Abstract
tRNA is essential for translation and decoding of the proteome. The yeast proteome responds to stress and tRNA biosynthesis contributes in this response by repression of tRNA transcription and alterations of tRNA modification. Here we report that the stress response also involves processing of pre-tRNA 3′ termini. By a combination of Northern analyses and RNA sequencing, we show that upon shift to elevated temperatures and/or to glycerol-containing medium, aberrant pre-tRNAs accumulate in yeast cells. For pre-tRNAUAUIle and pre-tRNAUUULys these aberrant forms are unprocessed at the 5′ ends, but they possess extended 3′ termini. Sequencing analyses showed that partial 3′ processing precedes 5′ processing for pre-tRNAUAUIle. An aberrant pre-tRNATyr that accumulates also possesses extended 3′ termini, but it is processed at the 5′ terminus. Similar forms of these aberrant pre-tRNAs are detected in the rex1Δ strain that is defective in 3′ exonucleolytic trimming of pre-tRNAs but are absent in the lhp1Δ mutant lacking 3′ end protection. We further show direct correlation between the inhibition of 3′ end processing rate and the stringency of growth conditions. Moreover, under stress conditions Rex1 nuclease seems to be limiting for 3′ end processing, by decreased availability linked to increased protection by Lhp1. Thus, our data document complex 3′ processing that is inhibited by stress in a tRNA-type and condition-specific manner. This stress-responsive tRNA 3′ end maturation process presumably contributes to fine-tune the levels of functional tRNA in budding yeast in response to environmental conditions.
Keywords: tRNA, Saccharomyces cerevisiae, tRNA end processing, stress response
INTRODUCTION
The unicellular organism, budding yeast, is able to adapt to sudden changes in environmental conditions by multiple levels of regulation of cell organization and gene expression. Most of these changes are caused by altered transcription efficiency, but RNA processing and decay are also affected. tRNA, the focus of this work, is transcribed by RNA polymerase III (Pol III). Control of tRNA transcription is mediated by a general negative regulator, Maf1 (Pluta et al. 2001). Under unfavorable growth conditions such as nonfermentable carbon sources, high temperature, nitrogen starvation, or stationary phase, Maf1 is dephosphorylated and binds to Pol III, leading to repression of transcription (Boguta 2013). Nutrient limitation and stress also down-regulate tRNA transcription by phosphorylation of Pol III subunit C53 (Lee et al. 2012).
Transcription by Pol III, followed by maturation of initial transcripts, generates a variety of stable and unstable tRNA species. Generally the first step in tRNA maturation is 5′ leader cleavage by RNase P, a multisubunit complex consisting in yeast of nine proteins and a single RNA encoded by the RPR1 gene (Xiao et al. 2001). The second step is 3′ end processing, which is more complex and involves multiple enzymes. In S. cerevisiae it requires Trz1, an essential endonuclease also known as RNase Z (Skowronek et al. 2014), and Rex1, a 3′-5′ exonuclease (van Hoof et al. 2000; Ozanick et al. 2009). Under some conditions, Rrp6, a 3′-5′ exonuclease that is a component of the nuclear exosome, performs tRNA 3′ end processing. It is suspected that other exonucleases may also contribute to tRNA maturation (Yoo and Wolin 1997; Copela et al. 2008). tRNA 3′ end processing also involves Lhp1, a homologue of human La protein. Lhp1 stabilizes pre-tRNAs by direct binding to the oligo U 3′ termini, generated upon Pol III termination (Stefano 1984; Lin-Marq and Clarkson 1995). Binding of Lhp1 maintains the proper structure of tRNA precursors, protects them from exonucleolytic digestion, and stimulates endonucleolytic maturation (Van Horn et al. 1997; Yoo and Wolin 1997; Chakshusmathi et al. 2003; Copela et al. 2008). Finally, a complex of Lsm proteins is also involved in tRNA end processing, by interaction with Lhp1. Inactivation of a single Lsm protein from this complex resulted in the accumulation of stable 5′ and 3′ end-extended intron-containing and 5′ end-matured 3′ end-extended tRNA precursors (Kufel et al. 2002). Following removal of the 3′ trailer from the original transcript, the ubiquitous C, C, A nucleotides are added to the 3′ terminus by nucleotidyltransferase Cca1 (Aebi et al. 1990). After end-maturation and nuclear export, splicing of intron-containing precursors occurs in the cytoplasm (Hopper 2013). Throughout all the maturation steps, modifications are added; the modified nucleotides serve numerous functions including an important role in maintaining proper tRNA structure and stability (Phizicky and Hopper 2010).
Turnover of tRNA maturation intermediates is controlled by the nuclear exosome. An indication that pre-tRNAs are normally degraded by the nuclear exosome came from the observation that their levels are increased in mutants lacking the nuclear exosome subunit Rrp6, and cofactor, Trf4 (Kadaba et al. 2004; Copela et al. 2008). Moreover, 3′ turnover by the nuclear exosome appears to compete with 3′ processing (Maraia and Lamichhane 2011). The inefficient pre-tRNA degradation in the exosome mutant facilitates pre-tRNA maturation; conversely, inefficient 3′ end maturation directs pre-tRNAs to the exosome for degradation (Gudipati et al. 2012).
It has recently become appreciated that tRNA modifications are affected by stress (Chan et al. 2010, 2012; Preston et al. 2013; Alings et al. 2015; Damon et al. 2015; Han et al. 2015), but there are no reports regarding whether there is regulation of the activities responsible of pre-tRNA end-maturation. Here we investigated early steps of tRNA maturation for yeast grown at different temperatures (23°C–39°C) and in media with different carbon sources (glucose and glycerol). We show that the pattern of tRNA precursors is differently affected depending on tRNA isotypes. tRNA-specific down-regulation of 3′ end processing by respiratory growth and elevated temperature was assessed by Northern analyses of pre-tRNAUAUIle, pre-tRNATyr, and pre-tRNAUUULys. Alteration of pre-tRNA 3′ end processing by stress was further confirmed by sequencing of 3′ trailers of pre-tRNAUAUIle. Moreover, our data suggest that the magnitude of inhibition of 3′ end processing correlates with the severity of the stress. The molecular mechanism responsible for control of pre-tRNA 3′ processing by environmental conditions was investigated and a model presenting the effect of stress conditions on end maturation of pre-tRNAUAUIle and pre-tRNATyr is proposed.
RESULTS
Aberrant tRNA precursors accumulate under stress conditions
Previous studies show that Maf1 regulates tRNA transcription in yeast upon changes in carbon sources (Ciesla et al. 2007). Here, we explore the effects of carbon source and temperature shifts upon tRNA end processing.
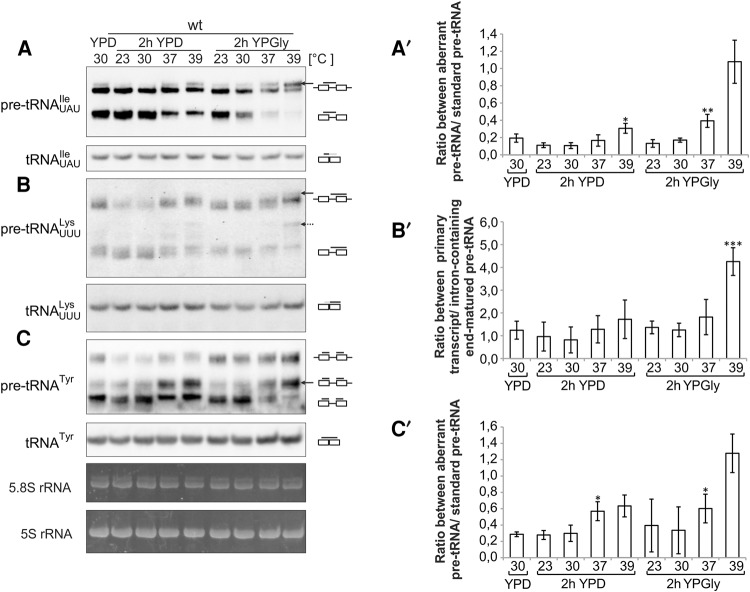
We first assessed intron-containing precursors of tRNAs by Northern analysis. RNAs were isolated from wild-type cells grown to exponential phase in rich glucose medium (YPD) and either shifted for 2 h to various temperatures (23°C, 30°C, 37°C, and 39°C) and/or to media with glycerol as the carbon source (YPGly). Each probe detected a characteristic set of processing intermediates for a given tRNA isotype (Fig. 1). For cells grown in YPD conditions, two major bands of tRNA precursors are usually detected which correspond, respectively, to the form containing the intron and extended 5′ and 3′ ends, generally assumed to be the primary transcript, and the end-matured, intron-containing form. Following the shift of cells to nonstandard conditions, the relative ratio of these two forms changes and unexpected aberrant forms become apparent. In cells exposed to elevated temperature, the amount of the end-matured, intron-containing form of pre-tRNAUAUIle decreased and a novel slower-migrating, intron-containing, end-extended form was detected (marked with the solid arrow, Fig. 1A). The relative abundance of this aberrant slowest migrating species increased when the stresses of high temperature and YPGly media were combined (Fig. 1A and A′).
FIGURE 1.
Patterns of tRNA precursors differ depending on growth conditions and tRNA isotype. (A–C) The wild-type yeast strain was grown in media with glucose as the carbon source (YPD) at 30°C to exponential phase and then shifted for 2 h to different temperatures (23°C–39°C) and/or to media containing glycerol as the carbon source (YPGly). RNAs were isolated and analyzed by Northern hybridization with probes specific to indicated tRNAs. Due to the high levels of mature tRNAs with respect to processing intermediates, different exposures were taken from blot hybridized with the same probe to assess pre-tRNAs and mature tRNAs in this and following figures. The major and minor forms of pre-tRNA precursors that accumulate under stress conditions are indicated by solid and dashed arrows, respectively. The following graphic representations for tRNA species were used in this and all subsequent figures: (–□–□–) tRNA precursor containing introns and unprocessed 5′ and 3′ends; (□–□–) tRNA precursor with intron and unprocessed 3′ end; (□–□) end-matured intron-containing pre-tRNA; (□□) mature tRNA. Positions of binding of probes to each form are marked with a black line above tRNA graphic representations. This and all succeeding Northern blots are representative of at least three independent biological replicates of experiments in which ethidium bromide–stained 5.8S and 5S rRNA serve as the loading controls. (A′–C′) Calculation of pre-tRNA ratios from at least three biological replicates. The bars represent the ratios between the levels of the following forms: for tRNAUAUIle the ratios between the slower migrating (aberrant) and faster migrating (standard) forms containing 5′ and 3′ termini and introns; for tRNAUUULys the ratio between end-extended intron-containing forms (primary transcript) and end-matured intron-containing form; and for tRNATyr the ratios of the aberrant 5′ processed 3′ end-containing (aberrant) form with respect to the end-matured intron-containing tRNA (standard). Standard deviation and P-value calculated as a two-tailed t-test are presented on the graphs. Asterisks above the bars designate, respectively, (*) P-value <0.05, (**) P-value <0.005, (***) P-value <0.0005.
Other tRNAs were also affected by temperature and carbon source stresses. For pre-tRNATyr, no additional slow-migrating, end-extended, intron-containing form was detected. However, an aberrant form of pre-tRNATyr that migrated faster than the initial transcript but slower than end-processed, intron-containing intermediates accumulated upon high temperature and carbon source stress (Fig. 1C and C′). For pre-tRNAUUULys, the precursors’ pattern resembled that observed for tRNAUAUIle; that is, at elevated temperatures and/or in glycerol-containing medium, a novel form of tRNAUUULys that migrated slower than the longest end-extended, intron-containing form detected in rich media at normal temperatures was identified (Fig. 1B, solid arrow). Moreover, as for tRNATyr under stress conditions, additional forms of pre-tRNAUUULys, which migrate between the primary transcript and the end-matured intron-containing forms, were detected (Fig. 1B, dashed arrow). However, when relative ratio between primary transcript and end-matured-intron containing forms was calculated, the only significant difference was observed in YPGly at 39°C (Fig. 1B′). Finally, subtle differences were visible for pre-tRNATrp and pre-tRNAPhe patterns upon the shift to a glycerol-containing medium (Supplemental Fig. S1). In sum, the results show that tRNA precursor forms are affected by environmental conditions, and the pattern of tRNA precursors changes in a tRNA-isotype and condition-specific manner.
Although the levels of initial tRNAs transcripts decreased upon stress treatments, the levels of mature tRNAs for given tRNAs, relative to 5S and 5.8S rRNA, as assessed by ethidium bromide staining, did not (Fig. 1, bottom panels). This is probably due to the fact that tRNAs are very stable and are not degraded during the 2 h exposure to stress and because rRNA transcription is likely coordinately decreased upon the stress conditions.
Alterations of pre-tRNA 3′ termini upon temperature and carbon source stress
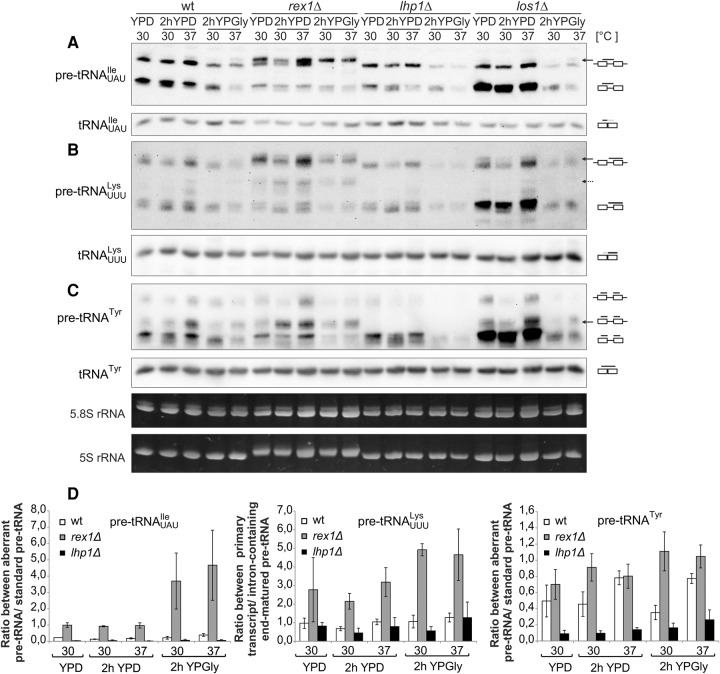
As the aberrant form of pre-tRNAUAUIle migrated slower than the accepted primary transcript, we explored the possibility that the slow form is the real primary transcript but that under favorable growth conditions it is rapidly partially processed. We compared the pre-tRNA species that accumulate in isogenic strains with mutations in genes that encode proteins that are involved in early steps of tRNA maturation. First, we examined the rex1Δ strain which lacks the 3′-5′ exoribonuclease that functions in maturation of the 3′ ends of 5S rRNA and some tRNAs (van Hoof et al. 2000; Copela et al. 2008; Ozanick et al. 2009; Skowronek et al. 2014). Second, we examined the lhp1Δ mutant lacking the Lhp1 protein that binds to the oligo U track of nascent pre-tRNAs. Lhp1 inhibits 3′ exonucleolytic processing of particular pre-tRNAs and thereby assists in endonucleolytic cleavage of their trailers (Yoo and Wolin 1997; Copela et al. 2008). As a control, we also employed the los1Δ mutant that has a defect in tRNA nuclear export resulting in accumulation of unspliced but end-matured pre-tRNAs (Hopper et al. 1980).
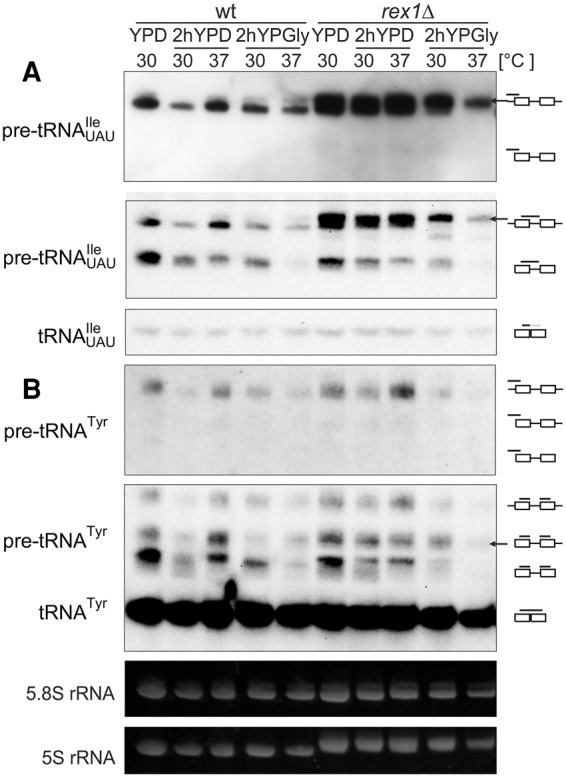
Analyses of pre-tRNAUAUIle in rex1Δ cells revealed the presence of an additional, slower migrating form even when grown in standard conditions (Fig. 2A,D), demonstrating, as anticipated, a defect in the 3′ end processing. This form of pre-tRNAUAUIle was similar in migration to the form that accumulated in wild-type cells subjected to carbon source and temperature stress and was the only form visible for rex1Δ in glycerol-containing media. The data indicate that the stress conditions might affect processing of pre-tRNAUAUIle 3′ trailers in a condition-specific manner. Also, the partially processed intron-containing form of pre-tRNATyr accumulated in rex1Δ cells (Fig. 2C,D). Moreover, both aberrant processing intermediates were observed for pre-tRNAUUULys in rex1Δ cells, even when grown in standard conditions (Fig. 2B,D). In contrast, aberrantly migrating forms of pre-tRNAUAUIle, pre-tRNAUUULys, and pre-tRNATyr were not detected in the lhp1Δ mutant (Fig. 2A–C). The observations suggest that the slowest migrating forms of pre-tRNAUAUIle and pre-tRNAUUULys detected in wild-type cells under stress conditions have extended 3′ ends that are protected by Lhp1, thus in lhp1Δ mutant these 3′ extensions are sensitive to exonucleases.
FIGURE 2.
Both exonucleolytic 3′ end maturation pathway and chaperone function of Lhp1 influence the precursor pattern of tested tRNAs. (A–C) Wild-type and indicated mutant strains were grown at 30°C to exponential phase and then shifted for 2 h to 30°C or 37°C and/or YPGly. RNA was isolated and analyzed by Northern hybridization with probes specific to tRNAUAUIle, tRNAUUULys, and tRNATyr. Ethidium bromide–stained 5.8S rRNA was used as a loading control. The major and minor forms of pre-tRNA precursors that accumulate under stress conditions are indicated by solid and dashed arrows, respectively. (D) Calculation of ratios between various forms of pre-tRNA (specified in the legend to Fig. 1) from at least three biological replicates.
As expected, the control los1Δ strain accumulated end-mature, intron-containing precursors for all tested tRNAs, but also the aberrant pre-tRNAUAUIle and pre-tRNATyr were present under stress conditions (Fig. 2A,C, solid arrows), further confirming that they are formed due to improper early maturation in the nucleus.
Processing of 3′ trailers consists of several steps, whereas the processing of 5′ leader in pre-tRNA is a single step catalyzed by the endoribonuclease, RNase P. We investigated whether the novel slow migrating form of pre-tRNAUAUIle observed under stress conditions contains a 5′ leader. Employing a probe specific to the 5′ leader of pre-tRNAUAUIle, both the conventional and the novel slow migrating forms were detected (Fig. 3A). Thus, the 5′ leader sequence is present in both forms of slow migrating pre-tRNAUAUIle. Therefore, the two forms of pre-tRNAUAUIle likely differ only at their 3′ termini and the faster migrating of the two intron-containing forms represents pre-tRNAUAUIle partially processed at the 3′ end. This partial 3′ end trimming occurring before 5′ processing is rapid under standard conditions and slowed-down under stress conditions.
FIGURE 3.
Species-specific tRNA processing intermediates under stress conditions: tRNAUAUIle contains the 5′ leader sequence whereas the aberrant tRNATyr species is processed at the 5′ end. Wild-type and rex1Δ mutant strains were grown in YPD at 30°C and then shifted for 2 h to 37°C and/or YPGly. RNA was isolated and analyzed by hybridization with probes specific to 5′ leaders of both tRNAUAUIle genes (A). and tRNATyr tY(GUA)D (B). Forms of tRNA precursors that accumulate under stress conditions are indicated by solid arrows. Hybridization with other specific probes recognizing mature and pre-tRNAs were employed as controls. The bottom panels present ethidium bromide–stained 5.8S and 5S rRNA used as the loading controls.
We extended the analyses to detection of the 5′ end of the aberrant form of pre-tRNATyr. We utilized short probe (17 nucleotides specific to the 5′ termini of pre-tRNATyr), which required optimization of conditions for specific detection of precursors containing 5′ leader sequence. Only primary transcripts and no other processing intermediates that accumulated under stress were observed (Fig. 3B). Therefore it appears that the aberrant form of pre-tRNATyr that accumulated under stress has been processed at the 5′ end and retains an unprocessed extended 3′ end. In support of this, we noted that a similarly migrating intermediate form of pre-tRNATyr accumulated under standard conditions in the rex1Δ mutant but was not detected in lhp1Δ cells (Fig. 2B). Altogether, the results of the Northern analysis indicated that the intermediates of tRNA processing which accumulate under stress differ at their 3′ ends and support the model that 3′ end processing of both, pre-tRNAUAUIle and pre-tRNATyr, is inhibited by stress and this inhibition involves Lhp1 binding to the 3′ termini.
We assessed whether the 3′ end-extended tRNA precursors observed under stress conditions could be further processed upon return to favorable growth conditions or instead are designated for degradation. Nonfunctional pre-tRNAs can be degraded by a nuclear TRAMP/exosome surveillance system (Gudipati et al. 2012; Schneider et al. 2012); therefore, we employed rrp6Δ mutant which impairs this quality control pathway. The fate of 3′-extended forms that accumulate in stress conditions (YPGly at 39°C) was monitored by Northern blotting at defined time points after transfer of cells back to standard conditions (YPD at 30°C) (Supplemental Fig. S2). The 3′ end-extended forms of tRNAUAUIle and pre-tRNATyr progressively decrease during incubation in YPD and the end-matured intron-containing form is generated. Moreover, the levels of aberrant forms are not increased in rrp6Δ mutant. The results suggest that the aberrant precursors that accumulate under stress are maturation intermediates that are rather not subjected to degradation but could be further processed when cells are shifted back to permissive conditions. However, due to continued transcription after the return to favorable growth conditions, we cannot exclude the contribution of newly synthetized transcripts to observed pre-tRNA pattern.
Defects of 3′ end processing of pre-tRNAUAUIle observed under stress conditions were further characterized by sequencing. RNA was isolated from wild-type cells grown under standard conditions or shifted to YPGly and incubated for 2 h at 37°C or 39°C. An oligonucleotide adapter was ligated to the 3′ ends of RNAs. The RNAs with ligated oligonucleotide were reverse transcribed using a primer specific to the adapter followed by PCR with primers specific to the intron of tRNAUAUIle and the adapter (Fig. 4A,B). Pre-tRNAUAUIle was chosen for analysis due to its relatively long 60 nucleotide (nt) intron, facilitating separation of PCR amplified products by gel electrophoresis. The products were then ligated into pGEM vector followed by transformation of E. coli. For each growth condition, we sequenced over 80 PCR fragments amplified from the resulting plasmids. All of the sequences were derived from pre-tRNAs prior to intron removal. As anticipated from the Northern analysis, the PCR fragments contained part of intron and 3′ exon but were heterogeneous at the 3′ end. End-matured pre-tRNAs became substrates of CCA nucleotidyltransferase and ended with C, CC, or CCA. Other sequences corresponded to unprocessed or partially processed 3′ ends (Fig. 4C; Supplemental Table S1).
FIGURE 4.
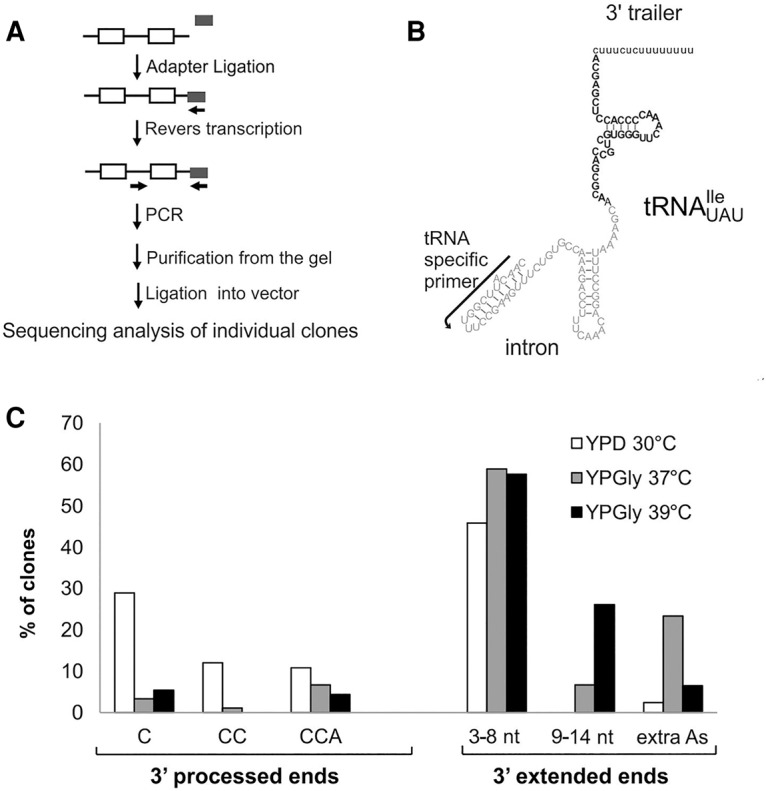
Sequencing of pre-tRNAUAUIle forms document 3′ end processing defects under stress conditions. (A) Outline of the strategy for linker ligation and sequencing of pre-tRNA 3′ ends. (B) Section of pre-tRNAUAUIle subjected to sequencing. (C) Summary of results obtained for pre-tRNAUAUIle sequencing of RNAs isolated from cells grown in YPD 30°C or shifted for 2 h to YPGly at 37°C or 39°C. Percent of pre-tRNAUAUIle molecules with 3′ processed or 3′ extended ends was calculated from the total number of sequenced clones that corresponded to genomic sequence and/or additional modification (A or CCA).
For cells grown in standard conditions, the majority of the clones (52%) represented 3′ end-matured pre-tRNAs; 29% of these had single C, 12% had CC, and 11% were fully modified to CCA (Fig. 4C; Supplemental Table S1). Under stress conditions, tRNA precursors had longer 3′ extensions that were not completely cleaved (Fig. 1A) and hence, fewer clones analyzed by sequencing contained the C, CC, or CCA (Fig. 4C).
The remaining 48% pre-tRNAUAUIle clones derived from cells grown in standard conditions possessed relatively short 3′ extensions of 3–8 nucleotides (Fig. 4C; Supplemental Table S1). For pre-tRNAUAUIle isolated from cells shifted to stress conditions, longer 3′ termini containing 9–14 nucleotides were detected (Fig. 4C). Moreover, the increase in the number of clones with longer trailers correlated with temperatures elevated from 30°C to 37°C and 39°C. The results show that the rate of 3′ processing of primary transcript is both temperature and carbon source dependent. The results are consistent with the hypothesis that 3′ processing of primary transcripts is very fast under standard conditions, but slows down under stress, resulting in the increase of longer forms of pre-tRNAUAUIle.
Additionally, mostly under stress conditions, some shorter pre-tRNAUAUIle have one or two extra adenines at their 3′ ends and oligoadenylation of longer form was detected only once among hundred sequenced clones (Fig. 4C; Supplemental Table S1). Low frequency of oligoadenylation supports our previous conclusion that the aberrant long pre-tRNAUAUIle form which accumulates under stress conditions is not subjected to degradation (Supplemental Fig. S2).
Rex1 is limiting for 3′ end trimming of particular pre-tRNAs under stress conditions
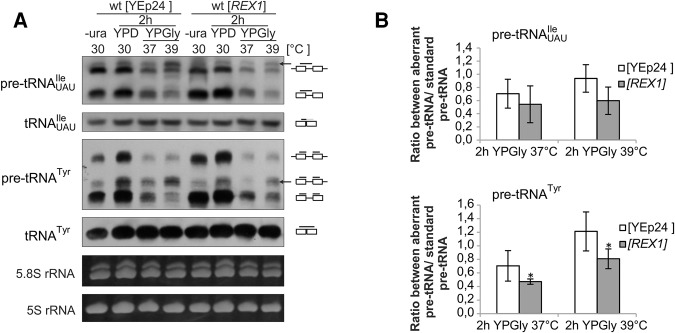
Decreased efficiency of tRNA 3′ end processing under stress could be caused by down-regulation of processing enzymes. To address this possibility, we assessed whether the levels of tRNA processing proteins respond to stress. Commercially available strains encoding tagged versions of Lhp1 and Trz1, grown at conventional or stress conditions, were examined by Western blotting. No effects on the levels of Lhp1 and Trz1 were observed (Supplemental Fig. S3). To explore whether REX1 expression is altered by stress, we created an HA-tagged Rex1 strain. However, Rex1-HA did not have normal catalytic activity as assessed by Northern analysis of tRNAs and 5S rRNA (Supplemental Fig. S4, forms marked with arrows).
To assess whether Rex1 level or function is reduced under stress conditions by another means, we monitored the ability of overexpressed Rex1 to suppress tRNA processing defects. We transformed yeast with a multicopy plasmid encoding the native Rex1 and analyzed tRNAs by Northern blotting (Fig. 5). As anticipated, this plasmid was functional because it complemented the rex1Δ defects of 5S rRNA and pre-tRNA processing (Supplemental Fig. S5, forms marked with arrows). When the blot was hybridized with the probe specific to pre-tRNAUAUIle, only a subtle effect of REX1 overexpression upon pre-tRNAUAUIle processing was observed (Fig. 5A) and differences were not statistically significant (Fig. 5B). However, the levels of aberrant form of pre-tRNATyr (intron-containing with unprocessed 3′ trailer) which accumulated under stress conditions were significantly decreased when Rex1 was overproduced (Fig. 5A). At the same time, the relative levels of end-mature intron-containing pre-tRNATyr increased upon REX1 overexpression (Fig. 5B).
FIGURE 5.
Effect of REX1 overexpression on 3′ end processing of pre-tRNAUAUIle and pre-tRNATyr under stress. (A) Wild-type strain transformed either with the multicopy vector or the vector containing REX1 was grown in minimal media to exponential phase and then shifted to YPD or YPGly at the indicated temperatures for 2 h. Isolated RNA was analyzed by Northern blotting with probes specific for tRNAUAUIle and tRNATyr. The bottom panels present ethidium bromide–stained 5.8S and 5S rRNA used as the loading controls. Forms of tRNA precursors that accumulate under stress conditions are indicated by solid arrows. (B) Calculation of RNA ratios from at least three biological replicates was performed as described in legend to Figure 1. Standard deviation and P-value calculated by a two-tailed t-test are presented on the graphs; (*) above the bars represent P-value = 0.05.
Additionally, we found that the examined defects of 3′ processing of pre-tRNAs were not suppressed by overproduction of Lhp1 or Trz1 (Supplemental Fig. S6). Northern analysis suggests further inhibition of 3′ processing by the excess of Lhp1 which probably limits the access of Rex1 and other nucleases to the 3′ end (Fig. 6A).
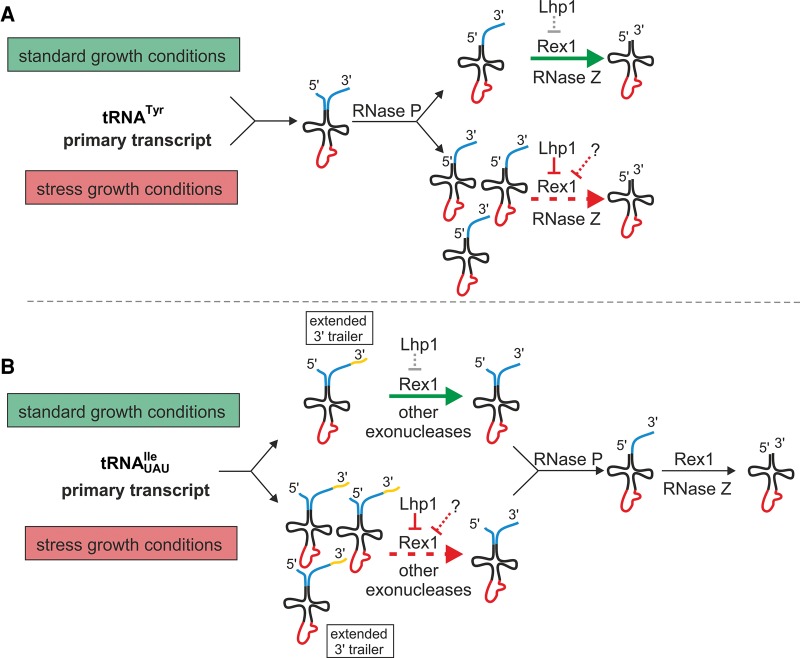
FIGURE 6.
Model presenting the effect of stress conditions on end-maturation of pre-tRNATyr and pre-tRNAUAUIle. (A) Under standard growth conditions, tRNATyr is first processed at the 5′ terminus by RNase P, then the 3′ end is matured by an exo- and/or endo-nucleolytic pathway. The step of 3′ removal is very fast under standard conditions (indicated by a green arrow), but under stress it is inhibited (marked with a red dashed arrow) by limited amount/activity of Rex1 and increased protection by Lhp1 and other putative factors. Thus, 5′-end-processed 3′-end-extended precursors of tRNATyr can be detected under standard conditions and they accumulate to higher levels under stress. (B) tRNAUAUIle has a longer primary transcript than previously assumed (3′ extension marked with yellow) and it is removed in two steps. The first step is very fast under standard conditions (indicated by a green arrow) and the 3′ end is partially trimmed before the 5′ end is removed. Inhibition of this step by stress due to a relative excess of Lhp1 and other putative factors (marked with a red dashed arrow) results in accumulation of pre-tRNAUAUIle unprocessed at both 3′ and 5′ termini and part of these accumulated pre-tRNAs have longer 3′ extensions. The second step of 3′ end removal is done in concert with 5′ end cleavage, thus precursors processed at one end are not detected.
These cumulative data provide compelling evidence in support of our conclusion that the pre-tRNA 3′ processing activity of Rex1 is limited under stress conditions.
DISCUSSION
The effect of repression of tRNA transcription under various types of stress in Maf1-dependent fashion has been thoroughly studied and well defined (Boguta 2013). The condition employed in this study—a nonfermentable carbon source together with an elevated temperature—is known to lower tRNA transcription (Ciesla et al. 2007). Also, the general level of tRNA modification was reported to change upon oxidative stress, DNA damage or hydroxyl peroxide and temperature (Chan et al. 2010, 2012; Alings et al. 2015). Furthermore, hypomodified tRNAs or tRNAs containing mutations that destabilize their structure were shown to be degraded at elevated temperatures via the RTD pathway (Chernyakov et al. 2008; Whipple et al. 2011). In addition, heat shock as well as methionine starvation, or oxidative stress, caused tRNA cleavage in the anticodon loop by Rny1, creating tRNA halves (Thompson et al. 2008; Thompson and Parker 2009). All those processes show different levels of response of cells to changes in environmental conditions.
However, there has not been a previous report that systematically examines the effects of growth conditions upon tRNA maturation, although differences in pre-tRNAs intermediates at elevated temperature were reported previously for some intron-containing yeast tRNA families (O'Connor and Peebles 1991; Kufel et al. 2002; Kufel and Tollervey 2003). Here, we employed Northern analyses and RNA sequencing to examine the effects of alterations of temperature and carbon source upon processing of several tRNA precursors.
Defects in tRNA maturation were detected by the accumulation of aberrant tRNA processing intermediates upon shift of cells to stress conditions—high temperature and/or medium with a nonfermentable carbon source. These intermediates can be categorized into two types. The first is represented by the slowest aberrantly migrating form that contains the 5′ leader, intron, and extended 3′ trailer. This form was most thoroughly examined for pre-tRNAUAUIle and its abundance was found to correlate with the severity of the stress. The second type of aberrant tRNA form observed for pre-tRNATyr at higher temperatures contained extended 3′ trailers and introns but processed 5′ termini. Both types of intermediates were detected for pre-tRNAUUULys when the stresses of high temperature and YPGly media were combined (Fig. 1B); however, these effects were subtle.
Similar aberrant precursor forms of pre-tRNAUAUIle and pre-tRNATyr were reported resulting from defects in 3′ end processing of pre-tRNA due to the rex1Δ mutation or to the combination of rex1Δ with partial inactivation of Trz1 endonuclease (Skowronek et al. 2014). Moreover, an earlier report detected slow migrating tRNAUAUIle and tRNAUUULys forms dependent on the presence of Lhp1 (Yoo and Wolin 1997). When the isogenic rex1Δ and lhp1Δ mutants were used as controls, we established that both types of the stress-related processing intermediates co-migrated with the precursors that are normally trimmed at the 3′ ends by Rex1. We thus propose that Rex1 activity is limiting under stress conditions. This idea is supported by partial suppression of the defect in tRNATyr maturation upon REX1 overexpression (Fig. 5A). Additionally, since accumulation of stress-related forms of pre-tRNAUAUIle and pre-tRNATyr was not detected in lhp1Δ mutant, we infer that Lhp1 is also involved in control of tRNA maturation by stress by protection of their 3′ extensions (Fig. 2). Protection by Lhp1 could be even stronger under the Pol III–repressive conditions when tRNA synthesis decreases leading to the increase of Lhp1:pre-tRNA ratio. Together, the results allow us to form a model in which defects of pre-tRNA processing observed under stress conditions are interconnected to inefficient processing of 3′ trailers (Fig. 6).
Formation of the 3′ end begins with termination of tRNA gene transcription in which intrinsic 3′ exonuclease activity is mediated by the C11 subunit of Pol III. Mutations in C11 that decrease cleavage activity resulted in increased 3′ oligo(U) length, but only by one or two nucleotides (Huang et al. 2005). Although it was not experimentally addressed, it is conceivable that stress conditions decrease the activity of endogenous C11. Thus, in addition to impaired 3′ processing, inefficient cleavage might have an impact upon the length of U tracts detected in 3′ trailers of tRNAUAUIle under stress conditions.
The C11 subunit forms a subcomplex C11/C53/C37 and altered activity of this complex results in read-through of termination signals (Huang et al. 2005; Landrieux et al. 2006; Rijal and Maraia 2013). The two tRNAUAUIle-encoding genes in the yeast genome possess the CTTTCTC sequence at their 3′ ends, followed by a tract of 9 or 10 T(s), respectively. Such long tracts of T(s) should function as strong transcription termination signals since 4–6 T(s) are sufficient for termination in eukaryotic cells (Hamada 2000; Braglia 2005). Theoretically, another T tract that could potentially serve for transcription termination is located about 50 nucleotides further downstream. Transcription terminated at this site would generate much longer transcripts than what was detected. Thus, transcription termination read-through appears not to cause the accumulation of slow migrating forms of tRNAUAUIle under stress conditions.
Interestingly, regardless of the growth conditions, for tRNAUAUIle we detect partial processing of 3′ trailer before the 5′ leader removal (Fig. 3A). Previous evidence has led to the generally accepted model that in yeast, pre-tRNA 5′ processing occurs prior to 3′ processing (Lee et al. 1991; O'Connor and Peebles 1991; Yoo and Wolin 1997). However, an exception was reported for tRNATrp for which the 3′ trailer is removed before the 5′ leader (Kufel and Tollervey 2003). The data reported here provide in vivo evidence supporting the model proposed by Copela et al. (2008), that is, partial 3′ end trimming can precede 5′ processing also for tRNAUAUIle (Fig. 6).
It is possible that the observed changes in processing of pre-tRNAUAUIle under stress conditions could be caused by lower availability of the enzymes involved in 3′ end processing. Pre-tRNAUAUIle has a relatively long trailer (up to 14 nt) and thus belongs to tRNA precursors which require both Trz1 and Rex1 for 3′ end processing (Ozanick et al. 2009; Skowronek et al. 2014). However, overexpression of REX1 is not sufficient to overcome the stress-induced defect in pre-tRNAUAUIle 3′ end processing that could be a result of increased protection of 3′ extension by Lhp1 (Fig. 5A). Especially since pre-tRNA levels are reduced under repressive conditions, a constant amount of Lhp1 (Supplemental Fig. S3) would lead to a larger fraction of pre-tRNAs to be bound by Lhp1. Moreover, LHP1 overexpression increases the accumulation of 3′ extended forms of pre-tRNAUAUIle (Supplemental Fig. S6). Another possible explanation for stress-mediated inhibition of 3′ trailer processing would be a structural change of pre-tRNAUAUIle that hinders enzyme access to the 3′ termini. In support of this notion, it has been reported that partial m5C modification at position 48 and m7G at position 64 can be introduced prior to 5′ end processing of pre-tRNAUAUIle (Ohira et al. 2009) and levels of these two modifications are increased under various stress conditions (Chan et al. 2010). It is conceivable that changes in folding/structure of pre-tRNAs could impact their 3′ end processing under stress. It has recently become appreciated that tRNA modifications that could alter the tRNA structure are affected by growth conditions. The effect of stress was reported for tRNA thiolation (Kamenski et al. 2007; Damon et al. 2015; Han et al. 2015), m5C methylation (Preston et al. 2013), and other modifications (Alings et al. 2015). It is worth noting that activities of at least two enzymes responsible for end processing, RNase P and RNase Z, depend not on substrate sequence but proper structure (Takaku et al. 2004; Mondragón 2013).
The form of pre-tRNATyr which accumulates under stress conditions is processed at its 5′ end. In this case, decreased activity of Rex1 exonuclease might account for inhibition of the processing of 3′ trailer. A role of endonucleolytic cleavage seems to be less probable because inactivation of Trz1 alone did not result in the accumulation of the tRNATyr intermediate that contained intron and 3′ trailer (Skowronek et al. 2014) and overproduction of Trz1 had no significant effect on the accumulation of aberrant pre-tRNA forms as well (Supplemental Fig. S6B).
Our study provides evidence for an environmental response of tRNA processing for several tRNA species highlighting the effects of temperature and/or carbon source stress. It is quite possible that additional processing steps in these and other tRNAs might be affected by various types of stress. It is known that effects of diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing (Kuhn et al. 2001; Pleiss et al. 2007; Bergkessel et al. 2011). Although the mechanism of adaptation and physiological function of the observed response require further clarification, our data document that the impact of environmental conditions also occurs for tRNA maturation.
MATERIALS AND METHODS
Strains, plasmids, and media
Yeast strains BY4741 (MATa his3Δ 1 leu2Δ 0 met15Δ0 ura3Δ0) and rex1Δ, lhp1Δ, and los1Δ derivatives were obtained from the yeast deletion collection (Open Biosystem). The plasmid harboring REX1 was obtained from M. Rosbach and is described in Abruzzi et al. (2007). YEp24 plasmid was used as the control vector.
YPD or YPGly complete media contained 1% yeast extract (Fisher Scientific), 1% peptone (BD), and 2% glucose (Fisher Scientific) or 2% glycerol (Sigma), respectively. Minimal medium (SC) contained 2% glucose and 0.67% yeast nitrogen base (BD). SC-ura and Sc-leu contained supplements required for growth (at 20 µg/mL each, except uracil or leucine, respectively).
LBA X-gal IPTG plates contained 1% bactotrypton, 0.5% yeast extract, 20 mg/mL Agar (all from BD), 0.5% NaCl, 1 mg/mL ampicillin, 1.5% X-Gal and, 0,8 M IPTG.
Northern blot analysis
Overnight yeast cultures were resuspended in fresh YPD media to OD600 = 0.15 and grown to exponential phase (OD600 = 0.5–0.8). The cells were collected by centrifugation and washed. Then the cultures were resuspended in YPGly media and/or shifted to different temperatures in the range of 23°C–39°C. After incubation under stress conditions for 2 h, the cells were harvested, washed, and resuspended in TSE buffer (0.01 M Tris, 0.01 M EDTA, 0.1 M NaCl, pH 7.5). TSE-saturated phenol was added to the samples and the mixture was incubated at 37°C with shaking for 1 h followed by 30 min incubation on ice, and then a second phenol extraction was performed, followed by ethanol precipitation for at least 1 h at −80°C. 3–5 μg of RNA was separated by electrophoresis on a 10% polyacrylamide, 8 M urea gel, transferred onto positively charged nylon membranes, and hybridized with DIG-labeled probes, as described previously (Wu et al. 2013). For some probes, the temperature of hybridization and washing protocol were modified to increase specificity of the signal. Some blots were developed using photographic films for higher resolution. Calculation of RNA signals was conducted using Total LabQuant software (Totallab).
For RNA hybridization, the following DIG-labeled oligonucleotides were used: JW0011 (tRNAUAUIle) 5′-GGCACAGAAACTTCGGAAACCGAATGTTGCTATAAGCACGAAGCTCTAACCACTGAGCTACA-3′; ND0060 (tRNAUUULys,) 5′-CTCCTCATAGGGGGCTCGAACCCCTGACATTTCGGTATCCTTGCTTAAGCAAATGCGCT-3′; SRIM15 (tRNATyr) 5′-GCGAGTCGAACGCCCGATCTCAAGATTTACAGTCTTGCGCCTTAAACCAACTTGGCTACC-3′; WHIT27 (tRNAUAUIle 5′ end) 5′-CTACACGAGCATTTTCGAAAGA-3′, tY(GUA)D (tRNATyr 5′ end of tY(GUA)D gene) 5′-GAGAGTCGATTTCTTGC-3′.
RNA 3′ adapter ligation and sequencing
700 ng of RNA from each sample was treated with DNase according to the manufacturer's instructions (TURBO DNase, Life Technologies) prior to ligation with Universal miRNA Cloning Linker (NEB). Ligation was conducted using truncated T4 RNA Ligase 2 (NEB) in the manufacturer's buffer supplemented with 12% PEG MW 8000; the reaction was incubated for 2 h at room temperature (RT) followed by phenol:chloroform extraction and ethanol precipitation. The entire sample was used for reverse transcription with Super Script II Reverse Transcriptase (Life Technologies), and performed according to the manufacturer's protocol with 2 pmol of primer specific to the linker (5′-ATTGATGGTGCCTACAG-3′) and incubation time for reverse transcription extended to 1.5 h. Negative controls were prepared using RNA without linkers for RT. cDNA was used for PCR with primers specific to the linker and pre-tRNA intron (5′-CAACATTCGGTTTCCG-3′). PCR product was analyzed by electrophoresis on 2.5% agarose gel. The proper size product was cut from the gel, purified with NucleoSpin Gel and PCR Clean-up (Macherey-Nagel), and ligated into pGEM-T Easy vector (Promega) followed by transformation of E. coli which was plated on LBA-X-gal IPTG selection plates. White colonies were selected and plasmids were isolated with QIAprep Spin Miniprep Kit (Qiagen) and sequenced at the laboratory of sequencing DNA and oligonucleotide synthesis IBB PAS. Sequencing results were analyzed with Sequence Scanner 2 (Applied Biosystem) and online Multi Align tool (Corpet 1988).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor J. Kufel for constructive discussion and critical reading of this manuscript. We thank Professor M. Rosbach for providing REX1 overexpression plasmid. We thank A.K.H. and M.B. laboratory members, especially Dr. R. Hurto, for valuable technical advice, Dr. E. Kramer, Dr. N. Dhungel, and Dr. T. Turowski for stimulating discussions, and D. Toczydłowska for laboratory assistance. We thank Dr. T. Ohira for scientific communication on pre-tRNAIle modifications and Dr. P. Nuc for technical advice. This study was supported by the Foundation for Polish Science within the International PhD Project “Studies of nucleic acids and proteins—from basic to applied research,” cofinanced by the European Union, Regional Development Fund Biology; National Science Center grant UMO-2012/04/A/NZ1/00052 to M.B. and grant 2013/09/N/NZ1/00129 to D.F.; and the National Institutes of Health, Medical Science grant GM27930 to A.K.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054973.115.
REFERENCES
- Abruzzi K, Denome S, Olsen JR, Assenholt J, Haaning LL, Jensen TH, Rosbash M. 2007. A novel plasmid-based microarray screen identifies suppressors of rrp6Delta in Saccharomyces cerevisiae. Mol Cell Biol 27: 1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, Abelson J. 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem 265: 16216–16220. [PubMed] [Google Scholar]
- Alings F, Sarin LP, Fufezan C, Drexler HCA, Leidel SA. 2015. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 21: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergkessel M, Whitworth GB, Guthrie C. 2011. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA 17: 1461–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M. 2013. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta 1829: 376–384. [DOI] [PubMed] [Google Scholar]
- Braglia P. 2005. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem 280: 19551–19562. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J 22: 6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. 2010. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. 2012. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 3: 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′–3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla M, Towpik J, Graczyk D, Oficjalska-Pham D, Harismendy O, Suleau A, Balicki K, Conesa C, Lefebvre O, Boguta M. 2007. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol Cell Biol 27: 7693–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL. 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damon JR, Pincus D, Ploegh HL. 2015. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol Biol Cell 26: 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild type cells. Mol Cell 48: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M. 2000. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J Biol Chem 275: 29076–29081. [DOI] [PubMed] [Google Scholar]
- Han L, Kon Y, Phizicky EM. 2015. Functional importance of 38 and 39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 21: 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA. 1980. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751. [DOI] [PubMed] [Google Scholar]
- Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. 2005. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol Cell Biol 25: 621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I. 2007. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell 26: 625–637. [DOI] [PubMed] [Google Scholar]
- Kufel J, Tollervey D. 2003. 3′-processing of yeast tRNATrp precedes 5′-processing. RNA 9: 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Allmang C, Verdone L, Beggs JD, Tollervey D. 2002. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol Cell Biol 22: 5248–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn KM, DeRisi JL, Brown PO, Sarnow P. 2001. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol Cell Biol 21: 916–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. 2006. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J 25: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR. 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol 11: 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moir RD, McIntosh KB, Willis IM. 2012. TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol Cell 45: 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Marq N, Clarkson SG. 1995. A yeast RNA binding protein that resembles the human autoantigen La. J Mol Biol 245: 81–85. [DOI] [PubMed] [Google Scholar]
- Maraia RJ, Lamichhane TN. 2011. 3′ Processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA 2: 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón A. 2013. Structural studies of RNase P. Annu Rev Biophys 42: 537–557. [DOI] [PubMed] [Google Scholar]
- O'Connor JP, Peebles CL. 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol 11: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira T, Miyauchi K, Sakaguchi Y, Suzuki T, Suzuki T. 2009. Precise analysis of modification status at various stage of tRNA maturation in Saccharomyces cerevisiae. Nucleic Acids Symp Ser 2004: 301–302. [DOI] [PubMed] [Google Scholar]
- Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT. 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res 37: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. 2007. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell 27: 928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M. 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol 21: 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, D'Silva S, Kon Y, Phizicky EM. 2013. tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae. RNA 19: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal K, Maraia RJ. 2013. RNA polymerase III mutants in TFIIFα-like C37 that cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res 41: 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. 2012. Transcriptome-wide analysis of exosome targets. Mol Cell 48: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek E, Grzechnik P, Späth B, Marchfelder A, Kufel J. 2014. tRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA 20: 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154. [DOI] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M. 2004. The N-terminal half-domain of the long form of tRNase Z is required for the RNase 65 activity. Nucleic Acids Res 32: 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R. 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. 1997. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3: 1434–1443. [PMC free article] [PubMed] [Google Scholar]
- Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev 25: 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang H-Y, Hopper AK. 2013. A rapid and sensitive non-radioactive method applicable for genome-wide analysis of Saccharomyces cerevisiae genes involved in small RNA biology. Yeast Chichester Engl 30: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Houser-Scott F, Engelke DR. 2001. Eukaryotic ribonuclease P: increased complexity to cope with the nuclear pre-tRNA pathway. J Cell Physiol 187: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.