Abstract
4,4′-Methylene diphenyl diisocyanate (herein 4,4′-MDI) is used in the production of polyurethane foams, elastomers, coatings, adhesives and the like for a wide range of commercial products. Occupational exposure to MDI levels above current airborne exposure limits can elicit immune mediated hypersensitivity reactions such as occupational asthma in sensitive individuals. To accurately determine exposure, there has been increasing interest in developing analytical methods to measure internal biomarkers of exposure to MDI. Previous investigators have reported methodologies for measuring MDI diamine metabolites and MDI-Lysine (4,4′-MDI-Lys) adducts. The purpose of this study was to develop and validate an ultra performance liquid chromatography isotope dilution tandem mass spectrometry (UPLC-ID/MS/MS) quantitation method via a signature peptide approach to enable biomonitoring of 4,4′-MDI adducted to human serum albumin (HSA) in plasma. A murine, anti-4,4′-MDI monoclonal IgM antibody was bound to magnetic beads and utilized for enrichment of the MDI adducted HSA. Following enrichment, trypsin digestion was performed to generate the expected 414 site (primary site of adduction) 4,4′-MDI-adducted HSA signature peptide that was quantified by UPLC-ID/MS/MS. An Agilent 6530 UPLC/quadrupole time of flight MS (QTOF) system was utilized for intact adducted protein analysis and an Agilent 6490 UPLC/MS/MS system operated in multiple reaction monitoring (MRM) mode was utilized for quantification of the adducted signature peptide biomarker both for in chemico and worker serum samples. Worker serum samples were initially screened utilizing the previously developed 4,4′-MDI-Lys amino acid method and results showed that 12 samples were identified as quantifiable for 4,4′-MDI-Lys adducts. The signature peptide adduct approach was applied to the 12 worker samples identified as quantifiable for 4,4′-MDI-Lys adducts. Results indicated no positive results were obtained above the quantification limit by the signature peptide approach. If the 414 site of lysine adduction accounted for 100% of the 4,4′-MDI adductions in the signature peptide adduct approach, the three highest quantifiable samples by the 4,4′-MDI-Lys method should have at least been detectable by the signature peptide method. Results show that although the 4,4′-MDI signature peptide approach is more selective, it is 18 times less sensitive than the 4,4′-MDI-Lys method, thus limiting the ability to detect adduct levels relative to the 4,4′-MDI-Lys amino acid method.
Keywords: MDI, Adduct, Mass spectrometry, Diisocyanates, Biomarkers
1. Introduction
In commerce, a majority of methylene diphenyl diisocyanate (MDI) is sold as “monomeric MDI” (typically containing greater than 95% of the 4,4′-MDI isomer) or “polymeric MDI” (pMDI). These diisocyanates are important industrial chemicals used in a variety of commercial and consumer applications such as thermoplastic polyurethanes, fibers, sealants, coatings, automotive bumpers, fenders and fascia, integral-skin plastics, tires and wheels, industrial wheels, shoe soles, recreational goods, and mechanical goods.
Occupational exposure to MDI levels above current airborne exposure limits can elicit immune mediated hypersensitivity reactions such as occupational asthma in sensitive individuals [1]. Hence, current industrial hygiene practices such as the utilization of personal protective equipment are designed to limit exposure below established exposure limits obviating potential health effects [1]. An important tool in the determination of worker exposure is the measurement of airborne MDI in the workplace. However, the external exposure measurements of MDI may not be representative of actual internal body burden levels, thus there has been increasing interest in developing analytical methods to measure internal biomarkers of exposure for MDI.
An understanding of the major metabolic pathways for MDI provides insight into potential biomarkers to measure internal exposure to MDI. Gledhill and coworkers [2] demonstrated that the major metabolites after inhalation exposure to 14C-MDI were the mono- and di-acetylated compounds with the methylene bridge oxidized to various states. This supports the most common method, measuring liberated hydrolysis products following acid digestion of protein conjugates in biological fluids. Some researchers have suggested that methylene diphenyl diamine (MDA) can be formed after exposure to MDI, and that the hydrolysis method could confound biomonitoring efforts for MDI as the metabolic products are the same whether there is MDI or MDA exposure [3], [4], [5], [6], [7], [8], [9], [10], [11]. Recent work by Wisnewski and coworkers [12] addresses alternative mechanisms associated with toluene diamine (TDI) metabolism, which is applicable to MDI. This work provides evidence that accounts for the formation of metabolites observed in the Gledhill et al. [2] study without the necessity of invoking the presence of free MDA as an intermediate. Most importantly, the hydrolysis method while tested [13] has not been fully validated, as incomplete release of MDA from protein conjugation may underestimate MDI exposure levels.
Beyond hydrolysis measurements, some investigators have moved to measuring MDI specifically adducted to circulating proteins. Groups have investigated 4,4′-MDI-hemoglobin adducts at the N-terminal valine [14]. Gries and Leng validated a method that measured hydantoin adducts levels in plasma which formed following acid treatment of 4,4′-MDI conjugated to the N-terminal amino acid [15]. Sufficient sensitivity was achieved to quantify the possible adduct levels, however poor analyte recoveries were observed following 2 M HCl hydrolysis (2 h at 80 °C) and solid phase extraction (SPE) cleanup of the biological samples (or spiked matrix samples). These low recoveries were likely a function of analyte degradation during the hydrolysis process or loss of the analyte during the SPE cleanup process. Additional work would be required to establish if technical modifications might make this methodology useful for 4,4′-MDI-hemoglobin adducts, and if this methodology would be sensitive enough to inform worker exposures.
More recently, Kumar and coworkers have identified and quantified 4,4′-MDI-Lys amino acid adducts in rats and humans [16], [17], [18]. Both 4,4′-MDI-Lys and acetylated 4,4′-MDI-Lys (AcMDI-Lys) adduct levels were quantified from human serum following human serum albumin (HSA) purification, pronase digestion, and SPE cleanup. However, due to the lack of a peptide sequence the amino acid adduct method does not provide HSA protein biomarker confirmation. As a consequence, the 4,4′-MDI-Lys and AcMDI-Lys quantitative results rely on the purity of the HSA fraction collected. Due to the non-specificity of the albumin purification method utilized, an overestimation in MDI albumin adducts levels could result due to adduct contributions from other proteins.
However, Wisnewski and coworkers [19] have identified 12 MDI lysine peptide conjugation sites with IgG on human albumin which when utilized as a biomarker of exposure could increase specificity by not relying on the purity of the HSA fraction. Recent advances in the development of diisocyanate-specific monoclonal antibodies have provided a new approach to detecting diisocyanate adducts [20], [21], [22]. The advantages of measuring protein adduct biomarkers are specificity for at least one diisocyanate adduct, a slow turnover (i.e., half-life of HSA is approximately 20 days) and therefore represents a viable potential tool for monitoring long-term exposure [23], [24].
Therefore, to overcome the current selectivity limitations associated with 4,4′-MDI-Lys amino acid biomarker quantification methods described above, we aimed to develop and validate a highly specific HSA signature peptide approach [25], [26], [27], [28] to identify and quantify 4,4′-MDI HSA protein adducts formed in chemico and sera from a human cohort population. We employ a highly specific IgM monoclonal antibody to capture the 4,4′-MDI adducted HSA proteins from sera, digest the captured adducted albumin with trypsin to produce the 4,4′-MDI adducted signature peptide biomarker and analyze with ultra performance liquid chromatography isotope dilution tandem mass spectrometry (UPLC-ID/MS/MS).
2. Methods
2.1. In chemico
2.1.1. Conjugation of 4,4′-MDI to HSA
To 1 mg/mL solutions of HSA, 0 mM, 0.01 mM, 0.1 mM and 1 mM of 4,4′-MDI were added and incubated with rotation at 37 °C for 2 h. Following incubation, each reaction was quenched with the addition of 0.5% acetic acid and vortex-mixed. Excess 4,4′-MDI was removed by centrifugation (10 min at 15,000 × g). Samples were transferred to autosampler vials for intact protein analysis via the Agilent 6530 UPLC/QTOF (Agilent, Santa Clara, CA) system as described below.
2.1.2. HSA-4,4′-MDI adduct stability
To determine stability of the 1 mM 4,4′-MDI adducted HSA, aliquots were stored at room temperature and −80 °C. Each sample was analyzed on days 1, 4 and 8 after initial analysis of freshly prepared adducted HSA by UPLC/QTOF as described below to determine stability.
2.1.3. LC/MS–MS conditions for intact HSA-adduct analysis
An Agilent 1290 UPLC system (Agilent, Santa Clara, CA) was utilized for intact adducted protein analysis. The analytical column utilized was a Zorbax rapid resolution 300SB-C18 (part number 863974-302, Agilent) 3 × 150 mm-i.d. (3.5 μm particle size). The aqueous mobile phase (A) was 0.1% acetic acid/water, and the organic phase (B) was 0.1% acetic acid/acetonitrile (ACN). After injection of 2 μL sample onto the column the sample was eluted at 400 μL/min from the column using a solvent gradient that initially consisted of 99% A and 1% B for 1 min, and then a 5.5% increase in B for the next 13.5 min to a final concentration of 75% B. The column eluent was introduced into an Agilent 6530 QTOF (Agilent) mass spectrometer with an electrospray ionization interface. The instrument, operated in full scan mode, was utilized for intact adducted protein analysis. The instrument was operated in the positive ion mode with a survey scan range from 800 to 3000 Da. Instrument parameters were as follows: gas temperature 350 °C, gas flow 10 L/min, nebulizer 60 psi, fragmentation voltage was 250 V and the capillary voltage was 3500 V. Instrument control and data processing were performed with the Mass Hunter software version for B.02.01 data acquisition and B.05.00 for qualitative analysis.
2.1.4. HSA adduct digestion
To determine the extent of adduction, the 1 mM 4,4′-MDI adducted HSA was digested with trypsin. Specifically, 5 μL of the 1 mM 4,4′-MDI adducted HSA and 45 μL of human plasma (sodium heparin, Bioreclamation, Westbury, NY) were transferred to a microfuge tube. Two hundred and fifty μL of 50 mM ammonium bicarbonate was added to adjust the pH to 7.8. Fifty μL of 0.1% Rapigest SF (Waters, Milford, MA) was added to the same microfuge tube and incubated for 5 min at 100 °C to solubilize the adducted protein and make it more susceptible to enzymatic cleavage without inhibiting enzyme activity. The sample was allowed to cool to room temperature and 20 μg (∼45 μL) of modified trypsin (Promega, Madison, WI, V5113) was added and incubated overnight at 37 °C to digest the adducted HSA into predictable trypsin cleaved peptides. The sample was subsequently removed from the incubator and allowed to cool to room temperature. One hundred μL of 175 mM HCl was added to adjust the pH to approximately 2 and incubated for an additional 30 min at 37 °C to break down the Rapigest surfactant into small molecules. The sample was removed from the incubator and allowed to cool to room temperature. Fifty five μL of a 1 μg/mL 13C 15N internal standard (ISTD, KVPQVSTPTLV(+6)EVSR) solution was added to the digest. Final volume was approximately 550 μL. The sample was transferred to an autosampler vial for UPLC-ID/MS/MS analysis.
The same process described above was followed for control HSA. Ten mL of mixed gender unfiltered human plasma (sodium heparin, Bioreclamation) aliquoted into 1 mL increments and stored at −80 °C. After the human plasma was thawed to room temperature, human plasma was digested with trypsin as described above.
2.1.5. MDA adducted amino acid/peptides syntheses
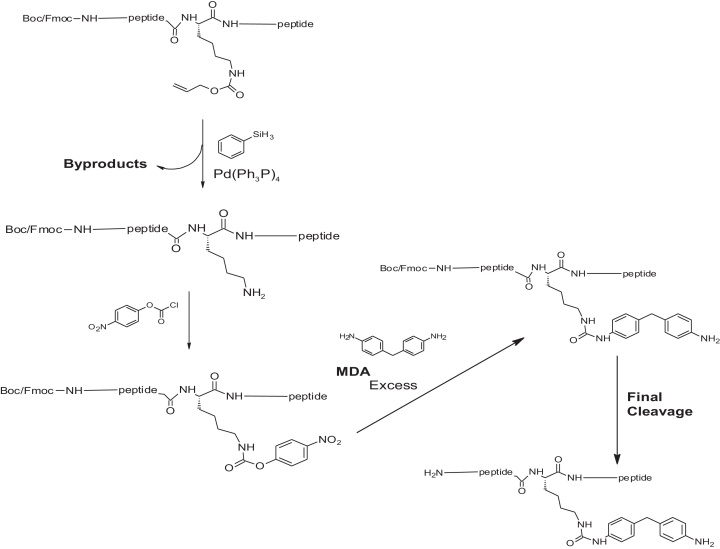
Adduction of lysine with 4,4′-MDI, results in the nucleophilic addition of an isocyanate group (N C O) that produces a methylene dianaline (MDA) urea adduct on the lysine amino acid side chain. To determine the primary site of adduction, identified peptides with a MDA urea adduct were synthesized (New England Peptide, Gardner, MA) to lysine positioned in the 234, 413 and or 414 on various HSA tryptic peptides to include one missed cleavage at the 413 and 414 sites of adduction. The unlabeled synthesized MDA adducted lysine amino acid and peptides are listed in Table 1 and the peptide adduct synthesis is shown in Fig. 1. Fig. 1 illustrates that following hydrogenation and de-protection during the synthesis process to produce an intermediate, MDA is introduced in excess and the final cleavage product, the peptide with an MDA adduct, is formed (resulting in a change in molecular weight of 224 Da).
Table 1.
4,4′-MDI HSA derived lysine amino acid and peptide adducts. Underlined lysine (K) indicates the site of MDI adduction.
| HSA site of adduction | MDI adducted amino acid or peptide | MW (g/mol) |
|---|---|---|
| Any, non-specific | K | 371.11 |
| 236 | AFKAWAVAR | 1242.67 |
| 413 | YTK | 634.22 |
| 414 | KVPQVSTPTLVEVSR | 1862.93 |
| 413 | KKVPQVSTPTLVEVSR | 2255.14 |
| 414 | KKVPQVSTPTLVEVSR | 2255.14 |
| 549 | KQTALAELVK | 1323.66 |
Fig. 1.
Synthesis process to adduct MDA to the lysine side chain on a peptide.
2.1.6. Monoclonal 4,4′-MDI IgM antibody production
The monoclonal antibody (mAb) 15D-1C4-1C3, an anti-aromatic and aliphatic diisocyanate IgM producing hybridoma cell line [20] was grown in complete DMEM over a period of three weeks. Culture supernatant was collected following the splitting of confluent cell lines. Approximately 2 L of hybridoma culture supernatant was collected, centrifuged, and filter sterilized. Protease activity was inhibited by the addition of Roche protease inhibitor per 500 mL culture supernatant. The IgM mAb 15D-1C4-1C3 was precipitated and concentrated using a saturated ammonium sulphate precipitation method. The mAb pellet was resuspended in NaHCO3 buffer, dialyzed against NaHCO3 buffer, and then lyophilized. The lyophilized mAb was reconstituted in Dulbecco's Phosphate-Buffered Saline (DPBS, Gibco, Life Technologies, Grand Island, NY) at 1 mg/mL and stored at −20 °C until use.
2.1.7. Immunoaffinity purification of MDI conjugated proteins
One mL of secondary coated IgM Dynabeads (110.39D Invitrogen, Carlsbad, CA) was washed twice with DPBS. To the washed secondary coated IgM Dynabeads, 60 μL primary 15D-1C4-1C3 IgM mAb murine monoclonal antibody was added along with 440 μL DPBS and allowed to incubated with rotation at room temperature for 2 h. After the 2 h binding period, the supernatant was removed and antibody-bead complex was washed twice with DPBS and stored in 1 mL DPBS solution at 4 °C until needed.
Five μL of the 1 mM 4,4′-MDI adducted human albumin was diluted 1:10, 1:100 and 1:1000 in human plasma. Dilutions were added to 200 μL of the above antibody bead complex and incubated at 37 °C with rotation overnight. After incubation, the supernatant was removed from the microfuge tubes and the remaining antigen–antibody bead bound complex was washed twice with 1 mL DPBS. The beads were resuspended in 50 μL of 0.1% Rapigest (Waters, Milford, MA) and 250 μL of 50 mM ammonium bicarbonate, heated at 100 °C for 5 min, allowed to cool to room temperature, and then 20 μg (∼45 μL) of sequencing grade modified trypsin was added (Promega, V5113). Samples were briefly vortex-mixed and subsequently incubated on a water bath at 37 °C overnight. After tryptic digestion, 100 μL of 175 mM HCl was added to adjust the pH to approximately 2 and an additional incubation at 37 °C for 30 min to break down the acid labile surfactant into small molecules was performed. The total final digest volume was ∼550 μL.
2.1.8. LC/MS–MS conditions for quantification of the HSA adducted peptide biomarker
MDI-peptide adduct standards were prepared in 50:50 water:acetonitrile with 0.1% formic acid from 1 ng/mL to 200 ng/mL. Matrix standards, with trypsin digested albumin from human serum (A8763, Sigma, St. Louis, MO) as the matrix, were prepared over the same range of concentrations. The analytical column utilized was a 300 SB RRHD (Agilent, part number 857750-902) 2.1 × 50-mm-i.d. reversed-phase C18 (1.8 μm particle size). The aqueous mobile phase (A) consisted of Milli-Q (18 MΩ) water with 0.1% formic acid, while the organic phase (B) was ACN with 0.1% formic acid. The autosampler was programmed to deliver a 10 μL injection. A gradient profile was utilized at a flow rate of 500 μL/min. The mobile phase was held for 1 min at of 95% A and 5% B and then B was increased at 19% per minute for the next 3.5 min to a final concentration of 28% A and 72% B, respectively.
An Agilent 6490 UPLC–MS/MS (Agilent) system operated in positive ESI multiple reaction monitoring (MRM) mode was utilized for quantification of the adducted peptide biomarker. The UPLC column eluent was introduced into a 6490 Agilent triple-quadrupole mass spectrometer with a jet spray interface. For multiple reaction monitoring, the instrument was operated in the positive ion mode with mass-to-charge (m/z) MRM transition pairs listed in Table 2. The collision energy for each transition was optimized. Instrument parameters were as follows: gas temperature 350 °C, gas flow 11 L/min, nebulizer 30 psi, sheath gas heater 400 °C, sheath gas flow 12, and the capillary voltage was 4000 V. Dwell time was 100 ms per transition and the fragmentation voltage was set to 380 V. Instrument control and data processing were performed with the Mass Hunter software version for B.04.01 data acquisition and version B.05.00 for both qualitative and quantitative analysis.
Table 2.
Multiple reaction monitoring transitions and corresponding collision energies for labeled and unlabeled adducted HSA peptides. Underlined lysine (K) indicates the site of MDI adduction.
| Adducted peptide | Monoisotopic transition pairs |
Optimized collision energies | |
|---|---|---|---|
| Precursor | Product | ||
| AFKAWAVAR | 622.3 (+2) | 673.1 | 16 |
| KQTALAELVK | 662.9 (+2) | 844.5 | 16 |
| YTK | 635.3 (+1) | 371.2 | 16 |
| KVPQVSTPTLVEVSR | 622.0 (+3) | 900.5 | 18 |
| 589.3 | 18 | ||
| 450.8 | 18 | ||
| KVPQVSTPTLVEV(+6)SR | 624.3 (+3) | 906.5 | 18 |
| 595.3 | 18 | ||
2.2. In vivo
2.2.1. Human sera screened utilizing 4,4′-MDI-Lys amino acid methodology
De-identified sera samples, approved by The Dow Chemical Company human subject review board, from human workers with known 4,4′-MDI exposures were obtained from the laboratory of Dr. Jean-Luc Malo at the Hôpital du Sacré-Coeur de Montréal. Demographic and classification information for sera samples is provided in Tables S1–S4. These samples were collected from workers undergoing specific inhalation challenge (SIC) in order to establish the diagnosis of diisocyanate related asthma as part of the worker compensation program. Workers were exposed to MDI in an enclosed-circuit inhalation apparatus, which maintained a constant concentration of diisocyanates below 20 ppb. The standardized protocol used was exposure of subjects for 1–4 min on the first day, 5–30 min on the second day, and 2 h on the third day, unless a decrease in FEV1 of 20% or more was observed immediately or 10 min after stopping exposure. The sensitivity and specificity of 4,4′-MDI HSA adduct determinations via the peptide method and to that of the previously published 4,4′-MDI-Lys method [17] were compared. Briefly, for the 4,4′-MDI-Lys method, 500 μL of human sera albumin (HSA) was purified through a HiTrap Blue column. The purified eluent containing the adducted HSA was subsequently centrifuged (15 min, 4000 × g) though a 30 kDa molecular weight cutoff filter. Two water washes were performed by centrifugation of the 30 kDa molecular weight cutoff filter. The concentration of the purified HSA in the retentate was determined by the bicinchoninic acid assay protein assay and 10 mg of purified albumin/mL was Pronase E digested for approximately 12 h at 37 °C. Five ng/mL of 13C 15N K ISTD (underlined K indicates MDI adducted lysine) was added prior to digestion. After digestion, samples were cleaned up utilizing SPE columns (Phenomenex, Strata-X 33u Polymeric Reversed Phase, 200 mg/3 mL, part number 8B-S100-FBJ). The column was activated with 3 mL of methanol and then equilibrated with distilled de-ionized water. The sample was applied to the SPE column and then washed with 3 mL of distilled de-ionized water, 3 mL of 90:10 distilled de-ionized water:methanol and 3 mL of 80:distilled de-ionized water–methanol. The sample was then eluted with 6 mL of 20:80 distilled de-ionized water: methanol. The SPE eluent was evaporated to dryness and reconstituted in 100 μL of 50 mM ammonium bicarbonate. Samples were vortex-mixed prior to analysis on an AB/Sciex 4000 QTRAP.
Supplementary Tables S1–S4 related to this article can be found, in the online version, at doi:10.1016/j.toxrep.2014.09.009.
MDI exposed worker demographics and classification criteria positive for occupational asthma.
MDI exposed worker demographics and classification criteria negative for asthma (non-asthma of any kind).
MDI exposed workers demographics and classification criteria negative for occupational asthma.
Non exposed worker control demographics and classification criteria.
2.2.2. Human sera screened utilizing the 4,4′-MDI signature peptide adduct methodology
Sera samples that were previously identified as containing 4,4′-MDI-Lys adducts [17] were subsequently screened with the modified signature peptide adduct method. To obtain definitive analyses of 4,4′-MDI signature peptide adducts, a 0.5 mL aliquot of each positive human sera was allowed to bind to the primary murine anti-diisocyanate IgM antibodies, trypsin digested and analyzed as described previously.
3. Results and discussion
3.1. In chemico
3.1.1. Exposure of 4,4′-MDI to HSA
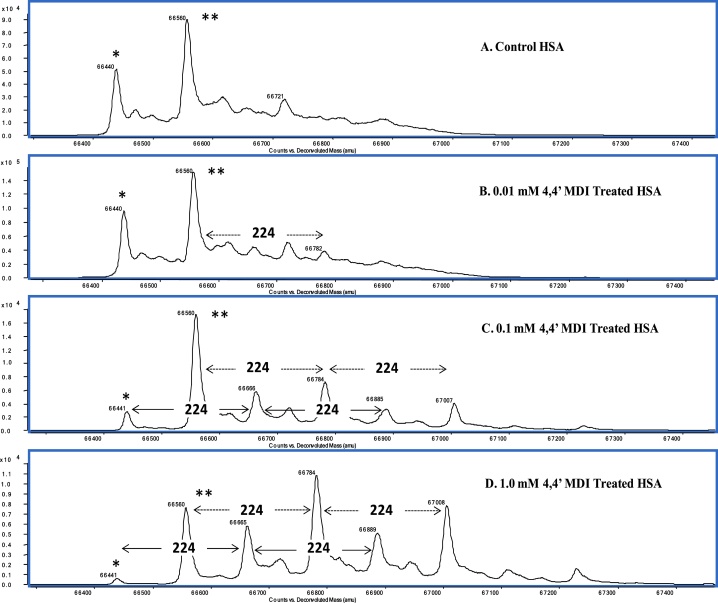
Fig. 2 contains deconvoluted full scan mass spectra taken in positive ion electrospray ionization mode of (A) HSA; (B) 0.01 mM 4,4′-MDI adducted HSA; (C) 0.1 mM 4,4′-MDI adducted HSA; and (D) 1 mM 4,4′-MDI adducted HSA. Fig. 2A demonstrated two major isoforms of HSA at [M+H]+ of 66,440 Da and 66,560 Da consistent with that previously reported [29], [30], [31]. When 0.01 mM of 4,4′-MDI was incubated with the HSA (Fig. 2B) adduction appeared to be evident and produced a mass difference of 224 indicative of an MDA adduction. At the 0.1 mM and the 1 mM 4,4′-MDI concentrations (Fig. 2C and D), MDA adduction were apparent on both isoforms, as well as multiple MDI additions to a single HSA molecule, which may represent adduction to multiple amino acid residues and/or polymerization onto a single nucleophilic site. Also evident were the decreased signal intensities of both isoforms and increased signal intensities at m/z ratios at the respective MDA adductions.
Fig. 2.
Deconvoluted full scan mass spectra taken in positive ion electrospray ionization mode of (A) control HSA; (B) 0.01 mM 4,4′-MDI adducted HSA; (C) 0.1 mM 4,4′-MDI adducted HSA; and (D) 1.0 mM 4,4′-MDI adducted HSA. The single asterisk indicates the first isomer of HSA while the double asterisk indicates a second isomer. The change in molecular weight is 224 Da.
3.1.2. HSA adduct stability
MDI-HSA adducts were analyzed on days 0, 1, 4 and 8 after preparation. Results demonstrated that the samples were stable over the entire 8 days when stored at either −80 °C or room temperature conditions (data not shown).
3.1.3. HSA adduct peptide identification
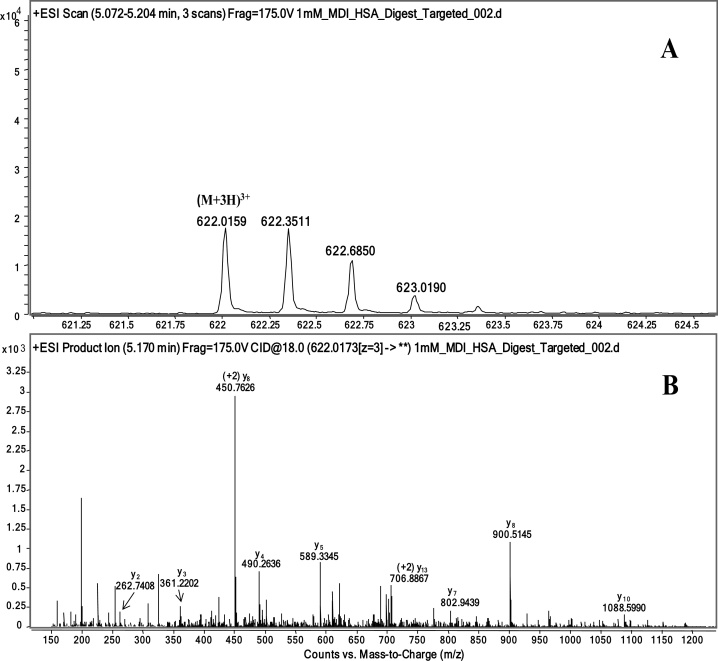
Targeted peptides from the 1 mM 4,4′-MDI adducted HSA trypsin digest were identified via data-dependent UPLC–MS/MS acquisition and subsequent analysis. Results in Fig. 3A show the adducted peptide KVPQVSTPTLVEVSR. A triply charged precursor (M+3H)3+ ion dominant at a m/z ratio of 622.0 of the MDI peptide adduct was observed. When the precursor ion was subjected to MS/MS fragmentation, the fragmentation pattern (Fig. 3B) confirmed the amino acid sequence of the adducted peptide. Precursor ions and fragmentation patterns were also confirmed for K, YTK, KQTALAELVK and AFKAWAVAR from MDI adducted HSA. A precursor (M+3H)3+ was identified for adducted peptides YTKKVPQVSTPTLVEVSR and YTKKVPQVSTPTLVEVSR, however ion abundance was insufficient for MS/MS fragmentation confirmation.
Fig. 3.
Adducted peptide KVPQVSTPTLVEVSR (A) full scan and (B) product ion scan.
For confirmation, tandem mass spectrometry analysis was performed on the HSA trypsin digests. Full scan and fragmentation data files utilized the Mascot database search engine (Matrix Science, Boston, MA) which employs probability base scoring, significance level, mass tolerances and sequence query scoring [32] to confirm identification of the MDI-derived trypsin digested HSA peptides from the non-redundant database of the National Center for Biotechnology Information (NCBI) for peptide and protein adduct identification. Results confirmed identification for three 4,4′-MDI HSA trypsin derived peptide adducts: AFKAWAVAR, KQTALAELVK and KVPQVSTPTLVEVSR (all peptides have one missed cleavage). The missed cleavage in each peptide is not unexpected due to the large 4,4′-MDI substituent present and likely hindering trypsin's ability to cleave at these sites. The identified MDI adducts with HSA were consistent with those reported by Wisnewski's research group [19].
3.1.4. Primary site of adduction
For external calibration purposes, matrix standards (trypsin digested HSA) were prepared from 1 ng/mL to 200 ng/mL. Quantification of the 1 mM 4,4′-MDI adducted HSA trypsin digest was performed. No quantifiable amounts resulted for K and YTK. Results showed that the KVPQVSTPTLVEVSR peptide had a concentration of 276 ng/mL. This resulted in a 10.3% recovery of the adducted peptide when 0.1 mg of HSA was digested. Recovery was 1.2% for the AFKAWAVAR peptide. This demonstrates that of the peptides monitored, the primary site of adduction appears to be the 414 site, KVPQVSTPTLVEVSR.
3.1.5. Sensitivity comparison with and without immunoaffinity purification
Five μL aliquots of the 1 mM 4,4′-MDI adducted human albumin (1 mg/mL) were diluted 1:10, 1:100 and 1:1000 in human plasma. Trypsin digestion was performed on samples with and without antibody capture. Results demonstrated in the absence of immunoaffinity purification adducted albumin could only be detected at the 1:10 dilution, while adducted albumin was non-quantifiable at the 1:1000 dilution. Capture efficiency was 99.5%, 70.8% and 69.4% when the 1 mM 4,4′-MDI adducted human albumin was diluted 1:10, 1:100 and 1:1000 in plasma respectively (n = 1 per protein level).
3.1.6. Suppression effects with peptide adduct methodology
Comparison of solvent standards to matrix standards showed that there is approximately a 30% suppression effect occurring in the matrix standards. When the internal standard was added at 100 ng/mL the area counts showed an average of 18,040 counts in solvent standards while in matrix standards the average internal standard area count was 12,624 demonstrating a suppression of 30%. A comparison of the signal-to-noise ratio was also performed of the 100 ng/mL matrix standard before and after a 10-fold dilution in solvent (50:50 water:acetonitrile with 0.1% formic acid). Results demonstrated that the signal-to-noise ratio of the 100 ng/mL matrix standard prior to dilution was approximately 114 and after the 10-fold dilution with solvent was approximately 29. However, the increased signal-to-noise ratio achieved by dilution of the matrix (2.5×) cannot overcome the absolute dilution of the analyte to enhance the peptide method detection limit.
3.1.7. Screening of serum samples by the 4,4′-MDI-Lys amino acid methodology
Human sera samples from workers with potential MDI exposure were purified for HSA, pronase digestion, SPE cleaned up and assayed for 4,4′-MDI-Lys as previously described (Kumar et al. [16]). 4,4′-MDI-Lys could be detected in 8 of the 15 MDI-asthmatic workers, but only 4 of the 15 had quantifiable levels (Supplementary Table S1) and maybe attributable to the half-life of albumin from when the date of last exposure took place to the time the actual sera samples were collected. 4,4′-MDI-Lys was detectable in a proportion of all workers, independent of disease status (Supplementary Tables S1–S4). Overall the 4,4′-MDI-Lys method could quantify adduct levels in 12 human sera samples from the human cohort above the limit of quantification (LOQ) for the method (Table 3). Concentrations of 4,4′-MDI-Lys ranged from 7.55 to 151 fmol adducted albumin/mg of albumin with the highest level of adduct observed in a non-exposed control. Sabbioni and coworker's [17] construction worker cohort identified 15% of the construction worker controls as positive with the 4,4′-MDI-Lys method. In the present human cohort, the presence of a high level of 4,4′-MDI-Lys observed in the non-exposed individual may be due to either a non-occupational exposure to MDI or lack of recognition of use of a MDI-containing product.
Table 3.
Summary demographic and classification information for all sera samples.
| Parameter | Exposed to diisocyanates workers with diisocyanate related asthma n = 15 | Exposed to diisocyanate workers non-asthmatic no diisocyante related asthma n = 10 | Exposed to diisocyanates workers asthmatic no diisocyanate related asthma n = 9 | Not exposed to diisocyanates non-asthmatic no diisocyante related asthma n = 7 |
|---|---|---|---|---|
| Gender | ||||
| Male | 13 | 8 | 8 | 1 |
| Female | 2 | 2 | 1 | 6 |
| Age (years) (mean ± SD) | 38.5 ± 9.9 | 36.5 ± 9.9 | 44.9 ± 7.5 | 37.9 ± 11.0 |
| Mean (±SD) duration of exposure to diisocyanates (months) | 66.8 ± 83.0 | 60.5 ± 54.3 | 152.5 ± 109.3 | Not applicable |
| Last exposure to diisocyanates prior to sample collection (mean days ± SD) | 94.8 ± 136.9 | 140.9 ± 113.1 | 170.9 ± 256.1 | Not applicable |
| Challenge test positive to MDI | Yes | No | No | No |
| Detect or non-detect | 4 detect | 4 detect | 3 detect | 1 detect |
| Sabbioni | 4 NQ | 1 NQ | 2 NQ | 0 NQ |
| Modified method | 7 ND | 5 ND | 4 ND | 6 ND |
3.1.8. Screening of positive serum samples with signature peptide adduct methodology
The 12 sera positive with quantifiable 4,4′-MDI-Lys adducts were subsequently screened by the 4,4′-MDI-signature peptide adduct method as described above. No 4,4′-MDI-signature peptide adduct positive results were obtained from the 12 samples (LOQ = 13.4 fmol adducted albumin/mg of albumin or matrix LOQ = 134 fmol adducted albumin/mg of albumin). If the 414 site accounted for 100% of the adduction, the three highest quantifiable samples by the 4,4′-MDI-Lys method should have at least been detectable by the signature peptide method. However an additional 5–19 exposure dose dependent lysine adduction sites have been demonstrated on HSA [20], [22].
4. Conclusions
The MDI adducted HSA is stable at both −80 °C and room temperature over 8 days. Of the possible HSA peptide adducts, the 414 adduct (KVPQVSTPTLVEVSR) appears to be a primary site of adduction. The peptide adduct method is 18 times less sensitive than the 4,4′-MDI-Lys method thus, limiting the ability to detect adduct levels relative to 4,4′-MDI-Lys method. While the adducted KVPQVSTPTLVEVSR peptide method is more selective and potentially the primary site of adduction, the additional 5–19 exposure dose dependent lysine adduction sites have been demonstrated on HSA, all of which would potentially contribute non-specifically to the total 4,4′-MDI-Lys measured following pronase digestion. Thus, while more specific, the 4,4′-MDI-signature peptide method's sensitivity appears to be inadequate for biomonitoring of workers’ exposure to MDI.
Biomonitoring is superior to many other exposure assessment methodologies since it allows assessment of the actual exposure on the individual level. While increased selectivity was achieved with the signature peptide method, it could not overcome sensitivity limitations discussed above. The Sabbioni method [17] appears to have the sensitivity to detect and quantify 4,4′-MDI-Lys adducts at the NIOSH and OSHA external occupational exposure limit of 0.2 mg/m3.
Transparency document
Acknowledgements
The authors extend their sincere appreciation to the International Isocyanate Institute, The Dow Chemical Company and the National Institute for Occupational Safety and Health for financial support of this work. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
Footnotes
Available online 4 October 2014
References
- 1.Allport D.C., Gilbert D.S., Outterside S.M., editors. MDI & TDI Safety, Health and the Environment. John Wiley & Sons; West Sussex: 2003. [Google Scholar]
- 2.Gledhill A., Wake A., Hext P., Leibold E., Shiotsuka R. Absorption, distribution, metabolism and excretion of an inhalation dose of [14C] 4,4′-methylenediphenyl diisocyanate in the male rat. Xenobiotica. 2005;35(3):273–292. doi: 10.1080/00498250500057591. [DOI] [PubMed] [Google Scholar]
- 3.Baur X. Isocyanates: occupational exposures and disorders. Pneumologie (Stuttgart, Germany) 2003;57(9):526–531. doi: 10.1055/s-2003-42221. [DOI] [PubMed] [Google Scholar]
- 4.Cocker J., Jones K., Morton J., Mason H.J. Biomonitoring at the UK Health and Safety Laboratory. Int. J. Hyg. Environ. Health. 2007;210(3–4):383–386. doi: 10.1016/j.ijheh.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Dalene M., Skarping G., Lind P. Workers exposed to thermal degradation products of TDI- and MDI-based polyurethane: biomonitoring of 2,4-TDA, 2,6-TDA, and 4,4′-MDA in hydrolyzed urine and plasma. Am. Ind. Hyg. Assoc. J. 1997;58(8):587–591. doi: 10.1080/15428119791012522. [DOI] [PubMed] [Google Scholar]
- 6.Pauluhn J. Critical analysis of biomonitoring endpoints for measuring exposure to polymeric diphenyl-methane-4,4′-diisocyanate (MDI) in rats: a comparison of markers of exposure and markers of effect. Arch. Toxicol. 2002;76(1):13–22. doi: 10.1007/s00204-001-0301-y. [DOI] [PubMed] [Google Scholar]
- 7.Pauluhn J., Lewalter J. Analysis of markers of exposure to polymeric methylene-diphenyl diisocyanate (pMDI) in rats: a comparison of dermal and inhalation routes of exposure. Exp. Toxicol. Pathol. 2002;54(2):135–146. doi: 10.1078/0940-2993-00242. [DOI] [PubMed] [Google Scholar]
- 8.Jin R., Day B.W., Karol M.H. Toluene diisocyanate protein adducts in the bronchoalveolar lavage of guinea pigs exposed to vapors of the chemical. Chem. Res. Toxicol. 1993;6(6):906–912. doi: 10.1021/tx00036a023. [DOI] [PubMed] [Google Scholar]
- 9.Schutze D., Sepai O., Lewalter J., Miksche L., Henschler D., Sabbioni G. Biomonitoring of workers exposed to 4,4′-methylenedianiline or 4,4′-methylenediphenyl diisocyanate. Carcinogenesis. 1995;16(3):573–582. doi: 10.1093/carcin/16.3.573. [DOI] [PubMed] [Google Scholar]
- 10.Sriramachari S., Chandra H. The lessons of Bhopal [toxic] MIC gas disaster scope for expanding global biomonitoring and environmental specimen banking. Chemosphere. 1997;34(9–10):2237–2250. doi: 10.1016/s0045-6535(97)00081-7. [DOI] [PubMed] [Google Scholar]
- 11.Sabbioni G., Hartley R., Henschler D., Hollrigi-Rosta A., Koeber R., Schneider S. Isocyanate-specific hemoglobin adduct in rats exposed to 4,4′-methylenediphenyl diisocyanate. Chem. Res. Toxicol. 2000;13:82–89. doi: 10.1021/tx990096e. [DOI] [PubMed] [Google Scholar]
- 12.Wisnewski A.V., Hettick J.M., Siegel P.D. Toluene diisocyanate reactivity with glutathione across a vapor/liquid interface and subsequent transcarbamoylation of human albumin. Chem. Res. Toxicol. 2011;24(10):1686–1693. doi: 10.1021/tx2002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabbioni G., Lamb J.H., Farmer P.B., Sepai O. Reactions of 4-methylphenyl isocyanate with amino acids. Biomarkers. 1997;2:223–232. doi: 10.1080/135475097231599. [DOI] [PubMed] [Google Scholar]
- 14.Sabbioni G., Hartley R., Henschler D., Hollrigl-Rosta A., Koeber R., Schneider S. Isocyanate-specific hemoglobin adduct in rats exposed to 4,4′-methylenediphenyl diisocyanate. Chem. Res. Toxicol. 2000;13(2):82–89. doi: 10.1021/tx990096e. [DOI] [PubMed] [Google Scholar]
- 15.Gries W., Leng L. Analytical determination of specific 4,4′-methylene diphenyl diisocyanate hemoglobin adducts in human blood. Anal. Bioanal. Chem. 2013;405(23):7205–7213. doi: 10.1007/s00216-013-7171-z. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A., Nagaraju D., Sabbioni G. New isocyanate-specific albumin adducts of 4,4′-methylenediphenyl diisocyanate (MDI) in rats. Chem. Res. Toxicol. 2009;22(12):1975–1983. doi: 10.1021/tx900270z. [DOI] [PubMed] [Google Scholar]
- 17.Sabbioni G., Dongari N., Kumar A. Determination of a new biomarker in subjects exposed to 4,4′-methylenediphenyl diisocyanate. Biomarkers. 2010;15(6):508–515. doi: 10.3109/1354750X.2010.490880. [DOI] [PubMed] [Google Scholar]
- 18.Sabbioni G., Hartley R., Schneider S. Synthesis of adducts with amino acids as potential dosimeters for the biomonitoring of humans exposed to toluene diisocyanate. Chem. Res. Toxicol. 2001;14:1573–1583. doi: 10.1021/tx010053+. [DOI] [PubMed] [Google Scholar]
- 19.Wisnewski A.V., Liu J., Redlich C.A. Antigenic changes in human albumin caused by reactivity with the occupational allergen diphenylmethane diisocyanate. Anal. Biochem. 2010;400:251–258. doi: 10.1016/j.ab.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemons A.R., Siegel P.D., Mhike M., Hettick J., Bledsoe T.A., Nayak A.P., et al. A murine monoclonal antibody with broad specificity for occupationally relevant diisocyanates. J. Occup. Environ. Hyg. 2014;11(2) doi: 10.1080/15459624.2013.843783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemons A.R., Bledsoe T.A., Siegel P.D., Beezhold D.H., Green B.J. Development of sandwich ELISAs for the detection of aromatic diisocyanate adducts. J. Immunol. Methods. 2013;397(1–2):66–70. doi: 10.1016/j.jim.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemons A.R., Siegel P.D., Mhike M., Hettick J.M., Bledsoe T.A., Nayak A.P., et al. A murine monoclonal antibody with broad specificity for occupationally relevant diisocyanates. J. Occup. Environ. Hyg. 2014;11(2):101–110. doi: 10.1080/15459624.2013.843783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannesson G., Sennbro C.J., Willix P., Lindh C.H., Jonsson B.A. Identification and characterisation of adducts between serum albumin and 4,4′-methylenediphenyl diisocyanate (MDI) in human plasma. Arch. Toxicol. 2004;78(7):378–383. doi: 10.1007/s00204-004-0555-2. [DOI] [PubMed] [Google Scholar]
- 24.Wisnewski A.V., Liu J., Redlich C.A. Connecting glutathione with immune responses to occupational methylene diphenyl diisocyanate exposure. Chem. Biol. Interact. 2013;205(1):38–45. doi: 10.1016/j.cbi.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F., Bartels M.J., Brodeur J.C., Woodburn K.B. Quantitative measurement of fathead minnow vitellogenin by liquid chromatography combined with tandem mass spectrometry using a signature peptide of vitellogenin. Environ. Toxicol. Chem. SETAC. 2004;23(6):1408–1415. doi: 10.1897/03-425. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F., Bartels M.J., Pottenger L.H., Schisler M.R., Grundy J.J., Gollapudi B.B. Quantitation of methylated hemoglobin adducts in a signature peptide from rat blood by liquid chromatography/negative electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2008;22(10):1455–1460. doi: 10.1002/rcm.3530. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F., Bartels M.J., Rick D.L., Krieger S., Hotchkiss J., Esenbrandt D. Quantitation of protein adducts of sulfuryl fluoride (vikane) with albumin protein in the male F344 rat. 46th SOT Annual Meeting & ToxEPO; Charlotte, North Carolina, USA, March; 2007. [Google Scholar]
- 28.Zhang F., Bartels M.J., Stott W.T. Quantitation of human glutathione S-transferases in complex matrices by liquid chromatography/tandem mass spectrometry with signature peptides. Rapid Commun. Mass Spectrom. 2004;18(4):491–498. doi: 10.1002/rcm.1364. [DOI] [PubMed] [Google Scholar]
- 29.Meloun B., Moravek L., Kostka V. Complete amino acid sequence of human serum albumin. FEBS Lett. 1975;58:134–137. doi: 10.1016/0014-5793(75)80242-0. [DOI] [PubMed] [Google Scholar]
- 30.Lawn R.M., Adelman J., Bock S.C., Franke A.E., Houck C.M., Najarian R.C., et al. The sequence of human serum albumin cDNA and its expression in E. coli. Nucleic Acids Res. 1981;9:6103–6114. doi: 10.1093/nar/9.22.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dugaiczyk A., Law S.W., Dennison O.E. Nucleotide sequence and the encoded amino acids of human serum albumin mRNA. Proc. Natl. Acad. Sci. U. S. A. 1982;79:71–75. doi: 10.1073/pnas.79.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappin D.J.C., Rahman D., Hansen H.F., Bartlet-Jones M., Jeffery W., Bleasby A.J. Chemistry, mass spectrometry and peptide-mass databases: evolution of methods for the rapid identification and mapping of cellular proteins. Mass Spectrom. Biol. Sci. 1996:135–160. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MDI exposed worker demographics and classification criteria positive for occupational asthma.
MDI exposed worker demographics and classification criteria negative for asthma (non-asthma of any kind).
MDI exposed workers demographics and classification criteria negative for occupational asthma.
Non exposed worker control demographics and classification criteria.