartmann D, Thum T, Laggerbauer B, Engelhardt S. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2012;52:13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 36.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao X, Wang J, Wang Z, Du J, Yuan X, Huang W, Meng J, Gu H, Nie Y, Ji B, Hu S, Zheng Z. MicroRNA profiling during rat ventricular maturation: A role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS letters. 2013;587:1548–55. doi: 10.1016/j.febslet.2013.01.075. [DOI] [PubMed] [Google Scholar]
- 38.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–6. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMullen JR, Shioi T, Zhang L, Tarnavski O, Sherwood MC, Kang PM, Izumo S. Phosphoinositide 3-kinase(p110alpha) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc Natl Acad Sci U S A. 2003;100:12355–60. doi: 10.1073/pnas.1934654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 41.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, Mills GB, Guan KL. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–91. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odashima M, Usui S, Takagi H, Hong C, Liu J, Yokota M, Sadoshima J. Inhibition of endogenous Mst1 prevents apoptosis and cardiac dysfunction without affecting cardiac hypertrophy after myocardial infarction. Circ Res. 2007;100:1344–52. doi: 10.1161/01.RES.0000265846.23485.7a. [DOI] [PubMed] [Google Scholar]
- 43.Del Re DP, Matsuda T, Zhai P, Maejima Y, Jain MR, Liu T, Li H, Hsu CP, Sadoshima J. Mst1 Promotes Cardiac Myocyte Apoptosis through Phosphorylation and Inhibition of Bcl-xL. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, Pu WT. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 2012;109:2394–9. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaulk SG, Lattanzi VJ, Hiemer SE, Fahlman RP, Varelas X. The Hippo pathway effectors TAZ/YAP regulate dicer expression and microRNA biogenesis through Let-7. J Biol Chem. 2014;289:1886–91. doi: 10.1074/jbc.C113.529362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, Ghoshal K, Jacob ST. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–58. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, Morrisey EE, Tucker PW. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–87. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Li S, Yuan L, Tian Y, Weidenfeld J, Yang J, Liu F, Chokas AL, Morrisey EE. Foxp1 coordinates cardiomyocyte proliferation through both cell-autonomous and nonautonomous mechanisms. Genes Dev. 2010;24:1746–57. doi: 10.1101/gad.1929210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai S, Kerppola TK. Opposing roles of FoxP1 and Nfat3 in transcriptional control of cardiomyocyte hypertrophy. Mol Cell Biol. 2011;31:3068–80. doi: 10.1128/MCB.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–76. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 52.Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, Lim DS, Isobe M, Sadoshima J. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–88. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013;113:676–89. doi: 10.1161/CIRCRESAHA.113.300226. [DOI] [PubMed] [Google Scholar]
- 54.Mooren FC, Viereck J, Kruger K, Thum T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am J Physiol Heart Circ Physiol. 2014;306:H557–63. doi: 10.1152/ajpheart.00711.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.