Abstract
Mammalian Mitochondrial ncRNA is a web-based database, which provides specific information on non-coding RNA in mammals. This database includes easy searching, comparing with BLAST and retrieving information on predicted structure and its function about mammalian ncRNAs.
Availability
The database is available for free at http://www.iitm.ac.in/bioinfo/mmndb/
Keywords: ncRNA, Database, BLAST
Background
An interesting aspect of non-coding RNAs (ncRNA) is the execution of functional RNA molecules in most of the cellular processes, specifically under illness and various disorders in human [1, 2]. Moreover, these regions are cell and tissue specific, confirming their roles in mitochondrial gene expression [3]. Hence, annotating these non-coding regions would reveal their specific roles in genomes [4]. In our preliminary study, we have reported some of the mitochondrial non-coding regions and their amenability for diverse ailments in human [5]. Consequently, we envisaged to predict the mammalian mitochondrial ncRNA types and their secondary structures. Further, the annotated information has been utilized to construct a database for facilitating accessibility to scientific community. It also has the feasibility to perform Local Alignment using BLAST with the collected mammalian ncRNA.
Methodology
The mitochondrial DNA sequences were retrieved from NCBI database and we extracted the non-coding segments based on the location of reported coding regions using a Perl script. From the extracted non-coding regions, the representative ncRNAs have been identified using the program RNAcon [6]. Further, secondary structures were generated for all the predicted ncRNA using ipKnot [7] and viewed using VARNA software [8]. The database has been developed and configured using LAMP (Linux, Apache, MySQL, and PHP) server.
Database content, functions and utility:
This database contains three major utilities: (1) The statistical information about the predicted non-coding RNA types and their structures in 319 mammalian species; (2) keyword search consisting of four identification categories (i.e., MND_ID, NCBI ID, TYPE and ncRNA TYPE); and (3) the BLAST implementation to perform search against the database with RNA query sequences (Figure 1). The BLAST 2.2.25+ version is utilized with BLSOUM62 matrix, Gap penalties of 11 and extension 1 for alignment. The window dimension for the multiple hits depends on the end user query, whereas the BLAST hit result is directly linked to the database.
Figure 1.
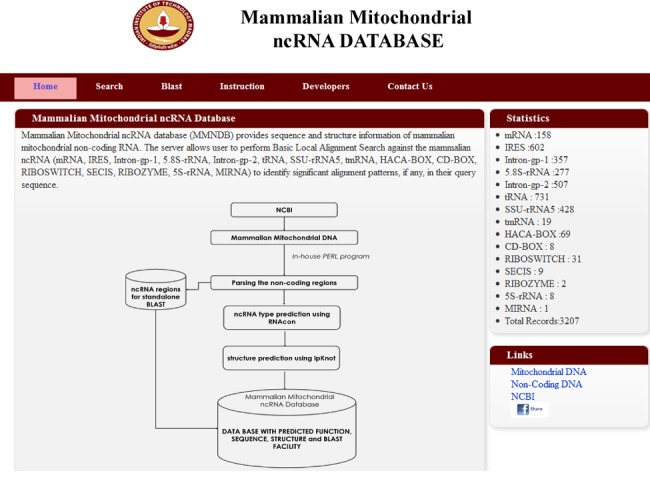
Snapshot of Mammalian Mitochondrial ncRNA Database
Future developments:
The current version of the database contains mammalian mitochondrial ncRNA related information. We plan to include the structural divergence of ncRNA in all other classes of whole genomes for data available in the NCBI database. We believe that the information available in the database will help in the functional annotation of mitochondrial ncRNA.
Acknowledgments
SA is grateful to University Grants Commission (UGC), Government of India for financial support (Award No. F.PDFSS-2014-15-SC-TAM-9117). The authors report no conflict of interest.
Footnotes
Citation:Anandakumar et al. Bioinformation 11(11): 512-513 (2015)
References
- 1.Eddy SR. Cell. 2002;109:137. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 2.Alexander RP, et al. Nat Rev Genet. 2010;8:559. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 3.Rackham O, et al. RNA. 2011;17:2085. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor RW, et al. Nat Rev Genet. 2015;6:389. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anandakumar S, et al. Mitochondrial DNA. 2014;20:1. [Google Scholar]
- 6.Panwar B, et al. BMC Genomics. 2014;15:127. doi: 10.1186/1471-2164-15-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato K, et al. Bioinformatics. 2011;27:i85. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darty K, et al. Bioinformatics. 2009;25:1974. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]


