Abstract
Prions derived from the prion protein (PrP) were first characterized as infectious agents that transmit pathology between individuals. However, the majority of cases of neurodegeneration caused by PrP prions occur sporadically. Proteins that self-assemble as cross-beta sheet amyloids are a defining pathological feature of infectious prion disorders and all major age-associated neurodegenerative diseases. In fact, multiple non-infectious proteins exhibit properties of template-driven self-assembly that are strikingly similar to PrP. Evidence suggests that like PrP, many proteins form aggregates that propagate between cells and convert cognate monomer into ordered assemblies. We now recognize that numerous proteins assemble into macromolecular complexes as part of normal physiology, some of which are self-amplifying. This review highlights similarities among infectious and non-infectious neurodegenerative diseases associated with prions, emphasizing the normal and pathogenic roles of higher-order protein assemblies. We propose that studies of the structural and cellular biology of pathological vs. physiological aggregates will be mutually informative.
I. Introduction
Adult-onset neurodegenerative diseases feature the accumulation of aggregated proteins in a variety of forms: neurofibrillary tangles, extracellular plaques, Lewy bodies, neuronal intranuclear inclusions, Hirano bodies, etc. With some rare exceptions (e.g. polyglutamine diseases like Huntington’s), such disorders are sporadic, arising from the aggregation of wild-type protein, or genetic, caused by dominantly inherited mutations. In genetic cases, causal mutations arise in the very same proteins that accumulate in aggregates, or are in proteins directly linked to them (e.g. presenilins in Alzheimer’s disease (AD)). Transmissible spongiform encephalopathies (TSEs), which are neurodegenerative diseases that can be passed between individuals, also feature the accumulation of protein amyloids. TSEs have held the attention of the scientific community for decades and continue to teach us about the pathogenesis of more common disorders. Carleton Gajdusek initially attributed the cause of Kuru, a TSE transmitted by ritual cannibalism among the Fore people in the New Guinea highlands, to a novel type of “slow virus” (Gajdusek et al., 1966). Stanley Prusiner ultimately determined that an infectious protein assembly causes this disease and other TSEs (Prusiner, 1998). He coined the word “prion” to describe this novel “proteinaceous infectious particle” (Prusiner, 1982) and termed the normal cellular form the “prion protein” (PrP) (Bolton et al., 1982). PrP has distinct native (cellular) and pathological (prion) structures (Pan et al., 1993). In their infectious conformation, PrP prions form self-amplifying beta-sheet rich amyloids that “seed” additional monomer to replicate. The idea of a life form that lacks nucleic acid and yet can somehow trigger predictable pathology via infection remains revolutionary. Nonetheless, while TSEs caused by PrP prions occur with an incidence of approximately 1 per 106 annually, virtually none of these cases is due to known infection (Wadsworth et al., 2003). Consequently, despite very rare cases of human-human transmission (mostly in the remote past, due to contaminated tissue grafts or surgical instruments, pituitary growth hormone extracts, tainted meat products, or cannibalism), human prion disease today is not typically an infectious disorder, and thus more closely resembles other common neurodegenerative diseases.
In recent years, beginning with studies of amyloid beta (Meyer-Luehmann et al., 2006; Petkova et al., 2005), α-synuclein (Desplats et al., 2009; Kordower et al., 2008; Li et al., 2008; Luk et al., 2009), and tau (Clavaguera et al., 2009; Frost et al., 2009a; 2009b), it has become clear that many amyloid-forming proteins exhibit essential properties of prions. In diverse inoculation paradigms, fibrillar assemblies induce unique pathologies depending on the biochemical characteristics of the initiating seed (Clavaguera et al., 2013; Heilbronner et al., 2013; Meyer-Luehmann et al., 2006; Sanders et al., 2014). Tau in particular clearly transmits unique amyloid structures that produce predictable pathology, whether in transmission from mother to daughter cells, or from one animal to another via serial inoculation (Sanders et al., 2014). Thus, in almost every respect these pathogenic proteins have properties that replicate what is known to be true for PrP prions, except for inter-organism transmission of pathology, i.e. “infection.” While some maintain that infection is the sine qua non for a true prion (Aguzzi and Rajendran, 2009; Brettschneider et al., 2015), we and others suggest that this property is not essential (Sanders et al., 2014; Walker and Jucker, 2015; Watts et al., 2013). As we consider myriad prion phenomena associated with mammalian proteins, we propose that the term “prion” and the biology of prions be considered beyond the fascinating and terrifying idea of the PrP prion as an infectious agent. Instead we define a prion as a protein assembly that communicates information stably via template-based amplification of a specific conformation.
The focus on prions as infectious particles has obscured a role for protein aggregates generally, and protein amyloids specifically, as stable carriers of information. Indeed, the ability of prions to faithfully transmit pathological information, producing distinct clinical and neuropathological phenotypes (an effect now linked to conformationally distinct “strains”) initially was used to argue that they must be viruses. Viruses encode within a genome the keys to their own pathological presentation: replication, host response, and cell-cell transmission. Prions similarly encipher these responses entirely within the structure of a simple protein assembly (Collinge and Clarke, 2007; Toyama and Weissman, 2011). In fact, longstanding studies of yeast (Alberti et al., 2009; Halfmann et al., 2012; True and Lindquist, 2000; True et al., 2004; Wickner, 1994), and more recent investigations in mammalian systems (Cai and Chen, 2014; Wu, 2013), suggest that self-replicating protein assemblies regulate diverse physiological processes that require catastrophic “all-or-none” responses. However, whether or not many of these proteins meet exactly the same criteria of a prion as defined above requires further investigation.
Fundamental experiments in cultured cells and animal models are consistent with trans-cellular propagation of protein aggregation as a mechanism for how self-amplification of amyloids, coupled with neural circuitry, could produce different neurodegenerative syndromes. This model predicts initial aggregation in a single cell or group of cells, but the precise events that nucleate an aggregate are unknown. Potential triggers could include spontaneous aggregation as a consequence of misfolding (Ross and Poirier, 2004), environmental exposure to toxins (Dauer and Przedborski, 2003), trauma (McKee et al., 2014), post-translational modifications (Alonso et al., 2008; Beyer and Ariza, 2013), liquid-to-solid phase transitions (Lin et al., 2015; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015; Zhang et al., 2015), or genetic mosaicism in post-mitotic neurons (e.g. copy number variation that increases steady-state levels past a critical threshold, or point mutations that predispose proteins to adopt a prion form) (Bushman et al., 2015; Frigerio et al., 2015). Presumably, a prion seed must encounter another appropriately presented cognate monomer to amplify its structure. This has generally been assumed to proceed by mass action, as it occurs in vitro, i.e. by a collision between an aggregate nucleus and monomer. This seems unlikely to occur efficiently within a cell, however, where most potential interactions will be with heterologous proteins.
In this light, mammalian cells have an amazing ability to faithfully reproduce and propagate pathological PrP prion structures from a proteopathic seed (Legname et al., 2006). This replication process requires considerable effort by investigators to accomplish efficiently in vitro (Deleault et al., 2007; Saborio et al., 2001), which suggests the existence of a cellular prion replication machinery. While seemingly absurd (after all, why would a healthy neuron participate in its own destruction?), this is in fact in keeping with most human diseases, which almost always result from perturbation of an underlying physiological process, e.g. dysregulated cell growth in cancer or glucose metabolism in diabetes. Cells undoubtedly have evolved mechanisms to degrade amyloid structures. For example, in inducible mouse models, pre-existing amyloids will disappear once further protein production is stopped (Mallucci et al., 2007; Polydoro et al., 2013; Walker et al., 2015; Yamamoto et al., 2000). Finally, following amplification the seed must escape to enter a new cell and propagate an aggregated state throughout the nervous system using physiologic cell uptake mechanisms (Holmes and Diamond, 2012).
This review will highlight our current knowledge about intracellular pathogenic amyloids such as tau and α-synuclein across multiple aspects of prion biology. We will also emphasize the importance of functional amyloids and other higher-order non-amyloid assemblies that are increasingly recognized to play a role in eukaryotic signaling (Cai and Chen, 2014; Wu, 2013), and which could conceivably carry and transmit information via conformationally distinct protein templates.
II. Cellular Mechanisms of Prion Propagation
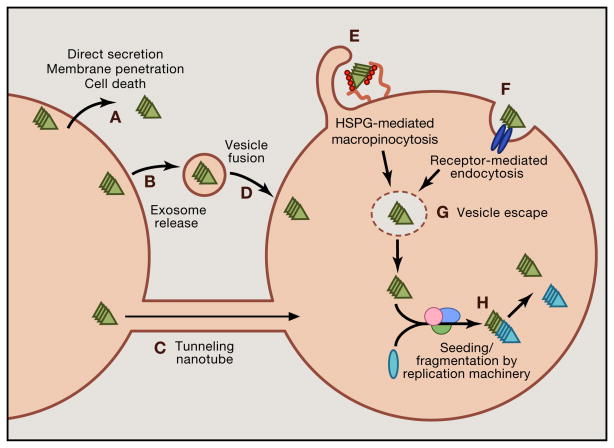
Trans-cellular propagation of prions requires specific events (Figure 1): escape from a neuron; adherence to the plasma membrane of a second cell; endocytosis; conversion of endogenous monomer to a pathogenic conformation; and amplification. Prions presumably escape a cell either by membrane breakdown or via active release into the extracellular space. Recent work has defined how aggregate uptake occurs, while mechanisms involved in intracellular seeding and aggregate release are still unclear (Holmes and Diamond, 2012). Early studies described “bulk endocytosis,” or more specifically, macropinocytosis, as the major mode of internalization for tau, α-synuclein and SOD1 prions (Frost et al., 2009a; Holmes et al., 2013a; Münch et al., 2011; Wu et al., 2013). Macropinocytosis, which accommodates even large protein aggregates, is a form of fluid-phase uptake that produces vesicles ranging from 0.5 – 10 μm in diameter (Holmes et al., 2013a; Mercer and Helenius, 2009). Macropinocytosis of extracellular aggregates requires heparan sulfate proteoglycans (HSPGs), which bind prions prior to their uptake (Holmes et al., 2013a). HSPGs are transmembane or glycolipid-anchored core proteins decorated with glycosaminoglycan (GAG) polysaccharides. Defined patterns of GAG sulfation specify interactions with extracellular ligands (Xu and Esko, 2014). Tau and α-synuclein prions both require HSPGs for binding, internalization, seeding, and trans-cellular propagation in cultured cell lines and primary neurons (Holmes et al., 2013a). HSPG inhibition also prevents neuronal entry of tau prions in vivo (Holmes et al., 2013a). Of note, PrP prions require HSPGs for productive infection of cultured cells, and Aβ and transthyretin monomer are also internalized via this pathway (Horonchik et al., 2005; Kanekiyo et al., 2011; Sousa and Saraiva, 2001). HSPGs could thus mediate aggregate propagation in multiple neurodegenerative diseases. The chemical composition of HSPGs may confer specificity for cellular uptake of different prions, but this has not been tested conclusively. Further, evidence suggests that tunneling nanotubes, filopodia-like extensions that connect the cytoplasm of neighboring cells, may play a role in the spread of PrP and other prions (Costanzo et al., 2013; Gousset et al., 2009), although additional work is required to confirm the importance of this pathway in vivo.
Figure 1. Steps in trans-cellular propagation.
Trans-cellular propagation of prions is likely to involve escape from a first-order cell, binding to a second-order cell, uptake into a second-order cell, seeding of native monomer, and fragmentation and amplification of the seeded aggregates. (A) Escape of prions from a first-order cell could occur by direct release into the extracellular space. This may be driven by unconventional secretion, cell death, or membrane penetration. (B) Alternatively, prions could escape in exosomes, or (C) could directly move to neighboring cells via tunneling nanotubes. (D) In the exosomal pathway, cell entry would presumably occur via vesicle fusion. (E) More likely, prions bind to heparan sulfate proteoglycans (HSPGs) to trigger macropinocytosis. (F) It is theoretically possible that prions gain entry by another form of receptor-mediated endocytosis, although there is not clear evidence for this. (G) Prions escape the lumen of vesicles to encounter cognate monomer. (H) The seed acts as a template for recruitment of monomer to amplify the prion structure. This likely involves a replication machinery that may also be involved in fibril fragmentation to amplify the number of seeds, which then repeat the cycle of propagation to other cells.
After internalization, prions serve as templates to convert monomer into an aggregate (Desplats et al., 2009; Frost et al., 2009a; Luk et al., 2009; Münch et al., 2011; Ren et al., 2009). Recombinant and AD-derived tau aggregates exhibit precise size requirements for cell surface binding, uptake, and seeding of intracellular protein (Mirbaha et al., 2015). Upon size fractionation, all tau assemblies, including monomer and dimer, bind cell surface HSPGs. However, only tau trimers and larger assemblies mediate cellular uptake and seeding (Mirbaha et al., 2015). These findings are consistent with a report that tau trimers are the minimal unit toxic for neuronal cells (Tian et al., 2013), and with molecular modeling studies which hypothesized that tau forms a three-stranded helical fiber based upon trimers (Ruben et al., 1991). PrP trimers also constitute the minimal apparent unit for infectivity (Bellinger-Kawahara et al., 1988; Govaerts et al., 2004; Wille et al., 2009), although PrP monomer in a pathogenic conformation may be toxic in certain conditions (Zhou et al., 2012b). Taken together, it seems likely that trimers are the smallest assemblies that can mediate trans-cellular propagation of the prion state. It is unknown whether smaller assemblies (monomer or dimer) could act as seeds or cause toxicity in certain circumstances. For example, two recent investigations suggested that tau monomer is competent to enter cells and in some cases seed (Falcon et al., 2015; Michel et al., 2014). However these studies relied on less stringent methods for tau monomer purification than those reported elsewhere (Mirbaha et al., 2015), which may have resulted in contamination by tau oligomers.
Prions may also require post-translational processing to become competent seeds. In a cell-based assay to detect seeded aggregation of α-synuclein, fibrils required digestion by cathepsin B to seed productively in cells (Tsujimura et al., 2014). Genetic or pharmacological inhibition of cathepsin B renders α-synuclein aggregates virtually inert, suggesting that α-synuclein fragments could act as templates, instead of full-length protein (Tsujimura et al., 2014). In these studies, α-synuclein prions were delivered directly to the cytosol via transfection reagents, and thus the precise role of cathepsin B remains unknown for physiological uptake and seeding. Multiple studies also support a central role for toxic tau fragments, based on proteolytic processing by cathepsin L, asparagine endopeptidase (AEP), and calpain, as inhibition of these proteases reduces tau phosphorylation, neurofibrillary tangle pathology, and neurodegeneration (Basurto-Islas et al., 2013; Rao et al., 2014; Wang et al., 2009; Zhang et al., 2014). Notably, AEP generates a tau fragment reminiscent of the tau repeat domain (Wischik et al., 1988) that is prone to fibril formation (Zhang et al., 2014).
The precise cellular sub-compartment where amplification of tau and α-synuclein prions occurs is unknown, as amyloid seeds enter a cell within the lumen of the macropinosome, whereas cognate monomer is generally localized in the cytosol. Prions may escape macropinosomes by rupturing the vesicular membrane. For example, internalized SOD1 aggregates are initially found in punctate macropinocytic vesicles (Münch et al., 2011). Within 30 minutes, however, the prions distribute diffusely, and acquire polyubiquitin chains, a post-translational modification that occurs only in the cytosol (Münch et al., 2011). Indeed, other macropinosome substrates such as viruses and the TAT peptide use this mode of entry into the cytosol (Mercer and Helenius, 2009; Wadia et al., 2008). Another study implicates the aggresome, a stress-induced perinuclear inclusion body, as the site of seeded aggregation, since tau aggregates induced by exogenous seeds co-localize with the aggresome markers γ-tubulin and vimentin (Santa-Maria et al., 2012). The alternate hypothesis, however, that seeded aggregation occurs within the lumen of a vesicle, has not been ruled out. Both tau and α-synuclein monomer can enter the endosomal-lysosomal system from the cytosol via chaperone-mediated autophagy (Cuervo and Wong, 2014). This model has been supported by microscopy studies of α-synuclein (Tsujimura et al., 2014). Meanwhile, some studies suggest PrP prions convert monomer into their pathogenic form within mutivesicular bodies (Marijanovic et al., 2009; Yim et al., 2015). However, simple co-localization studies may not sufficiently define where induced aggregation occurs, since coincidence may just represent sequestration of exogenous and endogenous aggregates to a common site of degradation. Subcellular resolution of initial intracellular seeding will require more quantitative and spatially precise methods.
Prions grow by nucleation of monomer at the ends of amyloid fibril templates (Collins et al., 2004). At a simplistic level, an amyloid fiber can be conceived as a seed with two ends for growth, and further fragmentation with produce more free ends that greatly amplify the rate at which free monomer converts to the prion state. Moreover, it is possible that smaller seeds could more readily spread the prion state between cells, although this remains to be tested. In budding yeast, the Hsp104 chaperone works as a two-tiered hexameric macromolecular complex that, along with additional chaperones, fragments amyloids into additional seeds in an ATP-dependent manner (Chernoff et al., 1995; Glover and Lindquist, 1998; Lee et al., 2003; Shorter and Lindquist, 2004). Presumably this mechanism has evolved to maintain balance between an aggregated and monomeric state (Shorter and Lindquist, 2004). Until recently, little evidence in metazoans existed for a disaggregase machinery to sever and amplify amyloid fibrils. Pioneering work suggested that a mixture of Hsp70, Hsp40, and Hsp110 may disassemble amorphous aggregates but not amyloids (Nillegoda et al., 2015; Rampelt et al., 2012; Shorter, 2011). A provocative recent paper has further observed that a similar three chaperone system (Hsc70, DNAJC1, and Hsp110) cooperates to rapidly disaggregate α-synuclein fibrils into smaller seeds and monomer when present at the right stoichiometry (Gao et al., 2015). Moreover, another study has demonstrated that the serine protease HTRA1 solubilizes tau amyloids in an ATP-independent manner in vitro (Poepsel et al., 2015). Conceivably, proteases in lysosomes could create additional seeds. Further work will be required to determine if these processes can generate additional seeding-competent prions in living organisms, and whether they are essential for trans-cellular propagation of the prion state.
To move between cells, prions must exit the cell, and multiple studies have documented release of tau and α-synuclein into the extracellular space (Chai et al., 2012; Danzer et al., 2011; de Calignon et al., 2012; Dujardin et al., 2014a; 2014b; Emmanouilidou et al., 2010; Karch et al., 2012; Kim et al., 2013a; 2010; Lee et al., 2011; 2005; 2012; Liu et al., 2012; Mohamed et al., 2014; Plouffe et al., 2012; Pooler et al., 2013; Saman et al., 2012; Simón et al., 2012; Yamada et al., 2011; 2014). Several mechanisms have been suggested for the trans-cellular propagation of prions, including secretion of soluble protein (Chai et al., 2012; Karch et al., 2012; Santa-Maria et al., 2012), exosomes (Fevrier et al., 2004; Saman et al., 2012), ectosomes (Dujardin et al., 2014a), neuronal activity (Pooler et al., 2013; Yamada et al., 2014), and membrane rupture in cell death (Hesse et al., 2001; Palmio et al., 2009). Clearly, further work must determine conditions that favor one release mechanism over another, and which play a role in trans-cellular propagation of aggregates in vivo. Finally, most published studies of extracellular pathogenic proteins have predominantly measured monomer. In the case of tau, monomer is unlikely to propagate pathology since it is not spontaneously internalized (Mirbaha et al., 2015), and aggregates are responsible for transmission of pathology from cell-to-cell and region-to-region in the brain (Holmes and Diamond, 2012). Direct sampling of the ISF in vivo (Yamada et al., 2011; 2014) to assess its seeding potential with sensitive aggregation biosensors (Holmes et al., 2014) might help resolve this issue. Ultimately multiple factors—the composition of cognate monomer, neural connections, extracellular matrix, glymphatics, and the degradation machinery of the cell—will influence the propensity for aggregate propagation. Despite so many variables, the prion hypothesis provides a model against which to test each of these potential mechanisms.
III. Network involvement in trans-synaptic spread of pathology
Studies of PrP prions first suggested a role for neural networks in pathogenesis, based on spread of pathology through the optic tract following intraocular inoculation (Fraser and Dickinson, 1985; Scott et al., 1992). This anticipated a later observation that neurofibrillary tau pathology in AD progresses along brain networks and correlates with the level of cognitive impairment in patients (Braak and Braak, 1991; 1995). Early in AD, tau aggregate deposition occurs in the transentorhinal region, then within the hippocampus, and finally in more distant regions of the neocortex (Braak et al., 2006). α-synuclein pathology also tracks along brain networks, beginning in the dorsal motor nuclei of the vagus and glossopharyngeal nerves, before involving the substantia nigra and finally neocortical areas (Braak et al., 2003). These cross-sectional pathological studies have highlighted the relationship of neuronal networks to pathogenesis, and anticipated the prion hypothesis for non-infectious neurodegenerative diseases. More recent studies have found that other tauopathies, synucleinopathies, and potentially amyotrophic lateral sclerosis-frontotemporal dementia (ALS-FTD) spectrum diseases may follow a similar hierarchical accumulation of misfolded proteins (Arnold et al., 2013; Brettschneider et al., 2013; Polymenidou and Cleveland, 2011; Ravits et al., 2007a; 2007b). These findings support the idea that toxic agents, e.g.prions, travel via neural networks to induce pathology.
Anatomical proximity of regions near the initial site of pathology may predict risk of involvement (Braak et al., 2006). However, more recent studies based on mapping patterns of degeneration to connectivity networks indicate relative functional and anatomical network connection strength, rather than geographical proximity per se, best predicts regions that are most vulnerable (Raj et al., 2012; Zhou et al., 2012a). Thus, the original site of pathology may determine the network involved and consequently the clinical presentation. As for PrP prion-containing homogenates (Fraser and Dickinson, 1985), inoculation of recombinant, cell, or patient-derived tau fibrils induces endogenous tau pathology in transgenic mice, and this pathology appears to spread through brain networks (Ahmed et al., 2014; Clavaguera et al., 2009; 2013; Iba et al., 2013; 2015; Peeraer et al., 2015; Sanders et al., 2014; Stancu et al., 2015). Injection of recombinant tau fibrils into the hippocampus has produced entorhinal cortex pathology, whereas injection into striatum and cortex caused pathology in the substantia nigra and thalamus (Iba et al., 2013). Injection of aggregated α-synuclein into the brain of wild-type or transgenic mice that express forms of human α-synuclein has produced similar results (Luk et al., 2012a; 2012b; Masuda-Suzukake et al., 2014; 2013; Recasens et al., 2014; Tran et al., 2014; Watts et al., 2013; Woerman et al., 2015). Inoculation studies into mice thus consistently demonstrate progressive accumulation of pathology along known anatomical networks.
These experiments support the prion model, but rely on introduction of exogenous protein aggregates to initiate pathology. Consequently, apparent trans-neural propagation may actually represent the traffic of an inoculum along axons to distant regions, flow along axon tracts, or local uptake by synaptic terminals that belong to distant neuronal cell bodies. Indeed, after just a few hours, α-synuclein oligomers injected into the olfactory bulb spread to distantly connected regions, including the amygdala and striatum (Rey et al., 2013). By contrast, recombinant tau fibrils injected into the hippocampus and cortex of tau transgenic mice reportedly remain at the injection site (Peeraer et al., 2015). Time, detection technique, and aggregate heterogeneity may influence the spread observed in these models. Since true trans-neuronal propagation in vivo itself remains conjecture, such models require rigorous validation, preferably by observation that endogenous prions spontaneously propagate aggregation through a network.
Others have attempted to model the trans-cellular propagation of tau and α-synuclein prions by overexpression of the proteins in specific regions of the nervous system. A substantial body of work suggests that these proteins can indeed move between cells in vivo. However, the role of aggregation (i.e. adoption of the prion state) in this phenomenon is unclear in many cases. Studies of tau protein began in lamprey models, in which plasmid-based expression was used to observe its movement across synapses (Kim et al., 2010). Other groups used the neuropsin promoter to drive tet-regulated mutant human tau primarily in the entorhinal cortex (de Calignon et al., 2012; Liu et al., 2012). These studies documented hyper-phosphorylated tau in regions such as CA1 and the dentate gyrus, and support a model of trans-synaptic propagation from the entorhinal cortex to the hippocampus by perforant pathway connections (de Calignon et al., 2012; Liu et al., 2012). However, while in situ analysis suggested this mouse model only expresses the transgene in the entorhinal cortex (de Calignon et al., 2012), analysis by qPCR of laser-captured dentate gyrus granule cells detected human tau mRNA (Liu et al., 2012). Further, recent work based on tet-induced LacZ expression now indicates that over time the neuropsin promoter activates gene expression fairly widely, including the hippocampus (Yetman et al., 2015). This casts some doubt on the use of this model to definitively test for trans-synaptic propagation. Nonetheless, crosses using tet-regulated GFP did not show the same spatial and temporal progression as was observed for tau (Polydoro et al., 2013), suggesting that protein movement itself, and not promoter activity, is the primary cause of the observations.
Viral inoculation models also support the idea that tau and α-synuclein traffic across synapses. Wild-type tau expressed in rat CA1 hippocampal region via lentivirus spread to distant sites after 8 months, while GFP expression remained restricted, with none in distant cell bodies (Dujardin et al., 2014b). Furthermore, human tau RNA was only detectable around the injection site and not at more distant regions that exhibited human tau protein accumulation, further supporting the hypothesis that tau protein can move between neurons (Dujardin et al., 2014b). Expression of α-synuclein in the rat vagus nerve via adeno-associated virus caused progressive α-synuclein pathology that moved rostrally to connected regions in the pons, midbrain and forebrain, whereas a control (GFP expression) remained restricted to the medulla (Ulusoy et al., 2013). These virus-based approaches may represent versatile tools for non-transgenic animal models of trans-cellular propagation, especially given detailed axonal projection maps developed for the mouse brain (Oh et al., 2014). However, further work will be required to determine which of these diverse models accurately represent the trans-cellular propagation of fibrillar prion assemblies.
Human studies using resting state functional connectivity MRI (fcMRI) also support a network-based propagation mechanism. fcMRI measures correlations in the blood-oxygen level dependent (BOLD) signal across brain regions. Synchronous BOLD activity often represents known anatomical networks or areas that commonly co-activate during a task (Greicius et al., 2009; Seeley et al., 2007; Vincent et al., 2007). Progressive atrophy observed in different tauopathies has been mapped onto known functional connectivity networks as defined in healthy controls (Seeley et al., 2009). This correlation of atrophy patterns with preexisting neural networks supports the model of trans-synaptic propagation. Further studies have found that connectivity to disease “epicenters” (i.e. sites with the most severe atrophy), predicted patterns of progressive atrophy based on the strongest functional path. This had the highest effect on variance, even controlling for absolute distances brain anatomy (Zhou et al., 2012a). Thus, the stronger the network connection between brain regions, the tighter the correlation with pathology. In related work, a network diffusion-based model was used to test whether particle movement could underlie the atrophy patterns in AD and behavioral variant frontotemporal dementia (bvFTD). Whole-brain tractography of diffusion MRI scans from healthy patients was used to model along neural connections. This closely matched the atrophy patterns that arise in AD and bvFTD (Raj et al., 2012), consistent with the idea that a toxic factor (e.g. a prion) diffuses along fibers and induces neuropathology within functional networks.
IV. Strains: amyloid structures that define pathology
The observation of disease “strains” defined by different incubation times, patterns of neuropathology, and behavior phenotypes, was one of the most perplexing aspects of TSEs, and hindered acceptance of the “protein-only” prion hypothesis. In experimental systems of serial inoculation, strain-specific phenotypes passage through multiple generations of mice, even over decades (Bruce, 1993; Collinge and Clarke, 2007; Dickinson and Meikle, 1969; Fraser and Dickinson, 1973). One explanation for this observation had been that different PrP prion agents contained their own self-replicating genomes and were merely acting as very unconventional viruses. Extensive searching, however, failed to find a nucleic acid that governed strain phenomena (Safar et al., 2005). Two pioneering studies instead suggested that strain variation derives from PrP amyloid conformers with distinct protease-resistant core regions (Bessen and Marsh, 1994; Telling et al., 1996). More extensive characterization of a panel of hamster PrP prion strains (Safar et al., 1998) solidified this hypothesis, and different PrP strains probably cause distinct human prion diseases (Collinge et al., 1996).
The precise mechanisms by which distinct amyloid structures produce different diseases, however, are unclear. The study of yeast prions has provided important clues to the relationship between amyloid structure and biological phenotype. Yeast prions switch between monomeric and aggregated forms, in which the latter is inherited by daughter cells as a stable epigenetic phenotype (Derkatch et al., 1996; Patino et al., 1996; Paushkin et al., 1996; Wickner, 1994). Sup35 is particularly well studied: aggregation blocks its activity as a transcription termination factor and is generally detrimental to the organism (McGlinchey et al., 2011), but may be advantageous in certain environmental situations (Halfmann et al., 2012; Holmes et al., 2013b; True and Lindquist, 2000; True et al., 2004). Like PrP, Sup35 prions form strains with distinct strengths, or degrees of aggregation (Tanaka et al., 2004). A strong yeast prion strain is analogous to a mammalian PrP prion strain with a short incubation period (Figure 2A). Two studies determined the physical and structural basis for these phenomena in several Sup35 prion strains. A combination of growth rate and amyloid fibril fragility defined the strength of the strain (Tanaka et al., 2006). Hydrogen-deuterium exchange indicated that strong strains amplify more readily because they contain a smaller amyloid-forming core that fragments more readily to produce additional seeds (Toyama et al., 2007). These studies suggested that biophysical parameters could account for most of the variance in strain strength. Consistent with these studies, the stability of multiple PrP prion strains in denaturant (GdHCl) strongly correlated with incubation time upon inoculation into transgenic PrP mice (Legname et al., 2006), suggesting that more fragile amyloid conformers produced shorter incubation periods (i.e. were stronger strains). While studies of sporadic Creutzfeldt-Jakob disease (sCJD) PrP strains initially seemed to support this idea (Kim et al., 2012), recent analysis of two versions of sCJD has suggested this is not the defining determinant of PrP prion pathogenicity, as more fragile fibrils cause longer disease duration in certain cases (Safar et al., 2015). Furthermore, the degree of interaction with chaperone protein clients, not just conformational stability, also determines the strength of certain Sup35 prion strains (Stein and True, 2014a; 2014b). Taken together, biophysical characteristics of the amyloid alone cannot explain the strength of a prion strain. Further structural examination of diverse prion strains as well as interrogation of their binding partners will help define how unique amyloid conformers produce distinct biological responses and disease phenotypes.
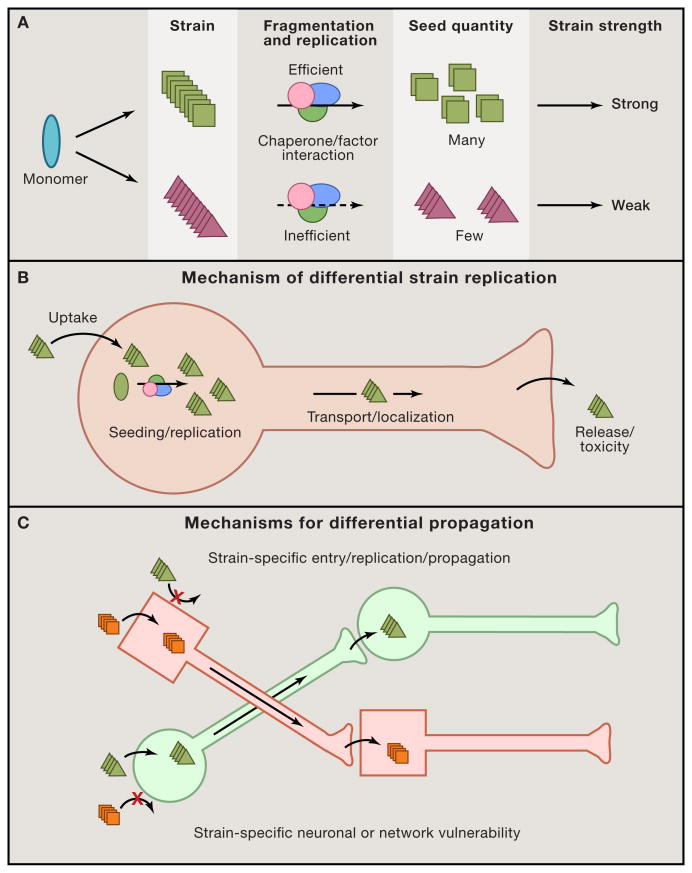
Figure 2. Prion replication mechanisms and network involvement link strains to biological phenotype.
The various steps in replication of a prion provide an explanation for how strains might exhibit different rates of progression, types of neuropathology, and involvement of specific networks. (A) Biophysical and biochemical parameters govern strain replication efficiency. A single protein monomer can adopt multiple potential amyloid configurations, illustrated by squares vs. triangles. Strain-specific fibril stability and the interaction with chaperone/replication machinery may determine the fragmentation rate and production of seeds. This likely determines the rate of spread and disease progression, with strong strains featuring more rapid phenotypes. (B) A strain must propagate pathology by entering a neuron and initiating seeding and subsequent replication of a particular structure. The newly formed aggregates will accumulate in certain regions of the cell, possibly through interaction with specific factors, and may be differentially transported in axons. Finally, strains may have differential rates of release and/or toxicity at the cell membrane/synapse that govern transfer to other cells. Each of these steps may be influenced by strain conformation. (C) Unique strains may target different cell types and neural networks to cause syndromic variation. Aggregate conformers (square vs. triangle) may target one cell type more readily than another. This could be based on parameters discussed above: cell entry, replication, and trans-cellular propagation rates. Distinct neuronal networks might thus be vulnerable to particular strains.
Like prionopathies, common neurodegenerative diseases such as tauopathies and synucleinopathies exhibit great phenotypic diversity (Halliday et al., 2011; Lee et al., 2001). AD, corticobasal degeneration, progressive supranuclear palsy, Pick’s disease, argyrophilic grain disease, among other tauopathies, have distinct clinical and neuropathological presentations (Lee et al., 2001). It is tempting to speculate that distinct tau prion strains could cause these syndromes. Similar to PrP, amyloid beta (Petkova et al., 2005), tau (Frost et al., 2009b), and α-synuclein (Bousset et al., 2013; Guo et al., 2013) form discrete conformers in vitro. Further, amyloid beta extracts from different mouse models induce unique amyloid deposition patterns upon in vivo inoculation, depending on the transgenic animal source of the protein seeds and the transgenic mouse host (Meyer-Luehmann et al., 2006). Likewise, inoculation of susceptible mice with brain extracts from different tauopathies has produced unique patterns of tau deposition, consistent with strains (Boluda et al., 2015; Clavaguera et al., 2013). Inoculation of mice with different preparations of a-synuclein fibrils has also produced variable degrees of pathology (Guo et al., 2013; Peelaerts et al., 2015).
However, to have clinical, pathological, or diagnostic significance, prion strains must faithfully replicate in vivo, producing predictable pathology upon serial inoculation. Multiple PrP strains have now been rigorously characterized, and can be maintained in constant passaging paradigms over decades (Bruce, 1993; Collinge and Clarke, 2007). This high fidelity of information transfer probably accounts for distinct syndromes that progress over years in humans. In human-derived amyloid beta samples, amplification of unique structures from different patients supports the idea that specific conformers predominate and stably propagate in individuals (Lu et al., 2013), although a causal link to distinct syndromes has not been established. Further, brain regions from a single patient contained a similar fibrillar amyloid conformation, consistent with the idea that a single conformation arises in one part of the brain and then spreads throughout the nervous system (Lu et al., 2013). Moreover, passage of amyloid beta in vivo has appeared to involve maintenance of unique structures, based on analysis with conformer-dependent amyloid dyes (Heilbronner et al., 2013).
A more thorough test of this idea would be to create unique amyloid conformers, replicate them in a biological system, and confirm preservation of their structure following passage in vivo. In cell culture, two tau prion strains have been defined based on their unique inclusion morphologies, biochemical characteristics, and ability to corrupt native monomer (Sanders et al., 2014). Distinct tau prion strains passed stably from mother to daughter cells over months of culture and produced unique pathologies in transgenic mice that propagated serially through multiple generations of animals, identical to PrP strains. More importantly, after three generations of passage in vivo, the conformations were unchanged when introduced back into the original reporter cells. These experiments indicated that, like PrP, tau prions have essential strain characteristics, and raised the question of whether human tauopathies might also be associated with distinct strains. Inoculation of brain lysate from five different tauopathy syndromes generated disease-associated tau prion strain patterns in a cell model (Sanders et al., 2014). These observations were consistent with prior studies based on brain homogenate inoculations into mice (Boluda et al., 2015; Clavaguera et al., 2013), and additional work based on biochemical analysis of aggregates from patient brain (Taniguchi-Watanabe et al., 2015). Furthermore, some patients appear to be afflicted by multiple tau prion strains (Sanders et al., 2014), reminiscent of previous findings regarding PrP prion strains isolated from single patients with CJD (Polymenidou et al., 2005; Puoti et al., 1999).
Isolation and characterization of tau strains suggest that tau (and by extension other amyloid proteins) has diverse amyloid structures that propagate with high fidelity. The existence of tau prion strains may explain the myriad tauopathies, but must be tested further with improved diagnostic assays. Finally, mechanisms that target particular prion strains to unique areas of the cell (to create characteristic inclusion morphologies), different cell types, and to discrete neuronal networks will require further elucidation (Figure 2B, 2C).
V. Prion-like assemblies in signaling cascades
Most, if not all, human disease results from normal physiology gone awry, and thus it is intriguing to speculate that formation and self-amplification of specific prion assemblies may encipher useful information. Recent work has suggested that this may be the case for certain signaling mechanisms (Cai and Chen, 2014; Wu, 2013). The catastrophic assembly of monomer into a fibril creates an “all or none” signaling mechanism, a binary switch for diverse innate immunity and cell death pathways (Wu, 2013). Signalosomes consist of higher-order, micron-sized fibrillar protein assemblies that link persistent upstream danger signals with irreversible downstream enzyme-driven cellular responses (Wu, 2013) (Figure 3A). Nucleation-dependent polymerization drives association of adapter proteins and enzymes that would not otherwise form active complexes. The switch to an aggregated state thus triggers activation of cell death pathways and downstream signaling responses (Cai and Chen, 2014). Three examples have especially informed our understanding of functional protein aggregates in signaling: MAVS filaments, which detect viral nucleic acids to drive interferon production and the innate immune response (Cai et al., 2014; Hou et al., 2011); ASC inflammasomes, which direct cytokine production (Cai et al., 2014; Lu et al., 2014); and necrosomes, which mediate programmed non-apoptotic cell death or necroptosis (Li et al., 2012).
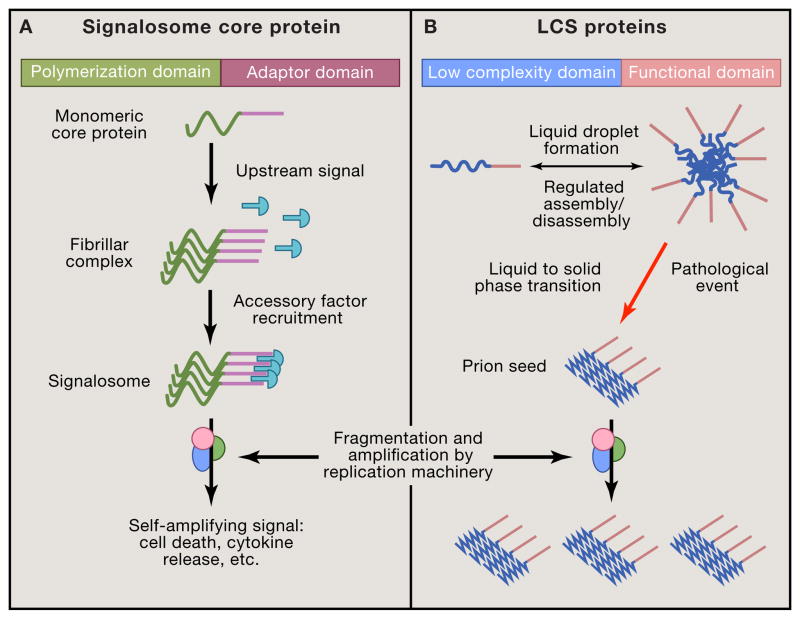
Figure 3. Formation of higher-order assemblies in signaling, RNA metabolism, and disease.
(A) Diverse proteins form micron-sized signalosomes to drive cellular responses. These structurally heterogeneous complexes rapidly assemble via polymerization domains in core proteins, and amplify an upstream signal. They form particles that recruit multiple accessory factors through adaptor domains. The resulting signalosome triggers all-or-none signaling responses such as cell death. The amplification and trans-cellular spread of signals may involve machinery similar to that used by prions. (B) Proteins with low-complexity sequences (LCS) undergo liquid-liquid phase separation to form large, dynamic membraneless organelles as part of RNA metabolism. Cell signals regulate their assembly and disassembly, which is based on self-association of the LCS to create a high concentration of the functional domain. In ALS and potentially other diseases, these liquid complexes may undergo an aberrant transition to a solid (i.e. fibrillar) form that becomes a self-propagating prion. Similar to other prions (e.g. PrP, tau), a cellular amyloid replication machinery may fragment these structures to produce additional seeds that spread between cells to drive progressive pathology.
MAVS exhibits catastrophic self-assembly when activated by the RNA helicase RIG-1, which is itself driven by binding to viral RNA (Hou et al., 2011). RIG-1 activates MAVS on the surface of mitochondria, directing interferon production and an antiviral response (Seth et al., 2005). Viral nucleic acid causes MAVS to switch from an inactive monomer to an active self-amplifying polymer (Hou et al., 2011). However it is unclear that MAVS is a true prion, as it is a three-stranded helical filament, lacks amyloid structure, and doesn’t undergo a major change in secondary structure upon assembly (Xu et al., 2014). Nonetheless, the Q/N rich prion domain of Sup35, which mediates amyloid formation, functionally substitutes for the non-amyloid CARD domain of MAVS, suggesting that formation of a higher-order polymer alone drives a prion-like switch (Cai et al., 2014).
Other innate immunity proteins with death domains form higher-order helical assemblies that behave similarly to prions. Inflammasome sensor proteins with pyrin domains seed the polymerization of the pyrin domain of an adaptor protein, ASC, which drives caspase-1 polymerization, inflammasome activation, and cytokine production (Lu et al., 2014). Like the MAVS CARD domain, the ASC pyrin domain also acts like a prion in yeast (Cai et al., 2014). Remarkably, ASC inflammasomes propagate their signal from cell to cell in vivo, as they are internalized to seed the polymerization of native ASC to communicate an immune response (Baroja-Mazo et al., 2014; Franklin et al., 2014). Thus, proteins in innate immunity may amplify a response within and between cells similar to those in neurodegenerative diseases, although the assemblies that form do not have amyloid structure.
Unlike MAVS and ASC polymers, the core constituents of necrosomes form true amyloids (Li et al., 2012). Upon induction of necroptosis in cells, the RIP1 and RIP3 kinases co-polymerize in thioflavin-positive assemblies that increase their respective enzymatic activities. Mutations in RIP1 and RIP3 that abolish amyloid formation also abrogate necrosome formation and downstream necroptosis (Li et al., 2012). Bioinformatics studies have suggested that RIP homotypic interaction motifs are homologous to polymerization motifs from HET-S (Kajava et al., 2014), a well-characterized fungal prion that mediates heterokaryon incompatibility-induced cell death (Coustou et al., 1997). Thus, these various all-or-none prion-like switches have been conserved from yeast to vertebrates. While it is clear that some assemblies can provide epigenetic inheritance from mother to daughter cells in yeast (Cai et al., 2014), the spectrum of prion-based signaling systems, and the degree to which aggregates in metazoans stably maintain or transfer information requires elucidation.
VI. Liquid-liquid phase separation in membraneless organelles
Like signalosomes, diverse micron-scale ribonucleoprotein (RNP) bodies concentrate biochemical reactions into membraneless, proteinaceous organelles within the cell (Figure 3B). RNP granules include, but are not limited to stress granules, processing bodies, splicing speckles, paraspeckles, and nucleoli (Kedersha et al., 2013; Sleeman and Trinkle-Mulcahy, 2014). These higher-order assemblies of protein and RNA perform specialized functions, but all undergo liquid-liquid phase separation (LLPS) to form condensed, spherical droplets whose assembly and size are controlled by physical constraints such as protein and solute concentrations, heterotypic interactions with other RNA-binding proteins, cell size, and post-translational modifications (Brangwynne, 2013; Hyman et al., 2014). Unlike pathogenic fibrillar inclusions, these droplets are only slightly denser than their milieu, rapidly exchange substrates with surrounding cytoplasm or nucleoplasm, and do not appear to undergo a conformational change to a self-propagating form or prion. Their dynamic properties concentrate reactions into membraneless compartments to spatially regulate nucleic acid biology (Brangwynne, 2013; Kedersha et al., 2013). Absence of a membrane may allow rapid assembly and disassembly depending upon environmental conditions, essential to regulated transfer of information within the cell (Kedersha et al., 2013).
Many of the core protein constituents of RNP granules contain vast, low-complexity sequences in disordered regions linked to modules that bind RNA and DNA (Han et al., 2012; Kato et al., 2012). The disordered regions are prone to self-interaction and drive the assembly of the higher-order, liquid-like complexes (Brangwynne et al., 2009; Elbaum-Garfinkle et al., 2015; Nott et al., 2015). These low complexity sequences are similar to Q/N-rich prion domains in yeast, which also mediate formation of higher-order assemblies (Alberti et al., 2009). Upon self-assembly, likely via tunable electrostatic interactions (Elbaum-Garfinkle et al., 2015; Nott et al., 2015), they form the structural constituents of granules and recruit other proteins with similar domains via heterotypic interactions (Kato et al., 2012; Lin et al., 2015). How proteins with similar domains are targeted to distinct membraneless compartments requires elucidation.
Several papers have demonstrated that higher-order assemblies of a specific RNA-binding protein, CPEB, may govern the maintenance of memory, possibly by converting to a self-propagating prion form that recalls synaptic activation (Fioriti et al., 2015; Majumdar et al., 2012; Si et al., 2010; 2003a; 2003b; Stephan et al., 2015). Aplysia CPEB (Si et al., 2003b) and mammalian CPEB3 (Stephan et al., 2015) have Q-rich low-complexity domains and that act like prions when overexpressed in budding yeast. Moreover, CPEB forms higher-order, SDS-resistant assemblies in Aplysia (Si et al., 2010), Drosophila (Majumdar et al., 2012), and mice (Fioriti et al., 2015) during memory formation. A single point mutation that blocks self-assembly is sufficient to disrupt long-term memory maintenance in Drosophila (Majumdar et al., 2012). Nevertheless, the biophysical characteristics of the native protein assemblies in vivo are unclear. As these are RNA-binding proteins with low-complexity sequences, it is possible that the higher-order structures that they form represent a maturation of phase-separated droplets to self-propagating amyloids. If so, other RNA-binding proteins that undergo LLPS and play a role in synapse maintenance and memory could form similar complexes.
VII. Liquid-to-solid phase transitions in disease pathogenesis
Numerous studies now link RNP granule dysfunction to ALS-FTD spectrum diseases associated with amyloid formation. These may result in part from impairment of information-transfer mechanisms such as mRNA maturation, RNA transport, and auto-inhibition of translation (Ramaswami et al., 2013). Mutations in over a dozen RNA- and DNA-binding proteins cause familial ALS and other less common motor neuron diseases (King et al., 2012; Li et al., 2013). Genetic analyses coupled with bioinformatics approaches to evaluate candidate RNA-binding proteins that share prion-like properties with TDP-43 and FUS in model organisms have identified additional RNA-binding proteins that, when mutated, cause ALS-FTD (Couthouis et al., 2012; 2011; Kim et al., 2013b). In contrast to their normal regulated assembly and disassembly, in pathological conditions these proteins undergo catastrophic aggregation to sequester soluble monomeric protein from the nucleus into large, stable cytoplasmic inclusions. Numerous studies indicate that pathogenic mutations in these proteins enhance their ability to form self-propagating pathogenic amyloids or amyloid-like structures (reviewed by (Li et al., 2013)) and disrupt trafficking and regulation of mRNP granules (Alami et al., 2014; Liu-Yesucevitz et al., 2014).
The precise cellular subcompartments that mediate initial amyloid formation within cells are unknown, and could vary for different proteins. Such compartments would require relatively high local concentration of protein to enable the formation of self-propagating prions. For example, pathogenic mutations that cause ALS-FTD can promote a phase transition from a liquid to a solid form) (Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). High local concentrations of these proteins, such as occur in LLPS, especially when combined with a pathogenic mutation that enhances self-assembly or beta-sheet structure, primes these proteins to undergo a nucleation event. It should be noted, however, that wild-type versions of diverse LCS proteins, not just those associated with disease, will form fibrils that nucleate out of droplets after sufficient time (Lin et al., 2015; Zhang et al., 2015). In fact, at high concentrations and low temperatures, many LCS proteins will form amyloid-like fibrils (Han et al., 2012; Kato et al., 2012; Kwon et al., 2013; 2014). Thus, whether fibrils play a normal physiological role in maintenance of these RNP bodies or are purely pathological is unknown. Regardless, it is likely that fibrils can form within the cell during LLPS, and must be cleared to prevent the formation of larger pathogenic aggregates that disrupt normal function of the associated proteins. Thus, study of proteins that undergo phase transitions will no doubt inform regulation and physiology of normal and abnormal protein assemblies. For example, VCP, which is associated with both ALS and multisystem proteinopathy (Johnson et al., 2010; Watts et al., 2004), is critical to both RNP granule disassembly and pathological aggregate clearance (Buchan et al., 2013; Ju et al., 2009; Meyer and Weihl, 2014). Moreover, RNA helicases such as Ddx6 may be critical to maintaining fluidity of these compartments, preventing the formation of pathological solids (Hubstenberger et al., 2015; 2013).
Together, these studies suggest that many ALS-FTD-associated proteins exist in a dynamic biophysical state, in which they can transition between function (liquid) and dysfunction (solid) due to rapid and physiological self-association and dissociation. More generally, a tendency for proteins to undergo transient self-association could initiate the formation of diverse self-propagating prions (Figure 3B). Further work will be required to determine why certain proteins that undergo LLPS form self-propagating prions that ultimately lead to disease whereas others do not.
VIII. Perspectives and Future Directions
Multiple diseases are now linked to the formation of prions comprised of proteins such as PrP, tau, and α-synuclein. Each of these proteins, and likely many others, can form higher-order assemblies that act as templates within a cell to corrupt native cognate protein, and recruit it to a growing amyloid fibril. According to the model of PrP prion propagation, these assemblies extend protein aggregation pathology throughout the nervous system via pre-existing neural networks. It is unknown whether distinct prion assembly structures, or strains, have a predilection for particular networks, and can thus account for distinct clinical syndromes. However, there is now evidence for a variety of strains within individual tauopathy syndromes, much as PrP prion disorders have been linked to similar constellations of strains. We predict that strains could represent novel ways to transmit information, whereby multiple assemblies of a single protein are used to maintain and communicate distinct (and possibly subtle) signaling outcomes, not just all-or-none responses.
While a biological role for prion strains in normal physiology remains conjecture, self-propagating protein aggregation is now fairly well established in several experimental systems, with or without formation of beta-sheet amyloid structures. Yeast prions clearly represent a well-characterized mode of epigenetic inheritance. In metazoans, the study of prion-like phenomena in signaling is nascent, and has mainly been linked to cell death pathways and regulation of RNA metabolism at synapses in conjunction with learning and memory. Moreover, it is becoming increasingly clear that proteins that undergo liquid-liquid phase separation to form dynamic ribonucleoprotein bodies as part of normal physiology are especially prone to forming fibrillar amyloids. An integrated “unification theory” that describes both amyloid-associated neurodegenerative diseases and the role of amyloids, non-amyloid fibrils, and other higher-order assemblies in normal biology will help guide research. This could bring together fields that span infectious prion diseases, sporadic non-infectious amyloid diseases, and functional higher-order assemblies in mammalian systems.
We anticipate that transient assemblies of proteins facilitate the formation of amyloids due to their high concentration. These might cause progressive disease by feeding into existing machinery that enables inappropriate replication and propagation along brain networks or by mediating local cell-to-cell transfer of pathology in other tissues. The underlying biophysical events are likely to be similar for different amyloid proteins, but with protein-specific players, and distinct cellular subcompartments. We speculate that common mechanisms will regulate the assembly and disassembly of functional and pathological assemblies alike. Thus, an integrated study of protein aggregation has the chance to create both a comprehensive understanding of disease mechanisms and an elucidation of new modes of signal transduction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Cooper J, Murray TK, Garn K, McNaughton E, Clarke H, Parhizkar S, Ward MA, Cavallini A, Jackson S, et al. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: the pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 2014;127:667–683. doi: 10.1007/s00401-014-1254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso ADC, Li B, Grundke-Iqbal I, Iqbal K. Mechanism of tau-induced neurodegeneration in Alzheimer disease and related tauopathies. Curr Alzheimer Res. 2008;5:375–384. doi: 10.2174/156720508785132307. [DOI] [PubMed] [Google Scholar]
- Arnold SE, Toledo JB, Appleby DH, Xie SX, Wang LS, Baek Y, Wolk DA, Lee EB, Miller BL, Lee VMY, et al. Comparative survey of the topographical distribution of signature molecular lesions in major neurodegenerative diseases. J Comp Neurol. 2013;521:4339–4355. doi: 10.1002/cne.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Mazo A, Martín-Sánchez F, Gomez AI, Martínez CM, Amores-Iniesta J, Compan V, Barberà-Cremades M, Yagüe J, Ruiz-Ortiz E, Antón J, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- Basurto-Islas G, Grundke-Iqbal I, Tung YC, Liu F, Iqbal K. Activation of asparaginyl endopeptidase leads to Tau hyperphosphorylation in Alzheimer disease. J Biol Chem. 2013;288:17495–17507. doi: 10.1074/jbc.M112.446070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger-Kawahara CG, Kempner E, Groth D, Gabizon R, Prusiner SB. Scrapie prion liposomes and rods exhibit target sizes of 55,000 Da. Virology. 1988;164:537–541. doi: 10.1016/0042-6822(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer K, Ariza A. α-Synuclein posttranslational modification and alternative splicing as a trigger for neurodegeneration. Mol Neurobiol. 2013;47:509–524. doi: 10.1007/s12035-012-8330-5. [DOI] [PubMed] [Google Scholar]
- Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- Boluda S, Iba M, Zhang B, Raible KM, Lee VMY, Trojanowski JQ. Differential induction and spread of tau pathology in young PS19 tau transgenic mice following intracerebral injections of pathological tau from Alzheimer’s disease or corticobasal degeneration brains. Acta Neuropathol. 2015;129:221–237. doi: 10.1007/s00401-014-1373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset L, Pieri L, Ruiz-Arlandis G, Gath J, Jensen PH, Habenstein B, Madiona K, Olieric V, Böckmann A, Meier BH, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–8. doi: 10.1016/0197-4580(95)00021-6. discussion278–84. [DOI] [PubMed] [Google Scholar]
- Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. 2006 doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rüb U, de Vos RAI, Jansen Steur ENH, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203:875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Lee VMY, Trojanowski JQ. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat Rev Neurosci. 2015;16:109–120. doi: 10.1038/nrn3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Toledo JB, Robinson JL, Irwin DJ, Grossman M, Suh E, Van Deerlin VM, Wood EM, Baek Y, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce ME. Scrapie strain variation and mutation. Br Med Bull. 1993;49:822–838. doi: 10.1093/oxfordjournals.bmb.a072649. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman DM, Kaeser GE, Siddoway B, Westra JW, Rivera RR, Rehen SK, Yung YC, Chun J. Genomic mosaicism with increased amyloid precursor protein (APP) gene copy number in single neurons from sporadic Alzheimer’s disease brains. Elife. 2015;4 doi: 10.7554/eLife.05116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen ZJ. Prion-like polymerization as a signaling mechanism. Trends Immunol. 2014;35:622–630. doi: 10.1016/j.it.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai X, Dage JL, Citron M. Constitutive secretion of tau protein by an unconventional mechanism. Neurobiol Dis. 2012;48:356–366. doi: 10.1016/j.nbd.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci USa. 2013 doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of “new variant” CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Abounit S, Marzo L, Danckaert A, Chamoun Z, Roux P, Zurzolo C. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci. 2013;126:3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USa. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USa. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer KM, Ruf WP, Putcha P, Joyner D, Hashimoto T, Glabe C, Hyman BT, McLean PJ. Heat-shock protein 70 modulates toxic extracellular α-synuclein oligomers and rescues trans-synaptic toxicity. Faseb J. 2011;25:326–336. doi: 10.1096/fj.10-164624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suárez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA, et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci USa. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USa. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AG, Meikle VM. A comparison of some biological characteristics of the mouse-passaged scrapie agents, 22A and ME7. Genet Res. 1969;13:213–225. doi: 10.1017/s0016672300002895. [DOI] [PubMed] [Google Scholar]
- Dujardin S, Bégard S, Caillierez R, Lachaud C, Delattre L, Carrier S, Loyens A, Galas MC, Bousset L, Melki R, et al. Ectosomes: a new mechanism for non-exosomal secretion of tau protein. PLoS ONE. 2014a;9:e100760. doi: 10.1371/journal.pone.0100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin S, Lécolle K, Caillierez R, Bégard S, Zommer N, Lachaud C, Carrier S, Dufour N, Aurégan G, Winderickx J, et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: relevance to sporadic tauopathies. Acta Neuropathol Commun. 2014b;2:14. doi: 10.1186/2051-5960-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CCH, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USa. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30:6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Cavallini A, Angers R, Glover S, Murray TK, Barnham L, Jackson S, O’Neill MJ, Isaacs AM, Hutton ML, et al. Conformation determines the seeding potencies of native and recombinant Tau aggregates. J Biol Chem. 2015;290:1049–1065. doi: 10.1074/jbc.M114.589309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci USa. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, et al. The Persistence of Hippocampal-Based Memory Requires Protein Synthesis Mediated by the Prion-like Protein CPEB3. Neuron. 2015;86:1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, et al. The adaptor ASC has extracellular and “prionoid” activities that propagate inflammation. Nat Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H, Dickinson AG. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- Fraser H, Dickinson AG. Targeting of scrapie lesions and spread of agent via the retino-tectal projection. Brain Res. 1985;346:32–41. doi: 10.1016/0006-8993(85)91091-1. [DOI] [PubMed] [Google Scholar]
- Frigerio CS, Lau P, Troakes C, Deramecourt V, Gele P, Van Loo P, Voet T, De Strooper B. On the identification of low allele frequency mosaic mutations in the brains of Alzheimer disease patients. Alzheimers Dement. 2015 doi: 10.1016/j.jalz.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009a;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Ollesch J, Wille H, Diamond MI. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J Biol Chem. 2009b;284:3546–3551. doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdusek DC, Gibbs CJ, Alpers M. Experimental transmission of a Kuru-like syndrome to chimpanzees. Nature. 1966;209:794–796. doi: 10.1038/209794a0. [DOI] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. Human Hsp70 Disaggregase Reverses Parkinson’s-Linked α-Synuclein Amyloid Fibrils. Mol Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J, et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol. 2009;11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USa. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, et al. Distinct α-Synuclein Strains Differentially Promote Tau Inclusions in Neurons. Cell. 2013;154:103–117. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, Holton JL, Revesz T, Dickson DW. Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 2011;122:187–204. doi: 10.1007/s00401-011-0852-9. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Heilbronner G, Eisele YS, Langer F, Kaeser SA, Novotny R, Nagarathinam A, Aslund A, Hammarström P, Nilsson KPR, Jucker M. Seeded strain-like transmission of β-amyloid morphotypes in APP transgenic mice. EMBO Rep. 2013;14:1017–1022. doi: 10.1038/embor.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse C, Rosengren L, Andreasen N, Davidsson P, Vanderstichele H, Vanmechelen E, Blennow K. Transient increase in total tau but not phospho-tau in human cerebrospinal fluid after acute stroke. Neurosci Lett. 2001;297:187–190. doi: 10.1016/s0304-3940(00)01697-9. [DOI] [PubMed] [Google Scholar]
- Holmes BB, Diamond MI. Cellular mechanisms of protein aggregate propagation. Curr Opin Neurol. 2012;25:721–726. doi: 10.1097/WCO.0b013e32835a3ee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, Devos SL, Kfoury N, Li M, Jacks R, Yanamandra K, Ouidja MO, Brodsky FM, Marasa J, Bagchi DP, et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci USa. 2013a;110:E3138–E3147. doi: 10.1073/pnas.1301440110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, Diamond MI. Proteopathic tau seeding predicts tauopathy in vivo. Proc Natl Acad Sci USa. 2014;111:E4376–E4385. doi: 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013b;153:153–165. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horonchik L, Tzaban S, Ben-Zaken O, Yedidia Y, Rouvinski A, Papy-Garcia D, Barritault D, Vlodavsky I, Taraboulos A. Heparan sulfate is a cellular receptor for purified infectious prions. J Biol Chem. 2005;280:17062–17067. doi: 10.1074/jbc.M500122200. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Cameron C, Noble SL, Keenan S, Evans TC. Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J Cell Biol. 2015;211:703–716. doi: 10.1083/jcb.201504044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Noble SL, Cameron C, Evans TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell. 2013;27:161–173. doi: 10.1016/j.devcel.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Iba M, Guo JL, McBride JD, Zhang B, Trojanowski JQ, Lee VMY. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J Neurosci. 2013;33:1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba M, McBride JD, Guo JL, Zhang B, Trojanowski JQ, Lee VM-Y. Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol. 2015 doi: 10.1007/s00401-015-1458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Klopffleisch K, Chen S, Hofmann K. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep. 2014;4:7436. doi: 10.1038/srep07436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo T, Zhang J, Liu Q, Liu CC, Zhang L, Bu G. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J Neurosci. 2011;31:1644–1651. doi: 10.1523/JNEUROSCI.5491-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Jeng AT, Goate AM. Extracellular Tau levels are influenced by variability in Tau that is associated with tauopathies. J Biol Chem. 2012;287:42751–42762. doi: 10.1074/jbc.M112.380642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Haldiman T, Surewicz K, Cohen Y, Chen W, Blevins J, Sy MS, Cohen M, Kong Q, Telling GC, et al. Small protease sensitive oligomers of PrPSc in distinct human prions determine conversion rate of PrP(C) PLoS Pathog. 2012;8:e1002835. doi: 10.1371/journal.ppat.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Ho DH, Suk JE, You S, Michael S, Kang J, Joong Lee S, Masliah E, Hwang D, Lee HJ, et al. Neuron-released oligomeric α-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat Commun. 2013a;4:1562. doi: 10.1038/ncomms2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013b;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lee S, Jung C, Ahmed A, Lee G, Hall GF. Interneuronal transfer of human tau between Lamprey central neurons in situ. J Alzheimers Dis. 2010;19:647–664. doi: 10.3233/JAD-2010-1273. [DOI] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14:504–506. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Baek SM, Ho DH, Suk JE, Cho ED, Lee SJ. Dopamine promotes formation and secretion of non-fibrillar alpha-synuclein oligomers. Exp Mol Med. 2011;43:216–222. doi: 10.3858/emm.2011.43.4.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Patel S, Lee SJ. Intravesicular localization and exocytosis of alpha-synuclein and its aggregates. J Neurosci. 2005;25:6016–6024. doi: 10.1523/JNEUROSCI.0692-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim W, Li Z, Hall GF. Accumulation of vesicle-associated human tau in distal dendrites drives degeneration and tau secretion in an in situ cellular tauopathy model. Int J Alzheimers Dis. 2012;2012:172837. doi: 10.1155/2012/172837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FTF. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Legname G, Nguyen HOB, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci USa. 2006;103:19105–19110. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Englund E, Holton JL, Soulet D, Hagell P, Lees AJ, Lashley T, Quinn NP, Rehncrona S, Björklund A, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K. Trans-synaptic spread of tau pathology in vivo. PLoS ONE. 2012;7:e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Lin AY, Ebata A, Boon JY, Reid W, Xu YF, Kobrin K, Murphy GJ, Petrucelli L, Wolozin B. ALS-linked mutations enlarge TDP-43-enriched neuronal RNA granules in the dendritic arbor. J Neurosci. 2014;34:4167–4174. doi: 10.1523/JNEUROSCI.2350-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm VM, Zhang B, O’Brien P, Trojanowski JQ, Lee VMY. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J Exp Med. 2012a;209:975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O’Brien P, Trojanowski JQ, Lee VMY. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012b;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Song C, O’Brien P, Stieber A, Branch JR, Brunden KR, Trojanowski JQ, Lee VMY. Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc Natl Acad Sci USa. 2009;106:20051–20056. doi: 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EML, Kannan K, Guo F, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Mallucci GR, White MD, Farmer M, Dickinson A, Khatun H, Powell AD, Brandner S, Jefferys JGR, Collinge J. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Marijanovic Z, Caputo A, Campana V, Zurzolo C. Identification of an intracellular site of prion conversion. PLoS Pathog. 2009;5:e1000426. doi: 10.1371/journal.ppat.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Suzukake M, Nonaka T, Hosokawa M, Kubo M, Shimozawa A, Akiyama H, Hasegawa M. Pathological alpha-synuclein propagates through neural networks. Acta Neuropathol Commun. 2014;2:88. doi: 10.1186/s40478-014-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Suzukake M, Nonaka T, Hosokawa M, Oikawa T, Arai T, Akiyama H, Mann DMA, Hasegawa M. Prion-like spreading of pathological α-synuclein in brain. Brain. 2013;136:1128–1138. doi: 10.1093/brain/awt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci USa. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AC, Daneshvar DH, Alvarez VE, Stein TD. The neuropathology of sport. Acta Neuropathol. 2014;127:29–51. doi: 10.1007/s00401-013-1230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- Michel CH, Kumar S, Pinotsi D, Tunnacliffe A, St George-Hyslop P, Mandelkow E, Mandelkow EM, Kaminski CF, Kaminski Schierle GS. Extracellular monomeric tau protein is sufficient to initiate the spread of tau protein pathology. J Biol Chem. 2014;289:956–967. doi: 10.1074/jbc.M113.515445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbaha H, Holmes BB, Sanders DW, Bieschke J, Diamond MI. Tau trimers are the minimal propagation unit spontaneously internalized to seed intracellular aggregation. J Biol Chem. 2015 doi: 10.1074/jbc.M115.652693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed NV, Plouffe V, Rémillard-Labrosse G, Planel E, Leclerc N. Starvation and inhibition of lysosomal function increased tau secretion by primary cortical neurons. Sci Rep. 2014;4:5715. doi: 10.1038/srep05715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH, et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron. 2015 doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc Natl Acad Sci USa. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmio J, Suhonen J, Keränen T, Hulkkonen J, Peltola J, Pirttilä T. Cerebrospinal fluid tau as a marker of neuronal damage after epileptic seizure. Seizure. 2009;18:474–477. doi: 10.1016/j.seizure.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USa. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. Embo J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Peelaerts W, Bousset L, Van der Perren A, Moskalyuk A, Pulizzi R, Giugliano M, Van den Haute C, Melki R, Baekelandt V. α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature. 2015 doi: 10.1038/nature14547. [DOI] [PubMed] [Google Scholar]
- Peeraer E, Bottelbergs A, Van Kolen K, Stancu IC, Vasconcelos B, Mahieu M, Duytschaever H, Ver Donck L, Torremans A, Sluydts E, et al. Intracerebral injection of preformed synthetic tau fibrils initiates widespread tauopathy and neuronal loss in the brains of tau transgenic mice. Neurobiol Dis. 2015;73:83–95. doi: 10.1016/j.nbd.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- Plouffe V, Mohamed NV, Rivest-McGraw J, Bertrand J, Lauzon M, Leclerc N. Hyperphosphorylation and cleavage at D421 enhance tau secretion. PLoS ONE. 2012;7:e36873. doi: 10.1371/journal.pone.0036873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poepsel S, Sprengel A, Sacca B, Kaschani F, Kaiser M, Gatsogiannis C, Raunser S, Clausen T, Ehrmann M. Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nat Chem Biol. 2015;11:862–869. doi: 10.1038/nchembio.1931. [DOI] [PubMed] [Google Scholar]
- Polydoro M, de Calignon A, Suárez-Calvet M, Sanchez L, Kay KR, Nicholls SB, Roe AD, Pitstick R, Carlson GA, Gómez-Isla T, et al. Reversal of neurofibrillary tangles and tau-associated phenotype in the rTgTauEC model of early Alzheimer’s disease. J Neurosci. 2013;33:13300–13311. doi: 10.1523/JNEUROSCI.0881-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 2005;4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- Pooler AM, Phillips EC, Lau DHW, Noble W, Hanger DP. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep. 2013;14:389–394. doi: 10.1038/embor.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USa. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti G, Giaccone G, Rossi G, Canciani B, Bugiani O, Tagliavini F. Sporadic Creutzfeldt-Jakob disease: co-occurrence of different types of PrP(Sc) in the same brain. Neurology. 1999;53:2173–2176. doi: 10.1212/wnl.53.9.2173. [DOI] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73:1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. Embo J. 2012;31:4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, McBrayer MK, Campbell J, Kumar A, Hashim A, Sershen H, Stavrides PH, Ohno M, Hutton M, Nixon RA. Specific calpain inhibition by calpastatin prevents tauopathy and neurodegeneration and restores normal lifespan in tau P301L mice. J Neurosci. 2014;34:9222–9234. doi: 10.1523/JNEUROSCI.1132-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravits J, Laurie P, Fan Y, Moore DH. Implications of ALS focality: rostral-caudal distribution of lower motor neuron loss postmortem. Neurology. 2007a;68:1576–1582. doi: 10.1212/01.wnl.0000261045.57095.56. [DOI] [PubMed] [Google Scholar]
- Ravits J, Paul P, Jorg C. Focality of upper and lower motor neuron degeneration at the clinical onset of ALS. Neurology. 2007b;68:1571–1575. doi: 10.1212/01.wnl.0000260965.20021.47. [DOI] [PubMed] [Google Scholar]
- Recasens A, Dehay B, Bové J, Carballo-Carbajal I, Dovero S, Pérez-Villalba A, Fernagut PO, Blesa J, Parent A, Perier C, et al. Lewy body extracts from Parkinson disease brains trigger α-synuclein pathology and neurodegeneration in mice and monkeys. Ann Neurol. 2014;75:351–362. doi: 10.1002/ana.24066. [DOI] [PubMed] [Google Scholar]
- Ren PH, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–225. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey NL, Petit GH, Bousset L, Melki R, Brundin P. Transfer of human α-synuclein from the olfactory bulb to interconnected brain regions in mice. Acta Neuropathol. 2013;126:555–573. doi: 10.1007/s00401-013-1160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Ruben GC, Iqbal K, Grundke-Iqbal I, Wisniewski HM, Ciardelli TL, Johnson JE. The microtubule-associated protein tau forms a triple-stranded left-hand helical polymer. J Biol Chem. 1991;266:22019–22027. [PubMed] [Google Scholar]
- Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, Riesner D. Search for a prion-specific nucleic acid. J Virol. 2005;79:10796–10806. doi: 10.1128/JVI.79.16.10796-10806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar JG, Xiao X, Kabir ME, Chen S, Kim C, Haldiman T, Cohen Y, Chen W, Cohen ML, Surewicz WK. Structural determinants of phenotypic diversity and replication rate of human prions. PLoS Pathog. 2015;11:e1004832. doi: 10.1371/journal.ppat.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, Jackson B, McKee AC, Alvarez VE, Lee NCY, et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. 2012;287:3842–3849. doi: 10.1074/jbc.M111.277061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, Devos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82:1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Maria I, Varghese M, Ksiezak-Reding H, Dzhun A, Wang J, Pasinetti GM. Paired helical filaments from Alzheimer disease brain induce intracellular accumulation of Tau protein in aggresomes. J Biol Chem. 2012;287:20522–20533. doi: 10.1074/jbc.M111.323279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JR, Davies D, Fraser H. Scrapie in the central nervous system: neuroanatomical spread of infection and Sinc control of pathogenesis. J Gen Virol. 1992;73(Pt 7):1637–1644. doi: 10.1099/0022-1317-73-7-1637. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003a;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003b;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Simón D, García-García E, Royo F, Falcón-Pérez JM, Avila J. Proteostasis of tau. Tau overexpression results in its secretion via membrane vesicles. FEBS Lett. 2012;586:47–54. doi: 10.1016/j.febslet.2011.11.022. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol. 2014;28:76–83. doi: 10.1016/j.ceb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Sousa MM, Saraiva MJ. Internalization of transthyretin. Evidence of a novel yet unidentified receptor-associated protein (RAP)-sensitive receptor. J Biol Chem. 2001;276:14420–14425. doi: 10.1074/jbc.M010869200. [DOI] [PubMed] [Google Scholar]
- Stancu I-C, Vasconcelos B, Ris L, Wang P, Villers A, Peeraer E, Buist A, Terwel D, Baatsen P, Oyelami T, et al. Templated misfolding of Tau by prion-like seeding along neuronal connections impairs neuronal network function and associated behavioral outcomes in Tau transgenic mice. Acta Neuropathol. 2015 doi: 10.1007/s00401-015-1413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, True HL. Extensive diversity of prion strains is defined by differential chaperone interactions and distinct amyloidogenic regions. PLoS Genet. 2014a;10:e1004337. doi: 10.1371/journal.pgen.1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, True HL. Structural variants of yeast prions show conformer-specific requirements for chaperone activity. Mol Microbiol. 2014b;93:1156–1171. doi: 10.1111/mmi.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. The CPEB3 Protein Is a Functional Prion that Interacts with the Actin Cytoskeleton. Cell Rep. 2015;11:1772–1785. doi: 10.1016/j.celrep.2015.04.060. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- Taniguchi-Watanabe S, Arai T, Kametani F, Nonaka T, Masuda-Suzukake M, Tarutani A, Murayama S, Saito Y, Arima K, Yoshida M, et al. Biochemical classification of tauopathies by immunoblot, protein sequence and mass spectrometric analyses of sarkosyl-insoluble and trypsin-resistant tau. Acta Neuropathol. 2015 doi: 10.1007/s00401-015-1503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- Tian H, Davidowitz E, Lopez P, Emadi S, Moe J, Sierks M. Trimeric tau is toxic to human neuronal cells at low nanomolar concentrations. Int J Cell Biol. 2013;2013:260787. doi: 10.1155/2013/260787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Weissman JS. Amyloid structure: conformational diversity and consequences. Annu Rev Biochem. 2011;80:557–585. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Tran HT, Chung CHY, Iba M, Zhang B, Trojanowski JQ, Luk KC, Lee VMY. A-synuclein immunotherapy blocks uptake and templated propagation of misfolded α-synuclein and neurodegeneration. Cell Rep. 2014;7:2054–2065. doi: 10.1016/j.celrep.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- Tsujimura A, Taguchi K, Watanabe Y, Tatebe H, Tokuda T, Mizuno T, Tanaka M. Lysosomal enzyme cathepsin B enhances the aggregate forming activity of exogenous α-synuclein fibrils. Neurobiol Dis. 2014;73C:244–253. doi: 10.1016/j.nbd.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Ulusoy A, Rusconi R, Pérez-Revuelta BI, Musgrove RE, Helwig M, Winzen-Reichert B, Di Monte DA. Caudo-rostral brain spreading of α-synuclein through vagal connections. EMBO Mol Med. 2013;5:1051–1059. doi: 10.1002/emmm.201302475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Wadia JS, Schaller M, Williamson RA, Dowdy SF. Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS ONE. 2008;3:e3314. doi: 10.1371/journal.pone.0003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth JDF, Hill AF, Beck JA, Collinge J. Molecular and clinical classification of human prion disease. Br Med Bull. 2003;66:241–254. doi: 10.1093/bmb/66.1.241. [DOI] [PubMed] [Google Scholar]
- Walker AK, Spiller KJ, Ge G, Zheng A, Xu Y, Zhou M, Tripathy K, Kwong LK, Trojanowski JQ, Lee VMY. Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP-43. Acta Neuropathol. 2015;130:643–660. doi: 10.1007/s00401-015-1460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LC, Jucker M. Neurodegenerative Diseases: Expanding the Prion Concept. Annu Rev Neurosci. 2015 doi: 10.1146/annurev-neuro-071714-033828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Krüger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GDJ, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Watts JC, Giles K, Oehler A, Middleton L, Dexter DT, Gentleman SM, DeArmond SJ, Prusiner SB. Transmission of multiple system atrophy prions to transgenic mice. Proc Natl Acad Sci USa. 2013;110:19555–19560. doi: 10.1073/pnas.1318268110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci USa. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischik CM, Novak M, Thøgersen HC, Edwards PC, Runswick MJ, Jakes R, Walker JE, Milstein C, Roth M, Klug A. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci USa. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerman AL, Stöhr J, Aoyagi A, Rampersaud R, Krejciova Z, Watts JC, Ohyama T, Patel S, Widjaja K, Oehler A, et al. Propagation of prions causing synucleinopathies in cultured cells. Proc Natl Acad Sci USa. 2015;112:E4949–E4958. doi: 10.1073/pnas.1513426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Herman M, Liu L, Simoes S, Acker CM, Figueroa H, Steinberg JI, Margittai M, Kayed R, Zurzolo C, et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J Biol Chem. 2013;288:1856–1870. doi: 10.1074/jbc.M112.394528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, la Cruz de MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Cirrito JR, Stewart FR, Jiang H, Finn MB, Holmes BB, Binder LI, Mandelkow EM, Diamond MI, Lee VMY, et al. In vivo microdialysis reveals age-dependent decrease of brain interstitial fluid tau levels in P301S human tau transgenic mice. J Neurosci. 2011;31:13110–13117. doi: 10.1523/JNEUROSCI.2569-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Holth JK, Liao F, Stewart FR, Mahan TE, Jiang H, Cirrito JR, Patel TK, Hochgräfe K, Mandelkow EM, et al. Neuronal activity regulates extracellular tau in vivo. J Exp Med. 2014;211:387–393. doi: 10.1084/jem.20131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Yetman MJ, Lillehaug S, Bjaalie JG, Leergaard TB, Jankowsky JL. Transgene expression in the Nop-tTA driver line is not inherently restricted to the entorhinal cortex. Brain Struct Funct. 2015 doi: 10.1007/s00429-015-1040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim YI, Park BC, Yadavalli R, Zhao X, Eisenberg E, Greene LE. The multivesicular body is the major internal site of prion conversion. J Cell Sci. 2015;128:1434–1443. doi: 10.1242/jcs.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song M, Liu X, Kang SS, Kwon IS, Duong DM, Seyfried NT, Hu WT, Liu Z, Wang JZ, et al. Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer’s disease. Nat Med. 2014;20:1254–1262. doi: 10.1038/nm.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012a;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Ottenberg G, Sferrazza GF, Lasmézas CI. Highly neurotoxic monomeric α-helical prion protein. Proc Natl Acad Sci USa. 2012b;109:3113–3118. doi: 10.1073/pnas.1118090109. [DOI] [PMC free article] [PubMed] [Google Scholar]