Abstract
A growing number of studies employ time-averaged experimental data to determine dynamic ensembles of biomolecules. While it is well known that different ensembles can satisfy experimental data to within error, the extent and nature of these degeneracies, and their impact on the accuracy of the ensemble determination remains poorly understood. Here, we use simulations and a recently introduced metric for assessing ensemble similarity to explore degeneracies in determining ensembles using NMR residual dipolar couplings (RDCs) with specific application to A-form helices in RNA. Various target ensembles were constructed representing different domain-domain orientational distributions that are confined to a topologically restricted (<10%) conformational space. Five independent sets of ensemble averaged RDCs were then computed for each target ensemble and a ‘sample and select’ scheme used to identify degenerate ensembles that satisfy RDCs to within experimental uncertainty. We find that ensembles with different ensemble sizes and that can differ significantly from the target ensemble (by as much as ΣΩ ~ 0.4 where ΣΩ varies between 0 and 1 for maximum and minimum ensemble similarity, respectively) can satisfy the ensemble averaged RDCs. These deviations increase with the number of unique conformers and breadth of the target distribution, and result in significant uncertainty in determining conformational entropy (as large as 5 kcal/mol at T = 298 K). Nevertheless, the RDC-degenerate ensembles are biased towards populated regions of the target ensemble, and capture other essential features of the distribution, including the shape. Our results identify ensemble size as a major source of uncertainty in determining ensembles and suggest that NMR interactions such as RDCs and spin relaxation, on their own, do not carry the necessary information needed to determine conformational entropy at a useful level of precision. The framework introduced here provides a general approach for exploring degeneracies in ensemble determination for different types of experimental data.
Keywords: Dynamics, helix-junction-helix (HJH) motifs, inter-domain orientation, RNA dynamics, topological constraints, entropy, spin relaxation, ensemble similarity
INTRODUCTION
There is great interest in developing methods for determining population-weighed dynamic ensembles of biomolecules using experimental data1–8. The ensemble describes the energetic preferences of the many conformations that are typically sampled by a biomolecule in solution. This information in turn is critical for achieving a quantitative understanding regarding processes such as folding9–10, enzymatic catalysis11–14, and adaptive recognition15–17. In particular, the ensemble description provides a route for determining the free energy and entropy contributions to such processes arising due to conformational changes in biomolecules18–22. Common approaches used to determine dynamic ensembles of biomolecules seek to identify not a single static structure, but rather, an ensemble of population-weighted conformations that reproduces time-averaged experimental data within error23. The data that has been used so far include several NMR interactions such as residual dipolar couplings (RDCs)24–30, the Nuclear Overhauser effect (NOE)31–32, paramagnetic relaxation enhancements (PRE)33, chemical shifts (CS)34–36, as well as X-ray diffraction37 and small angle X-ray scattering (SAXS) and gold-SAXS data38–39. These data can have distinct spatial and temporal sensitivities and have been applied to determine ensembles for biomolecules including globular well-folded proteins, DNA, RNA, and intrinsically disordered proteins40–43. In particular, studies have examined the utility of RDCs in determining population-weighted ensembles of nucleic acids7,24,28 and protein1,2,25. These ensembles have provided important insights into functionally important motions including the role of dynamics in molecular recognition7,28,25. Despite advances in using such experimental approaches for determining dynamic ensembles of biomolecules, the source and magnitude of the uncertainties in ensemble determination are not fully understood.
Despite advances in experimental approaches for determining dynamic ensembles of biomolecules, the source and magnitude of uncertainties in ensemble determination are not fully understood. Rather, many ‘degenerate’ ensembles can typically satisfy data to within experimental error. Such degeneracies have not yet been rigorously explored for ensemble determination though they have long been recognized as a major source of uncertainty even when determining single-static structures by NMR spectroscopy and X-ray crystallography44–45. Bayesian approaches46 make it possible to estimate uncertainties in the populations of conformers in an ensemble, but they require a prior distribution, and can be computationally expensive. Understanding the nature of the degeneracies associated with a given type of experimental data is not only important to avoid over interpreting data, but also, to guide the design of approaches for optimally addressing these sources of uncertainty.
One of the challenges in determining degeneracies in ensemble determination, and more generally, in evaluating the accuracy with which ensembles can be determined using various methods, is the lack of metrics that can be used to quantitatively compare the similarity between two ensembles, for example, a target ensemble that is used to predict experimental data, and ensembles that are generated that satisfy the data. Commonly used metrics such as Jensen-Shannon divergence (Ω2)46–48 and the S-score49 and measure the extent of overlap between two ensemble distributions but fail to quantify the extent of structural similarity between non-overlapping parts48. Moreover, these measures of ensemble similarity can vary significantly depending on the size of the bin used to create the histogram distribution48. Recently, we introduced an approach for overcoming this problem that relies on summing the square root of Jensen-Shannon divergence (Ω) over variable bin size48. In particular, the normalized value of Ω summed over variable bin sizes (ΣΩ) is a metric that varies between 0 and 1 for perfect and zero similarity, respectively between two ensembles (see Methods). Here, we use ΣΩ and simulations to quantitatively examine degeneracies in determining ensembles with the use of NMR residual dipolar couplings (RDCs)50–51 measured in partially aligned systems.
There has been great interest in recent years to harness the broad time-scale and rich spatial sensitivity of RDCs in determining dynamic ensemble of biomolecules24–30. Here, we focus specifically on the problem of using RDCs to determine ensembles defining inter-domain orientation distributions. Domain-domain motions can significantly reorganize a biomolecule and play important roles in catalysis, ordered assembly of complexes, and adaptive recognition52–54. While our study will focus on RNA A-form helices and RDCs, the conclusions deduced equally apply to any chiral domain and extend to other anisotropic interactions such as residual chemical shift anisotropies (RCSA)55.
The determination of inter-helical ensembles for locally rigid A-form domains represents a best-case-scenario ensemble determination problem. First, one can pool together a large number of RDCs measured for various bond vectors within a domain to characterize what amounts to only three Euler angles describing the relative orientation of the two domains (Figure 1A). This is in contrast to determining an atomic resolution ensembles in which RDCs are parsed to determine distributions for many local degrees of freedom46, 49. Second, a realistic and highly restricted range of inter-helical orientations can be defined a priori in an unbiased manner based on simple topological constraints encoded by the junctions linking helices56–59. This obviates the need to rely on conformational pools derived from other methods such as molecular dynamics (MD) simulation in which correlations between various degrees of freedom can affect conformational sampling and data analysis. In addition, the topologically allowed conformational space is restricted to <10% of the total Euler space. This reduced conformational space captures realistic stereochemical constraints that serve to minimize degeneracies in ensemble determination. Our simulations also assume the theoretical maximum of five independent RDC data sets60–63, originating from five alignment tensors that are assumed to be known a priori, and decoupling limit in which one domain fully dominates alignment, thus avoiding any complications arising due to correlations between internal motions and overall reorientation7, 28. This framework allows us to hone in on the maximum information contained within second rank anisotropic interactions, and specifically, the twenty-five Wigner rotation elements that describe the information carried by five independent sets of RDCs in locally rigid domains64.
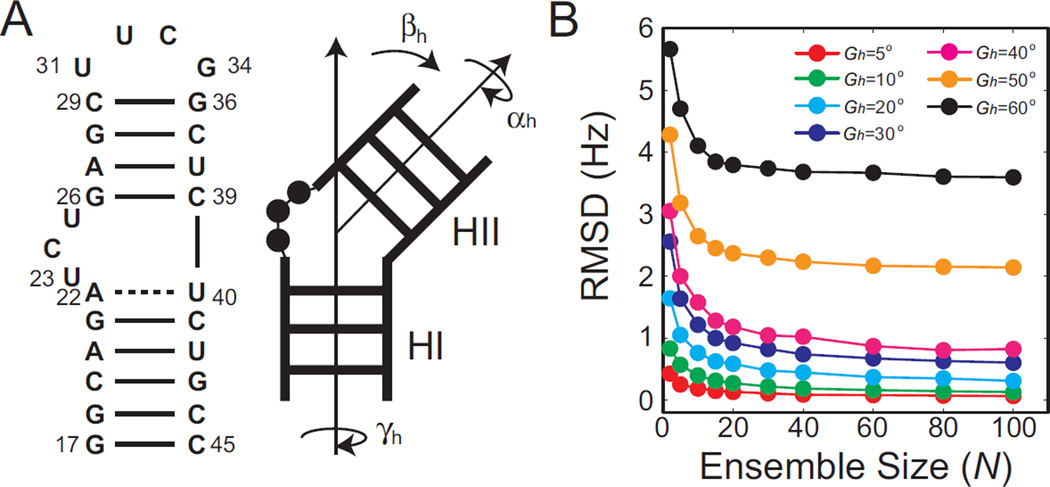
Figure 1. Defining an optimal grid size for binning histogram distributions of inter-helical angles.
(A) Secondary structure of the HIV-1 TAR used to model the two A-form helices linked by a bulge HJH motif and the inter-helical angles (αh βh γh) describing the relative orientation of the two A-form helices. (B) RMSD between five sets of RDCs calculated for ensembles derived from an MD simulation of HIV-1 TAR with variable size (N) and the RDC computed for corresponding grid-approximated conformational ensembles binned using variable grid size (Gh).
Our results reveal that ensembles with very different ensemble sizes and that can differ significantly from the target ensemble (by as much as ΣΩ ~ 0.4) can satisfy the ensemble averaged RDCs. These deviations increase with number of conformers in the target ensemble size, breadth of the distribution, and result in significant uncertainty in determining conformational entropy (as large as 5 kcal/mol at T = 298 K). Nevertheless, the RDC-degenerate ensembles are biased towards populated regions of the target ensemble, and capture other essential features of the distribution, including the shape.
METHODS
Conformational pool
All simulations for reconstructing ensembles employed a conformational pool of RNA inter-helical orientation consisting of two idealized A-form helices that are tethered together with a trinucleotide bulge helix-junction-helix (HJH) motif. The ‘lower’ helix was fixed and aligned with its helix axis oriented along the molecular z-direction. The ‘upper’ helix that was pivoted by placement of the phosphorus atom of the first nucleotide in the 3’ strand to the origin of the coordinate frame 57–59 (Figure 1A). The orientation of the upper helix relative to the lower helix is then specified using three inter-helical Euler angles (αh, βh, γh) (Figure 1A). αh and γh specify a rotation around the helical axis of the upper and lower helices respectively, and βh is the inter-helical bend angle (Figure 1A). The topological pool of allowed inter-helical conformations was constructed as described previously58 and consisted of all allowed inter-helical orientations subject to two topological constraints: i) helices cannot sterically collide with one another and ii) O3′(i) and P(i + 1) in the 3′ and 5′ helices across the bulge, respectively, cannot exceed the bulge linker length (4.9 Å per nucleotide)57–59. First, a non-degenerate Euler grid is generated by incrementing each of the three angles by 5° (i.e. each conformation differs from its closest neighbor by a 5° change in one of the three Euler angles)57–59. Note that each point on the grid represents a unique conformation. Next, all inter-helical orientations that fail to satisfy the above two topological constraints are discarded. For the trinucleotide bulge, only <10% of the space is topologically allowed. Note that for more complex RNA HJH motifs, the coarse grain model TOPRNA65 can be used to construct the typologically allowed inter-helical conformations. Additional simulations were carried out using an unrestricted conformational pool (see DISCUSSION and Supplementary Materials). This conformational pool is constructed by Euler angles (αh, βh, γh) ranging from −180° to 180° without degenerate Euler degeneracy (αh±180, ±βh, γh±180)57–59.
Grid Size
An optimum grid size of 5° for the conformational pool was selected that yields the lowest number of total conformations in the pool needed to reproduce a given set of RDC to within experimental uncertainty (<0.5 Hz). It was selected as following. A target ensemble containing N conformations was constructed by randomly selecting conformations from a 8.2µs MD simulation of HIV-1 TAR28 (see Identifying RDC-satisfying ensembles using sample and select). Next, each conformer in the selected ensemble was binned to a 3D Euler grid with grid size Gh (see Measuring similarity between RDC-satisfying and target ensembles). The binning is preformed as described previously48 by calculating the amplitude of single axis rotation that transforms the inter-helical orientation in a given conformer and every point on the grid, and the conformer is binned to the grid point with minimum amplitude single axis rotation i.e. to the grid point that is most similar to the selected conformer. In the case that more than one grid points show identical amplitude of single axis rotations, the grid point is set to be the one with smallest value of α2+β2+γ2. For both target and grid-approximated ensemble, five sets of ensemble averaged RDCs were then computed (see Computing RDCs) assuming five independent sets of hypothetical alignment tensors anchored on the lower helix48. The root-mean-square-deviation (RMSD) between the ensemble averaged RDCs for the target and grid-approximation was then computed and compared to RDC experimental uncertainty.
Building target ensembles
We examined RDC-satisfying ensembles for four types of target ensembles (i) Gaussian ensembles which are generated by restricting all three Euler angles in an Gaussian distribution with a specified standard deviation (σ= 10° or 30°) applied equally to all three Euler angles (ii) Random ensembles in which conformations are selected randomly from the topological pool (iii) discrete ensembles with N≤4 and (iv) ensemble of HIV-1 TAR inter-helical orientations obtained from an 8.2 µs MD trajectory generated using the CHARMM3666 force field on Anton supercomputer67. Ensembles (i)-(iii) were constructed using the topologically allowed inter-helical orientations for a 3-0 bulge58. The Gaussian ensembles were constructed by randomly picking a conformation (αm, βm, γm) as the mean conformation and then randomly selecting conformers from the pool and accepting them with a probability calculated using three Gaussian distributions on three Euler angles respectively: in which x=α, β, γ and xm=αm, βm, γm and σ is the pre-defined width of the Gaussian target ensemble. A random number r varying between 0 and 1 is then generated and compared to all three Gaussian probabilities. The randomly selected conformation is accepted to construct the Gaussian target ensemble if and only if r is smaller than all three Gaussian probabilities. While the Gaussian target ensemble allows different conformations to carry different population weights, the random ensemble consists of distinct conformations with equal population weight.
Computing RDCs
Five synthetic RDC data sets corresponding to five perfectly orthogonal alignment tensors64 were computed for all C-H vectors in the two helices of HIV-1 TAR assuming a standard A-form helical geometry68. The five orthogonal alignment tensors were fixed on the reference lower helix and generated using the Gram-Schmidt procedure 64. For each of the five alignment tensors, RDCs were computed using equation 1.
| (1) |
where γi and γj are the gyromagnetic ratios of spin i and j respectively; µ0 is the magnetic permittivity of vacuum; h is the Plank’s constant; rij is the separation between spin i and j, and θ represents the angle between the internuclear vector connecting spin i and j and the external magnetic field; Skl (k, l = x, y, z) are the elements of the alignment tensor represented in Cartesian form; ϕn is the angle between the bond vector and the corresponding nth axis (n = x, y, z).
For all ensembles used in this study, RDCs were computed for each conformation in an ensemble and averaged over all conformations to generate ensemble-averaged RDCs. The averaged RDCs were then error-corrupted by adding to each RDC a number randomly selected from a normal distribution with standard deviation equal to the assumed RDC error.
Identifying RDC-satisfying ensembles using sample and select
We use the computed RDCs and sample and select (SAS) approach8,24 to determine population-weighted inter-helical ensembles that satisfy RDCs to within experimental uncertainty. Here, RDCs were used to guide selection of N distinct conformations from a topological pool containing thousands of conformations such to identify ensembles that satisfy the measured RDCs. The initial N distinct conformations are randomly selected from the pool and the agreement between measured and predicted RDCs, χ2, is computed using equation 2.
| (2) |
where L is the number of RDCs. Next, one of the conformations is replaced randomly by another conformation from the remaining conformations in the pool, and the agreement with measured RDCs is re-examined and the newly selected conformation is either accepted or rejected based on the Metropolis criteria: at each step (k) of the selection procedure, the change from step k to k+1 is accepted if χ2 (k+1) < χ2 (k); if χ2 (k+1) ≥ χ2 (k) with a probability P=exp((χ2 (k)-χ2 (k+1))/T), where T is an effective temperature that is linearly decreased using a simulated-annealing scheme8,24. The initial effective temperature is set sufficiently high so that >99% of the conformations can be replaced and slowly decreased until the acceptance probability is smaller than 10−5. At each effective temperature, 200,000 steps were implemented followed by a decrease of effective temperature using Ti+1=0.9Ti. Using such a simulated annealing based approach, many SAS iterations are carried out until the penalty function shown in equation 2 is minimized, defined as achieving the best agreement with the measured RDCs at ensemble size N. The population-weighted ensemble is then constructed by combining the sub-ensembles from all SAS iterations.
Note that in the SAS analysis, a given conformation can only be selected once such that the reconstructed ensemble consists of equally populated conformations. Since the grid (Gh=5°) is significantly smaller than the RDC-distinguishable resolution (Figure 1B), ensembles with asymmetric population distributions can be obtained naturally by varying the number of neighboring conformations selected around a specific grid point. We avoid selecting the same conformation more than once to maximize sampling efficiency.
Measuring similarity between RDC-satisfying and target ensembles
The accuracy of RDC-determined ensembles was evaluated using a recently developed REsemble method48. In this approach, one computes the similarity between a target and RDC-determined ensembles at different bin sizes using the square root of Jensen-Shannon divergence (JSD), Ω, as shown in equation 346–48.
| (3) |
in which represent the population weights for the ith bin in ensemble T and P, respectively for a given bin size, m. S(wi)=−Σwi(m)log2wi(m) in equation 3 is the information entropy. Ω varies between 0 and 1 for maximum and minimum similarity, and is equal to zero if and only if . The sum of population overlap over all bin sizes normalized relative to values expected for worst prediction (e.g. random selection without guidance by RDCs) provides a convenient single-value measure of population overlap and structural similarity which we refer to as ΣΩ(wT, wP) that ranges between 0 and 1 for perfect and zero similarity, respectively,
| (4) |
in which K is the normalization factor. Note that ΣΩ(wT, wP) is also a metric, and therefore symmetric ΣΩ(wT, wP) = ΣΩ(wP, wT) and equal to zero if and only if two distributions are identical at all bin sizes or {wT}={wP}.
To measure the similarity between two ensembles, one has to have the ability to measure the similarity between distinct conformations within each ensemble. In the Euler conformational space, the straightforward Cartesian distance, ((αhA- αhB)2 + (βhA- βhB)2 + (γhA- γhB)2)1/2 between two sets of Euler angles A and B defining two distinct inter-helical orientations does not provide a measure of structural similarity between the two conformations and in general, the Cartesian distance between Euler angles can be smaller than, equal to or larger than the actual difference between two conformations48. Therefore we used the amplitude of single axis rotation to bin inter-helical orientations together and measure similarity between ensembles.
The binning grid points are constructed by picking a binning origin, defined by minimum value of each of the three Euler angle in the two ensembles upon comparison, and then incrementing each Euler angle by an amount defined by the bin size to cover the entire non-degenerate 3D Euler space. Next, the amplitude of a single axis rotation (ω) connecting a given conformation in the ensemble defined by Euler angles (αh1, βh1, γh1) and a point on the grid (αh2, βh2, γh2) is computed,
| (5) |
in which O(x, y, z, ω) represents a single axis rotation about a unit vector (x, y, z) with amplitude (ω) and the rotation amplitude ω is given by,
| (6) |
in which O11, O22 and O33 are the three diagonal elements of O(x, y, z, ω).
In this manner, the amplitude of the single axis rotation connecting a given conformation in an ensemble to every grid point is computed, and the conformation is binned to the grid point that leads to the minimum single axis rotation amplitude ω. In the case that more than one grid points show identical amplitude of single axis rotations, the grid point is set to be the one with the smallest value of α2 + β2 + γ2. The population of each grid point is then calculated to be the number of conformations binned divided by the total number of conformations in the ensemble. In our case, binning of the target and the predicted ensemble led to two population distributions on the same set of binning grids for a given bin size, and the value of Ω between the two ensembles at the given bin size (bin size ranges from 0° to 180° in increments of 10°) is then calculated using equation 3.
Calculating ensemble conformational entropy
The conformational entropy S for population-weighted ensembles can be computed using equation 7:
| (7) |
where R=1.987*10−3 kcal/mol/K is the gas constant and pi is the population of the ith conformation in the ensemble.
RESULTS
Defining an experimentally informed grid size for binning histogram distributions
It is generally not feasible to enumerate each and every conformation in an ensemble and experimentally determine its population weight. Two distinct conformations in the ensemble may not be distinguishable experimentally either because they differ in features that are not sensed by the data or because they do not differ sufficiently so as to give rise to experimental observations that differ within measurement uncertainty. Most ensemble determination approaches approximate the complex distribution into a discrete histogram distribution43,46,48. Here, the histogram bin size should be small enough to ensure that it properly captures the target ensemble distributions in so far as accounting for the time-averaged experimental data. Bin sizes that are larger than optimum can result in rough approximations that fail to capture features of the ensemble that is sensed by the experimental data whereas smaller bin sizes push the resolution limit beyond that achievable by the data and unnecessarily increases the conformational space that needs to be sampled.
In the context of defining the relative orientation of two domains using RDCs, we can define a 3D Euler grid covering all of the unique and topologically allowed (see Methods) inter-helical orientations56–59 (Figure 1A). The three Euler angles are incremented by a grid size given by Gh. Conformers in a given arbitrary ensemble can then be binned into their nearest points on the 3D grid, defined as the conformer that differs by the smallest amplitude single axis rotation48 (see Methods). We then asked the question; how small of an Euler grid size is needed when binning inter-helical ensembles to account for an ideal set of five independent sets of RDCs? We computed five sets of independent ensemble averaged RDCs for target ensembles with varying size (number of unique conformations, N = 1 – 100) that were randomly selected from an 8.2µs MD simulation for HIV-1 TAR28. We then binned all conformers in the ensemble to a set of grids with varying grid size Gh (grid-approximated ensemble, see Methods) and computed the expected RDCs for the grid-approximated ensemble. We then examined whether the RDCs for the target and grid-approximated ensembles are distinguishable based on typical RDC experimental uncertainty. These simulations assume alignment levels of Szz ~10−3.
As expected, the RMSD between the target and grid-approximated ensemble RDCs decreases with the decreasing grid size. Thus, as the grid size decreases, the grid-approximated ensemble becomes more similar to the actual target ensemble resulting in similar ensemble-averaged RDCs. Conversely, as the grid size increases, the grid-approximated ensemble differs considerable from the target ensemble resulting in increasing RDC RMSD values. Note that large increase in RMSD between Gh = 40° and Gh > 40° can be attributed to a rapid drop in number of unique conformations that are used to bin the ensemble for this particular target ensemble (Figure 1B).
The RDC RMSD between the target and grid-approximated ensembles also decreases with increasing number of unique conformations in the target ensemble regardless of the size of Gh (Figure 1B). This is because as N increases, any perturbation in the predicted RDCs for a given grid-approximated conformer can be compensated for in part by opposite deviations in a second conformer. This mutual cancelation arises due to the fact that different conformers will effectively experience independent perturbations when binned into the grid, resulting in near random perturbations to the RDCs that increasingly cancel with increasing N resulting in a lower RDC RMSD. Therefore a much larger Gh can be afforded for a larger ensemble size, and the ‘resolution’ with which the ensemble can be determined using RDCs decreases. This result underscores an important inverse relationship between the number of unique conformations that can be used to approximate an ensemble and the corresponding bin size that can be afforded to create the histogram distribution.
For the simulations that follow, we assume a grid size of Gh=5°. This guarantees a sufficiently small bin size that ensures we are working well below the theoretical resolution limit of RDCs.
RDC-satisfying ensembles with predefined size
Next, we used the SAS scheme to identify the family of degenerate ensembles that reproduce five sets of independent RDCs, computed for a variety of target ensembles, to within experimental error. One of the unknowns during an ensemble determination is the size of the target ensemble defined as the number of unique conformations given some assumed bin size. In general, there can be a wide range of ensembles that differ in size that can satisfy experimental data. We initially avoided such size-related degeneracies and examined the degeneracies that arise when assuming the target ensemble size is known. Here, the ensemble size used in the SAS scheme was set equal to the target ensemble size from which the RDCs were derived. In this manner, we asked the question, how similar are the ensembles satisfying the RDCs to the target ensemble when restricting the size of the ensemble to be equal that of the target ensemble? In all cases, the pool grid size was set to Gh=5°.
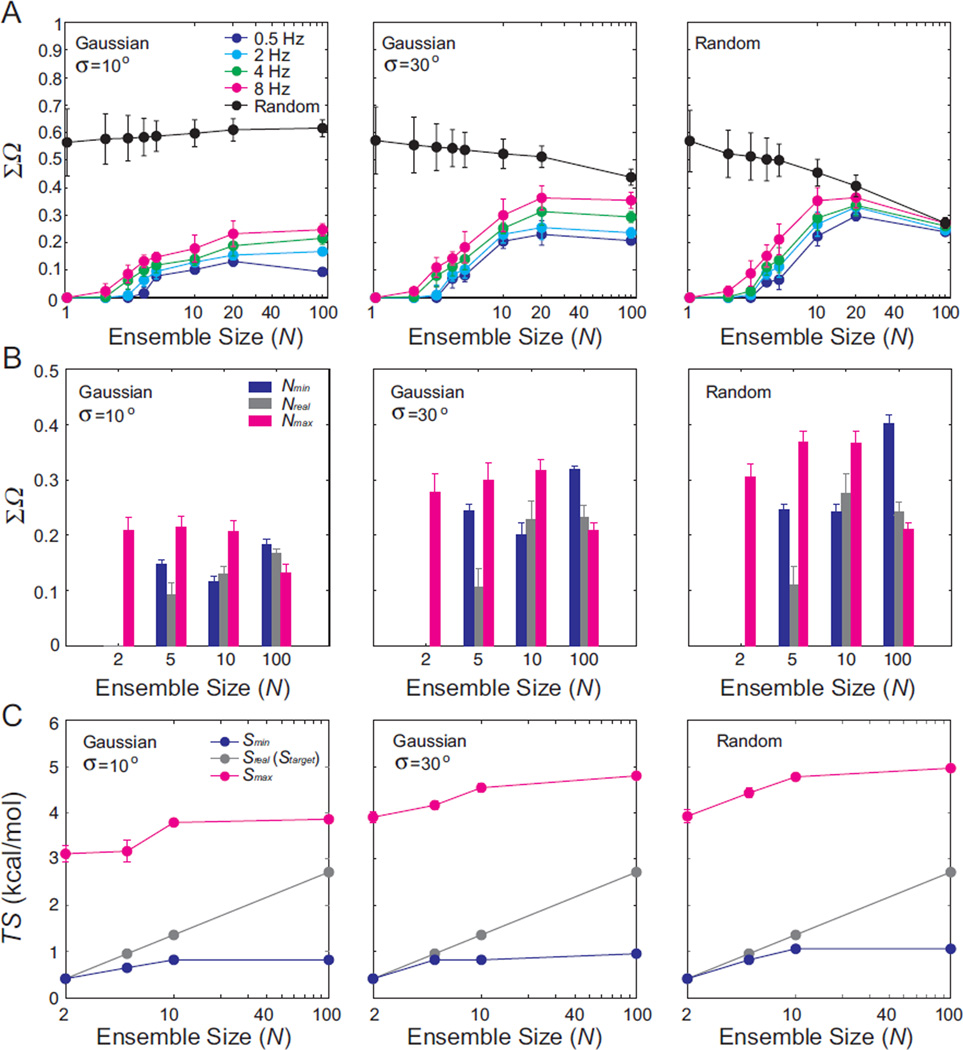
We carried out these simulations for Gaussian and random target ensemble distributions containing a variable number of conformations (N = 1–100) derived from the topologically allowed pool (see Methods). The RDC-satisfying ensembles were then compared to the target ensemble using the REsemble approach48. In particular, we computed ΣΩ which ranges between 0 and 1 for maximum and minimum similarity, respectively. For comparison, we also computed ΣΩ when comparing the similarity between the target ensemble and an ensemble of equal size that is selected randomly from the pool without guidance from the RDCs.
Shown in Figure 2 are the ΣΩ values for three different target ensembles. The results shown in Figure 2 represent averages and standard deviations over 100 independent SAS runs. Note that the similarity between RDC-satisfying ensembles obtained from independent SAS runs are well captured by the mean and standard deviation relative to the target ensemble. Overall we find good agreement between the RDC-satisfying and target ensembles with ΣΩ < 0.3 when assuming RDC uncertainty of 2 Hz, which can be compared to a 30° structural difference in a single structure (Figure S1). For comparison, ΣΩ > 0.3 and as large as ~ 0.6 when selecting ensembles randomly from the pool (Figure 2A, in black). The similarity between the RDC-satisfying and target ensemble generally decreases with increasing target ensemble size, conformational breadth, and RDC uncertainty. In some cases, we observe a decrease in ΣΩ in going from N=20 to N=100 particularly as the target ensembles increases in conformational breadth (Figure 2A, Table S1). This can be attributed to the fact that for target ensembles with broad distributions, there are fewer ensembles that can accomplish a given set of RDC averaging when restricting N to be large. Here, restricting the size of N increases the similarity between the RDC-satisfying and target ensembles (see section below). Note that sizeable ΣΩ values on the order of 0.3 are observed even when assuming negligible RDC uncertainty (<0.2Hz, data not shown). This indicates that at least some of the degeneracy manifests at levels far smaller than the typical RDC uncertainty.
Figure 2. Comparison of RDC-satisfying ensembles with target ensembles.
The similarity between RDC-satisfying ensembles with (A) prefixed and (B) variable ensemble size and target ensembles for three distinct distributions: Gaussian target ensemble with σ =10°, 30° and random target ensembles with RDC uncertainty. The values of ΣΩ represent the average over 100 simulations and the error bar represents the standard deviation. The similarity between the target ensemble and a randomly selected ensemble of equal size is also shown in black. (B) Results are shown for RDC-satisfying ensembles predicted using different ensemble sizes: Nmin (blue), Nreal(gray) and Nmax (magenta) using 2Hz RDC uncertainty. The values of ΣΩ represent the average over 100 simulations and the error bars represent the standard deviation. (C) Comparison of conformational entropy for RDC-satisfying ensembles determined using Nmin (blue), Nreal (gray) and Nmax (magenta) and for the target ensemble. Values represent the average of 100 simulations and error bars indicate the standard deviation.
RDC-satisfying ensembles without predefined size
In principle, ensembles with variable size can satisfy a given set of experimental data. In the SAS approach, one often selects the smallest sized ensemble (Nmin) that can satisfy experimental data to construct the ensemble and larger ensemble sizes are usually ignored due to the risk of overfitting of the experimental data2,26. However, the choice of Nmin is arbitrary and larger N values often satisfy experimental data within uncertainty. These RDC degenerate families of ensembles cannot be ruled out on the basis of the data, and therefore, represent another set of degeneracies that need to be considered.
We therefore repeated the SAS analysis for some of the target ensembles shown in Figure 2 to identify the degenerate family of RDC-satisfying ensembles with variable size that reproduce RDCs within the assumed 2 Hz uncertainty. We identified RDC-satisfying ensembles with the following sizes: Nmin which represents the smallest value of N that reproduces RDCs within uncertainty; Nreal which represents the real target ensemble size; and Nmax arbitrarily chosen to be 500 so long as it reproduces RDCs to within uncertainty. Note that for each target ensemble, the predicted ensemble using Nmin was constructed from more SAS runs compared to the ensemble predicted using Nmax so that the Nmin and Nmax RDC-satisfying ensembles have the same or very similar number of conformations. This avoids biased comparisons between Nmin and Nmax due to the different number of conformations.
Surprisingly, for all target ensembles examined, we were able to identify ensembles that satisfy the RDCs with Nmax = 500. This was the case even for relatively small target ensembles with N=1 or 2 (Figure 2B and Figure S2, in this Figure we show all of the RDC RMSD plots). Conversely, ensembles with small Nmin = 2–6 could be identified that satisfy RDCs for large sized target ensembles with N=100 (Figure 2B, S2). It should be noted that the variable sized ensembles reproduced the RDCs equally well with no obvious differences in frequency of outliers (Figure S3) in the commonly used “leave-out” cross-validation analysis23,27,43. Here, four out of the five RDC data sets are used to generate ensembles and the RMSD between measured and predicted RDCs determined for the left out dataset (Figure S4).
Interestingly, the variability in N only slightly increases the maximum ΣΩ values (Figure 2B, Table S2) relative to the ΣΩ values observed when N is pre-fixed (Figure 2A). However, it results in large ΣΩ values even for small target ensemble sizes where the ΣΩ values were much smaller in the case that the ensemble size as assumed known (Figure 2A, B). In general, we find that for small target ensembles with N ≤ 10, the larger ΣΩ values arise due to Nmax RDC-satisfying ensembles. Conversely, for target ensembles with N > 10, the largest ΣΩ values are generally observed with the smaller Nmin satisfying ensembles. For these cases, lack of knowledge of N results in large uncertainty in the RDC-satisfying ensembles. The deviations generally increase with increasing conformational breadth of the target ensembles. This increase in ensemble uncertainty with breadth of the distribution has been noted previously46,64. Interestingly, in some cases, especially for relatively large target ensembles (N=10, 100), the Nreal RDC-satisfying ensembles do not best resemble the target ensemble. This can be attributed to the combinatorial increase of distinct ensembles that have to be sampled with large Nreal and existence of degenerate ensembles that can satisfy RDCs to within experimental error. It should be noted that Nmax was arbitrarily set to 500 and that the similarity may decrease even further with larger Nmax values.
Conformational entropy
Next, we asked how well do the RDC-satisfying ensembles reproduce the conformational entropy of the target ensemble. We calculated the conformational entropy S of the ensembles (see Methods) predicted using Nmin (Smin), Nreal (Sreal), and Nmax (Smax), and compared these values to the entropy of the target (Starget). As shown in Figure 2C, the uncertainty in ensemble size described above results in a very large uncertainties in ensemble entropy. As expected, the Nmax RDC-satisfying ensembles yield Smax that are significantly larger than Starget especially for small target ensembles whereas Smin is generally smaller particularly for larger target ensembles. Overall, the values of Smin and Smax yield large differences of TS values that range between 0.4–5.0 kcal/mol at T = 298K (Figure 2C, Table S3).
RDC-satisfying ensembles capture key feature of target ensembles
Next, we examined how the RDC-satisfying ensembles capture various features of the target ensembles. Shown in Figure 3 are representative examples of Nmin and Nmax RDC-satisfying ensembles for target ensembles with N=100 Gaussian σ = 10° (Figure 3A), σ = 30° (Figure 3B) and randomly selected target ensemble (Figure 3C). Shown are 1D populated weighted distributions comparing target ensemble, RDC-satisfying ensembles, the topological pool along with corresponding 2D distributions with color-coded population weights.
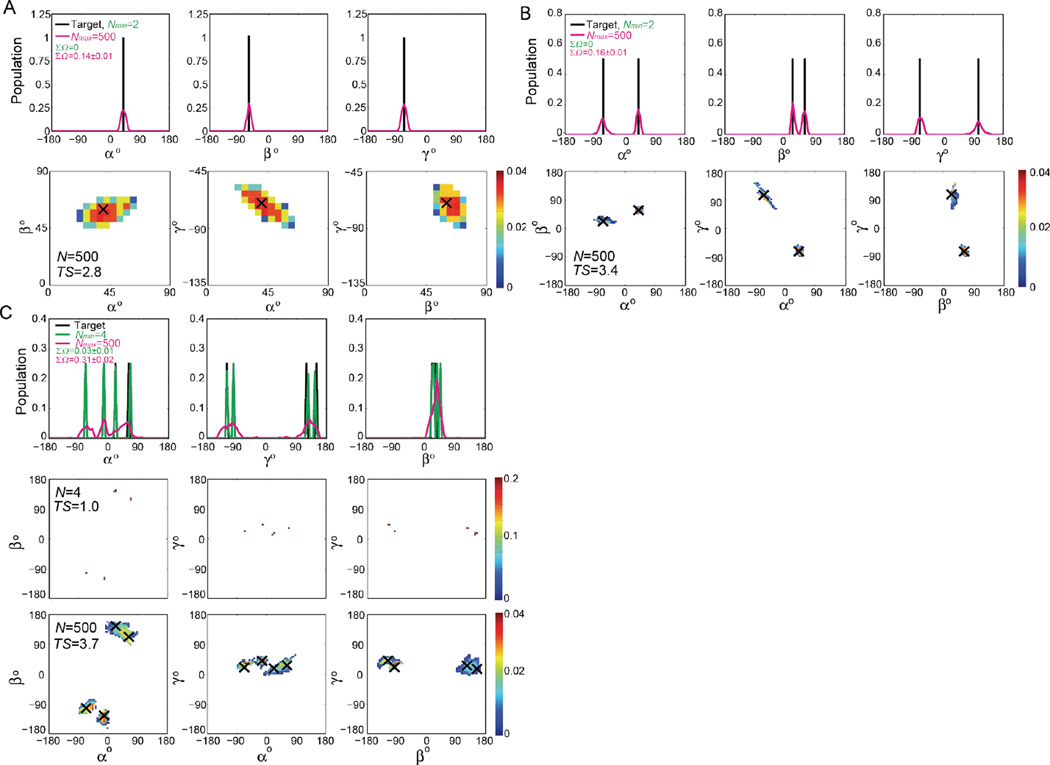
Figure 3. Comparison of RDC-satisfying and target ensembles: Gaussian and random target distributions.
1D and 2D population-weighted distributions of inter-helical angles ( αh βh γh) for Nmin (green) and Nmax (magenta) RDC-satisfying ensembles and corresponding target ensembles consisting of a Gaussian distribution with (A) σ = 10°, (B) σ = 30° and (C) randomly selected conformers from the pool. The value of ΣΩ comparing the RDC-satisfying and target ensemble and the TS values (in kcal/mol) are shown in the 2D distributions for each ensemble.
The Nmin RDC-satisfying ensembles give rise to discretized ensembles that feature a smaller number of conformations in and around the most populated conformations in the target. Nevertheless, many conformers are erroneously lowly populated and there are also small differences in the population-distributions in and around the dominant target conformations. On the other hand, the Nmax RDC-satisfying ensembles are broader, and while the most populated conformers are near the most populated conformers in the target ensemble, many conformers are also erroneously populated albeit typically to low levels that fall outside the target. On a quantitative level, the differences between the Nmin and Nmax RDC-satisfying ensembles ΣΩ(Nmin-Nmax) range between 0.2 and 0.4 and are comparable to the ΣΩ values observed when compared against the target ensembles. Visual inspection of these distributions shows that despite their obvious differences, both Nmin and Nmax RDC-satisfying ensembles recapitulate key features of the target distribution, including its breadth and shape (Figure 3).
We also carried out detailed simulations for more discrete smaller sized target ensembles with N=1, 2 and 4 conformations (Figure 4). In general, the RDC-satisfying ensembles are more similar to the target ensemble as compared to the ensembles with large size (Figure 4). The Nmin ensembles nearly perfectly reproduce the target ensemble while Nmax erroneous broadening around the dominant conformations (Figure 4). Interestingly, this broadening is not as significant as observed with target ensembles with much larger size (Figure 3) and there are fewer significantly populated conformations outside the dominant conformations. As expected, the differences between the Nmin and Nmax RDC-satisfying ensembles with ΣΩ(Nmin-Nmax) ranging between 0.1 and 0.3 are comparable to the ΣΩ values observed when compared against the target ensembles. The RDC satisfying ensemble capture salient feature of the distributions, including for example, whether the target ensemble is unimodal (Figure 4A), bimodal (Figure 4B) or tetramodal (Figure 4C).
Figure 4. Comparison of RDC-satisfying and target ensembles: Discrete distributions.
1D and 2D population-weighted distributions of inter-helical angles (αh βh γh) for Nmin (green) and Nmax (magenta) RDC-satisfying ensembles and corresponding target ensembles consisting of a discrete distributions with size (A)N=1, (B)N=2, and (C)N=4. The value of ΣΩ comparing the RDC-satisfying and target ensemble is shown in each case. The Nmin RDC-satisfying ensembles perfectly overlap with the target ensembles and therefore only the Nmax RDC-satisfying ensembles are shown in the 2D distributions with target conformations indicated using black crosses. The value of ΣΩ comparing the RDC-satisfying and target ensemble and the TS values (in kcal/mol) are shown in the 2D distributions for each ensemble.
It is important to note that even for N=1 target ensemble, the SAS scheme identifies an Nmax = 500 ensemble that reproduces the RDCs within error (Figure S3). This ensemble features broadening around the conformer in the target resulting in standard deviation in β ~10° (Figure 4A). This highlights how RDCs can be misinterpreted in terms of dynamics. In particular, deviations in σ(β) < 10° are likely difficult to characterize using RDCs with uncertainty 2Hz and alignment levels of Szz ~ 10−3.
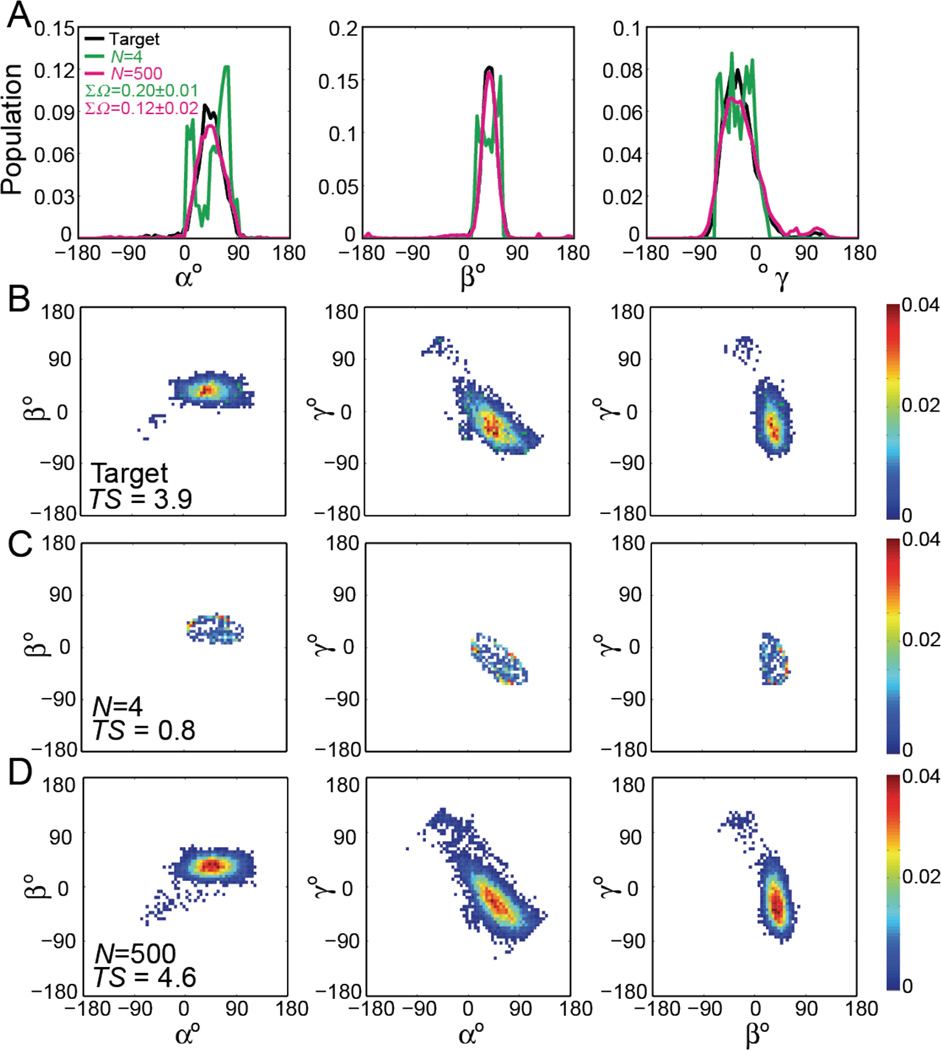
Finally, we examined a more realistic target ensemble corresponding to 8000 conformations obtains from an 8.2 µs MD trajectory of HIV-1 TAR28 generated using the CHARMM36 force field66 and the Anton supercomputer67 (see Methods). RDC-satisfying ensembles could be identified with Nmin=4 and Nmax=500 (Figure S5). The Nmax=500 RDC-satisfying ensemble is more similar to the target ensemble (ΣΩ=0.12±0.02) as compared to the Nmin=4 RDC-satisfying ensemble (ΣΩ=0.20±0.01) (Figure 5A). Despite these differences, the Nmin=4 and Nmax=500 capture salient features and dominant conformations in the target MD ensemble. As shown in Figure 5B–D, both ensembles capture the most populated region of the target ensemble that is not captured by the starting 3-0 junction-topology conformational pool although observable differences exist in lowly populated regions. While the Nmin=4 ensemble is focused on the mostly populated conformations, the Nmax=500 ensemble is broadened and samples many conformations around the populated regions. The target MD ensemble has an entropy TS = 3.9 kcal/mol at T= 298K. By comparison, the ensembles reconstructed using Nmin=4 was 0.8 kcal/mol and that of Nmax=500 was 4.6 kcal/mol at T = 298K. This uncertainty in free energy, particularly for larger and broader target ensembles, is way too large to allow meaningful insights into the role of conformational entropy in basic biological processes.
Figure 5. Comparison of RDC-satisfying and target ensembles: MD trajectory of HIV-1 TAR.
1D and 2D population-weighted distributions of the inter-helical angles (αh βh γh) for Nmin (green) and Nmax (magenta) RDC-satisfying ensembles and the corresponding target ensemble constructed by selecting conformers from an MD trajectory of HIV-1 TAR. The value of ΣΩ comparing the RDC-satisfying and target ensemble and the TS values (in kcal/mol) are shown in the 2D distributions.
It is worth noting that although our analysis assumes a topologically restricted inter-helical conformational space, similar results were obtained when using an unbiased unrestricted conformational pool spanning the entire Euler angle space (Figure S6). Although the ΣΩ values of the simulations using unrestricted pool are slightly larger than for the case using topologically restricted pool (Figure S6), the RDC-satisfying ensemble obtained from the entire pool capture salient features of different target ensembles (Figure S6). This is again consistent with degeneracies representing various degrees of broadening around the target conformations rather than having distinct minima along the landscape. Nevertheless, we note that the topologically allowed conformational pool may help reduce degeneracies in ensemble determination when using less than five independent RDC datasets and improve sampling particularly for systems with many inter-helical junctions.
Sparse RDC datasets
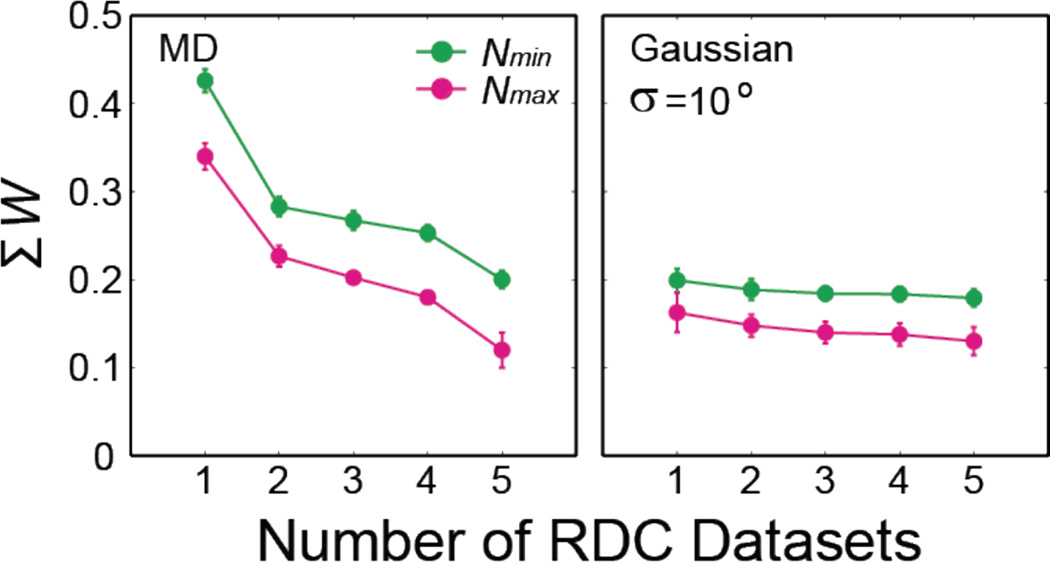
While our simulations assume a theoretical maximum of five independent datasets, in practice, such measurements are not easily accessible28. We therefore also explored the more realistic scenario involving a smaller number of RDC datasets. As expected, reducing the number of independent RDC datasets resulted in RDC-satisfying ensembles that can differ even more significantly from target ensembles. However, in general, this decrease in similarity with decreasing number of RDC datasets was more significant for broad target ensembles such as the MD trajectory of HIV-1 TAR, where ΣΩ from Nmax increases from 0.12±0.02 with five RDC datasets to 0.34±0.01 with only one RDC dataset as compared to the narrower Gaussian target ensemble with σ =10° where the corresponding ΣΩ increase is from 0.13±0.01 to 0.16±0.02 (Figure 6). These results indicate that it may be possible to determine inter-helical ensemble with reasonable accuracy for rigid molecules such as mutants of TAR16 and ligand-bound states of TAR7,69–70 with experimentally accessible RDCs. Further simulations showed that even for flexible RNAs such as HIV-1 TAR, the ensembles predicted using Nmin and Nmax and experimentally accessible RDCs sample the ligand-bound states within experimental uncertainty, although ensembles predicted using Nmin and Nmax have different subtle details of conformational distributions (Figure S7).
Figure 6. Prediction of target ensembles using different number of independent RDC datasets.
Shown are the accuracy (ΣΩ) of predicted ensembles using Nmin (green) and Nmax(magenta) for the MD trajectory of HIV-1 TAR (left) and Gaussian target ensemble with σ = 10° (right).
DISCUSSION
For decades, structural biologists have struggled with the problem that while it may be feasible to find a structural model that satisfies experimental data obtained by NMR or X-ray diffraction, it is very difficult to enumerate all other models that satisfy the data equally well. Not too surprisingly, this problem is exacerbated when determining dynamic ensembles of biomolecules, given the large number of ensemble combinations that have to be tested. These degeneracies have not yet been actively examined in ensemble determination. Understanding the nature of the degeneracies is not only key to avoid over interpreting data, but also, to define approaches that can help improve ensemble determination, for example, by suitable combination of complimentary data. Prior studies28,71 have estimated uncertainty in ensembles by using Monte Carlo type approaches. These approaches primarily provide an estimate of precision in ensemble determination and may not fully capture ensemble accuracy or the range of degenerate ensembles that equally satisfy the data such as the degenerate ensembles with different size and entropy uncovered in this work. In this work, we sought to develop a general framework for unveiling such ensemble degeneracies with specific application to NMR RDCs.
We designed our simulations to represent a ‘best-case-scenario’ ensemble determination problem where RDCs are calculated and predicted using five linearly independent alignment tensors. This allowed us to identify the degeneracies and sources of uncertainty that restrict the theoretical limits with which ensembles can be defined using the information contained in anisotropic interactions such as RDCs. We find that even under these ideal conditions, RDC-satisfying ensemble can vary significantly in size and differ from the target ensemble by ΣΩ ~ 0.4. This degeneracy increases with the breadth of the target ensemble and makes it difficult to extract quantitative thermodynamic information of interest. For example, the uncertainty translates into large errors in ensemble entropy, as high as ~5 kcal/mol or ~1.7 kcal/mol per degree of freedom. One can in principle gauge the amplitude of inter-helical motions and obtain estimates of the uncertainty in ensemble determination by using the RDC data to determine the generalized degree of order (GDOint), which is the ratio of the degree of order measured in each of the two domains64. Like any order parameter, the GDOint varies between 0 (maximum motions) to 1 (minimum motions). As might be expected, systems with lower GDOint represent broader ensembles, which are more susceptible to uncertainty including due to uncertainty in the ensemble size. Indeed, we find a good correlation between the uncertainty in the entropy Smax -Smin and the GDOint of the target ensemble (data not shown). Therefore, the GDOint can be used to obtain a priori information regarding the breadth of the distribution and potential uncertainties in parameters such as conformational entropy. In practice, the accuracy of inter-helical RNA ensembles determined with RDCs as the only source of constraints are likely to be less reliable, not only due to more limited RDCs, broader conformational space, but also because of inherent approximations that have to be assumed when computing RDCs for a candidate conformer24,28. On the other hand, inclusion of RDCs on linker residues i.e. the helix junction motifs, may help further constrain the linker conformation and thereby facilitate definition of inter-helical ensemble28. Further studies are needed to examine these effects and to evaluate how inclusion of other experimental data may help further improve determination of RNA ensembles.
In prior studies, we reported ensembles of HIV-1 TAR with size N=37 and N=2024, 28. In both cases, the ensembles come close to sampling seven distinct ligand bound conformations of TAR7,24. This is consistent with our findings here that salient features of ensembles can be determined despite the uncertainty due to ensemble size. For completion, we also generated RDC-satisfying ensembles for HIV-1 TAR with Nmin = 3 and Nmax = 500. The ensembles sample similar conformational regions and come close to sampling ligand bond states (Figure S7).
In general, the RDC-satisfying ensembles are biased toward the populated regions of the target ensemble distribution, and capture other features, including the breadth and shape. We did not find any evidence for RDC-satisfying ensembles that do not strongly resemble the target ensemble. Rather, the degeneracies are better described as random broadening around the target conformations. Nevertheless, the RDC-satisfying ensembles can populate conformers that are never sampled in the real target ensemble or conversely, can result in depopulation of regions that are clearly populated in the target ensemble. These results emphasize the need to account for such uncertainty during the ensemble determination process.
The ensemble degeneracy, particularly due to variability in ensemble size, contributes significantly to uncertainty in conformational entropy. In this regard, it should be noted that spin relaxation data measured on amide backbones and methyl groups in proteins, which are based on the same basic NMR anisotropic interactions, have been used extensively to characterize conformational entropy in proteins18–22. This represents a significantly smaller amount of data than used here. In particular, the five orthogonal sets of RDCs measured on locally rigid domains carries information described by 25 ensemble-averaged Wigner rotation elements7,64. Here, all 25 Wigner elements are effectively applied in the ensemble determination. On the other hand, in studies that rely on measurements of spin relaxation data, one typically relies on a single Lipari-Szabo order parameter for one bond or methyl group - which effectively represents only one independent Wigner element18–22. While our results suggest that it should not be feasible to determine entropy based on such parameters, it is possible that unique attributes of motions in proteins are such to permit estimation of entropy from such reduced parameters. Nevertheless, our study emphasizes the need to carefully evaluate the relationship between entropy and order parameters for different systems that may exhibit different dynamic attributes including nucleic acids studied here.
It is very likely the case that the ensemble degeneracies uncovered here will be common in other types of time-averaged experimental data. What is less clear is whether these degeneracies will be orthogonal, so that combining data together can help resolve uncertainties including those related to ensemble size. The framework introduced here can be generally applied to test intrinsic degeneracies in ensemble determination, and can be immediately applied to explore ideal combinations of data that can help overcome these sources of uncertainty.
CONCLUSION
Our results reveal that ensembles with variable size (or number of unique conformations) can satisfy the RDCs to within experimental uncertainty. This results in significant uncertainty in determining thermodynamic parameters of interest, such as conformational entropy. Nevertheless, the RDC-satisfying ensemble did not differ from the target ensembles by more than ΣΩ ~ 0.4 and are significantly more similar to the target ensemble as compared to those selected randomly from the pool. Future studies should quantitatively evaluate how inclusion of other types of experimental data including on the helix-junction-helix motif linkers can improve the accuracy and uniqueness with which ensembles can be determined. The framework introduced here can readily be applied to evaluate other types of data.
Supplementary Material
Acknowledgments
We thank members of the Al-Hashimi laboratory for critical comments on the manuscript. This work was supported by the US National Institutes of Health (R01AI066975 and PO1GM0066275).
Footnotes
Supporting information
A full set of ensemble prediction and cross-validation results. This material is available free of charge via the Internet at http://pubs.acs.org.
Competing financial interests
H.M.A. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company. The research reported in this article was performed by the Duke University faculty, research associate and was funded by US National Institute of Health contract to H.M.A.
References
- 1.Ravera E, Salmon L, Fragai M, Parigi G, Al-Hashimi H, Luchinat C. Insights into Domain-Domain Motions in Proteins and RNA from Solution NMR. Acc. Chem. Res. 2014;47:3118–3126. doi: 10.1021/ar5002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter B, Gsponer J, Varnai P, Salvatella X, Vendruscolo M. The Mumo (Minimal under-Restraining Minimal over-Restraining) Method for the Determination of Native State Ensembles of Proteins. J. Biomol.NMR. 2007;37:117–135. doi: 10.1007/s10858-006-9117-7. [DOI] [PubMed] [Google Scholar]
- 3.Beauchamp KA, Pande VS, Das R. Bayesian Energy Landscape Tilting: Towards Concordant Models of Molecular Ensembles. Biophys. J. 2014;106:1381–1390. doi: 10.1016/j.bpj.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Showalter SA, Johnson E, Rance M, Bruschweiler R. Toward Quantitative Interpretation of Methyl Side-Chain Dynamics from NMR by Molecular Dynamics Simulations. J. Am. Chem. Soc. 2007;129:14146–14147. doi: 10.1021/ja075976r. [DOI] [PubMed] [Google Scholar]
- 5.Bertini I, Luchinat C, Nagulapalli M, Parigi G, Ravera E. Paramagnetic Relaxation Enhancement for the Characterization of the Conformational Heterogeneity in Two-Domain Proteins. Phys. Chem. Chem. Phys. 2012;14:9149–9156. doi: 10.1039/c2cp40139h. [DOI] [PubMed] [Google Scholar]
- 6.Salmon L, Pierce L, Grimm A, Ortega Roldan JL, Mollica L, Jensen MR, van Nuland N, Markwick PR, McCammon JA, Blackledge M. Multi-Timescale Conformational Dynamics of the Sh3 Domain of Cd2-Associated Protein Using NMR Spectroscopy and Accelerated Molecular Dynamics. Angew. Chem. Int. Ed. Engl. 2012;51:6103–6106. doi: 10.1002/anie.201202026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing Spatially Correlated Dynamics That Directs RNA Conformational Transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Campbell SL, Dokholyan NV. Deciphering Protein Dynamics from NMR Data Using Explicit Structure Sampling and Selection. Biophys. J. 2007;93:2300–2306. doi: 10.1529/biophysj.107.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furtig B, Buck J, Manoharan V, Bermel W, Jaschke A, Wenter P, Pitsch S, Schwalbe H. Time-Resolved NMR Studies of RNA Folding. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 10.Chu VB, Lipfert J, Bai Y, Pande VS, Doniach S, Herschlag D. Do Conformational Biases of Simple Helical Junctions Influence RNA Folding Stability and Specificity? RNA. 2009;15:2195–2205. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple Native States Reveal Persistent Ruggedness of an RNA Folding Landscape. Nature. 2010;463:681–684. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris DA, Rueda D, Walter NG. Local Conformational Changes in the Catalytic Core of the Trans-Acting Hepatitis Delta Virus Ribozyme Accompany Catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 13.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A Conformational Switch Controls Hepatitis Delta Virus Ribozyme Catalysis. Nature. 2004;429:201–5. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 14.Shi X, Mollova ET, Pljevaljcic G, Millar DP, Herschlag D. Probing the Dynamics of the P1 Helix within the Tetrahymena Group I Intron. J. Am. Chem. Soc. 2009;131:9571–9578. doi: 10.1021/ja902797j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehr DD, Nussinov R, Wright PE. The Role of Dynamic Conformational Ensembles in Biomolecular Recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA Dynamics by Design: Biasing Ensembles Towards the Ligand-Bound State. Angew. Chem. Int. Ed. Engl. 2010;49:5731–5733. doi: 10.1002/anie.201000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Structures of RNA Switches: Insight into Molecular Recognition and Tertiary Structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 18.Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational Entropy in Molecular Recognition by Proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee AL, Sharp KA, Kranz JK, Song XJ, Wand AJ. Temperature Dependence of the Internal Dynamics of a Calmodulin-Peptide Complex. Biochemistry. 2002;41:13814–13825. doi: 10.1021/bi026380d. [DOI] [PubMed] [Google Scholar]
- 20.Kasinath V, Sharp KA, Wand AJ. Microscopic Insights into the NMR Relaxation-Based Protein Conformational Entropy Meter. J. Am. Chem. Soc. 2013;135:15092–15100. doi: 10.1021/ja405200u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzeng SR, Kalodimos CG. Protein Activity Regulation by Conformational Entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 22.Karplus M, Ichiye T, Pettitt BM. Configurational Entropy of Native Proteins. Biophys. J. 1987;52:1083–1085. doi: 10.1016/S0006-3495(87)83303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmon L, Yang S, Al-Hashimi HM. Advances in the Determination of Nucleic Acid Conformational Ensembles. Annu. Rev. Phys. Chem. 2013;65:293–316. doi: 10.1146/annurev-physchem-040412-110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank AT, Stelzer AC, Al-Hashimi HM, Andricioaei I. Constructing RNA Dynamical Ensembles by Combining Md and Motionally Decoupled NMR Rdcs: New Insights into RNA Dynamics and Adaptive Ligand Recognition. Nucleic Acids Res. 2009;37:3670–3679. doi: 10.1093/nar/gkp156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition Dynamics up to Microseconds Revealed from an RDC-Derived Ubiquitin Ensemble in Solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 26.Nodet G, Salmon L, Ozenne V, Meier S, Jensen MR, Blackledge M. Quantitative Description of Backbone Conformational Sampling of Unfolded Proteins at Amino Acid Resolution from NMR Residual Dipolar Couplings. J. Am. Chem. Soc. 2009;131:17908–17918. doi: 10.1021/ja9069024. [DOI] [PubMed] [Google Scholar]
- 27.Jensen MR, Markwick PR, Meier S, Griesinger C, Zweckstetter M, Grzesiek S, Bernado P, Blackledge M. Quantitative Determination of the Conformational Properties of Partially Folded and Intrinsically Disordered Proteins Using NMR Dipolar Couplings. Structure. 2009;17:1169–1185. doi: 10.1016/j.str.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Salmon L, Bascom G, Andricioaei I, Al-Hashimi HM. A General Method for Constructing Atomic-Resolution RNA Ensembles Using NMR Residual Dipolar Couplings: The Basis for Interhelical Motions Revealed. J. Am. Chem. Soc. 2013;135:5457–5466. doi: 10.1021/ja400920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and Dynamics of Micelle-Associated Human Immunodeficiency Virus Gp41 Fusion Domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 30.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR Evidence for Slow Collective Motions in Cyanometmyoglobin. Nat. Struc. Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 31.Vogeli B, Orts J, Strotz D, Chi C, Minges M, Walti MA, Guntert P, Riek R. Towards a True Protein Movie: A Perspective on the Potential Impact of the Ensemble-Based Structure Determination Using Exact Noes. J. Magn. Reson. 2014;241:53–59. doi: 10.1016/j.jmr.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Blackledge MJ, Bruschweiler R, Griesinger C, Schmidt JM, Xu P, Ernst RR. Conformational Backbone Dynamics of the Cyclic Decapeptide Antamanide. Application of a New Multiconformational Search Algorithm Based on NMR Data. Biochemistry. 1993;32:10960–10974. doi: 10.1021/bi00092a005. [DOI] [PubMed] [Google Scholar]
- 33.Salmon L, Nodet G, Ozenne V, Yin G, Jensen MR, Zweckstetter M, Blackledge M. NMR Characterization of Long-Range Order in Intrinsically Disordered Proteins. J. Am. Chem. Soc. 2010;132:8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 34.Frank AT, Horowitz S, Andricioaei I, Al-Hashimi HM. Utility of (1)H NMR Chemical Shifts in Determining RNA Structure and Dynamics. J. Phys. Chem. B. 2013;117:2045–2052. doi: 10.1021/jp310863c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M. Using NMR Chemical Shifts as Structural Restraints in Molecular Dynamics Simulations of Proteins. Structure. 2010;18:923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Jensen MR, Salmon L, Nodet G, Blackledge M. Defining Conformational Ensembles of Intrinsically Disordered and Partially Folded Proteins Directly from Chemical Shifts. J. Am. Chem. Soc. 2010;132:1270–1272. doi: 10.1021/ja909973n. [DOI] [PubMed] [Google Scholar]
- 37.Fenwick RB, van den Bedem H, Fraser JS, Wright PE. Integrated Description of Protein Dynamics from Room-Temperature X-Ray Crystallography and NMR. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E445–E454. doi: 10.1073/pnas.1323440111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang X, et al. An Unusual Topological Structure of the Hiv-1 Rev Response Element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi X, Herschlag D, Harbury PA. Structural Ensemble and Microscopic Elasticity of Freely Diffusing DNA by Direct Measurement of Fluctuations. Proc Natl Acad Sci U S A. 2013;110:E1444–E1451. doi: 10.1073/pnas.1218830110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sterckx YG, et al. Small-Angle X-Ray Scattering- and Nuclear Magnetic Resonance-Derived Conformational Ensemble of the Highly Flexible Antitoxin Paaa2. Structure. 2014;22:854–865. doi: 10.1016/j.str.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Jensen MR, Ruigrok RW, Blackledge M. Describing Intrinsically Disordered Proteins at Atomic Resolution by NMR. Curr. Opin. Struct. Biol. 2013;23:426–435. doi: 10.1016/j.sbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Sibille N, Bernado P. Structural Characterization of Intrinsically Disordered Proteins by the Combined Use of NMR and Saxs. Biochem Soc Trans. 2012;40:955–62. doi: 10.1042/BST20120149. [DOI] [PubMed] [Google Scholar]
- 43.Clore GM, Schwieters CD. Amplitudes of Protein Backbone Dynamics and Correlated Motions in a Small Alpha/Beta Protein: Correspondence of Dipolar Coupling and Heteronuclear Relaxation Measurements. Biochemistry. 2004;43:10678–10691. doi: 10.1021/bi049357w. [DOI] [PubMed] [Google Scholar]
- 44.Aboul-ela F, Karn J, Varani G. Structure of Hiv-1 Tar Rna in the Absence of Ligands Reveals a Novel Conformation of the Trinucleotide Bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DePristo MA, de Bakker PI, Blundell TL. Heterogeneity and Inaccuracy in Protein Structures Solved by X-Ray Crystallography. Structure. 2004;12:831–838. doi: 10.1016/j.str.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 46.Fisher CK, Huang A, Stultz CM. Modeling Intrinsically Disordered Proteins with Bayesian Statistics. J. Am. Chem. Soc. 2010;132:14919–14927. doi: 10.1021/ja105832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindorff-Larsen K, Ferkinghoff-Borg J. Similarity Measures for Protein Ensembles. PLoS One. 2009;4:e4203. doi: 10.1371/journal.pone.0004203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang S, Salmon L, Al-Hashimi HM. Measuring Similarity between Dynamic Ensembles of Biomolecules. Nat. Methods. 2014;11:552–554. doi: 10.1038/nmeth.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Simone A, Richter B, Salvatella X, Vendruscolo M. Toward an Accurate Determination of Free Energy Landscapes in Solution States of Proteins. J. Am. Chem. Soc. 2009;131:3810–3811. doi: 10.1021/ja8087295. [DOI] [PubMed] [Google Scholar]
- 50.Tjandra N, Bax A. Direct Measurement of Distances and Angles in Biomolecules by Nmr in a Dilute Liquid Crystalline Medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 51.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear Magnetic Dipole Interactions in Field-Oriented Proteins - Information for Structure Determination in Solution. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shapiro YE. Nmr Spectroscopy on Domain Dynamics in Biomacromolecules. Prog. Biophys. Mol. Biol. 2013;112:58–117. doi: 10.1016/j.pbiomolbio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Hoffman RM, Sykes BD. Disposition and Dynamics: Interdomain Orientations in Troponin. Adv. Exp. Med. Biol. 2007;592:59–70. doi: 10.1007/978-4-431-38453-3_7. [DOI] [PubMed] [Google Scholar]
- 54.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional Complexity and Regulation through RNA Dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cornilescu G, Bax A. Measurement of Proton, Nitrogen, and Carbonyl Chemical Shielding Anisotropies in a Protein Dissolved in a Dilute Liquid Crystalline Phase. J. Am. Chem. Soc. 2000;122:10143–10154. [Google Scholar]
- 56.Bailor MH, Mustoe AM, Brooks CL, 3rd; Al-Hashimi HM. Topological Constraints: Using RNA Secondary Structure to Model 3d Conformation, Folding Pathways, and Dynamic Adaptation. Curr. Opin. Struct. Biol. 2011;21:296–305. doi: 10.1016/j.sbi.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailor MH, Mustoe AM, Brooks CL, 3rd; Al-Hashimi HM. 3d Maps of RNA Interhelical Junctions. Nat. Protoc. 2011;6:1536–1545. doi: 10.1038/nprot.2011.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mustoe AM, Bailor MH, Teixeira RM, Brooks CL, 3rd; Al-Hashimi HM. New Insights into the Fundamental Role of Topological Constraints as a Determinant of Two-Way Junction Conformation. Nucleic Acids Res. 2012;40:892–904. doi: 10.1093/nar/gkr751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailor MH, Sun X, Al-Hashimi HM. Topology Links RNA Secondary Structure with Global Conformation, Dynamics, and Adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 60.Tolman JR. A Novel Approach to the Retrieval of Structural and Dynamic Information from Residual Dipolar Couplings Using Several Oriented Media in Biomolecular NMR Spectroscopy. J. Am. Chem. Soc. 2002;124:12020–12030. doi: 10.1021/ja0261123. [DOI] [PubMed] [Google Scholar]
- 61.Briggman KB, Tolman JR. De Novo Determination of Bond Orientations and Order Parameters from Residual Dipolar Couplings with High Accuracy. J. Am. Chem. Soc. 2003;125:10164–10165. doi: 10.1021/ja035904+. [DOI] [PubMed] [Google Scholar]
- 62.Meiler J, Prompers JJ, Peti W, Griesinger C, Bruschweiler R. Model-Free Approach to the Dynamic Interpretation of Residual Dipolar Couplings in Globular Proteins. J. Am. Chem. Soc. 2001;123:6098–6107. doi: 10.1021/ja010002z. [DOI] [PubMed] [Google Scholar]
- 63.Peti W, Meiler J, Bruschweiler R, Griesinger C. Model-Free Analysis of Protein Backbone Motion from Residual Dipolar Couplings. J. Am. Chem. Soc. 2002;124:5822–5833. doi: 10.1021/ja011883c. [DOI] [PubMed] [Google Scholar]
- 64.Fisher CK, Zhang Q, Stelzer A, Al-Hashimi HM. Ultrahigh Resolution Characterization of Domain Motions and Correlations by Multialignment and Multireference Residual Dipolar Coupling NMR. J. Phys. Chem. B. 2008;112:16815–16822. doi: 10.1021/jp806188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mustoe AM, Al-Hashimi HM, Brooks CL., 3rd Coarse Grained Models Reveal Essential Contributions of Topological Constraints to the Conformational Free Energy of RNA Bulges. J. Phys. Chem. B. 2014;118:2615–2627. doi: 10.1021/jp411478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacKerell AD, Banavali N, Foloppe N. Development and Current Status of the Charmm Force Field for Nucleic Acids. Biopolymers. 2000;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 67.Shaw DE, et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 68.Musselman C, Pitt SW, Gulati K, Foster LL, Andricioaei I, Al-Hashimi HM. Impact of Static and Dynamic a-Form Heterogeneity on the Determination of RNA Global Structural Dynamics Using NMR Residual Dipolar Couplings. J. Biomol. NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 69.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-Arginine Complex by NMR Spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 70.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. Argininamide Binding Arrests Global Motions in Hiv-1 TAR RNA: Comparison with Mg2+-Induced Conformational Stabilization. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang J, Warner LR, Sanchez C, Frank G, Tobias M, Mackereth CD, Sattler M, Blackledge M. Transient Electrostatic Interactions Dominate the Conformational Equilibrium Sampled by Multidomain Splicing Factor U2AF65: A Combined NMR and SAXS Study. J. Am. Chem. Soc. 2014;136:7068–7076. doi: 10.1021/ja502030n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.