Abstract
Although molecular tests for drug-resistant TB perform well on culture isolates, their accuracy using clinical samples, particularly from TB and HIV-endemic settings, requires clarification. The MTBDRplus and MTBDRsl line probe assays were evaluated in 181 sputum samples and 270 isolates from patients with culture-confirmed drug-sensitive-TB, MDR-TB, or XDR-TB. Phenotypic culture-based testing was the reference standard. Using sputum, the sensitivities for resistance was 97.7%, 95.4%, 58.9%, 61.6% for rifampicin, isoniazid, ofloxacin, and amikacin, respectively, whereas the specificities were 91.8%, 89%, 100%, and 100%, respectively. MTBDRsl sensitivity differed in smear-positive vs. smear-negative samples (79.2% vs. 20%, p < 0.0001 for ofloxacin; 72.9% vs. 37%, p = 0.0023 for amikacin) but not by HIV status. If used sequentially, MTBDRplus and MTBDRsl could rule-in XDR-TB in 78.5% (22/28) and 10.5% (2/19) of smear-positive and smear-negative samples, respectively. On culture isolates, the sensitivity for resistance to rifampicin, isoniazid, ofloxacin, and amikacin was 95.1%, 96.1%, 72.3% and 76.6%, respectively, whereas the specificities exceeded 96%. Using a sequential testing approach, rapid sputum-based diagnosis of fluoroquinolone or aminoglycoside-resistant TB is feasible only in smear-positive samples, where rule-in value is good. Further investigation is required in samples that test susceptible in order to rule-out second-line drug resistance.
Tuberculosis (TB) remains a leading cause of morbidity and mortality in developing countries42. National and global TB control efforts are undermined by the emergence of drug-resistant TB. MDR-TB is defined as resistance to rifampicin [RIF] and isoniazid [INH], and extensively drug-resistant TB (XDR-TB) is defined as MDR-TB plus resistance to a fluoroquinolone [FLQ] and a second-line injectable drug, such as amikacin [AMK], or capreomyin [CAP]2,3. DR-TB is associated with high mortality4,5,6,7, is a threat to healthcare workers8,9, and results in unsustainable costs that destabilise national TB control programmes (NTPs)42,10. If patients are placed on effective treatment earlier11,12,13, which can be facilitated by rapid genotypic rather than the slower phenotypic diagnostic testing, transmission will be reduced and the clinical prognosis of these patients will likely be improved.
In contrast to testing methods like the nitrate reductase assay14 and Microscopic Observation Drug Susceptibility (MODS)15, the MTBDRplus line probe assay (LPA; Hain Lifescience, Nehren, Germany), which is approved by the World Health Organisation (WHO) for the detection of RIF and INH resistance, is a same day test with a short relatively rapid within laboratory turn-around-time (~5 hours). It has an estimated sensitivity and specificity of 98.1% and 98.7% for both RIF and INH, when performed on culture isolates16, however, there are limited data about its performance using sputum. Several recent reports have reported a sensitivity of ~95% in smear-positive samples and ~65% in smear-negative samples17,18,19,20,21. Another LPA, MTBDRsl, was developed to diagnose XDR-TB by detecting mutations in the gyrA and rrs genes, thereby determining susceptibility to the FQs (ofloxacin, moxifloxacin, levofloxacin), and the second line injectable drugs (SLIDs; amikacin, kanamycin, and capreomycin). The assay displays a sensitivity and specificity of 85.1% and 98.2% to detect FQ resistance when performed directly (using smear-positive sputum samples)22, and 83.1% and 97.7, respectively when performed indirectly (using culture isolates)22. MTBDRsl exhibited sensitivities and specificities of 76.9% and 99.5% for the SLID class when performed indirectly (using isolates) and 94.4% and 98.2%, respectively when performed using smear-positive sputum samples (directly).
MTBDRsl makes the rapid same-day diagnosis of XDR-TB possible when it is used in combination with rapid tests for MDR-TB such as MTBDRplus. However, there are several gaps in our knowledge before such a strategy can be applied in appropriate settings. There are few data from a small number of cases about the performance of MTBDRsl using clinical samples23,24,25, none of which were smear-negative, and there are no studies evaluating the impact of HIV on accuracy. Moreover, a sequential testing strategy to inform clinical practice (e.g. MTBDRplus followed by MTBDRsl), and determinants of performance in this context, has hitherto not been evaluated. To address these considerations we evaluated the comparative diagnostic accuracy of the MTBDRplus and MTBDRsl assays using smear-positive and smear-negative sputum samples, and culture isolates obtained from patients in a TB and HIV endemic setting.
Methods
Study site and population
Sputum was collected from 234 patients enrolled in Brooklyn Chest Hospital, in Cape Town, South Africa, or undergoing routine testing at a centralised testing reference laboratory (National Health Laboratory Services [NHLS] at Groote Schuur Hospital). Brooklyn Chest Hospital is the designated provincial treatment centre for XDR-TB in the Western Cape. Brooklyn Chest Hospital also enrols patients with other types of drug-resistant TB. We accessed sputum or culture isolates from patients with culture-confirmed MDR-TB, or XDR-TB. The sputum samples and culture isolates came from different patients. Patients with confirmed drug susceptible isolates were sourced from the NHLS at Groote Schuur Hospital. DR-TB were based on phenotypical DST results. Patients were on anti-TB treatment at the time of specimen collection. In addition to the specimen collected for LPA testing, we collected a paired, second specimen that was used for microscopy and liquid culture. A HIV test was performed after appropriate counselling. This study was approved by the University of Cape Town Faculty of Health Sciences Ethics Committee and the methods were carried out in accordance with the approved guidelines. All patients provided written informed consent.
Specimen processing
Samples were processed using the standard NALC-NaOH method (final NaOH concentration 1%26). Smear microscopy was performed using Ziehl-Neelsen staining. The WHO-recommended critical concentrations for RIF (1 μg/mL), INH (1 μg/mL), AMK (1 μg/mL) and ofloxacin (OFX) (2 μg/mL), were used for DST using the MGIT 960 liquid culture system (BD Bioscience, Erebodegem, Belgium27).
MTBDRplus and MTBDRsl line probe assays
The MTBDRplus assay (version 1) and the MTBDRsl assay (version 1) were performed directly on a single sputum sediments. Sputum from 181 culture-positive patients received both tests. MTBDRplus and MTBDRsl were also performed indirectly on culture isolates (MTBDRplus and MTBDRsl; n = 270 received both tests) according to the manufacturer’s instructions (Hain Lifescience, Germany). The person performing the tests was blinded to the reference standard results. Manufacturer-recommended polymerase (HotStarTaq; Qiagen) was used for both LPAs, and the PCR on DNA from culture isolates was performed using the following parameters: 95 °C for 15 min, 95 °C for 30 s, 58 °C for 2 min (10 cycles), 95°C for 25 s, 53 °C for 40 s, 70 °C for 40 s (20 cycles) and final extension at 70 °C for 8 min (the parameters used for detecting DNA from sputum using the LPAs used 30 cycles of elongation). A valid LPA result was defined by a Mycobacterium tuberculosis complex-specific control (TUB), conjugate controls (CC) and amplification control (AC) bands in conjunction with the target gene locus control.
Discrepant analysis
Sequencing was performed on the inhA promoter, rpoB, katG, gyrA and rrs genes, from isolates that were discrepant between phenotypic DST and either of the LPAs. The sequences of the primers used can be found in supplementary Table S1.
Statistics
The sensitivity and specificity were calculated for each drug compared to the gold standard of culture-based DST. Patients whose paired sputum specimen was culture-negative were excluded. Statistical analyses were performed using Graphpad Prism (version 6.0; GraphPad Software, USA, www.graphpad.com), and STATA SE (version 12; StataCorp, USA). P-values less than 0.05 were considered significant. Fisher’s exact test with mid-P correction was used for comparisons between proportions.
Results
Patients and samples
The demographic data of the patients enrolled in the study is shown in Table 1. Demographic data was unavailable for 7/234 patients because of technical problems accessing the electronic NHLS records.
Table 1. Demographic data of the cohorts used in study.
| Demographic data | Study Cohort (%) (n = 227)* |
|---|---|
| Age | |
| Mean years (range) | 37 (18–111) |
| Sex | |
| Male | 110 (48) |
| Female | 117 (52) |
| Race | |
| Black | 109 (48) |
| Mixed | 118 (52) |
| HIV-infected | |
| Yes | 107 (47) |
| No | 113 (50) |
| Unknown | 7 (3) |
| CD4 count (cells/mL) rangeΨ | 308 (2–983) |
*Demographic data for 7 patients was missing.
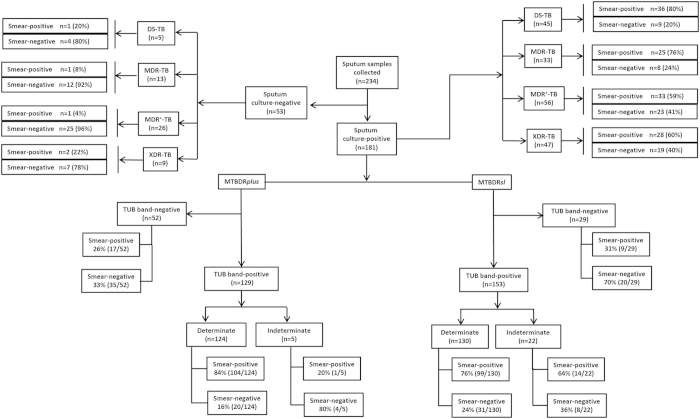
Figure 1 shows the study plan of the 234 patients tested using the LPAs. Fifty three patients were excluded because they were culture-negative. Of the 181 culture-positive samples, 45, 33, 56 and 47 were, according to phenotypic DST, DS-, MDR, MDR+ (MDR-TB and resistance to a FQ or SLID but did not meet the criteria for XDR-TB) and XDR, respectively.
Figure 1. Study plan showing the number of sputum samples tested directly using MTBDRplus (version 1) or MTBDRsl (version 1) according to patients’ diagnoses and smear-status.
The diagnosis was obtained using phenotypic liquid culture-based DST on a specimen collected at the same time as the specimen used for the line probe assays. A test is classified as positive for the Mycobacterium tuberculosis complex by the presence of the M. tb complex band (TUB), while a test is classified as negative by the absence of the M. tb complex band (TUB). Indeterminate results are those which are TUB-band positive yet are missing controls bands for gene specific loci. TB = tuberculosis; DS = drug sensitive, MDR = multi drug resistant, MDR+ = MDR-TB but with additional resistance to OFX, KAN or INH. XDR = extensively drug resistant.
MTBDRplus performance
Direct testing of sputum samples by MTBDRplus
Accuracy: The diagnostic accuracy of MTBDRplus is shown in Table 2. The accuracy (sensitivity, specificity) for RIFR and INHR were (97.7%, 91.8%) and (95.4%, 89%), respectively. When the discrepant results were resolved by sequencing, the accuracy (sensitivity, specificity %) to detect RIFR and INHR increased to (100%, 100%) and (97.7%, 97.4%), respectively.
Table 2. Diagnostic accuracy of MTBDRplus and MTBDRsl for the direct detection of drug resistance in sputum samples using phenotypic culture-based susceptibility testing as a reference standard.
| All sputum samples |
Smear-positive sputum |
Smear-negative sputum |
|||||
|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | ||
| MTBDRplus‡(v 1.0) | RIFR* | 97.7 (86/88) | 91.8 (34/37) | 97.1 (69/71) | 91.2 (31/34) | 100 (17/17) (p = 0.484) | 100 (3/3) |
| INHR* | 95.4 (84/88) | 89 (33/37) | 95.6 (68/71) | 88.2 (30/34) | 94.1 (16/17) (p = 0.768) | 100 (3/3) | |
| MTBDRsl†(v 1.0) | OFXR | 58.9 (43/73) | 100 (38/38) | 79.2 (38/48) | 100 (34/34) | 20 (5/25) (p < 0.001) | 100 (4/4) |
| AMKR | 61.6 (45/73) | 100 (38/38) | 72.9 (35/48) | 100 (34/34) | 37 (10/27) (p < 0.001) | 100 (4/4) | |
‡ 7.4% (9/122) MTBDRplus results from smear-positive samples were indeterminate, compared to 17% (10/59) from smear-negative (p = 0.049).
†1.6% (2/122) MTBDRsl results from smear-positive samples were indeterminate, compared to 5.1% (3/59) from smear-negative (p = 0.185). Refer to the materials and methods for a description of what defines an indeterminate result. P-values are for comparisons between smear statuses.
*When the discrepant results were resolved by sequencing the sensitivities and specificities of MTBDRplus were 100% and 100% to detect RIFR, respectively and 97.7% and 97.4% to detect INHR, respectively. RIFR = rifampicin resistance, INHR = isoniazid resistance, OFXR = ofloxacin resistance, AMKR = amikacin resistance.
Indeterminate rate: The indeterminate rates are shown in Fig. 1. Amongst the MTBDRplus-TUB-positive sputum samples, 4% (5/129) were indeterminate. Twenty percent and 80% were smear-positive and smear-negative, respectively (p = 0.058).
Impact of HIV: Table 3 shows the sensitivities and specificities amongst samples from HIV-infected or -uninfected patients. The sensitivities and specificities to detect RIFR or INHR did not change according to HIV status.
Table 3. Diagnostic accuracy of MTBDRplus and MTBDRsl for the direct detection of drug resistance in sputum samples according to HIV status compared to phenotypic culture-based susceptibility testing (reference standard).
| All sputum samples |
Smear-positive sputum |
Smear-negative sputum |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-infected |
HIV-uninfected |
HIV-infected |
HIV-uninfected |
HIV-infected |
HIV-uninfected |
||||||||
| Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | Sensitivity(%) | Specificity(%) | ||
| MTBDRplus(v 1.0) | RIFR | 96.7 (29/30) | 90 (9/10) | 98 (49/50) (p = 0.712) | 88.2 (15/17) (p = 0.888) | 100 (23/23) | 100 (8/8) | 97.7 (43/44) (p = 0.466) | 100 (14/14) | 100 (7/7) | 100 (1/1) | 100 (8/8) | 100 (1/1) |
| INHR | 93.3 (28/30) | 90 (9/10) | 96 (48/50) (p = 0.596) | 88.2 (15/17) (p = 0.888) | 91.6 (22/24) | 100 (8/8) | 97.6 (41/42) (p = 0.264) | 50 (2/4) | 100 (7/7) | 100 (1/1) | 87.5 (7/8) (p = 0.333) | 100 (1/1) | |
| MTBDRsl(v 1.0) | OFXR | 48.1 (13/27) | 94.4 (17/18) | 64.1 (25/39) (p = 0.197) | 83.3 (25/30) (p = 0.260) | 69 (11/16) | 100 (9/9) | 82.1 (23/28) (p = 0.308) | 100 (17/17) | 18.2 (2/11) | 100 (2/2) | 18.2 (2/11) | 100 (1/1) |
| AMKR | 59.2 (16/27) | 100 (18/18) | 64.1 (25/39) (p = 0.690) | 90 (27/30) (p = 0.166) | 62.5 (10/16) | 100 (9/9) | 78.6 (22/28) (p = 0.250) | 100 (17/17) | 50 (6/12) | 100 (2/2) | 27.3 (3/11) (p = 0.265) | 100 (1/1) | |
P-values are for comparisons between HIV statuses.
RIFR = rifampicin resistance, INHR = isoniazid resistance, OFXR = ofloxacin resistance, AMKR = amikacin resistance.
Indirect testing of the culture isolates
Accuracy: The diagnostic accuracy of MTBDRplus for the culture isolates is shown in Table 4. The LPA had a sensitivity and specificity to detect RIFR of 95.1% (95% CI 92.2% to 98.1%) and 100%, respectively and a sensitivity and specificity of 96.1% (93.5% to 98.7%) and 96.1% (90.8% to 100%), respectively to detect INHR.
Table 4. Diagnostic accuracy of MTBDRplus and MTBDRsl for the detection of drug resistance in culture isolates compared to phenotypic culture-based susceptibility testing (reference standard).
| Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|
| MTBDRplus(v 1.0) | RIFR | 95.1% (196/206) (p = 0.117) | 100% (51/51) (p = 0.039) |
| INHR | 96.1% (198/206) (p = 0.495) | 96.1% (49/51) (p = 0.698) | |
| MTBDRsl (v 1.0) | OFXR | 72.3% (115/159) (p = 0.042) | 99.0% (100/101) (p = 0.538) |
| AMKR | 76.6% (125/157) (p = 0.004) | 98.0% (99/101) (p = 0.382) | |
P-values are for comparisons between direct testing on specimens (both smear-positive and smear-negative; data shown in Table 2).
Indeterminate rate: The study plan for the 270 culture isolates is shown in Figure S1. From the 270 culture isolates tested, 95.2% (257/270) were TUB band-positive and all were MTBDRplus determinate. The indeterminate rate amongst the direct testing of the sputum samples (5/129) was significantly different to the indirect testing of the culture isolates (257/257; p < 0.001).
MTBDRsl performance
Direct testing of sputum samples by MTBDRsl
Accuracy: The diagnostic accuracy for MTBDRsl is shown in Table 2. Overall the LPA exhibited suboptimal sensitivity for OFXR (58.9% [95% CI 47.6% to 70.2%]) and AMKR (61.6% [50.4% to 72.8%]). However, sensitivity was higher in smear-positive sputum samples (OFXR: 79.2% [95% CI 67.7% to 90.7%] and AMKR: 72.9% [60.3% to 85.5%], respectively; p = 0.473) compared to smear-negative sputum samples (OFXR: 20% [4.3% to 35.7%; p < 0.001] and AMKR: 37%; [18.8% to 55.2%; p < 0.001]), respectively. MTBDRsl displayed excellent specificities of 100% to detect OFXR and AMKR in sputum. Furthermore, the sensitivities and specificities of MTBDRsl to detect OFXR and AMKR did not significantly change when the discordant results were resolved by sequencing.
Impact of HIV: The diagnostic accuracy of MTBDRsl when stratified according to HIV status is shown in Table 3. Similarly to MTBDRplus, the sensitivities and specificities of MTBDRsl amongst the samples from HIV-infected versus HIV-uninfected patients were not significantly different at 48.1% versus 64.1% (p = 0.197) and 94.4% versus 83.3% (p = 0.26) for OFXR and 59.2% versus 64.2% (p = 0.690) and 100% versus 90% (p = 0.166) for AMKR.
Indeterminate rate: Figure 1 shown that indeterminate rates for MTBDRsl. From the 153 TUB-positive sputum samples tested by MTBDRsl, 14.4% (22/153) were indeterminate, of which 64% (14/22) and 36% (8/22) were smear-positive and smear-negative, respectively (p = 0.070). The overall indeterminate rates are depicted in Table 2. Of the MTBDRsl TUB-positive results from smear-positive samples 1.6% (2/122) were indeterminate, compared to 5.1% (3/59) from smear-negative samples (p = 0.185).
Indirect testing of the culture isolates
Accuracy: The accuracy of MTBDRsl for the culture isolates is shown in Table 4. Indirect testing of the culture isolates by MTBDRsl showed a sensitivity and specificity of 72.3% (115/159) and 99% (100/101) for OFXR, respectively. For AMKR the sensitivity and specificity was 76.6% (125/157) and 98% (99/101), respectively.
Indeterminate rate: The study plan of the culture isolates is shown in Figure S1. From the 270 culture isolates tested indirectly by MTBDRsl, 97% (262/270) were TUB band-positive and all were determinate. The proportion of indeterminate results for MTBDRsl between direct testing of sputum (14.4% [22/153]) and indirect testing of isolates (0% [262/262]; p < 0.001) was statistically different.
Comparison of direct and indirect testing for MTBDRsl
A comparison of the accuracy for the culture isolates versus the sputum samples using MTBDRsl is shown in Tables 4 and 2, respectively. MTBDRsl had increased sensitivity to detect OFXR indirectly on the culture isolates (72.3% [115/159]) compared to direct testing of the sputum samples (58.9% [43/73]; p = 0.042). MTBDRsl had an improved sensitivity to detect AMKR indirectly (76.6% [125/157] versus directly on sputum (61.6% [54/73]; p = 0.004).
XDR-TB diagnosis by sequential use of MTBDRplus and MTBDRsl on sputum samples
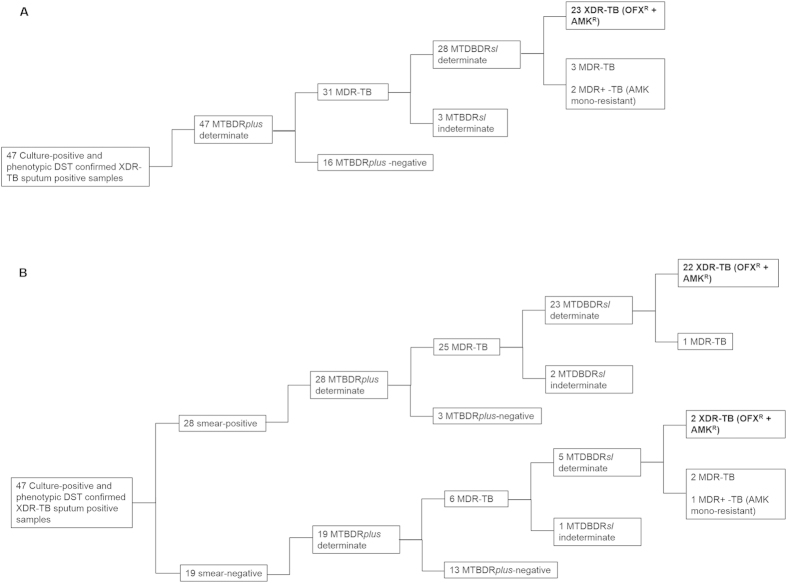
OVERALL: Fig. 2A, depicts the ability of MTBDRsl to detect XDR-TB when performed directly and in conjunction with MTBDRplus. From the 47 culture-positive and phenotypically-confirmed XDR sputum samples, all 47 were determinate for MTBDRplus and 31 of these were detected as MDR-TB (RIF and INH resistant). Of these 31, 93.3% (28/30) were MTBDRsl-determinate and 23 were detected as XDR-TB (resistance to OFX and AMK). When used sequentially on sputum samples, MTBDRplus and MTBDRsl could thus rule in 49% (23/47 [95% CI 34.71% to 63.29%) of XDR-TB samples. The sequential use of MTBDRplus and MTBDRsl to detect XDR-TB when stratified according to smear-positive or smear-negative samples is shown in Fig. 2B. This testing strategy could rule in 78.5% (22/28) of smear-positive XDR-TB samples and 10.5% (2/19; p < 0.001) of smear-negative XDR-TB samples. The lower rule-in value in smear-negative samples is due to the high indeterminate rate relative to the smear-positive specimens.
Figure 2.
The testing pathway for the diagnosis of XDR-TB overall (A) or according to smear-status (B) in the clinical sputum specimens, when MTBDRplus and MTBDRsl were used sequentially. From the 234 sputum specimens tested, 47 culture-positive samples were diagnosed as XDR-TB by phenotypic DST.
Discussion
We evaluated the diagnostic accuracy of MTBDRplus and MTBDRsl using sputum samples and culture isolates. There are hardly any data comparing the accuracy of MTBDRsl in smear-positive and smear-negative samples or interrogating the impact of HIV status. We show that MTBDRplus has excellent sensitivity for both RIFR and INHR using smear-positive and smear-negative sputum samples. By contrast, MTBDRsl showed modest sensitivity for OFX and AMK resistance in sputum samples. However, sensitivity was markedly reduced in smear-negative versus smear-positive sputum samples and indeterminate rates were elevated. Both LPAs had high specificity for the detection of drug-specific resistance, and thus a positive result for resistance can be treated with confidence.
This is the first study to evaluate the diagnostic accuracy of MTBDRsl directly on smear-negative clinical sputum samples. This information is critical to initiate rapid treatment and minimise transmission in areas with high HIV and TB co-infection where most patients are smear-negative28. Furthermore, although Xpert MTB/RIF can predict smear status29,30,31, the initial smear status of patients is often unknown and it can be unclear what DR-TB testing modality is suitable for sputum.
Sensitivities for OFXR and AMKR in sputum were lower than that published previously in our setting32, however, our study was the first to use smear-negative specimens. The reduced sensitivity of MTBDRsl amongst smear-negative samples indicate that, when used directly MTBDRsl, will likely only be useful in smear-positive sputum. The low sensitivity in smear-negative sputum can be explained by low concentrations of bacilli in the sputum, which are below the detection limit of the LPA33.
When performed indirectly on the culture isolates no MTBDRplus and MTBDRsl-TUB band positive results were indeterminate. However, when performed directly on the sputum samples, 4% of the MTBDRplus-TUB band positive results were indeterminate. By contrast, MTBDRsl had a high number of indeterminate results when performed directly at 14.4% (22/153). This is higher than has been reported in other studies22 and is explained by the testing of smear-negative samples, which harboured the bulk of the indeterminate (36%) readouts34.
This is the first study to evaluate a sequential testing strategy. The data is shown in Figs S2 and 2. We show that when MTBDRplus and MTBDRsl are used sequentially on DST culture-confirmed MDR+ or XDR-TB samples, the assays could rule-in 60%, 62.5% and 49% of OFX mono-resistant, AMK mono-resistant, and XDR-TB samples, respectively. When used sequentially on smear-positive XDR-TB samples, the assays could rule-in 78.6% of cases. However, the ability of the assay to accurately rule-in XDR-TB samples amongst the smear-negative sputum samples (10.5%) was substantially lower.
Overall our data indicate that MTBDRsl is likely a useful tool to rapidly diagnose MDR+ and XDR-TB, but only in smear-positive sputum samples. This is useful from a clinical and public health perspective as it enables a more rapid diagnosis (potentially by several weeks) thus likely minimising patient morbidity and mortality32, and most importantly ongoing transmission in the community. Transmission of DR-TB explains almost 80% of new cases in South Africa35 and has led to the emergence and transmission of resistance beyond XDR-TB36,37,38. We are of the view that the MTBDRsl assay should be used routinely in high MDR-TB burden programmatic settings in patients with rifampicin resistance. However, we acknowledge that further studies are urgently needed in different settings to confirm our findings so that global recommendations can be made that apply to high MDR-TB settings including South Africa and Eastern Europe. Our study represents the first step in this direction.
There are several limitations to our work. MTBDRplus version 1 and MTBDRsl version 1 were used, which have recently been succeeded by a new iteration (version 2)39,40. Nevertheless, diagnostic accuracy of MTBDRplus was excellent in the clinical sputum samples and similar to recent studies where version 2 of MTBDRplus was shown to have comparable accuracy to Xpert MTB/RIF in smear-negative sputum39,41. Both MTBDRplus and MTBDRsl were not performed at initial diagnosis, however, we collected a paired specimen for culture in order to control for the viability of the bacilli. Our samples size, particularly of the smear-negative group were small, yet substantially more than what has been reported elsewhere21,22,23,24,25. We also lacked data on the duration of treatment for each patient. A further limitation to the study was that both LPAs were only tested on culture-positive and not culture-negative sputum samples. There were 5 smear-positive culture-negative samples and these were excluded from the analysis given that they did not meet the reference standard definition. Finally, we did not test the impact of using MTBDRplus and MTBDRsl on treatment outcomes, cure, and death of the patients. However, our work lays the foundation for confirmatory and impact studies to now be undertaken.
In conclusion, even though MTBDRsl had suboptimal diagnostic sensitivity for OFXR and AMKR, it remains an important rule-in tool to rapidly detect XDR-TB and MDR+ TB using smear-positive clinical samples, given that alternative tests have a prolonged within-laboratory turn-around-time and are technically challenging. Negative results, however, require further investigation as resistance to second line drugs may still be present but undetected by the assay. Smear-negative sputum specimens should be cultured prior to MTBDRsl testing.
Additional Information
How to cite this article: Tomasicchio, M. et al. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Sci. Rep. 6, 17850; doi: 10.1038/srep17850 (2015).
Supplementary Material
Footnotes
Author Contributions Conceived and designed the experiments: K.D., G.T., R.W. and P.V.H. Performed the experiments: E.S., D.S.J. and R.W. Analysed the data: M.T., G.T., E.P., E.S., D.S.J., R.W. and K.D. Contributed to reagents/materials/analysis tools: K.D., G.T., R.W. and P.V.H. Wrote the paper: M.T., G.T. and K.D.
References
- World Health Organisation. Tuberculosis Fact Sheet Number 104 (2014). Available at: www.who.int/mediacentre/factsheets/fs104/en (Accessed: 5th June 2014).
- World Health Organization. WHO Global Task Force outlines measures to combat XDR-TB worldwide (2014). Available at: http://www.who.int/mediacentre/news/notes/2006/np29/en/ (Accessed on: 5th June 2014).
- Centers for Disease Control and Prevention. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs worldwide, 2000–2004. MMWR Morb Mortal Wkly Rep. 55, 301–305 (2006). [PubMed] [Google Scholar]
- Ahuja S. D. et al. Multidrug resistant pulmonary tuberculosis treatment regimens and patient outcomes: an individual patient data meta-analysis. PLoS med. 9, e1001300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K. et al. Early treatment outcomes and HIV status of patients with extensively drug-resistant tuberculosis in South Africa: a retrospective cohort study. Lancet. 375, 1798–1807 (2010). [DOI] [PubMed] [Google Scholar]
- O'Donnell M. R. et al. Treatment outcomes for extensively drug-resistant tuberculosis and HIV co-infection. Emerg Infect Dis. 19, 416–424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotgiu G. et al. Epidemiology and clinical management of XDR-TB: a systematic review by TBNET. Eur Respir J. 33, 871–881 (2009). [DOI] [PubMed] [Google Scholar]
- Baussano I. et al. Tuberculosis among health care workers. Emerg Infect Dis. 17, 488–494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M. R. et al. High incidence of hospital admissions with multidrug-resistant and extensively drug-resistant tuberculosis among South African health care workers. Ann Intern Med. 153, 516–522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooran A., Pieterson E., Davids M., Theron G. & Dheda K. What is the cost of diagnosis and management of drug resistant tuberculosis in South Africa? PloS one. 8, e54587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrahan C. F. et al. The impact of expanded testing for multidrug resistant tuberculosis using genotype [correction of geontype] MTBDRplus in South Africa: an observational cohort study. PloS one. 7, e49898 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K. R. et al. Implementation of genotype MTBDRplus reduces time to multidrug-resistant tuberculosis therapy initiation in South Africa. Clin Infect Dis. 56, 503–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipiani M. et al. Significant Clinical Impact of a Rapid Molecular Diagnostic Test (Genotype MTBDRplus Assay) to detect Multidrug-Resistant Tuberculosis. Clin Infect Dis (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. et al. The nitrate reductase assay for the rapid detection of isoniazid and rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. J Antimicrob Chemother. 62, 56–64 (2008). [DOI] [PubMed] [Google Scholar]
- Moore D. A. et al. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 355, 1539–1550 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling D. I., Zwerling A. A. & Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J. 32, 1165–1174 (2008). [DOI] [PubMed] [Google Scholar]
- Anek-Vorapong R. et al. Validation of the GenoType MTBDRplus assay for detection of MDR-TB in a public health laboratory in Thailand. BMC Infect Dis. 10, 123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentz M., Sorensen B., Horne D. J. & Walson J. L. Systematic review of the performance of rapid rifampicin resistance testing for drug-resistant tuberculosis. PloS one 8, e76533 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen M. N. et al. Validation of the GenoType MTBDRplus assay for diagnosis of multidrug resistant tuberculosis in South Vietnam. BMC Infect Dis. 10, 149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J. et al. GenoType(R) MTBDRplus assay detection of drug-resistant tuberculosis in routine practice in Korea. Int J Tuberc Lung Dis. 17, 120–124 (2013). [DOI] [PubMed] [Google Scholar]
- Yadav R. N. et al. Comparative evaluation of GenoType MTBDRplus line probe assay with solid culture method in early diagnosis of multidrug resistant tuberculosis (MDR-TB) at a tertiary care centre in India. PloS one. 8, e72036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron G. et al. The diagnostic accuracy of the GenoType((R)) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev (10), 4–123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillemann D., Rusch-Gerdes S. & Richter E. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 47, 1767–1772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoma A. et al. GenoType MTBDRsl for molecular detection of second-line-drug and ethambutol resistance in Mycobacterium tuberculosis strains and clinical samples. J Clin Microbiol. 50, 30–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto P. et al. GenoType MTBDRsl performance on clinical samples with diverse genetic background. Eur resp j. 40, 690–698 (2012). [DOI] [PubMed] [Google Scholar]
- Kent PT K. G. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services, Publication No. 86-216546. Atlanta, GA: Centers for Disease Control (1985).
- World Health Organization. Policy Guidance on Drug-Susceptibility Testing (DST) of Second-Line Antituberculosis Drugs (2014). Available at: whqlibdoc.who.int/hq/2008/WHO_HTM_TB_2008.392_eng.pdf.WHO/HTM/TB/2008.392 (Accessed: 5th June 2014). [PubMed]
- Hoek K. G., Van Rie A., van Helden P. D., Warren R. M. & Victor T. C. Detecting drug-resistant tuberculosis: the importance of rapid testing. Mol diagn ther. 15, 189–194 (2011). [DOI] [PubMed] [Google Scholar]
- Theron G. et al. The Use of an Automated Quantitative Polymerase Chain Reaction (Xpert MTB/RIF) to Predict the Sputum Smear Status of Tuberculosis Patients. Clin. Infect. Dis. 54, 384–388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrahan C. F. et al. Xpert MTB/RIF as a measure of sputum bacillary burden: variation by HIV status and immunosuppression. Am. J. Respir. Crit. Care Med. 189, 1426–1434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore R. et al. A Multisite Assessment of the Quantitative Capabilities of the Xpert MTB/RIF Assay. Am. J. Respir. Crit. Care Med. 184, 1076–1084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard M. et al. Genotype MTBDRsl line probe assay shortens time to diagnosis of extensively drug-resistant tuberculosis in a high-throughput diagnostic laboratory. Am. J. Respir. Crit. Care Med. 186, 1298–1305 (2012). [DOI] [PubMed] [Google Scholar]
- Campbell P. J. et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 55, 2032–2041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foundation for Innovative New Diagnostics. Drug resistant TB and new diagnostics for people living with HIV: emerging results from FIND 2009 (2009). Available at: www.stoptb.org/wg/tb_hiv/assets/documents/DRUGRE1.PDF (Accessed: 9th September 2014).
- Streicher E. M. et al. Emergence and treatment of multidrug resistant (MDR) and extensively drug-resistant (XDR) tuberculosis in South Africa. Infect Genet Evol. 12, 686–694 (2012). [DOI] [PubMed] [Google Scholar]
- Dheda K. Better treatment of XDR tuberculosis needed in South Africa - Author's reply. Lancet. 384, 582 (2014). [DOI] [PubMed] [Google Scholar]
- Dheda K. et al. Global control of tuberculosis: from extensively drug-resistant to untreatable tuberculosis. The Lancet Respir Med. 2, 321–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen E. et al. Long-term outcomes of patients with extensively drug-resistant tuberculosis in South Africa: a cohort study. Lancet. 383, 1230–1239 (2014). [DOI] [PubMed] [Google Scholar]
- Crudu V. et al. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J Clin Microbiol. 50, 1264–1269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliani E. et al. Diagnostic Performance of the new version of GenoType MTBDRsl (V2.0) Assay for detection of resistance to Fluoroquinolones and Second Line Injectable Drugs: a Multicenter study. J Clin Microbiol. 53, 2961–2999. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard M. et al. The diagnostic performance of the GenoType MTBDRplus version 2 line probe assay is equivalent to that of the Xpert MTB/RIF assay. J Clin Microbiol. 50, 3712–3716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K., Barry C.E. & Maartens G. Tuberculosis. Lancet. [Epub ahead of print].Sept 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.