Abstract
Pre-harvest sprouting (PHS) is a worldwide problem for wheat production and transgene antisense-thioredoxin-s (anti-trx-s) facilitates outstanding resistance. To understand the molecular details of PHS resistance, we analyzed the metabonomes of the transgenic and wild-type (control) wheat seeds at various stages using NMR and GC-FID/MS. 60 metabolites were dominant in these seeds including sugars, organic acids, amino acids, choline metabolites and fatty acids. At day-20 post-anthesis, only malate level in transgenic wheat differed significantly from that in controls whereas at day-30 post-anthesis, levels of amino acids and sucrose were significantly different between these two groups. For mature seeds, most metabolites in glycolysis, TCA cycle, choline metabolism, biosynthesis of proteins, nucleotides and fatty acids had significantly lower levels in transgenic seeds than in controls. After 30-days post-harvest ripening, most metabolites in transgenic seeds had higher levels than in controls including amino acids, sugars, organic acids, fatty acids, choline metabolites and NAD+. These indicated that anti-trx-s lowered overall metabolic activities of mature seeds eliminating pre-harvest sprouting potential. Post-harvest ripening reactivated the metabolic activities of transgenic seeds to restore their germination vigor. These findings provided essential molecular phenomic information for PHS resistance of anti-trx-s and a credible strategy for future developing PHS resistant crops.
Pre-harvest sprouting (PHS) is characterized by the premature germination of seeds occurring in the spikes and becomes a serious problem in many wheat-growing areas worldwide with prolonged pre-harvest rainfall and high humidity1. PHS leads to reduced yields, lower end-product quality of the grains and hence great economic losses. In China, for instance, about 25 million hectares of wheat are affected by PHS every year accounting for 83% of the whole wheat-growing area. Therefore, PHS resistant varieties are developed to solve the problem especially for the wheat-producing areas where long period of rainfall occurs frequently in the harvest season.
Pre-harvest sprouting is a complex phenotype resulting from interactions between wheat genotypes2 with biotic and/or abiotic environmental factors3. The untimely breakdown of seed dormancy is considered to be the major event for PHS to occur and therefore improvement of PHS resistance is often accompanied with prolonged seed dormancy to pass the harvest stage4.
Thioredoxin h (trx-h) gene is important for wheat seed germination and thus PHS5. Trx-h initially found in wheat kernels in 19796 is now found widely present in the higher plants. This gene acts as an important regulator for seed germination by facilitating the reduction of intramolecular disulfide bonds in storage proteins of cereals, such as wheat and barley. During seed germination, trx-h also promotes the activation of α-amylase, pullulanase and proteases by weakening the inhibitive effect of inhibitor proteins on amylases and proteases7. Overexpressing trx-h gene in barley accelerated germination of the embryos and activated both α-amylase and starch pullulanase8,9. On the other hand, underexpressing trx h9 gene in wheat lowered the activities of Trx protein, α-amylase and pullulanase slowing seed germination10. It is particularly worth-noting that the transgenic wheat underexpressing trx h9 gene has also shown outstanding PHS resistance10.
Trx-s is another member of the thioredoxin gene family initially cloned from Phalaris coerulescens5. In fact, trx-s and trx-h have more than 90% homology in their cDNA sequences and similar biological functions for their expression products11. By using pollen-tube pathway, antisense thioredoxin s (anti-trx-s) was transferred into wheat to have successfully developed transgenic wheat with PHS resistance as well12. It was reported that anti-trx-s inhibited the endogenous trx-h expression and lowered α-amylase activity between day-30 post anthesis and 10 days post-harvest ripening resulting in high PHS resistance in the transgenic wheat13,14,15. It was also found that the introduction of anti-trx-s gene inactivated starch hydrolases and slowed hydrolysis of storage proteins16,17,18 in seeds imbibed for 3 to 4 days.
Systems biology approaches offer excellent opportunity to understand pre-harvest sprouting in terms of protein expressions and metabolism in a more holistic manner. Proteomic analyses already showed that transferred anti-trx-s gene caused down-regulation of many proteins in wheat seed kernels involving protein biosynthesis/degradation, starch degradation, gene expression regulation, lipid and energy metabolisms19. Anti-trx-s also caused up-regulation of proteins in kernels involving α-amylase activity suppression and disulfide bond formation compared to wild-type19. Several proteins related to stress resistance (such as antioxidant and disease resistance) were further up-regulated in the transgenic wheat kernels19. Moreover, transgenic wheat showed differential gene expression in trx-h, serpin, heat shock protein 70 (hsp70), and WRKY transcription factor 6 (WRKY6) compared to wild-type19. These results also imply that anti-trx-s gene may induce comprehensive metabolic changes in the transgenic wheat seeds. However, it remains unknown what metabolic changes such transgene causes, at which seed development stages and how these transgenic effects on seed metabolic activities are related to PHS.
Metabonomics ought to be a useful approach for understanding the dynamic metabolic changes since metabonomic analysis measures the metabolite composition (metabonome) of a given biological system and its dynamic responses to both endogenous and exogenous factors20,21,22. Such approach has been proven to be powerful in disease diagnosis23, in understanding metabolic variation between different rice varieties24 and metabolic responses to gene modifications25. Metabonomic analysis has increasingly become a powerful approach in understanding the effects of biotic and abiotic stressors on plant physiology and biochemistry26,27,28,29. So far, however, there have been no reports about the effects of anti-trx-s on the wheat seed metabonome, to the best of our knowledge, though these effects are expected to be insightful for developing PHS resistant wheat. It is also conceivable that PHS and its resistance ought to be associated with the development dependence of wheat seed metabolic phenome since sprouting of wheat seeds generally go through four grain filling periods including milk stage, dough development stage, mature seeds and post-harvest ripening period15.
In this study, we analyzed the seed metabonomic phenotypes (metabotypes) of transgenic wheat with anti-trx-s and wild-type at four different time-points of seed development (milk stage, dough development stage, mature seed and post-harvest ripeness period) using NMR spectroscopy in conjunction with multivariate statistical analysis. We also analyzed the developmental dependence of the fatty acid composition of these seeds using GC-FID/MS method. We further conducted integrative analysis on the metabonomic and proteomic differences between the PHS susceptible and resistant seeds. Our objectives are (1) to define the metabonomic changes induced by introduction of anti-trx-s and (2) to understand the molecular aspects of the PHS resistance acquired through introduction of such gene which will offer important information for further development of PHS-resistant wheat varieties.
Results
Taking into consideration of the developmental periods of wheat seeds15, in this study, we analyzed metabonomic features of wheat seeds harvested at about day-20 post anthesis (20-dpa, milk stage), day-30 post anthesis (30-dpa, dough development stage), day-40 post anthesis (40-dpa, mature seed) and 30 days post-harvest ripening (30-dpr), respectively. We also considered the metabonomic data in an integrative manner with our proteomic results. Our results showed that this anti-trx-s transgenic wheat seeds had lower trx-h expression and α-amylase activity but higher PHS resistance compared with the corresponding wild-type wheat seeds as reported previously13,14,15. Our proteomic analysis results also showed that anti-trx-s gene introduction led to significant expressional changes of a number of metabolism-related proteins in wheat seeds19 at 40-dpa. This was highlighted by up-regulation of protein disulfide isomerase (PDI), serpin, xylanase inhibitor protein I precursor (XIP-I) but down-regulation of glucose-6-phosphate isomeraseprecursor (GPI), aldolase (ALD), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoenolpyruvate carboxylase (PEPC), glutamate dehydrogenase (GDH), malate dehydrogenase (MDH), pullulanase, aspartate aminotransferase (AST), adenylylsulfate kinase (APK), α- and β-amylase19.
Seed metabolite composition for transgenic and wild-type wheat
1H NMR spectra of seed extracts (Fig. 1) showed rich but obvious different metabolite profiles for anti-trx-s transgenic and wild-type wheat seeds at 30- and 40-dpa, respectively. Both 1H and 13C signals were unambiguously assigned to individual metabolites (Table S1) based on the literature data30,31,32, in-house databases and publicly available databases33 with further confirmation using a series of 2D NMR spectral data. More than 40 metabolites were identified including 16 amino acids and their metabolites (Ala, Thr, Val, Ile, Leu, Glu, Gln, Asp, Asn, Phe, Tyr, Trp, GABA, His, Arg and 4-guanidinobutyrate), 10 organic acids (trans-aconitate, citrate, α-ketoglutarate, succinate, malate, fumarate, formate, acetate, lactate, isobutyrate), 2 plant secondary metabolites namely 4-hydroxy-3-methoxyphenylacetate (HMPA) and ferulate, 6 nucleotide metabolites (uridine, adenosine, guanosine, allantoin, NAD+ and ATP), 5 choline metabolites (choline, ethanolamine, betaine, dimethylamine and trimethylamine), 4 sugars (sucrose, glucose, raffinose, D-ribose-5-phosphate) and ethanol (Fig. 1, Table S1 and Table 1). 15 fatty acids were also detected and quantified from these seeds (Table 2). Visual inspection showed that compared with wild-type, transgenic seeds had higher levels of fumarate and GABA at 30-dpa (Fig. 1a,b) but lower levels of Trp and Ala at 40-dpa (Fig. 1c,d). To obtain more detailed metabolic changes induced by anti-trx-s gene, multivariate data analyses were performed on the NMR data of these seeds.
Figure 1.
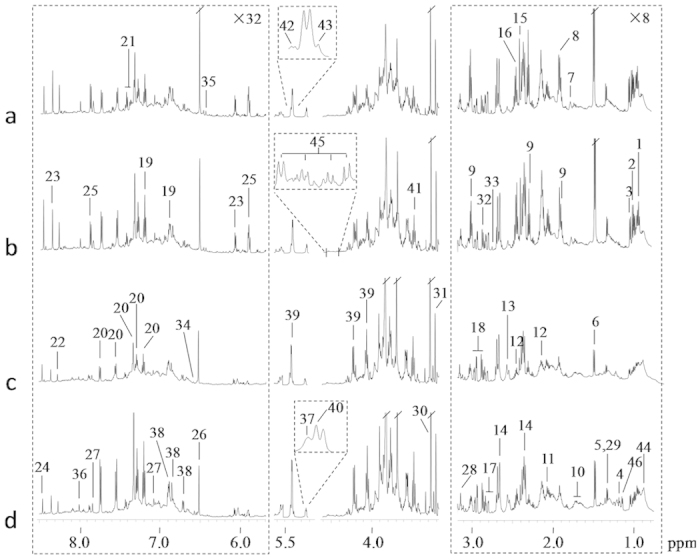
Average 1H NMR spectra of seed extracts from (a) transgenic wheat with anti-trx-s at 30-dpa, (b) wild type wheat at 30-dpa, (c) transgenic wheat with anti-trx-s at 40-dpa, and (d) wild type wheat at 40-dpa. The regions δ 0.75-3.19 and δ 5.68-8.50 were vertically expanded 8 times and 32 times, respectively. Keys: 1, Isoleucine (Ile); 2, Leucine (Leu); 3, Valine (Val); 4, Ethanol; 5, Threonine (Thr); 6, Alanine (Ala); 7, 4-Guanidinobutyrate (4-GB); 8, Acetate; 9, γ-Aminobutyrate (GABA); 10, Arginine (Arg); 11, Glutamate (Glu); 12, Glutamine (Gln); 13, Citrate; 14, Malate; 15, Succinate; 16, α-Ketoglutarate; 17, Aspartate (Asp); 18, Asparagine (Asn); 19, Tyrosine (Tyr); 20, Tryptophan (Trp); 21, Phenylalanine (Phe); 22, Adenosine triphosphate (ATP); 23, Adenosine; 24, Formate; 25, Uridine; 26, Fumarate; 27, Histidine (His); 28, Ethanolamine; 29, Lactate; 30, Betaine; 31, Choline; 32, Trimethylamine; 33, Dimethylamine; 34, trans-Aconitate; 35, Ferulate; 36, Guanosine; 37, D-Ribose-5-phosphate; 38, 4-Hydroxy-3-methoxyphenylacetate (HMPA); 39, Sucrose; 40, α-Glucose; 41, β-Glucose; 42, Raffinose; 43, Allantoin; 44, Lipids; 45, Nicotinamide adenine dinucleotide (NAD+); 46, Isobutyrate.
Table 1. Significantly differential seed metabolites between anti-trx-s transgenic (TG) and wild type wheat (WT)a.
| Metabolites (no.) | Coefficient (r)b |
||
|---|---|---|---|
| TG vsWT at 30-dpa | TG vs WT at 40-dpa | TG vs WT at 30-dpr | |
| Amino acids & metabolites | |||
| GABA (9) | 0.77 | ||
| Asp (17) | 0.66 | 0.53 | |
| Glu (11) | −0.57 | −0.60 | |
| Gln (12) | −0.57 | −0.58 | |
| Arg (10) | −0.57 | ||
| Ala (6) | −0.74 | ||
| Phe (21) | 0.61 | ||
| Trp (20) | −0.70 | ||
| Val (3) | −0.61 | 0.59 | |
| Leu (2) | −0.58 | ||
| Ile (1) | −0.60 | ||
| Thr (5) | −0.70 | ||
| 4-GB (7) | 0.69 | ||
| Sugars | |||
| Sucrose (39) | −0.54 | −0.90 | 0.66 |
| Glucose (40, 41) | −0.92 | ||
| Raffinose (42) | 0.88 | ||
| TCA Cycle Metabolites | |||
| Fumarate (26) | 0.66 | ||
| Citrate (13) | −0.69 | 0.68 | |
| Nucleotide Metabolites | |||
| Adenosine (23) | −0.74 | ||
| Uridine (25) | −0.77 | ||
| Guanosine (36) | −0.75 | −0.54 | |
| Allantoin (43) | −0.86 | ||
| NAD+ (45) | −0.69 | 0.68 | |
| ATP (22) | −0.71 | ||
| Choline Metabolites and Others | |||
| Ethanolamine (28) | −0.65 | 0.75 | |
| Choline (31) | −0.59 | 0.69 | |
| Betaine (30) | −0.65 | 0.79 | |
| HMPA (38) | −0.61 | 0.88 | |
aThe level of malate was lower in transgenic wheat than that in wild type seeds at 20-dpa (t-test, p = 0.037).
bThe coefficients were obtained from OPLS-DA results, and positive and negative signs indicate positive and negative correlation in the concentrations, respectively.
Table 2. Seed fatty acid composition for wild type (WT) and transgenic (TG) wheat with anti-trx-s gene.
| Fatty acids (μmol/g) | WT at 40-dpa | TG at 40-dpa | WT at 30-dpr | TG at 30-dpr |
|---|---|---|---|---|
| C14:0 | 0.08 ± 0.01 | 0.07 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.10 |
| C15:0 | 0.13 ± 0.01 | 0.12 ± 0.01 | 0.12 ± 0.02 | 0.16 ± 0.02* |
| C16:0 | 12.56 ± 0.72 | 11.72 ± 0.46 | 12.16 ± 0.67 | 14.99 ± 1.47** |
| C16:1n7 | 0.04 ± 0.01 | 0.02 ± 0.01** | 0.04 ± 0.01 | 0.03 ± 0.01 |
| C18:0 | 1.35 ± 0.10 | 1.21 ± 0.10 | 1.39 ± 0.10 | 1.30 ± 0.12 |
| C18:1n9 | 7.01 ± 0.75 | 6.20 ± 0.82 | 6.32 ± 0.75 | 8.85 ± 0.97** |
| C18:2n6 | 40.09 ± 1.67 | 34.06 ± 3.18** | 38.04 ± 2.45 | 46.05 ± 5.13* |
| C18:3n3 | 3.20 ± 0.13 | 2.54 ± 0.18** | 2.98 ± 0.29 | 3.37 ± 0.45 |
| C20:0 | 0.07 ± 0.01 | 0.06 ± 0.01 | 0.08 ± 0.01 | 0.07 ± 0.01 |
| C20:1n9 | 0.32 ± 0.03 | 0.31 ± 0.04 | 0.28 ± 0.06 | 0.47 ± 0.08** |
| C20:2n6 | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.07 ± 0.01 |
| C20:4n6 | 0.16 ± 0.01 | 0.13 ± 0.02* | 0.12 ± 0.02 | 0.14 ± 0.01* |
| C22:0 | 0.09 ± 0.01 | 0.07 ± 0.01 | 0.08 ± 0.01 | 0.08 ± 0.01 |
| C22:1n9 | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.06 ± 0.01 | 0.08 ± 0.02 |
| C24:0 | 0.10 ± 0.01 | 0.08 ± 0.01 | 0.10 ± 0.01 | 0.10 ± 0.01 |
| Total FAs | 65.3 ± 3.21 | 56.71 ± 4.66* | 61.90 ± 4.06 | 75.87 ± 8.13** |
| SFAs | 14.37 ± 0.75 | 13.35 ± 0.46* | 14.01 ± 0.61 | 16.80 ± 1.60** |
| UFAs | 50.93 ± 2.48 | 43.36 ± 4.23* | 47.89 ± 3.46 | 59.06 ± 6.58** |
| MUFAs | 7.42 ± 0.77 | 6.59 ± 0.86 | 6.70 ± 0.80 | 9.43 ± 1.07** |
| PUFAs | 43.51 ± 1.78 | 36.77 ± 3.38** | 41.19 ± 2.67 | 49.63 ± 5.57* |
| PUFA/MUFA | 5.89 ± 0.40 | 5.60 ± 0.24 | 6.18 ± 0.34 | 5.27 ± 0.20** |
| PUFA/UFA | 0.85 ± 0.01 | 0.85 ± 0.01 | 0.86 ± 0.01 | 0.84 ± 0.01 |
| SFA% | 22 ± 1 | 23 ± 1 | 23 ± 1 | 22 ± 1 |
| UFA% | 78 ± 1 | 76 ± 1 | 77 ± 1 | 78 ± 1 |
| MUFA% | 11 ± 1 | 12 ± 1 | 11 ± 1 | 12 ± 1 |
| n3 | 3.20 ± 0.13 | 2.54 ± 0.18** | 2.98 ± 0.29 | 3.37 ± 0.45 |
| n6 | 40.31 ± 1.68 | 34.24 ± 3.20** | 38.21 ± 2.46 | 46.26 ± 5.14* |
| n6/n3 | 12.59 ± 0.39 | 13.49 ± 0.38* | 12.88 ± 0.87 | 13.76 ± 0.50 |
Data expressed as mean ± SD. *p < 0.05, **p < 0.01. SFAs, saturated fatty acids; MUFAs, monounsaturated fatty acids; PUFA, polyunsaturated fatty acids; n3, n3-type polyunsaturated fatty acids; n6, n6-type polyunsaturated fatty acids.
Anti-trx-s gene induced metabolic changes in transgenic wheat seeds
PCA (principal component analysis) scores plots showed clear development-associated metabonomic changes for both transgenic and wild-type wheat seeds reflected by a metabolic trajectory from 20-dpa, 30-dpa, 40-dpa and 30-dpr (Figure S1). Some obvious metabonomic differences were also observable between the transgenic and wild-type wheat seeds at 30-dpa, 40-dpa and 30-dpr (Figure S2B–D) although such difference appeared to be less prominent at 20-dpa (Figure S2A).
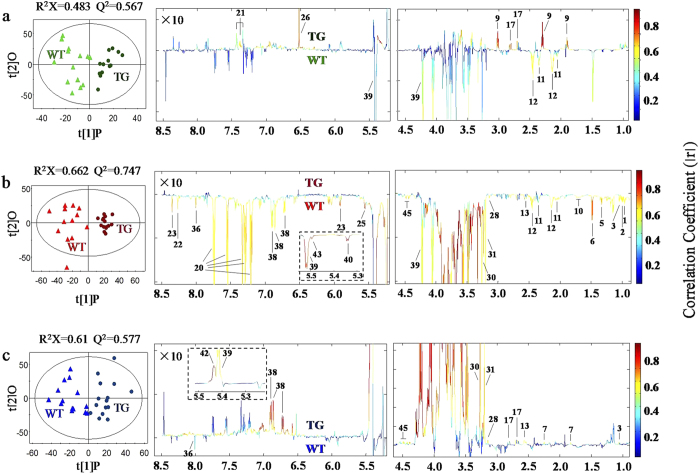
To obtain the detailed information on significant metabolic alterations induced by anti-trx-s, pairwise OPLS-DA (orthogonal projection to latent structure-discriminant analysis) was conducted between the extracts of transgenic wheat and wild-type seeds harvested at 20-dpa, 30-dpa, 40-dpa and 30-dpr, respectively; significantly differential metabolites between these two groups were tabulated in Table 1. At 20-dpa, OPLS-DA model parameters indicated only some limited metabolic differences were present between transgenic wheat and wild-type seeds. Careful analysis of all variables individually34 showed that at this development stage, only malate in the transgenic wheat seeds had significant level difference (p = 0.037) compared to the wild type (Table 1).
At the other three time points of seed development (i.e., 30-, 40-dpa and 30-dpr), in contrast, OPLS-DA model parameters (Fig. 2, Table 1) all showed significant seed metabonomic differences between the transgenic and wild-type wheat. The coefficient-coded loadings plots indicated that introduction of anti-trx-s caused significant elevation of seed fumarate, Asp, GABA and Phe at 30-dpa together with level decreases for sucrose, Glu and Gln compared with wild-type wheat (Fig. 2a, Table 1). At 40-dpa, anti-trx-s transgenic wheat seeds had significantly lower levels for 2 sugars (sucrose and glucose), 9 amino acids (Ala, Thr, Val, Ile, Leu, Glu, Gln, Trp and Arg), a TCA intermediate (citrate) and a secondary metabolite (HMPA), 3 choline metabolites (ethanolamine, choline and betaine) and 6 nucleotide derivatives (uridine, adenosine, guanosine, allantoin, NAD+ and ATP) than the wild type (Fig. 2b, Table 1). Furthermore, after 30 days post-harvest ripening (30-dpr), the transgenic wheat seeds had significantly lower level of guanosine but higher contents of sucrose, Val, citrate, HMPA, choline metabolites (ethanolamine, choline and betaine) and NAD+, being in contrast to the seed metabonomic features at 40-dpa. Moreover, introduction of anti-trx-s caused elevation of wheat seed raffinose, Asp and 4-guanidinobutyrate compared with the wild-type (Fig. 2c, Table 1).
Figure 2. OPLS-DA scores plots (left) and coefficient-coded loadings plots (right) showing time-dependence of anti-trx-s gene effects on wheat seed metabolism.
These models were derived from transgenic wheat (TG) and wild type (WT) seeds (a) at 30-dpa (p = 0.0007), (b) 40-dpa (p = 0.000001) and (c) 30-dpr (p = 0.0003). Metabolite keys are the same as in Fig. 1 and Table S1.
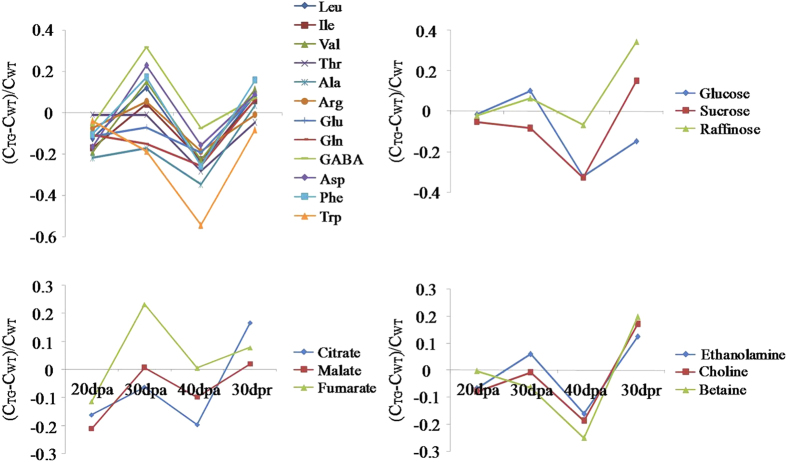
We further calculated the ratios of concentration changes for the transgene-altered metabolites (against the wild-type wheat) at four different time points. The results indicated that compared with the wild type, a clear metabolic switch for the transgenic wheat occurred between 40-dpa and 30-dpr. At 40-dpa, for instance, the levels of Trp, Ala, Thr, Gln and Phe in the transgenic wheat seeds showed about 54%, 35%, 28%, 26% and 26% level decrease, respectively, compared with the wild-type (Fig. 3). In contrast, after 30 days post-harvest ripening, the levels of most seed amino acids in transgenic wheat were similar to these in wild type with exceptions for Asp and Val whose levels were higher in transgenic wheat (Fig. 3). Furthermore, much more significant differences between sucrose and glucose levels in transgenic wheat and wild-type were observed at 40-dpa than at the other three time-points; their levels were more than 30% lower in transgenic wheat seeds than in wild type (Fig. 3). After 30 days post-harvest ripening, in contrast, the levels of sucrose and raffinose were about 15% and 34% higher in transgenic wheat seeds than in wild-type (Fig. 3). Moreover, at-40 dpa, citrate and malate levels were lower in transgenic wheat seeds than wild type whereas, at 30-dpr, the levels of these metabolites and fumarate were higher in transgenic wheat seeds (Fig. 3). Ethanolamine, choline and betaine levels also showed a decrease at 40-dpa but an elevation at 30-dpr (Fig. 3) in transgenic wheat seeds compared with wild type.
Figure 3. The ratios of concentration changes for metabolites in transgenic wheat seeds against that in wheat of wild type seeds.
Metabolites with statistically significant variations are listed in Fig. 2 and Table 1.
Anti-trx-s gene induced fatty acid changes in transgenic wheat seeds
Composition of wheat seed fatty acids was dominated by palmitate (C16:0), stearate (C18:0) and their unsaturated forms. Linoleic acid (C18:2n6) was the most abundant wheat seed fatty acid (34.1–46.1 μmol/g fresh weight) accounting for about 60% of total fatty acids (Table 2). Palmitic acid (C16:0, 11.7–15.0 μmol/g) and oleic acid (C18:1n9, 6.2–8.9 μmol/g) accounted for about 20% and 10% of total seed fatty acids, respectively. Linolenic acid (C18:3n3, 2.5–3.4 μmol/g) and stearic acid (C18:0, 1.2–1.4 μmol/g) were about 5% and 2% of total fatty acids, respectively.
Results showed outstanding dynamic changes in fatty acids induced by introduction of anti-trx-s with an obvious switch after 40-dpa. Compared with wild type seeds, the contents of total fatty acids (ToFAs), saturated fatty acids (SFAs), unsaturated fatty acids (UFAs), monounsaturated fatty acids (MUFAs), polyunsaturated fatty acids (PUFAs) and n6-type fatty acids were all decreased in transgenic wheat seeds at 40-dpa whereas the contrary was observed at 30-dpr (Table 2). Specifically, at 40 dpa, transgenic wheat seeds had significantly lower contents of C16:1n7 and PUFAs (C18:2n6, C18:3n3 and C20:4n6) than the wild-type (Table 2). At 30 dpr, in contrast, the concentration of C15:0, C16:0, C18:1n9, C20:1n9, C18:2n6 and C20:4n6 were significantly higher in transgenic wheat seeds than wild type (Table 2). Furthermore, at 40-dpa, the n6-to-n3 ratio for fatty acids was higher in transgenic wheat seeds than in wild type whereas the PUFA-to-MUFA ratio in transgenic wheat was lower than in wild type at 30-dpr (Table 2).
Discussion
Wheat seeds with anti-trx-s gene expression showed excellent resistance to pre-harvest sprouting (PHS)35,36. The PHS resistant wheat differed significantly from the PHS susceptible wild-type in the seed metabonomic phenotypes at several seed development stages (Fig. 2 and Table 2). Such differences were outstanding especially when seeds reached maturity (at 40-dpa) and post-harvest ripening for 30 days (30-dpr). It is known that the period from day-5 pre-mature to day-10 post-mature is critical for wheat PHS to occur15. Our results showed that whilst most of the seed metabolic activities were inactivated at harvest time (40-dpa), these activities were restored after post-harvest ripening for a month (Fig. 4). This indicates that appropriate regulation and control of the seed metabolic activity during seed development is critical for pre-harvest sprouting to occur and thus for development of PHS resistant wheat.
Figure 4. Anti-trx-s gene-induced metabolic changes in transgenic wheat compared with wild type wheat.
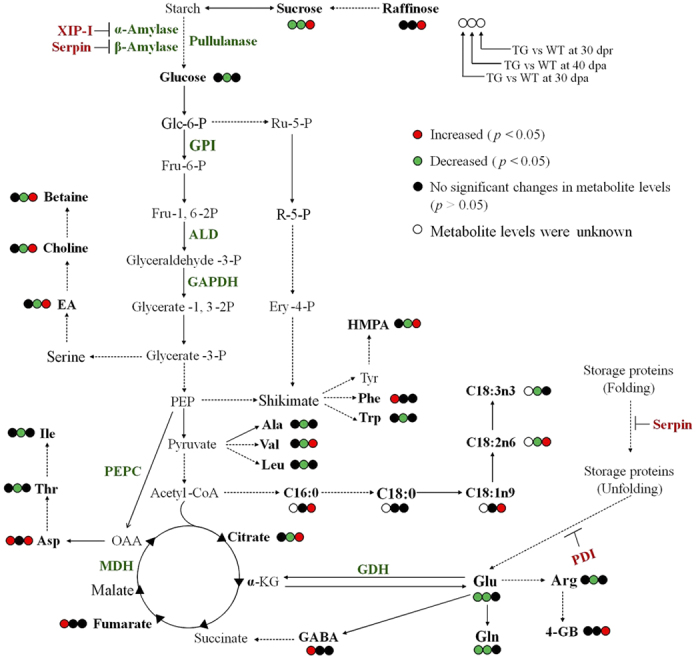
Results for protein expressional changes were derived from Guo et al. (ref 19) at 40-dpa. Dark red bold letters and dark green bold letters denoted significant increases and decreases in protein levels respectively in transgenic wheat seeds compared with wild type. TG, transgenic wheat; WT, wild type wheat; 30-dpa, 30 days post anthesis; 40-dpa, 40 days post anthesis; 30-dpr, 30 days post-harvest ripening; Glc-6-P, glucose-6-phosphate; Fru-6-P, fructose-6-phosphate; Fru-1,6-2P, fructose-1,6-biphosphate; Ru-5-P, ribulose-5-phosphate; R-5-P, ribose-5-phosphate; Ery-4-P, erythrose-4-phosphate; Glyceraldehyde-3-P, Glyceraldehyde-3-phosphate; Glycerte-1,3-2P, Glycerte-1,3-biphosphate; Glycerte-3-P, Glycerte-3-phosphate; EA, ethanolamine; PEP, phosphoenolpyruvate; α-KG, α-ketoglutarate; HMPA, 4-hydroxy-3-methoxyphenylacetate; OAA, oxalacetate; GABA, γ-aminobutyrate; 4-GB, 4-guanidinobutyrate; XIP-1, xylanase inhibitor protein 1; GPI, glucose-6-phosphate isomerase; ALD, aldolase; PEPC, phosphoenolpyruvate carboxylase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GDH, glutamate dehydrogenase; MDH, malate dehydrogenase; PDI, protein disulfide isomerase.
This current study showed that introduction of anti-trx-s led to different metabonomic changes at different development stages in wheat seeds. In the early seed developmental stage (20-dpa or milk stage), only malate level in anti-trx-s transgenic wheat differed from that in wild type (Fig. 3). At this stage, the expression level of trx-h was only about 15.6% lower in the transgenic wheat than in wild type (Yumai-70)15. When anti-trx-s was transferred to another wild type cultivar Yangmai-5, no difference in α-amylase activity was observable between the transgenic and wild type wheat seeds at 20-dpa37. This implies that, at 20-dpa, the effects of introduction of anti-trx-s have only limited impacts on seed metabolism simply because the major metabolic events at this stage are programmed towards biosynthesis of materials needed for seed development. The transgenic effects on seed malate level may indicate that Krebs cycle was slower to some extent in the transgenic wheat during this early grain fill period.
When seeds reached dough development stage (at 30-dpa), much more metabolic differences were observable between the transgenic and wild type wheat seeds. GABA, Asp, Phe and fumarate were significantly up-regulated in the transgenic line whereas Glu, Gln and sucrose were significantly down-regulated (Fig. 2, Table 1). At this stage, about 23% decreases in the trx-h expression level were observed in the anti-trx-s transgenic wheat seeds compared with the control15. This indicated that biosynthesis of storage proteins and consumption of biogenic Glu was faster in transgenic wheat than in wild type offering nitrogen source for protein biosynthesis. Significant lower level of sucrose in transgenic seeds implied that anti-trx-s slowed the starch degradation to favor starch storage compared with wild type. At the dough development stage, therefore, the major metabolic events for both transgenic and wild type wheat seeds appeared to be biosynthesis of storage matters including starch and proteins although anti-trx-s expression seemed to accelerate such biosyntheses to some extent.
When wheat seeds reached maturity at 40-dpa, germinating ratio of transgenic wheat seeds was significantly lower than wild type36. Significant lower levels for 26 seed metabolites in the transgenic wheat than in wild type (Fig. 2, Tables 1,2) indicated comprehensive reduction of metabolic activities in anti-trx-s transgenic wheat seeds. This is agreeable with 36.5% decreases in α-amylase activity in anti-trx-s transgenic wheat seeds compared with the wild type control Yangmai-5 at 40-dpa37. These results are also consistent with the proteomic results for this wheat with significant up-regulations of XIP-I and PDI but down-regulations of GAPDH, GPI, ALD and PEPC19. This is reasonable since XIP-I suppresses α-amylase activity in wheat seeds38 whilst GAPDH, GPI and ALD are enzymes involved in glycolysis and thioredoxin can activate GAPDH39. Therefore, the markedly reduction of sucrose and glucose levels (Table 1, Fig. 2), XIP-I up-regulations and down-regulations of GAPDH, GPI and ALD19 suggests that both the seed starch degradation and glycolysis were slowed down in transgenic wheat at this particular stage (Fig. 4). PEPC catalyzing conversion of oxaloacetate into TCA intermediates can be hydrolyzed by enzymes having disulfide bonds40. At 40-dpa, expression of anti-trx-s gene in the transgenic wheat lowered thioredoxin h protein expression hence down-regulation of PEPC19. This and significantly lower citrate levels in the transgenic wheat seeds than in controls (Table 1) also suggested that TCA cycle was slowed down at 40-dpa in the transgenic wheat (Fig. 4).
Significant level decreases for many amino acids (Glu, Gln, Arg, Ala, Trp, Val, Leu, Ile and Thr) in transgenic wheat at 40-dpa suggest that expression of anti-trx-s gene affected protein metabolism. This and up-regulations of serpin and PDI19, two important proteins inhibiting breakdown of seed proteins, in transgenic wheat (Fig. 4) indicated that anti-trx-s expression slowed degradation of reserve proteins in seeds at this stage. Furthermore, down-regulations of GDH, AST and APK19 in the transgenic wheat (relative to wild type) indicated that anti-trx-s expression also slowed protein biosynthesis since these were essential enzymes for sulfydryl-dependent protein biosynthesis41,42.
Lower choline and ethanolamine levels in transgenic wheat seeds than in wild type are probably related to changes in membrane biosynthesis with choline and ethanolamine as the main fragments of membrane phospholipids in plants43. These two fragments can be produced through phospholipid hydrolysis catalyzed by phospholipase D (PLD). In fact, PLD antagonist did inhibit seed germination in Arabidopsis44. Our observation of choline and ethanolamine decreases resulting from introduction of anti-trx-s implies probable PLD inactivation in transgenic wheat to inhibit untimely seed germination (i.e., PHS) and/or down-regulation of glycerate-3-phosphate due to slowed glycolysis (Fig. 4). The decreases of adenosine, guanosine and uridine in transgenic wheat seeds were probably associated with the reduced overall metabolic activities of seeds including nucleic acid degradation and de novo biosynthesis purines and pyrimidines. The level reduction of allantoin further supports this notion with it as a catabolic metabolite of purines. Since wheat seed germination accompanies initiation of DNA and RNA biosynthesis45, it is reasonable that the decreased adenosine, guanosine and uridine levels in transgenic wheat seeds observed here are related to inhibition of potential seed germination. The change of HMPA associated with introduction of anti-trx-s indicates some roles of plant secondary metabolism in seed germination and such clearly warrants further investigations. Lower levels of main fatty acids in transgenic wheat seeds than in wild type (Table 2) are probably also related to slower overall metabolic activities in transgenic seeds although slowing glycolysis is one of the major reasons since de novo biosynthesis of fatty acids in plants is critically dependent on production of acetyl-CoA from glycolysis.
After 30 days postharvest ripening (30-dpr), 17 metabolites in anti-trx-s transgenic wheat seeds showed significant increase compared with wild type (Fig. 2, Tables 1,2) including NAD+, a TCA intermediate (citrate), two sugars (glucose, raffinose), three amino acid metabolites (Val, Asp, 4-guanidinobutyrate), three choline metabolites (choline, ethanolamine, betaine), a secondary metabolite (HMPA) and six fatty acids (C15:0, C16:0, C18:1n9, C18:2n6, C20:1n9 and C20:4n6). This indicates that break of seed dormancy occurs and transgenic wheat seeds start having more metabolic activities than corresponding wild type. Interestingly, 10 metabolites with lower levels in transgenic wheat at 40-dpa had higher levels at 30-dpr than the wild type controls (Tables 1,2). Nevertheless, at 30-dpr, the α-amylase activity together with the ratio and rates of germination were all similar for anti-trx-s transgenic and wild type seeds36,37 indicating both lines had similar starch degradation potentials required for germination. Compared with wild type, therefore, transgenic wheat seeds activate their metabolism to reduce the inhibiting effects of anti-trx-s gene so as to break dormancy and to initiate germination. Furthermore, 30 days postharvest ripening was sufficient for transgenic wheat seeds to recover the germination vigor by recovering the required metabolic activity of seeds.
To sum up, PHS-resistance of transgene anti-trx-s resulted largely from reprogramming seeds metabolism especially at later seed development (dough development, seed maturing) and post-harvest ripening stages. The expression of antisense thioredoxin s gene suppressed the overall metabolic activities of seeds when seeds reach maturity (40-dpa) (Fig. 4) so as to prevent from seed germination (i.e., pre-harvest sprouting). This metabolic reprogramming is critically important at about 40-dpa to prepare the seed metabolic activities since wheat seeds are most susceptible PHS at this stage15. During 30-days post-harvest ripening, however, the transgenic seeds re-adjusted their metabolic activities by up-regulating a number of pathways to overcome the anti-trx-s effects. This re-adjustment of metabolic activities is also critically important for seeds to break dormancy and germinate. Although anti-trx-s gene expressed throughout wheat growing period, its effects on seed metabolism was less obvious at milk (20-dpa) probably because biosyntheses of proteins, starch and nucleic acids were the dominant events in seed metabolism at such stage. These results offered essential biochemistry information for pre-harvest sprouting of wheat and for PHS resistance of anti-trx-s.
Methods
Plant materials
Seeds from the transgenic wheat with antisense trx-s gene and wild-type (Yumai 18) were harvested, respectively, at day-20 post anthesis (20-dpa, milk stage), day-30 post anthesis (30-dpa, dough development stage) and day-40 post anthesis (40-dpa) when seeds reached complete maturity. Wheat seeds harvested at 20-dpa and 30-dpa were snap-frozen in liquid nitrogen and stored at −80 °C until further analysis. Seeds at 40-dpa were divided into two groups with one group snap-frozen in liquid nitrogen followed with storage at −80 °C (designated as 40-dpa in this study). The other group of samples were kept at room temperature for 30 more days to have post-harvest ripening (and designated as 30-dpr here) followed with snap-frozen in liquid nitrogen and storage at −80 °C. Nine to eleven independent biological replicates were employed for 30-dpa, 40-dpa and 30-dpr whereas only six biological replicates were possible for samples harvested at 20-dpa.
Seed metabolite extraction
Each sample was individually ground to fine power in liquid nitrogen with a mortar and a pestle followed with extraction with an optimized method for seeds46. In brief, above seed powder (150 ± 3 mg) was individually weighted into an Eppendorf tube followed with addition of 600 μL aqueous methanol (66%, v/v) and 3 mm tungsten carbide bead (Qiagen, Germany). The mixture was subjected to homogenized with a tissuelyser (Qiagen, Germany) followed with 15 min intermittent sonication in an ice bath (1 min sonication and 1 min break, repeated for 15 times). After centrifugation for 10 min (16099 × g, 4 °C), the supernatant was collected. This extraction process was further repeated twice and these three supernatants obtained were combined. After removal of methanol under vacuum, samples were lyophilized. The freeze-dried extracts were re-dissolved into 600 μL phosphate buffer (0.1 M, 50% D2O, pD 7.4) prepared from K2HPO4/NaH2PO447 containing 0.002% TSP-d4. Following 10 min centrifugation (16099 × g, 4 °C), 550 μL of such solution from each sample was then transferred into a 5 mm NMR tube for metabolite analysis.
NMR Measurements
All 1H NMR spectra were recorded at 298 K on a Bruker AVIII 600 spectrometer (600.13 MHz for 1H) equipped with an inverse detection cryogenic probe (Bruker Biospin, Germany). A standard noesypr1d pulse sequence (RD-90o-t1-90o-tm-90o-acquisition) was used to record one-dimensional 1H NMR spectra with the 90o pulse length of about 10 μs and t1 of 3 μs. Water peak was saturated with a continuous wave irradiation during the recycle delay (RD, 2s) and mixing time (tm, 80 ms). A total of 64 transients were collected with 32 k data points over a spectral width of 12 kHz. For resonance assignment purposes, a set of 2D NMR spectra were recorded and processed as previously reported48,49 for selected samples including 1H-1H COSY, 1H-1H TOCSY, 1H-JRES, 1H-13C HSQC and 1H-13C HMBC spectra.
Spectral processing and multivariate date analysis
An exponential line-broadening factor of 1 Hz was applied to each free induction decay (FID) prior to Fourier transformation (FT), and the spectra were phase- and baseline-corrected followed by referenced to TSP at δ 0.00. NMR spectral region between 0.5 and 8.5 ppm was divided into segments with width of 0.003 ppm using AMIX (v3.9.2, Bruker Biospin) whilst both methanol region at δ 3.339-3.384 and water region at δ 4.610-5.220 were removed.
The spectral areas of all buckets were normalized to the fresh weight of wheat seed power so that resultant data represented absolute metabolite concentration in the form of “peak intensity per weight unit of seeds”. PCA (principal component analysis) and OPLS-DA (orthogonal projection to latent structure-discriminant analysis)50 were carried out on the normalized NMR data using SIMCA-P+ software (v11.0, Umetrics, Sweden). In OPLS-DA models, one orthogonal and one predictive component were calculated using the unit-variance (UV) scaled NMR data as X-matrix and the class information as Y-matrix. The model qualities were described by the explained variances for X-matrix (R2X values) and the model predictability (Q2 values) with further assessment with CV-ANOVA approach51 where intergroup differences were considered as significant with p < 0.05.
The results were displayed in the forms of scores plots showing group clustering and loadings plots indicating variables (metabolites) contributing to inter-group differences. In these plots, loadings were back-transformed52 and the variables were color-coded according to the absolute values of the correlation coefficients (|r|)53 using an in-house developed script. In such plots, variable (i.e., metabolites) with hot colors (e.g., red) have more significant contributions to the group classification than the cold colored ones (e.g., blue). In this study, the metabolites exhibiting statistically significant changes were obtained at the level of p < 0.05. The ratios of changes for metabolites at four time-points were also calculated against their levels in wild-type wheat seeds in the form of (CTG –CWT)/CWT, where CTG stands for the metabolite concentrations in transgenic wheat seeds whereas CWT stands for the metabolite concentrations in wild-type wheat seeds.
GC-FID/MS analysis of fatty acids in the wheat seeds
Fatty acid composition in wheat seeds were measured in the methylated forms with a previously reported method54,55 with some minor modifications. About 20 mg of seed powder of wheat harvested at 40-dpa and 30-dpr was used with addition of 500 μL HPLC-grade CH3OH and one 3 mm tungsten carbide bead (Qiagen, Germany). Seeds at 20- and 30-dpa were not measured here due to limitation of samples availability. After vortex mixing, the mixture was homogenized with a tissuelyser (Qiagen, Germany) at 20 Hz for 90 s. Such treatment was repeated thrice with 3 min rest between each homogenization. 100 μL of homogenized mixture from each sample in a Pyrex tube was added with 20 μL internal standards in hexane (containing 1 mg/mL methyl heptadecanoate, 0.5 mg/mL methyl tricosanate and 2 mg/mL 3,5-di-tert-butyl-4-hydroxytoluene BHT) , then 1 mL mixture of methanol and hexane (4:l v/v). After cooling down the sample tubes above in a home-made liquid nitrogen bath for 10 min, 100 μL of precooled acetyl chloride was added carefully. Tubes were screw-capped and kept at room temperature in the dark for 24 hours followed with cooling down in an ice-bath for 4 min. 2.5 mL of 6% K2CO3 solution was then added (with gentle shaking) for neutralization. 200 μL hexane was added to extract the methylated fatty acids thrice and the combined supernatants were evaporated to dryness. The extracts were re-dissolved in 100 μL hexane for GC–FID/MS analysis.
Methylated fatty acids were quantified on a Shimadzu 2010 Plus GC-MS spectrometer. A flame ionization detector (FID) was used for quantification and a mass spectrometer with an electron impact (EI) ion source was employed for identification purpose. A DB-225 capillary GC column (10 m × 0.1 mm × 0.1 μm) was used with helium as carrier gas with 1 μL sample injected. The injection port and detector temperatures were set to 230 °C. The oven temperature was increased from 55 °C to 205 °C at 25 °C per min, kept at 205 °C for 3 min and then increased to 225 °C at 10 °C per min. The temperature was then kept at 225 °C for further 3 min.
All MS spectra over the m/z range of 45–450 were acquired with EI at 70 eV. Methylated fatty acids were identified by comparing retention times and mass spectra of 37 fatty acid standards. The results were expressed as micromole fatty acids per gram wheat seed fresh weight (FW).
Proteomics analysis
Seed proteomics analysis for both transgenic and wild type wheat was done at 40-dpa as previously reported19.
Additional Information
How to cite this article: Liu, C. et al. Reprogramming of Seed Metabolism Facilitates Pre-harvest Sprouting Resistance of Wheat. Sci. Rep. 6, 20593; doi: 10.1038/srep20593 (2016).
Supplementary Material
Acknowledgments
The authors would like to thank Dr Yixuan Duan for metabolite assignments and helpful discussions. We acknowledge financial supports from the National Natural Science Foundation of China (31200208, 21175149 and 21375144), and the Ministry of Agriculture of China (2009ZX08012-023B).
Footnotes
Author Contributions Y.W. and H.T. conceived the project. C.L., M.Y., X.W., Z.Z., H.G., Y.J. and F.D. performed experiments. H.T., C.L., F.H. and H.L. analyzed data. C.L., Y.W. and H.T. wrote the manuscript.
References
- Sharma S., Dhaliwal H., Multani D. & Bains S. Inheritance of preharvest sprouting tolerance in Triticum aestivum and its transfer to an amber-grained cultivar. J. Hered. 85, 312–314 (1994). [Google Scholar]
- Groos C. et al. Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white × red grain bread-wheat cross. Theor. Appl. Genet. 104, 39–47 (2002). [DOI] [PubMed] [Google Scholar]
- Walker-Simmons M. ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84, 61–66 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derera N. & Bhatt G. Germination inhibition of the bracts in relation to pre-harvest sprouting tolerance in wheat. Cereal Res. Commun. 8, 199–201 (1980). [Google Scholar]
- Li X., Nield J., Hayman D. & Langridge P. Cloning a putative self-incompatibility gene from the pollen of the grass Phalaris coerulescens. Plant Cell 6, 1923–1932 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G., Wagner W. & Follmann H. NADPH-dependent thioredoxin reductase and a new thioredoxin from wheat. Z Naturforsch 34, 214–221 (1979). [Google Scholar]
- Montrichard F., Renard M., Alkhalfioui F., Duval F. D. & Macherel D. Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol. 132, 1707–1715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. J. et al. Overexpression of thioredoxin h leads to enhanced activity of starch debranching enzyme (pullulanase) in barley grain. Proc. Natl. Acad. Sci. USA 96, 14641–14646 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. H. et al. Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc. Natl. Acad. Sci. USA 99, 16325–16330 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. C. et al. The level of expression of thioredoxin is linked to fundamental properties and applications of wheat seeds. Mol. Plant 2, 430–441 (2009). [DOI] [PubMed] [Google Scholar]
- Li X., Nield J., Hayman D. & Langridge P. Thioredoxin activity in the C terminus of Phalaris S protein. Plant J. 8, 133–138 (1995). [DOI] [PubMed] [Google Scholar]
- Yin J., Yu G., Ren J., Li L. & Song L. Transforming anti-trx S gene into wheat by means of pollen tube pathway and ovary injection. Acta Botanica Boreali-Occidentalia Sinica 24, 776–780 (2004). [Google Scholar]
- Zhou S., Yin J., Ren J. & Zhang R. Study on molecular identification and pre-harvest sprouting characteristic of the transgenic anti-trxs-gene wheat line 00T89. Chinese Journal of Biotechnology 22, 438–444 (2006). [PubMed] [Google Scholar]
- Ren J., Yin J., Niu H., Wang X. & Li Y. Effects of antisense-thioredoxin s gene on expression of endogenous thioredoxin h gene in transgenic wheat seed. Journal of Plant Physiology and Molecular Biology 33, 325–332 (2007). [PubMed] [Google Scholar]
- Ren J. P. et al. Expression of endogenous thioredoxin h gene and a-amylase activity in transgenic wheat with antisense- thioredoxins gene at grain filling stage. Journal of Triticeae Crops 27, 893–897 (2007). [Google Scholar]
- Guo H. & Yin J. Effects of antisense TRX s gene on hydrolysis of storage proteins in germination of transgenic wheat (Triticum aestivum L.). Acta Agronomica Sinica 33, 885–890 (2007). [Google Scholar]
- Guo H., Yin J. & Ren J. Effects of antisense trxs on starch hydrolases of wheat (Triticum aestivum L.) seeds in germination. Plant Physiology Communications 43, 273–277 (2007). [Google Scholar]
- Guo H. & Yin J. Effects of antisense Trx s gene on α-amylase in germinating seeds of wheat (Triticum aestivum L.). Acta Agriculturae Jiangxi 21, 1–3 (2009). [Google Scholar]
- Guo H. et al. Identification of changes in wheat (Triticum aestivum L.) seeds proteome in response to anti–trx s gene. PloS one 6, e22255. doi: 10.1371/journal.pone.0022255 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J. K., Lindon J. C. & Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 (1999). [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002). [PubMed] [Google Scholar]
- Tang H. R. & Wang Y. L. Metabonomics: a revolution in progress. Prog. Biochem. Biophys. 33, 401–417 (2006). [Google Scholar]
- Wang Y. et al. Rapid diagnosis and prognosis of de novo acute myeloid leukemia by serum metabonomic analysis. J. Proteome Res. 12, 4393–4401 (2013). [DOI] [PubMed] [Google Scholar]
- Hu C. Y. et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 4, 5067. doi: 10.1038/srep05067 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. Metabonomic Changes Associated with Atherosclerosis Progression for LDLR−/− Mice. J. Proteome Res. 14, 2237–2254 (2015). [DOI] [PubMed] [Google Scholar]
- Gargallo-Garriga A. et al. Opposite metabolic responses of shoots and roots to drought. Sci. Rep. 4, 6829. doi: 10.1038/srep06829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E., Juszczak I., Lee Y. P., Baier M. & Hincha D. K. Time-dependent deacclimation after cold acclimation in Arabidopsis thaliana accessions. Sci. Rep. 5, 12199. doi: 10.1038/srep12199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. Revealing different systems responses to brown planthopper infestation for pest susceptible and resistant rice plants with the combined metabonomic and gene-expression analysis. J. Proteome Res. 12, 6774–6785 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Du Y., Chen S. & Tang H. Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. J. Proteome Res. 10, 1704–1714 (2011). [DOI] [PubMed] [Google Scholar]
- Fan T. W. M. Metabolite profiling by one-and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Mag. Res. Sp. 28, 161–219 (1996). [Google Scholar]
- Fan T. W. M. & Lane A. N. Structure-based profiling of metabolites and isotopomers by NMR. Prog. Nucl. Mag. Res. Sp. 52, 69–117 (2008). [Google Scholar]
- Dai H., Xiao C., Liu H. & Tang H. Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza Bunge induced by water depletion. J. Proteome Res. 9, 1460–1475 (2010). [DOI] [PubMed] [Google Scholar]
- Cui Q. et al. Metabolite identification via the Madison metabolomics consortium database. Nat. Biotechnol. 26, 162–164 (2008). [DOI] [PubMed] [Google Scholar]
- Duan Y. et al. Multiple univariate data analysis reveals the inulin effects on the high-fat-diet induced metabolic alterations in rat myocardium and testicles in the preobesity state. J. Proteome Res. 12, 3480–3495 (2013). [DOI] [PubMed] [Google Scholar]
- Guo J. J. et al. Gene expression difference of antisense trx s transgenic wheat during seed development. Acta Agriculturae Boreali-occidental is Sinica 20, 60–64 (2011). [Google Scholar]
- Zhou S. et al. Study on the pre-harvest sprouting resistance in the transgenic wheat 00TY5 lines with anti-trx sgene. Journal of Triticeae Crops 26, 46–50 (2006). [Google Scholar]
- Zhou S. M. et al. Relationship between -SH content of protein ingredients and a-amylase activity in grains of transgenic anti-trx s gene wheat. Acta Agriculturae Sinica 32, 821–826 (2006). [Google Scholar]
- Payan F. et al. The dual nature of the wheat xylanase protein inhibitor XIP-I. J. Biol. Chem. 279, 36029–36037 (2004). [DOI] [PubMed] [Google Scholar]
- Sparla F., Pupillo P. & Trost P. The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J. Biol. Chem. 277, 44946–44952 (2002). [DOI] [PubMed] [Google Scholar]
- Saze H., Ueno Y., Hisabori T., Hayashi H. & Izui K. Thioredoxin-mediated reductive activation of a protein kinase for the regulatory phosphorylation of C4-form phosphoenolpyruvate carboxylase from maize. Plant Cell Physiol. 42, 1295–1302 (2001). [DOI] [PubMed] [Google Scholar]
- Miflin B. J. & Habash D. Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987 (2002). [DOI] [PubMed] [Google Scholar]
- Kochkina V. Isolation, purification, and crystallization of aspartate aminotransferase from wheat grain. Biochemistry (Moscow) 69, 897–900 (2004). [DOI] [PubMed] [Google Scholar]
- Morris A. J., Engebrecht J. & Frohman M. A. Structure and regulation of phospholipase D. Trends Pharmacol. Sci. 17, 182–185 (1996). [DOI] [PubMed] [Google Scholar]
- Gardiner J., Collings D. A., Harper J. D. I. & Marc J. The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol. 44, 687–696 (2003). [DOI] [PubMed] [Google Scholar]
- Rejman E. & Buchowicz J. The sequence of initiation of RNA, DNA and protein synthesis in the wheat grains during germination. Phytochemistry 10, 2951–2957 (1971). [Google Scholar]
- Wu X., Li N., Li H. & Tang H. An optimized method for NMR-based plant seed metabolomic analysis with maximized polar metabolite extraction efficiency, signal-to-noise ratio, and chemical shift consistency. Analyst 139, 1769–1778 (2014). [DOI] [PubMed] [Google Scholar]
- Jiang L., Huang J., Wang Y. & Tang H. Eliminating the dication-induced intersample chemical-shift variations for NMR-based biofluid metabonomic analysis. Analyst 137, 4209–4219 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. Metabonomic analysis reveals efficient ameliorating effects of acupoint stimulations on the menopause-caused alterations in mammalian metabolism. Sci. Rep. 4, 3641. doi: 10.1038/srep03641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci. Rep. 5, 8421. doi: 10.1038/srep08421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J. & Wold S. Orthogonal projections to latent structures (O‐PLS). J. Chemometr. 16, 119–128 (2002). [Google Scholar]
- Eriksson L., Trygg J. & Wold S. CV‐ANOVA for significance testing of PLS and OPLS® models. J. Chemometr. 22, 594–600 (2008). [Google Scholar]
- Wang Y. et al. Metabonomic investigations of aging and caloric restriction in a life-long dog study. J. Proteome Res. 6, 1846–1854 (2007). [DOI] [PubMed] [Google Scholar]
- Cloarec O. et al. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal. Chem. 77, 517–526 (2005). [DOI] [PubMed] [Google Scholar]
- An Y. et al. High-fat diet induces dynamic metabolic alterations in multiple biological matrices of rats. J. Proteome Res. 12, 3755–3768 (2013). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Combined NMR and GC-MS analyses revealed dynamic metabolic changes associated with the carrageenan-induced rat pleurisy. J. Proteome Res. 12, 5520–5534 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.