Abstract
The immune response is highly active in Alzheimer's disease (AD). Identification of genetic risk contributed by immune genes to AD may provide essential insight for the prognosis, diagnosis, and treatment of this neurodegenerative disease. In this study, we performed a genetic screening for AD-related top immune genes identified in Europeans in a Chinese cohort, followed by a multiple-stage study focusing on Complement Factor H (CFH) gene. Effects of the risk SNPs on AD-related neuroimaging endophenotypes were evaluated through magnetic resonance imaging scan, and the effects on AD cerebrospinal fluid biomarkers (CSF) and CFH expression changes were measured in aged and AD brain tissues and AD cellular models. Our results showed that the AD-associated top immune genes reported in Europeans (CR1, CD33, CLU, and TREML2) have weak effects in Chinese, whereas CFH showed strong effects. In particular, rs1061170 (Pmeta=5.0 × 10−4) and rs800292 (Pmeta=1.3 × 10−5) showed robust associations with AD, which were confirmed in multiple world-wide sample sets (4317 cases and 16 795 controls). Rs1061170 (P=2.5 × 10−3) and rs800292 (P=4.7 × 10−4) risk-allele carriers have an increased entorhinal thickness in their young age and a higher atrophy rate as the disease progresses. Rs800292 risk-allele carriers have higher CSF tau and Aβ levels and severe cognitive decline. CFH expression level, which was affected by the risk-alleles, was increased in AD brains and cellular models. These comprehensive analyses suggested that CFH is an important immune factor in AD and affects multiple pathological changes in early life and during disease progress.
Introduction
Late-onset Alzheimer's disease (AD, OMIM 104300, 104310) is the most common neurodegenerative disorder and leads to a progressive cognitive decline and dementia in the elderly (Alzheimer's Association, 2013; Querfurth and LaFerla, 2010). The major histological features of the disease include the presence of neurofibrillary tangles, extracellular amyloid β peptide (Aβ) deposition, synaptic dysfunction, and loss of neuronal integrity (Querfurth and LaFerla, 2010). The underlying cause of the disease is unclear in most cases, but numerous genetic alterations have been identified as being associated with Alzheimer's risk (Bertram et al, 2007; Karch and Goate, 2015; Lambert et al, 2013). Immune-related genes, especially complement genes such as complement receptor 1 (CR1) and clusterin (CLU; Bertram et al, 2007), have been identified as the top AD susceptibility genes in European populations or of European origin, and the complement system has been reported to be involved in the initiation and development of AD (Crehan et al, 2012).
The complement regulator, Complement Factor H (CFH, OMIM 134370), has a key role in inhibiting complement activation and inflammation. CFH was recognized as the major genetic risk factor for age-related macular degeneration (AMD; Klein et al, 2005), which is another age-related neurodegenerative disease and shares similar risk factors and pathological features with AD (Sivak, 2013). CFH protein was suggested to be a potential top serum biomarker for AD (Hye et al, 2006, 2014; Thambisetty et al, 2008). However, the involvement of CFH in AD is contentious.
We performed a genetic screening in a Han Chinese cohort with AD for five immune genes (CR1, CR2, CLU, CD33, and TREML2) that were identified as the top AD susceptibility genes for Europeans (Bertram et al, 2007; Lambert et al, 2013). After the screening, a multiple-stage genetic association study focusing on the CFH gene was performed. We aimed to answer two key questions: (1) Do genetic variants in these immune genes, especially CFH, confer risk to AD in Han Chinese? and (2) How does CFH function in AD? The involvement of CFH in functional and structural brain changes, as well as AD biomarker (cerebrospinal fluid (CSF) tau and Aβ levels) alterations, were explored using data from the Alzheimer's Disease Neuroimaging Initiative (ADNI) project (Weiner et al, 2010). Moreover, the effect of AD-related CFH SNPs on morphological changes of hippocampus and entorhinal cortex, which were recognized as the most and the first affected regions of the brain with AD (Harris et al, 2010; Khan et al, 2014), respectively, was measured in healthy young adults at genetic risk by magnetic resonance imaging (MRI) scan. The effects of CFH genotypes and expression changes were analyzed in aged and AD brain tissues and in AD cellular models. Our collective data indicated that CFH is an important AD susceptibility gene and may affect the structure and function of the brain and alter the immune response as the disease progresses.
Materials and methods
Subjects
A two-stage cohort of 2041 Han Chinese with and without AD was analyzed. In stage 1, 380 patients (AD1, 45.8% men, mean age 76.5±9.6 years, mean onset age 70.9±9.7 years) and 475 healthy individuals from the general populations (PC1) were recruited from East China. In stage 2, we recruited 345 patients (AD2), 337 healthy individuals from the general populations (PC2), and 504 healthy longevity individuals (LC, age 93±2.6 years; as another control) from Southwest China. Most of these AD patients had been analyzed for other risk loci in our recent studies (Bi et al, 2014, 2015; Wang et al, 2014). In brief, patients were diagnosed following the DSM-IV and the NINCDS-ADRDA criteria independently by at least two senior clinicians. The healthy controls were confirmed to have normal cognitive ability. Informed consents conforming to the tenets of the Declaration of Helsinki were obtained from all participants, or the supervisors of patients, after being given a complete description of the study. The institutional review board of the Kunming Institute of Zoology, Chinese Academy of Sciences, approved this study.
To confirm the results of the two-stage study, we performed jointed comparisons with multiple world-wide sample sets. Additional cohorts from East and Southwest China: 2460 individuals from Shanghai, 1549 individuals from Sichuan Province, and 2751 individuals from Yunnan Province (Zhang et al, 2014), which were enrolled for other genetic association analyses, were included in this analysis to enlarge the population controls. All of these subjects were collected from the general populations with normal cognitive ability and no history of dementia. Individuals with genotype data of rs800292 and rs1061170 available were included in our jointed comparison. Genetic data from ADNI (http://adni.loni.usc.edu/; Weiner et al, 2010) were also retrieved for re-analysis. Subjects with available genotype data from all stages of the ADNI 1/GO/2 were included in our analyses. These samples contain 760 individuals in the ADNI1 cohort (180 probable AD patients, 363 mild cognitive impairment (MCI) patients, and 214 cognitively normal aging controls) and 430 individuals in the ADNI GO/2 cohort (29 probable AD patients, 275 MCI patients, and 126 cognitively normal aging controls). Because of the limited sample size of probable AD, AD and MCI participants in these two cohorts were pooled as the patients' group. Previously reported data regarding the association of rs1061170 with AD (Hamilton et al, 2007; Le Fur et al, 2010; Proitsi et al, 2012; Zetterberg et al, 2008) were re-analyzed together with the data from our current samples. In total, 719 patients and 6217 population controls from China, and 845 patients and 345 controls of European origin were analyzed for rs800292; 713 patients and 6747 controls from China and 3604 patients and 10 048 controls of European origin were analyzed for rs1061170.
SNP Genotyping and Association Analysis
We genotyped 17 SNPs of five immune genes (CR1, CR2, CLU, CD33, and TREML2) that were identified as the top Alzheimer's susceptibility genes in Europeans (Bertram et al, 2007; Lambert et al, 2013) and 11 SNPs of the CFH gene in our stage 1 cohort from East China for the preliminary screening. Previously reported genome-wide association study (GWAS) top hits, tagging SNPs and potentially functional SNPs of these genes were selected for genotyping. The selection criteria and details for selected SNPs were described in the Supplementary Methods and Supplementary Table 1. APOE ɛ4 was determined as previously described (Bi et al, 2014, 2015; Wang et al, 2014).
Association analysis was carried out using PLINK (Purcell et al, 2007). Allelic (Table 1) and genotypic (Supplementary Table 2) comparisons with 2 d.f. genotypic, Cochran-Armitage trend, dominant, and recessive models were conducted for individual SNPs. All available samples from the general populations were pooled as a combined sample for Chinese (termed ‘Combined Chinese', Table 2) and Europeans (termed ‘Combined Europeans', Table 2), respectively. Comparison of the genotype counts between the combined case and control populations was estimated by the χ2 test. Meta-analysis for the association of CFH SNPs with AD in the two combined sample sets was performed by using Review manager (RevMan 5.2, http://tech.cochrane.org/revman), with the Cochran–Mantel–Haenszel method under a fixed effect. The heterogeneity was measured by the I2 index (Higgins and Thompson, 2002; Table 2).
Table 1. Association of CFH Variants with AD in Han Chinese Populations (N=2041).
| SNP | Allele |
Stage 1 (SH:AD1
vs
PC1) |
Stage 2-1 (SC:AD2
vs
PC2) |
Stage 2-2 (SC:AD2
vs
LC) |
Combined (AD1+AD2
vs
PC1+PC2) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FAD1 | FPC1 | OR (95%CI) | PA | FAD2 | FPC2 | OR (95%CI) | PA | FLC | OR (95%CI) | PA | FAD | FPC | OR (95%CI) | PA | PCorrected | ||
| rs800292 | T/C | 0.362 | 0.402 | 0.844 (0.693–1.028) | 0.092 | 0.376 | 0.441 | 0.764 (0.615–0.949) | 0.015 | 0.426 | 0.811 (0.664–0.989) | 0.038 | 0.369 | 0.418 | 0.812 (0.702–0.940) | 5.1 × 10−3 | 0.019 |
| rs1061170 | C/T | 0.066 | 0.039 | 1.749 (1.130–2.705) | 0.011 | 0.073 | 0.043 | 1.749 (1.091–2.805) | 0.019 | 0.037 | 2.047 (1.320–3.174) | 0.001 | 0.069 | 0.041 | 1.759 (1.277–2.422) | 4.7 × 10−4 | 0.005 |
| rs10801555 | A/G | 0.044 | 0.040 | 1.098 (0.682–1.769) | 0.699 | 0.057 | 0.043 | 1.345 (0.822–2.201) | 0.237 | 0.052 | 1.105 (0.721–1.693) | 0.648 | 0.050 | 0.041 | 1.225 (0.871–1.722) | 0.242 | 0.409 |
| rs10922096 | T/C | 0.122 | 0.128 | 0.943 (0.706–1.260) | 0.692 | 0.109 | 0.110 | 0.990 (0.704–1.393) | 0.954 | 0.115 | 0.942 (0.691–1.284) | 0.705 | 0.116 | 0.121 | 0.954 (0.765–1.189) | 0.674 | 0.823 |
| rs2019727 | T/A | 0.073 | 0.092 | 0.781 (0.549–1.110) | 0.167 | 0.051 | 0.071 | 0.703 (0.449–1.102) | 0.123 | 0.060 | 0.847 (0.552–1.300) | 0.446 | 0.063 | 0.083 | 0.736 (0.558–0.971) | 0.030 | 0.065 |
| rs10733086 | A/T | 0.057 | 0.060 | 0.948 (0.630–1.425) | 0.796 | 0.069 | 0.064 | 1.086 (0.708–1.666) | 0.705 | 0.070 | 0.986 (0.672–1.446) | 0.940 | 0.063 | 0.062 | 1.019 (0.759–1.367) | 0.900 | 0.900 |
| rs10737680 | C/A | 0.395 | 0.430 | 0.868 (0.715–1.055) | 0.154 | 0.452 | 0.435 | 1.071 (0.865–1.327) | 0.528 | 0.433 | 1.078 (0.887–1.311) | 0.449 | 0.422 | 0.432 | 0.962 (0.833–1.110) | 0.595 | 0.817 |
| rs1410996 | T/C | 0.391 | 0.428 | 0.858 (0.706–1.042) | 0.122 | 0.431 | 0.433 | 0.992 (0.800–1.230) | 0.942 | 0.433 | 0.993 (0.816–1.208) | 0.940 | 0.410 | 0.430 | 0.921 (0.797–1.063) | 0.260 | 0.409 |
| rs11582939 | C/T | 0.443 | 0.494 | 0.816 (0.673–0.988) | 0.037 | 0.461 | 0.510 | 0.819 (0.662–1.013) | 0.066 | 0.496 | 0.867 (0.714–1.054) | 0.152 | 0.489 | 0.492 | 0.988 (0.857–1.138) | 0.863 | 0.900 |
| rs426736 | T/C | 0.532 | 0.458 | 1.345 (1.110–1.629) | 0.002 | 0.494 | 0.503 | 0.965 (0.780–1.194) | 0.745 | 0.489 | 1.021 (0.840–1.240) | 0.836 | 0.520 | 0.474 | 1.199 (1.040–1.382) | 0.012 | 0.034 |
Abbreviations: AD1, patients with AD of stage 1 from Shanghai (SH); AD2, patients with AD of stage 2 from Sichuan (SC); CI, confidence interval; Fx, minor allele frequency of the ‘x' population; LC, longevity controls of stage 2 from Sichuan; OR, odds ratio; PA, allelic association P-value; PCorrected, adjusted P-value (combined PA) for multiple testing by Benjamini-Hochberg FDR (FDR_BH) control algorithm; PC1, population controls of stage 1 from Shanghai; PC2, population controls of stage 2 from Sichuan. The P-values less than 0.05 were marked in italic. Results of genotypic associations and genotype counts distribution were shown in Supplementary Table 2.
Table 2. Validating the Associations of rs800292 and rs1061170 with AD in Enlarged Sample Sets.
| SNP | Population |
Case |
Control |
P-value | OR | ||
|---|---|---|---|---|---|---|---|
| N | MAF | N | MAF | ||||
| rs800292 | Shanghaia | 377 | 0.362 | 1917 | 0.410 | 1.6 × 10−2 | 0.82 |
| Sichuanb | 342 | 0.376 | 1549 | 0.419 | 3.7 × 10−2 | 0.84 | |
| Combinedc | 719 | 0.369 | 3466 | 0.414 | 2.0 × 10−3 | 0.83 | |
| Combined Chinesed | 719 | 0.369 | 6217 | 0.415 | 7.0 × 10−4 | 0.82 | |
| ADNI_1e | 543 | 0.236 | 214 | 0.304 | 6.0 × 10−3 | 0.71 | |
| ADNI_GO2e | 302 | 0.207 | 126 | 0.246 | 2.1 × 10−1 | 0.78 | |
| European (ADNI) | 845 | 0.225 | 340 | 0.282 | 3.0 × 10−3 | 0.74 | |
| Meta-analysisf | 1564 | — | 6557 | — | 1.3 × 10−5 | 0.80 | |
| rs1061170 | Shanghaia | 377 | 0.066 | 2460 | 0.055 | 2.2 × 10−1 | 1.21 |
| Sichuanb | 336 | 0.073 | 1542 | 0.043 | 3.0 × 10−3 | 1.65 | |
| Combinedc | 713 | 0.069 | 4002 | 0.051 | 6.0 × 10−3 | 1.38 | |
| Combined Chinesed | 713 | 0.069 | 6747 | 0.057 | 5.5 × 10−2 | 1.24 | |
| European (Zetterberg et al, 2008) | 800 | 0.427 | 1265 | 0.394 | 3.9 × 10−2 | 1.14 | |
| European (Le Fur et al, 2010) | 701 | 0.357 | 6990 | 0.361 | 7.7 × 10−1 | 0.98 | |
| European (Proitsi et al, 2012) | 2103 | 0.385 | 1793 | 0.375 | 3.4 × 10−1 | 1.05 | |
| Combined Europeansg | 3604 | 0.389 | 10048 | 0.368 | 1.0 × 10−3 | 1.09 | |
| Meta-analysish | 4317 | — | 16795 | — | 5.0 × 10−4 | 1.10 | |
Abbreviation: MAF, minor allele frequency.
Enlarged general population controls including the case-matched controls and available general individuals with normal cognitive ability from Shanghai (author's unpublished data).
Enlarged controls including the case-matched controls and other available general individuals with normal cognitive ability from Sichuan (author's unpublished data).
Samples combining both Shanghai and Sichuan subjects.
Chinese samples combining Shanghai, Sichuan, and Yunnan subjects with available genotype data as the general population control. For rs800292: heterogeneity among combined Chinese populations, χ2=0.03, d.f.=1 (P=0.86), I2=0%, overall meta-analysis effect Z=3.21 (P=0.001). For rs1061170: heterogeneity among combined Chinese populations, χ2=2.34, d.f.=1 (P=0.13), I2=57%, overall meta-analysis effect Z=3.05 (P=0.002).
Data retrieved from the ADNI (Alzheimer's Disease Neuroimaging Initiative) project. For rs800292: heterogeneity among combined European populations, χ2=0.32, d.f.=1 (P=0.57), I2=0%, overall meta-analysis effect Z=2.95 (P=0.003); genotype data of rs1061170 is not available in the ADNI subjects.
Meta-analysis for rs800292 in combined Chinese and Europeans; heterogeneity: χ2=0.91, d.f.=1 (P=0.34), I2=0% overall meta-analysis effect Z=4.32 (P<0.0001).
All three available European sample sets were pooled together as a combined European population, with the original genotype counts measured by χ2 test; heterogeneity among populations, χ2=3.04, d.f.=2 (P=0.22), I2=34%, overall meta-analysis effect Z=1.48 (P=0.14).
Meta-analysis for rs1061170 in combined Chinese and Europeans, heterogeneity: χ2=0.93, d.f.=1 (P=0.34), I2=0%, overall effect Z=3.49 (P=0.0005). The P-values less than 0.05 were marked in italic.
The genetic associations were explored further by estimating the significance of SNP–SNP interaction using the multifactor-dimensionality reduction (MDR) method (Ritchie et al, 2001) or the ‘—epistasis' command in PLINK (Purcell et al, 2007).
Neuroimaging Analysis for the Effects of CFH Variants on Structural and Functional Brain Changes
We recruited 360 healthy young adults (age 19.4±1.1 years; 51.7% men) to study the effects of the Alzheimer's risk CFH SNPs on morphological changes of the brain. These samples were described in our previous study and were effective to identify risk alleles affecting brain structure variations (Li et al, 2015; Zhang et al, 2015). MRI scans were performed on a MR750 3.0 Tesla magnetic resonance scanner (GE Healthcare, detailed parameters in Supplementary Methods). The protocol was approved by the Ethics Committee of School of Life Science and Technology at University of Electronic Science and Technology of China.
First, we performed a whole-brain voxel-based morphometry analysis for volume and density of the gray matter. Second, we detected the effect of the AD-risk CFH SNPs on total intracranial volume (ICV) and hippocampus volume changes. Finally, we tested the effect of the AD-risk CFH SNPs on the thickness of the entorhinal cortex. To test the effect of CFH genotypes on brain morphological changes, we applied a general linear regression model adjusted for gender, age, education year, and ICV. To correct for multiple comparisons for the entorhinal cortex, the statistical significance level was set as P<0.005 (0.05/10 [5 SNPs × 2 hemisphere], Bonferroni correction). Details regarding the imaging process and statistics were described in Supplementary Methods.
Detecting the Effects of AD-Risk SNPs on AD Endophenotypes
To confirm our results and investigate further the role of CFH in AD pathogenesis, we obtained genetic, neuroimaging, and biomarker data from the ADNI project (http://adni.loni.usc.edu/; Weiner et al, 2010). Effect of the top AD-risk CFH SNP (only rs800292 was available) on AD endophenotypes, eg, CSF tau and Aβ levels, cognitive score, entorhinal regional atrophy rate, and entorhinal volume, was analyzed using PLINK (Purcell et al, 2007).
Expression Quantitative Trait Loci (eQTL) Analysis
To investigate the effect of CFH variants on CFH mRNA expression level, we performed eQTL analysis in 10 brain regions of 134 neuropathologically normal individuals. Details were shown in Supplementary File and the brain eQTL database (http://www.braineac.org/; Ramasamy et al, 2014). The eQTL effect of the CFH variants was validated by using the Genotype-Tissue Expression project (GTEx, http://www.gtexportal.org/home/) database, which provides a comprehensive atlas of gene expression and regulation across multiple human tissues (The GTEx Consortium, 2013). The tibial nerve tissue (n=102) was used in the analysis; the other brain tissues, such as cortex or hippocampus, had a sample size less than 30 and was not considered (http://www.gtexportal.org/home/). For the effect of the CFH variants on CFH protein level, we used an earlier GWAS data of plasma CFH levels (Ansari et al, 2013).
CFH mRNA Expression Alterations in Aged and AD Brains and AD Cellular Models
A total of 49 hippocampal samples of Rattus norvegicus at 5 age points (3, 6, 9, 12, and 23 months, GSE9990; Kadish et al, 2009) and 30 postmortem frontal cortex of normal individuals at 26–106 years of age (GSE1572; Lu et al, 2004) were used to assess CFH mRNA expression changes during brain aging. Expression data of 272 human postmortem dorsolateral prefrontal cortex (DLPFC) of normal subjects across the lifespan from the BrainCloud (http://braincloud.jhmi.edu/; Colantuoni et al, 2011) were also included to investigate the expression pattern of CFH with aging. In all, 22 hippocampal samples from postmortems showing AD at different stages of severity (GSE1297; Blalock et al, 2004), and entorhinal cortex neurons containing neurofibrillary tangles from 10 mid-stage patients (GSE4757; Dunckley et al, 2006) were used to assess CFH mRNA expression changes during disease processing. Expression differences between groups were measured by Student's t-test using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, USA). The expression data were retrieved through the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/sites/GDSbrowser). Correlation analysis was performed for the mRNA expression of CFH and APP in hippocampus of AD patients (GSE1297; Blalock et al, 2004).
U251 cells (a glioma cell line) with Aβ1–42 treatment or with stable overexpression of APP mutant (APPMut, APP-p.M671L) and PSEN1 mutant (PSEN1Mut, PSEN1-p.M139V/M146L/H163R) were used as AD cellular models to test CFH expression changes in response to these stimuli. Quantitative real-time PCR was used to determine the relative mRNA level of the CFH gene in the AD cellular models. Detailed information was shown in the Supplementary Materials.
Results
Genetic Screening of the AD-Related Immune Genes and CFH in Han Chinese Patients with AD
SNPs within the five AD-related top immune genes (CR1, CR2, CLU, CD33, and TREML2) identified in Europeans showed no association with AD in our stage 1 Chinese samples (Supplementary Table 1). We observed positive associations of CFH SNPs rs426736 (OR=1.345, allelic P=2.4 × 10−3, genotypic P=1.2 × 10−2) and rs1061170 (p.Y402H, OR=1.749, allelic P=1.1 × 10−2, genotypic P=1.3 × 10−2) with AD, whereas rs800292 (p.V62I) and rs11582939 showed a marginal significance (Table 1 and Supplementary Table 2) in our stage 1 samples.
We replicated the association of CFH with AD in stage 2 cohort. Associations of both rs1061170 (OR=1.749, allelic P=1.9 × 10−2, genotypic P=3.4 × 10−2) and rs800292 (OR=0.764, allelic P=1.5 × 10−2, genotypic P=6.5 × 10−4) with AD were confirmed. When the cases were compared with the healthy longevity controls (>90 years old), rs1061170 (OR=2.047, allelic P=1.1 × 10−3, genotypic P=9.3 × 10−4) showed a much stronger association with AD, and the association of rs800292 (OR=0.811, allelic P=0.038, genotypic P=3.0 × 10−3) with AD remained significant. We combined the two independent samples and found that rs1061170 (OR=1.759, PFDR_BH=5.2 × 10−3) and rs800292 (OR=0.812, PFDR_BH=1.9 × 10−2) showed strong associations even after correcting for multiple testing (false discovery rate (Benjamini Hochberg), FDR_BH). No SNP–SNP interaction among CFH variants and between CFH and APOE SNPs (rs429358 and rs7412 that defining the ɛ4 status) was observed, suggesting that CFH was involved in AD independently of APOE.
Validating the Association of CFH with AD in the Enlarged Sample Sets
We validated the association of the most robust CFH SNPs rs800292 and rs1061170 with AD in enlarged world-wide sample sets (Table 2). Compared with the pooled larger population controls (n=3466) from East and Southwest China, the association of rs800292 with AD remained robust (P=2.0 × 10−3). When all the population controls (n=6217) were considered, the association was even stronger (P=7.0 × 10−4). Analysis of the ADNI data showed that rs800292 was also associated with AD in Europeans (845 cases vs 340 controls; P=3.0 × 10−3). Meta-analysis combining all Chinese and European samples (1564 cases vs 6557 controls), which would increase the statistical power and had no significant study heterogeneity (cf. the footnote of Table 2), further validated the association of rs800292 with AD (Pmeta=1.3 × 10−5, OR=0.80).
Similarly, we validated the association of rs1061170 with AD in combined population controls (n=4002) from East and Southwest China (P=6.0 × 10−3), but this effect turned out to be marginally significant (P=5.5 × 10−2) when all Han Chinese controls (n=6747) were considered. Combined analysis of previously reported data showed a positive association of rs1061170 with AD in Europeans (3604 cases vs 10048 controls; P=1.0 × 10−3). When all combined Han Chinese and European samples (4317 cases vs 16795 controls) were used for meta-analysis, we observed a significant association between rs1061170 and AD (Pmeta=5.0 × 10−4, OR=1.10; Table 2).
Effects of the AD-Risk CFH SNPs on Structural Brain Changes in Young Adults
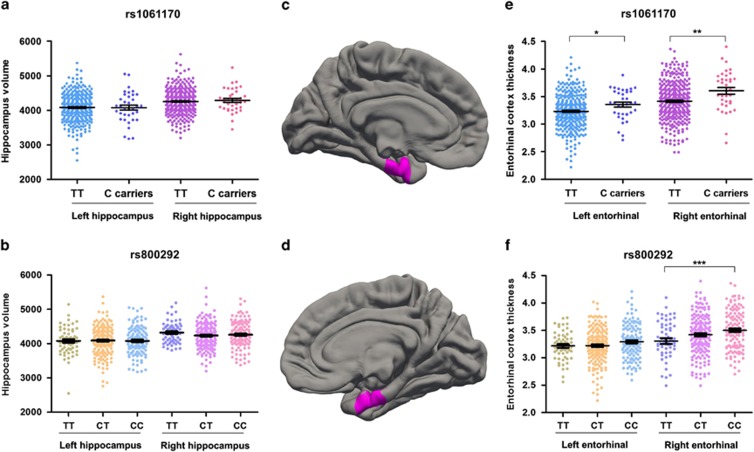
We detected the effects of the AD-risk CFH SNPs on morphological changes of the brain in young individuals using structural MRI scan. The AD-related CFH SNPs had no apparent effect on estimated total ICV (Supplementary Figure 2) and hippocampal volume (Figure 1 and Supplementary Table 3) in our pilot screening. However, all suggestive AD-associated CFH SNPs showed a trend of association with the entorhinal thickness (Supplementary Table 4). Both rs1061170 (P=2.5 × 10−3) and rs800292 (P=4.7 × 10−4) were significantly associated with the entorhinal thickness, especially for the right entorhinal cortex (Figure 1). Intriguingly, the AD-risk allele carriers have increased thickness of the entorhinal cortex in the right hemispheres (Figure 1) in their early age. It was reported that trans-synaptic progression of Aβ-induced cortex dysfunction and cortical spread was driven and initiated from the entorhinal cortex in preclinical Alzheimer's disease (Harris et al, 2010; Khan et al, 2014). Interference of the entorhinal cortex may contribute to the development of AD.
Figure 1.
Risk allele carriers have similar hippocampal volume and thicker entorhinal cortex compared with non-risk allele carriers at young age. Regression analysis was conducted to detect the associations of rs1061170 (Y402H) and rs800292 (V62I) with bilateral hippocampal volume (a and b) and entorhinal cortical thickness (e and f). The left entorhinal cortex (c) and the right entorhinal cortex (d) were labeled in FreeSurfer. *P<0.05, ** P<0.01, ***P<0.001, linear regression analyses. Data represent mean±SEM.
Effects of the AD-Risk CFH SNPs on AD Endophenotypes and CFH Expression
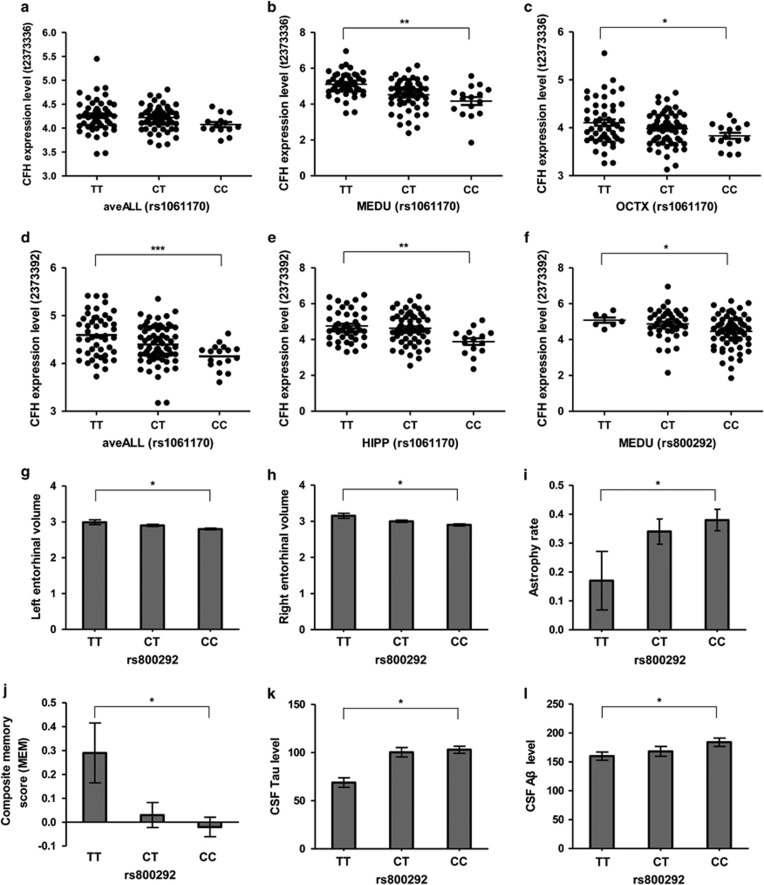
Our MRI scan analyses showed that individuals at risk of AD had a thicker entorhinal cortex in early life, suggesting a potential compensatory effect. Indeed, we observed a decreased volume of the entorhinal cortex in AD patients with risk allele of rs800292 (Figure 2g, h), indicating a higher atrophy rate of risk allele carriers as confirmed in our regional atrophy rate analysis (Figure 2i). In addition, risk allele carriers of rs800292 showed a marginally significant (P<0.05) decrease of cognitive score (Figure 2j), and increase of CSF tau (Figure 2k) and Aβ (Figure 2l) levels. These observations added more support for the contribution of CFH variants to AD susceptibility and development. Note that we also found a positive association of CFH variant with MCI patients using the ANDI data (Supplementary Table 5), which suggested that an analysis for the association between AD stage and CFH genotype might be rewarding.
Figure 2.
Effects of AD-risk CFH SNPs on CFH expression and AD endophenotypes. eQTL effect of AD-risk CFH SNPs (rs1061170 and rs800292) on CFH mRNA expression level was investigated in brain tissues using Affymetrix Human Exon 1.0 ST Array data from the UK Brain Expression Consortium (UKBEC; Ramasamy et al, 2014). We retrieved the genotyping and expression data from the UKBEC web server (http://www.braineac.org/; Ramasamy et al, 2014). Affymetrix ID t2373336 (a–c), CFH transcript probe; Affymetrix ID 2373392 (d–f), CFH exon-specific probe (chr1: 196712667–196712698). aveALL, average expression level among the 10 available brain regions; MEDU, the inferior olivary nucleus (sub-dissected from the medulla); OCTX, occipital cortex; HIPP, hippocampus. The potential effects of AD-risk CFH SNP rs800292 on AD-related endophenotypes, eg, entorhinal volume (g–h), entorhinal regional atrophy rate (i), composite memory score (MEM, j), and CSF tau (k) and Aβ (l) levels, were analyzed using data retrieved from the ADNI project (http://adni.loni.usc.edu/; Weiner et al, 2010). Data represent mean±SEM. *P<0.05, **P<0.01, ***P<0.001, one-way ANOVA for eQTL analysis; linear regression analyses for SNP rs800292 on AD-related endophenotypes.
Besides their effects on AD neuroimaging and biomarker endophenotypes, the risk alleles of rs800292 and rs1061170 were associated with lower CFH mRNA level (Figure 2). In particular, the CFH mRNA level was significantly decreased in the inferior olivary nucleus (MEDU, P<0.01) and occipital cortex (OCTX, P<0.05) in carriers with the risk allele C of rs1061170 (Figure 2a–c). The most significant genotype-affected CFH mRNA changes were observed for an exon-specific probe 2373392, which showed strong associations in all 10 brain regions (aveALL, P<0.001, Figure 2d) and hippocampus (HIPP, P<0.01, Figure 2e). In addition, carriers of rs800292 risk allele showed a significantly decreased CFH mRNA expression level in MEDU (Figure 2f). The significant decrease of CFH mRNA level associated with rs800292 and rs1061170 risk alleles could be validated in the tibial nerve tissues using the GTEx data (Supplementary Figure 3). Note that our results were in agreement with a reported GWAS study showing a significant association of lower serum CFH protein level with the AD-risk allele of rs1061170 (Ansari et al, 2013).
CFH mRNA Expression in Aged and Alzheimer's Brain Tissues and Cellular Assays
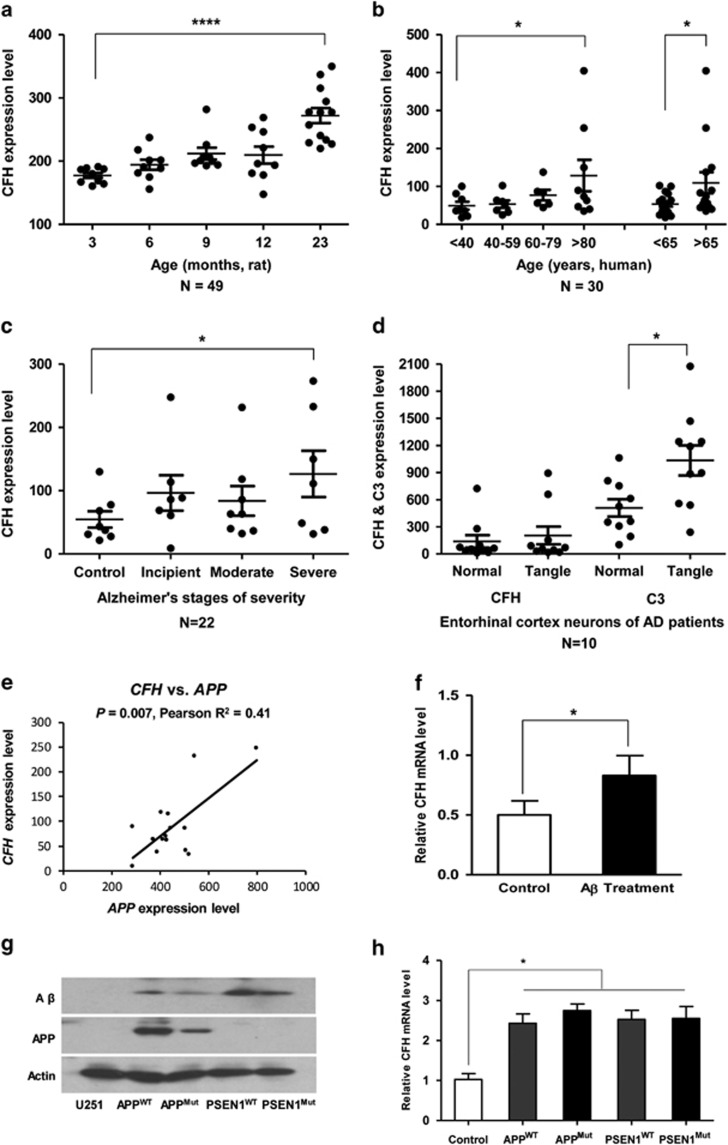
As we have observed a decrease of CFH expression levels in risk allele carriers, we further evaluated the alteration of CFH mRNA expression in aged and AD brains and AD cellular models. There was an increase of Cfh mRNA level with age in the rat hippocampus (Figure 3a). A similar pattern of upregulated CFH mRNA during aging was observed in human frontal cortex samples from 30 normal individuals of age 26–106 years (Figure 3b). The significant increase of CFH expression level with aging was confirmed in human prefrontal cortex using the BrainCloud data (Supplementary Figure 4). Moreover, CFH mRNA expression level increased in hippocampus as the severity of the disease worsened (Figure 3c). We also observed an increase of CFH mRNA level in entorhinal neurons containing neurofibrillary tangles compared with normal neurons, although the increase was not significant. The expression of C3, the central component of the complement system, was strongly elevated in tangled entorhinal neurons (Figure 3d). Considering its anti-inflammatory role, the increase of CFH may be the result of aging and balanced CFH level may have a protective effect on aging.
Figure 3.
CFH expression in aged and AD brains and cellular assays. (a) Cfh mRNA expression changes in the hippocampus from 49 Rattus norvegicus across the adult lifespan. (b) CFH mRNA expression of the postmortem frontal cortex from 30 normal individuals from 26 to 106 years of age. (c) CFH mRNA expression in brain hippocampus from 22 postmortem subjects with AD at different stages of severity. (d) CFH and C3 mRNA expression levels in entorhinal cortex neurons containing neurofibrillary tangles were increased relative to those of normal neurons from the same brain region in 10 mid-stage AD patients. (e) Correlation between CFH mRNA level (213800_at) and APP mRNA level (probe 211277_x_at) in hippocampus of AD patients (N=15) with incipient and moderate stages of severity. (f) Increase of CFH mRNA expression in U251 cells with Aβ1–42 treatment. (g) The APP and Aβ levels in cells with stable overexpression of APP (wild-type (APPwt) and APP-p.M671L mutant (APPMut)) and PSEN1 (wild-type (PSEN1wt) and PSEN1-p.M139V/M146L/H163R (PSEN1Mut). (h) CFH mRNA expression level was increased in cells with stable overexpression of APP and PSEN1. Data represent mean±SEM. *P<0.05, ****P<0.0001, Student's t-test.
In Aβ1–42-treated U251 cells, we observed a significant increase of CFH mRNA level (Figure 3f). Expression level of CFH was also increased in cells with stable overexpression of APP mutant (APPMut, APP-p.M671L) and PSEN1 mutant (PSEN1Mut, PSEN1-p.M139V/M146L/H163R; Figure 3h). Consistent with the results of cellular assays, CFH mRNA level was positively correlated with APP mRNA level in hippocampus of AD patients (Pearson R2=0.41, P=0.007; Figure 3e). This significant correlation disappeared in control sample or patient–control combined sample (Supplementary Figure 5). Taken together, our results indicated a protective role of increased CFH level in brain aging and AD development, whereas the CFH risk alleles were associated with lower CFH level, resulting in an insufficient protection of this immune regulator.
Discussion
Increased activity of the complement system has been reported to be involved in the initiation and development of AD (Crehan et al, 2012). Immune genes, especially complement genes, were identified as the top Alzheimer's susceptibility genes in Europeans (Bertram et al, 2007; Lambert et al, 2013). However, our analysis showed that these genes (CR1, CLU, CD33, and TREML2) had very weak effects in Han Chinese. Intriguingly, we found that CFH, the most important genetic factor for AMD (Klein et al, 2005), acts as an important AD susceptibility gene in Han Chinese patients and has multiple roles in AD pathology.
CFH Variants are Associated with Brain Changes and Confer Alzheimer's Susceptibility
By a comprehensive analysis of the CFH SNPs in Han Chinese with and without AD, and a meta-analysis of world-wide published data, we found that several SNPs, especially rs1061170 (a well-known causal risk SNP for AMD; Klein et al, 2005) and rs800292, showed robust associations with AD (Table 1). This result clarified the previous conflicting observations (Hamilton et al, 2007; Le Fur et al, 2010; Proitsi et al, 2012; Zetterberg et al, 2008). It is to be noted the risk allele C of rs1061170 presents with a marked regional distribution (7% in the East Asian Ancestry population, 28% in the African Ancestry population, 41% in the European Ancestry population; data from the 1000 genome (http://www.1000genomes.org; Abecasis et al, 2012) and this might account for the different patterns of association between different populations. The higher risk allele frequency in Europeans might interpret partially the higher prevalence of AMD (Wong et al, 2014) and AD (cf. Alzheimer's Disease International, World Alzheimer Report 2009: The Global Prevalence of Dementia, http://www.alz.co.uk/research/world-report-2009) in Europeans than in Asians, although the effect size of the risk allele was smaller in Europeans than in Asians.
Intriguingly, our neuroimaging analysis showed that the Alzheimer's risk alleles were associated with an increased right entorhinal thickness in young adults (Figure 1). The brain immune cell glia, the most abundant cells in brain, was previously reported to contribute to half of brain volume changes and would be overactive in neuro-inflammation (DiBattista et al, 2014). It is thus possible that the increase in entorhinal cortex thickness might be due to a deficit in CFH risk allele carriers to control neuro-inflammation in the brain, as CFH serves an anti-inflammatory component. In addition, it has been reported that increased entorhinal cortex volume during the brain development could indicate a deficit in neural efficiency (DiBattista et al, 2014; Gogtay et al, 2004). Although the molecular and cellular mechanisms responsible for the increased entorhinal thickness in young CFH risk allele carriers remained to be elucidated, this result is not unexpected as the entorhinal cortex has an essential role in AD (Khan et al, 2014). There are further lines of evidence supporting an enhanced entorhinal structure or activity in healthy adults with young age and a higher atrophy rate as the disease progresses for those risk allele carriers. For instance, healthy APOE ɛ4 carriers showed a thicker right entorhinal cortex as compared with the left hemisphere (Donix et al, 2013) and a thinner left entorhinal cortex in APOE ɛ4 carriers than in non-carriers could be identified in children and adolescents (Shaw et al, 2007). Meanwhile, APOE ɛ4 may lead to an increased activity but greater atrophy in right hemisphere in healthy young subjects (O'Dwyer et al, 2012). The Alzheimer's risk BDNF genotype (Val/Val of Val66Met) carriers had a thicker entorhinal thickness in early adult life and a higher rate of entorhinal atrophy in elderly (Voineskos et al, 2011). These observations indicated that young healthy individuals at risk may have altered entorhinal thickness and improved brain activity. It might reflect a compensatory hypothesis (Filippini et al, 2009) wherein disease risk individuals appear to require additional effort to achieve comparable performance levels to overcome potential preclinical neural dysfunction.
Indeed, our data showed that AD-related CFH variant rs800292 not only altered the brain structure (eg, entorhinal cortex) in early life, but also affected the atrophy rate of the entorhinal cortex, CSF tau and Aβ levels, and memory decline as the disease progresses (Figure 2). These results suggest that CFH is actively involved in the onset and development of AD by promoting structural and functional brain changes. These observations indicated a role of immune genes in neuroimaging alterations in early age.
Alteration of CFH Expression is Involved in AD
Consistent with previous reports that CFH protein has the potential to be a biomarker for AD (Hye et al, 2006, 2014) and the above genetic association results, we found that CFH mRNA level in the hippocampus increases with age, suggesting an active role of CFH in the brain aging process (Figure 3). Moreover, there was a positive correlation between CFH mRNA level and severity of AD (and APP mRNA level) in brain tissues, and this result was consistent with the previous finding of increased CFH protein in the AD brain (Honda et al, 2000; Strohmeyer et al, 2000, 2002). Note that there are some controversies regarding serum CFH level in AD: an increase of CFH level was observed in the serum of AD patients (Hye et al, 2006, 2014); however, serum CFH level was reported to be significantly downregulated in patients with AD and mild cognitive impairment (Gezen-Ak et al, 2013). The exact reason for this discrepancy remains unknown. Based on our results, we speculate that higher brain CFH levels may be related to AD development, supported by the observed increase of CFH mRNA levels in our cellular assays in response to Aβ1–42 treatment or with stable overexpression of APP mutant and PSEN1 mutant (Figure 3).
Implications of CFH in the Pathogenesis of AD
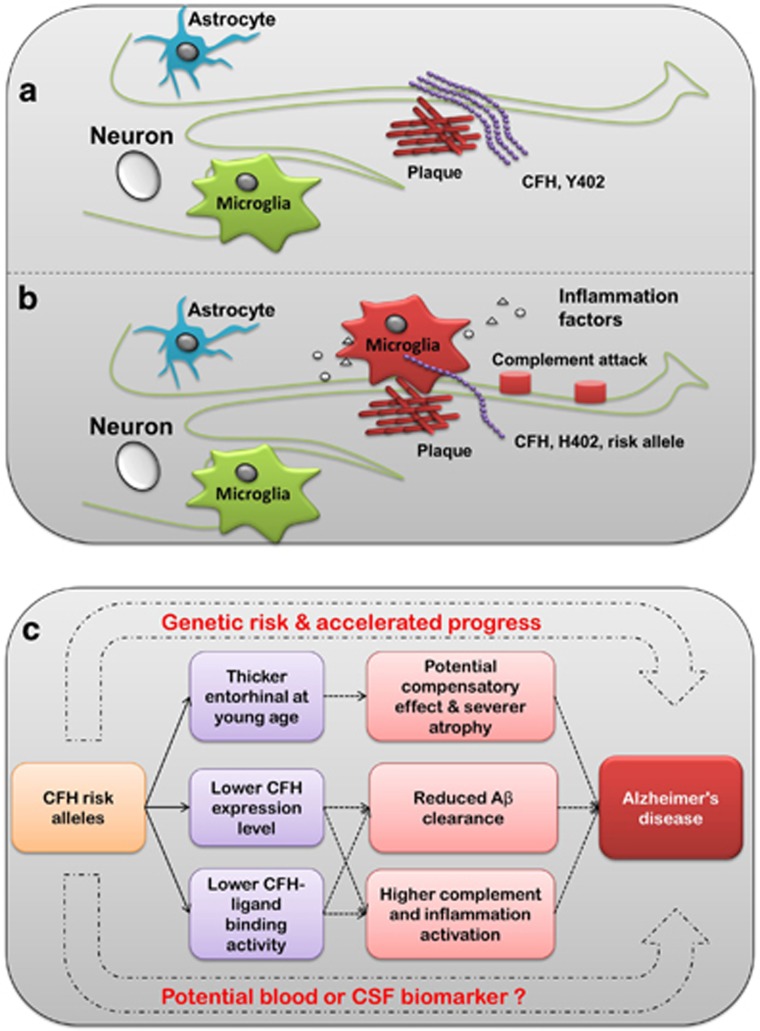
Recent studies have demonstrated that the risk allele 402H (allele C of rs1061170) interacts less well (compared with 402Y (allele T)) with the binding sites in CFH ligands within the macula, resulting in complement activation and inflammation that may contribute to the accumulation of drusen, thus leading to the initiation and progression of AMD (Clark et al, 2010). There may be a similar mechanism by which Y402H may contribute to progression of AD. It is now known that Aβ plaques and local inflammation are central to the pathogenesis of AD (Kamer, 2010; Wyss-Coray, 2006). In addition to the anti-inflammation role, CFH acts as an extracellular matrix component and interacts with a wide selection of ligands (Ferreira et al, 2010), such as the C-reactive protein (Strang et al, 2012), heparin (Bergamaschini et al, 2009), zinc (Suh et al, 2000), and sialic acid (Patel et al, 2006). All these ligands may be involved in the accumulation of senile plaque in the AD brain. It has been reported that the risk allele 402H presented a reduced affinity to these ligands (Ormsby et al, 2008). Hence, patients harboring risk allele 402H might bind less CFH in amyloid plaques, resulting in an impaired regulation of complement activation and local inflammation that may contribute to the accumulation of deposits and neuron loss in the development of AD (Figure 4). Furthermore, patients harboring the risk allele 402H might have decreased CFH levels in their brains according to our eQTL analysis. With impaired CFH levels and activity, extracellular deposition in the nidus may lead to inappropriate complement activation and thus contribute to the progression of clinical disease (Figure 4). Plaques in the AD brain, drusen in the eyes of patients with AMD, drusen-like deposits in the kidney of patients with kidney diseases, and even in the arteries of patients with atherosclerosis may also be the result of such processes. Further in vivo experiments using AD animal models are warranted to confirm our speculation.
Figure 4.
A simplified schematic profile for modeling the effect of CFH in AD. CFH acts as an extracellular matrix component and interacts with a wide range of ligands. The Alzheimer's risk allele 402H (rs1061170 C) was associated with lower CFH expression level and activity, thus presents a reduced affinity for these ligands. Patients harboring the risk allele 402H (b) may bind less CFH in the Aβ plaque compared with the wild-type (a), resulting in impaired regulation of complement activation (eg, membrane attack complex) and chronic local inflammation (inflammatory recruitment) in 402H carriers, which finally contributes to the accumulation of deposits and neuron loss during the development of AD. (c) Summary of the potential role of CFH in AD pathogenesis.
It is worth mentioning that CFH was identified as the most important genetic factor for AMD (Klein et al, 2005) and its SNPs associated with AD were found to be risk SNPs for AMD; are we really bringing these two diseases, which share similar pathological characteristics and environmental risk factors (Keenan et al, 2014; Sivak, 2013), much closer based on this study? Significant cognitive impairment and subsequent occurrence of AD in AMD patients have been reported (Baker et al, 2009; Kaarniranta et al, 2011; Pham et al, 2006; Sivak, 2013; Woo et al, 2012), although the conclusion remains controversial (Kaarniranta et al, 2011; Keenan et al, 2014). The sequential occurrence of brain and retinal damage needs to be clarified. Our current observations added more evidence to the notion that AMD and AD, to some extent, share some common pathological features such as chronic oxidative stress and inflammation, active complement involvement, and intra- and extracellular deposits (Kaarniranta et al, 2011). These two diseases likely represent two related but distinct parallel amyloidopathies that might benefit from common targeted therapeutic approaches.
The presence of the CFH risk alleles in AD and AMD poses an evolutionary paradox during human evolution, as the diseases may have negative effects on fitness, but the risk alleles have not been eliminated by natural selection and persist within global populations. Why the deleterious allele 402H (rs1061170 C) was retained in populations with a relatively high frequency? We performed a positive selection analysis on the CFH region to look for an evolutionary explanation of this phenomenon. Two online tools for detecting positive selection in human genome were used: Haplotter (Voight et al, 2006; using the HapMap data) and CMS viewer (http://www.broadinstitute.org/mpg/cmsviewer/, using the 1000 genome data; Supplementary Figure 6). Positive selection was observed in a region next to the CFH gene cluster in African and Asian populations, which contains the ASPM (abnormal spindle-like microcephaly associated) gene. This gene controls brain development and was reported to have evolved rapidly in human (Zhang, 2003). However, it is the CFH region itself that showed evidence of positive selection in the European (CEU) population, consistent with the fact that the CEU population also has a higher prevalence of AD and AMD. These observations indicate an evolutionary imprint on this region that may affect AD. The risk allele 402H may provide an advantageous effect against pathogens, which can evade complement attack by recruitment of CFH (Ferreira et al, 2010). Therefore, it is possible that the derived 402H allele was retained during evolution to limit immune evasion by potential pathogens. Because of this trade-off effect, the retained allele may contribute a deleterious effect on common diseases (such as AMD, AD, uremia, and atherosis), which commonly affect the elderly in our modern world.
In summary, our results showed that CFH may contribute to AD development by affecting neuroimaging endophenotypes and biomarkers as well as immune response. Most of all, CFH affects structural change of the entorhinal cortex in early life and atrophy rate during AD progression, indicating a multifaceted role of immune regulators. Population-based longitudinal analyses focusing on neuroimaging (eg, memory task-based functional MRI), biomarker indicators, and AD Braak stage progression in risk allele carriers might provide more support and benefit clinical research and applications. The biological implication of CFH in AD needs further characterization.
Funding and disclosure
This work was supported by the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (XDB02020003) and Sichuan Province (2014JZ0004). The authors declare no conflict of interest.
Acknowledgments
We thank Ian Logan for language editing and helpful comments and the three anonymous reviewers for their critical comments on the early version of the manuscript.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer's Association (2013). 2013 Alzheimer's disease facts and figures. Alzheimers Dement 9: 208–245. [DOI] [PubMed] [Google Scholar]
- Ansari M, McKeigue PM, Skerka C, Hayward C, Rudan I, Vitart V et al (2013). Genetic influences on plasma CFH and CFHR1 concentrations and their role in susceptibility to age-related macular degeneration. Hum Mol Genet 22: 4857–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Wang JJ, Rogers S, Klein R, Kuller LH, Larsen EK et al (2009). Early age-related macular degeneration, cognitive function, and dementia: the Cardiovascular Health Study. Arch Ophthalmol 127: 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschini L, Rossi E, Vergani C, De Simoni MG (2009). Alzheimer's disease: another target for heparin therapy. Sci World J 9: 891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE (2007). Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 39: 17–23. [DOI] [PubMed] [Google Scholar]
- Bi R, Zhang W, Yu D, Li X, Wang HZ, Hu QX et al (2015). Mitochondrial DNA haplogroup B5 confers genetic susceptibility to Alzheimer's disease in Han Chinese. Neurobiol Aging 36: 1604.e7. [DOI] [PubMed] [Google Scholar]
- Bi R, Zhao L, Zhang C, Lu W, Feng JQ, Wang Y et al (2014). No association of the LRRK2 genetic variants with Alzheimer's disease in Han Chinese individuals. Neurobiol Aging 35: 444.e5. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW (2004). Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA 101: 2173–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Perveen R, Hakobyan S, Morgan BP, Sim RB, Bishop PN et al (2010). Impaired binding of the age-related macular degeneration-associated complement factor H 402H allotype to Bruch's membrane in human retina. J Biol Chem 285: 30192–30202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT et al (2011). Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crehan H, Hardy J, Pocock J (2012). Microglia, Alzheimer's disease, and complement. Int J Alzheimers Dis 2012: 983640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista AM, Stevens BW, Rebeck GW, Green AE (2014). Two Alzheimer's disease risk genes increase entorhinal cortex volume in young adults. Front Hum Neurosci 8: 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donix M, Burggren AC, Scharf M, Marschner K, Suthana NA, Siddarth P et al (2013). APOE associated hemispheric asymmetry of entorhinal cortical thickness in aging and Alzheimer's disease. Psychiatry Res 214: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Beach TG, Ramsey KE, Grover A, Mastroeni D, Walker DG et al (2006). Gene expression correlates of neurofibrillary tangles in Alzheimer's disease. Neurobiol Aging 27: 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira VP, Pangburn MK, Cortes C (2010). Complement control protein factor H: the good, the bad, and the inadequate. Mol Immunol 47: 2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM et al (2009). Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A 106: 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gezen-Ak D, Dursun E, Hanagasi H, Bilgic B, Lohman E, Araz OS et al (2013). BDNF, TNFalpha, HSP90, CFH, and IL-10 serum levels in patients with early or late onset Alzheimer's disease or mild cognitive impairment. J Alzheimers Dis 37: 185–195. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC et al (2004). Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci USA 101: 8174–8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G, Proitsi P, Williams J, O'Donovan M, Owen M, Powell J et al (2007). Complement factor H Y402H polymorphism is not associated with late-onset Alzheimer's disease. Neuromolecular Med 9: 331–334. [DOI] [PubMed] [Google Scholar]
- Harris JA, Devidze N, Verret L, Ho K, Halabisky B, Thwin MT et al (2010). Transsynaptic progression of amyloid-beta-induced neuronal dysfunction within the entorhinal-hippocampal network. Neuron 68: 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG (2002). Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- Honda S, Itoh F, Yoshimoto M, Ohno S, Hinoda Y, Imai K (2000). Association between complement regulatory protein factor H and AM34 antigen, detected in senile plaques. J Gerontol A Biol Sci Med Sci 55: M265–M269. [DOI] [PubMed] [Google Scholar]
- Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, Byers HL et al (2006). Proteome-based plasma biomarkers for Alzheimer's disease. Brain 129: 3042–3050. [DOI] [PubMed] [Google Scholar]
- Hye A, Riddoch-Contreras J, Baird AL, Ashton NJ, Bazenet C, Leung R et al (2014). Plasma proteins predict conversion to dementia from prodromal disease. Alzheimers Dement 10: 799–807, e792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta K, Salminen A, Haapasalo A, Soininen H, Hiltunen M (2011). Age-related macular degeneration (AMD): Alzheimer's disease in the eye? J Alzheimers Dis 24: 615–631. [DOI] [PubMed] [Google Scholar]
- Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM et al (2009). Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci 29: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer AR (2010). Systemic inflammation and disease progression in Alzheimer disease. Neurology 74: 1157. [DOI] [PubMed] [Google Scholar]
- Karch CM, Goate AM (2015). Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan TD, Goldacre R, Goldacre MJ (2014). Associations between age-related macular degeneration, Alzheimer disease, and dementia: record linkage study of hospital admissions. JAMA Ophthalmol 132: 63–68. [DOI] [PubMed] [Google Scholar]
- Khan UA, Liu L, Provenzano FA, Berman DE, Profaci CP, Sloan R et al (2014). Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer's disease. Nat Neurosci 17: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C et al (2005). Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fur I, Laumet G, Richard F, Fievet N, Berr C, Rouaud O et al (2010). Association study of the CFH Y402H polymorphism with Alzheimer's disease. Neurobiol Aging 31: 165–166. [DOI] [PubMed] [Google Scholar]
- Li J, Cui Y, Wu K, Liu B, Zhang Y, Wang C et al (2015). The cortical surface area of the insula mediates the effect of DBH rs7040170 on novelty seeking. Neuroimage 117: 184–190. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J et al (2004). Gene regulation and DNA damage in the ageing human brain. Nature 429: 883–891. [DOI] [PubMed] [Google Scholar]
- O'Dwyer L, Lamberton F, Matura S, Tanner C, Scheibe M, Miller J et al (2012). Reduced hippocampal volume in healthy young ApoE4 carriers: an MRI study. PLoS One 7: e48895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormsby RJ, Ranganathan S, Tong JC, Griggs KM, Dimasi DP, Hewitt AW et al (2008). Functional and structural implications of the complement factor H Y402H polymorphism associated with age-related macular degeneration. Invest Ophthalmol Vis Sci 49: 1763–1770. [DOI] [PubMed] [Google Scholar]
- Patel D, Henry J, Good T (2006). Attenuation of beta-amyloid induced toxicity by sialic acid-conjugated dendrimeric polymers. Biochim Biophys Acta 1760: 1802–1809. [DOI] [PubMed] [Google Scholar]
- Pham TQ, Kifley A, Mitchell P, Wang JJ (2006). Relation of age-related macular degeneration and cognitive impairment in an older population. Gerontology 52: 353–358. [DOI] [PubMed] [Google Scholar]
- Proitsi P, Lupton MK, Dudbridge F, Tsolaki M, Hamilton G, Daniilidou M et al (2012). Alzheimer's disease and age-related macular degeneration have different genetic models for complement gene variation. Neurobiol Aging 33: 1843.e9. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM (2010). Alzheimer's disease. N Engl J Med 362: 329–344. [DOI] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al (2014). Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17: 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF et al (2001). Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet 69: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D et al (2007). Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol 6: 494–500. [DOI] [PubMed] [Google Scholar]
- Sivak JM (2013). The aging eye: common degenerative mechanisms between the Alzheimer's brain and retinal disease. Invest Ophthalmol Vis Sci 54: 871–880. [DOI] [PubMed] [Google Scholar]
- Strang F, Scheichl A, Chen YC, Wang X, Htun NM, Bassler N et al (2012). Amyloid plaques dissociate pentameric to monomeric C-reactive protein: a novel pathomechanism driving cortical inflammation in Alzheimer's disease? Brain Pathol 22: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeyer R, Ramirez M, Cole GJ, Mueller K, Rogers J (2002). Association of factor H of the alternative pathway of complement with agrin and complement receptor 3 in the Alzheimer's disease brain. J Neuroimmunol 131: 135–146. [DOI] [PubMed] [Google Scholar]
- Strohmeyer R, Shen Y, Rogers J (2000). Detection of complement alternative pathway mRNA and proteins in the Alzheimer's disease brain. Brain Res Mol Brain Res 81: 7–18. [DOI] [PubMed] [Google Scholar]
- Suh SW, Jensen KB, Jensen MS, Silva DS, Kesslak PJ, Danscher G et al (2000). Histochemically-reactive zinc in amyloid plaques, angiopathy, and degenerating neurons of Alzheimer's diseased brains. Brain Res 852: 274–278. [DOI] [PubMed] [Google Scholar]
- Thambisetty M, Hye A, Foy C, Daly E, Glover A, Cooper A et al (2008). Proteome-based identification of plasma proteins associated with hippocampal metabolism in early Alzheimer's disease. J Neurol 255: 1712–1720. [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006). A map of recent positive selection in the human genome. PLoS Biol 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineskos AN, Lerch JP, Felsky D, Shaikh S, Rajji TK, Miranda D et al (2011). The brain-derived neurotrophic factor Val66Met polymorphism and prediction of neural risk for Alzheimer disease. Arch Gen Psychiatry 68: 198–206. [DOI] [PubMed] [Google Scholar]
- Wang HZ, Bi R, Hu QX, Xiang Q, Zhang C, Zhang DF et al (2014). Validating GWAS-identified risk loci for Alzheimer's disease in Han Chinese populations. Mol Neurobiol. (doi:10.1007/s12035-014-9015-z). [DOI] [PubMed]
- Weiner MW, Aisen PS, Jack CR Jr, Jagust WJ, Trojanowski JQ, Shaw L et al (2010). The Alzheimer's disease neuroimaging initiative: progress report and future plans. Alzheimers Dement 6: 202–211 e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY et al (2014). Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2: e106–e116. [DOI] [PubMed] [Google Scholar]
- Woo SJ, Park KH, Ahn J, Choe JY, Jeong H, Han JW et al (2012). Cognitive impairment in age-related macular degeneration and geographic atrophy. Ophthalmology 119: 2094–2101. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T (2006). Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med 12: 1005–1015. [DOI] [PubMed] [Google Scholar]
- Zetterberg M, Landgren S, Andπersson ME, Palmer MS, Gustafson DR, Skoog I et al (2008). Association of complement factor H Y402H gene polymorphism with Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet 147B: 720–726. [DOI] [PubMed] [Google Scholar]
- Zhang DF, Wang D, Li YY, Yao YG (2014). Mapping genetic variants in the CFH gene for association with leprosy in Han Chinese. Genes Immun 15: 506–510. [DOI] [PubMed] [Google Scholar]
- Zhang J (2003). Evolution of the human ASPM gene, a major determinant of brain size. Genetics 165: 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yu JT, Li J, Wang C, Tan L, Liu B et al (2015). Bridging Integrator 1 (BIN1) genotype effects on working memory, hippocampal volume, and functional connectivity in young healthy individuals. Neuropsychopharmacology 40: 1794–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.