Abstract
The zinc finger protein tristetraprolin (TTP) promotes translation repression and degradation of mRNAs containing AU-rich elements (AREs). Although much attention has been directed toward understanding the decay process and machinery involved, the translation repression role of TTP has remained poorly understood. Here we identify the cap-binding translation repression 4EHP-GYF2 complex as a cofactor of TTP. Immunoprecipitation and in vitro pull-down assays demonstrate that TTP associates with the 4EHP-GYF2 complex via direct interaction with GYF2, and mutational analyses show that this interaction occurs via conserved tetraproline motifs of TTP. Mutant TTP with diminished 4EHP-GYF2 binding is impaired in its ability to repress a luciferase reporter ARE-mRNA. 4EHP knockout mouse embryonic fibroblasts (MEFs) display increased induction and slower turnover of TTP-target mRNAs as compared to wild-type MEFs. Our work highlights the function of the conserved tetraproline motifs of TTP and identifies 4EHP-GYF2 as a cofactor in translational repression and mRNA decay by TTP.
Keywords: AU-rich elements, tristetraprolin, translation repression, mRNA turnover, ARE-mediated decay
INTRODUCTION
The translation and degradation of mRNAs are central, interrelated steps in the control of gene expression. Both processes are impacted by general mRNA-associated factors. The cap-binding translation initiation complex eIF4F serves to promote translation initiation, while preventing access to the mRNA cap for the DCP2 decapping complex (von der Haar et al. 2004). Cytoplasmic poly(A)-binding protein (PABPC) stimulates translation initiation while impacting mRNA deadenylation through repression or stimulation of deadenylase complexes (Goss and Kleiman 2013). Consistent with this relation between translation initiation and mRNA stability, many RNA-binding proteins (RBPs) that promote mRNA degradation also repress translation initiation (Wharton et al. 1998; Pillai et al. 2004; Pfeiffer and Brooks 2012). However, the underlying mechanisms remain poorly understood.
A subset of translation repressors inhibits translation at the initiation step by interfering with the eIF4F complex. This includes eIF4E-binding proteins (4EBPs) that bind to eIF4E in competition with eIF4G, thereby repressing translation initiation by inhibition of eIF4F complex formation (Pause et al. 1994; Marcotrigiano et al. 1999). 4E-T is another eIF4E-binding protein, which inhibits translation initiation by a less well-defined mechanism (Dostie et al. 2000). Furthermore, a class of Saccharomyces cerevisiae RGG-domain proteins associate with eIF4G to represses translation (Rajyaguru et al. 2012). Another factor that interferes with cap-dependent translation is 4EHP (also known as eIF4E2). 4EHP is a homolog of eIF4E and is thought to repress translation by competition with the eIF4F complex for binding to the mRNA cap (Rom et al. 1998; Joshi et al. 2004). Drosophila 4EHP was identified as a cofactor of the RNA-binding protein Bicoid required for translation repression of caudal mRNA (Cho et al. 2005). Mammalian 4EHP has been reported to form translation repression complexes with GYF1/GYF2 (also known as GRB10 interacting GYF protein 1/2) cofactors (Morita et al. 2012), but mRNA-specific recruitment of 4EHP-GYF complexes has not been previously identified in mammals.
Tristetraprolin (TTP; also known as ZFP36 or Tis11) is an RBP that represses translation and activates degradation of mRNAs containing 3′ UTR AU-rich elements (AREs) (Brooks and Blackshear 2013). TTP plays an important role in attenuating cytokine expression during immune responses via repression and degradation of ARE-containing cytokine mRNAs (Carballo et al. 1998, 2000; Stoecklin et al. 2008). In the mouse, TTP knockout results in auto- and hyperimmunity due to overproduction of the cytokine TNFα, which is produced from an ARE-containing mRNA (Taylor et al. 1996). TTP binds AREs through its tandem Zn-finger RNA-binding domain, and recruits the DCP2 decapping complex (Fenger-Gron et al. 2005; Lykke-Andersen and Wagner 2005), the CCR4-NOT deadenylase complex (Lai et al. 2003; Lykke-Andersen and Wagner 2005; Sandler et al. 2011; Fabian et al. 2013), as well as exonucleases (Chen et al. 2001; Lehner and Sanderson 2004; Lykke-Andersen and Wagner 2005). The activity of TTP is controlled through phosphorylation by the p38-MK2 pathway, which inhibits recruitment of the CCR4-NOT deadenylase complex and prevents cytokine mRNA degradation during early stages of an immune response (Chrestensen et al. 2004; Stoecklin et al. 2004; Hitti et al. 2006; Clement et al. 2011). In addition to activation of mRNA decay, TTP also promotes translation repression, which appears to be the dominant mechanism of repression by TTP under certain conditions (Schott et al. 2014). The helicase DDX6 (also called Rck/p54) was recently implicated in translation repression by TTP (Qi et al. 2012), but the specific mechanism of TTP-mediated translation repression has remained poorly understood.
The RNA-binding zinc finger domain and a C-terminal CCR4-NOT interaction motif (CIM) are highly conserved regions of TTP. Another evolutionary conserved feature of TTP is its tetraproline (PPPPG) motifs, which served as the basis for the naming of the protein (Lai et al. 1990). However, the functional significance of these motifs has remained unknown. Two paralogs of TTP exist in the human genome, BRF1 (also known as ZFP36L1 and Tis11b) and BRF2 (ZFP36L2, Tis11d), which, like TTP, promote degradation of ARE-containing mRNAs (Ciais et al. 2004; Hodson et al. 2010; Zhang et al. 2013). These proteins share the conserved zinc finger and CIM domains with TTP, but lack the tetraproline motifs characteristic of TTP.
In this study, we identify the translation repression complex 4EHP-GYF2 as a TTP cofactor. Our interaction studies in vitro and in cells demonstrate that TTP associates with the 4EHP-GYF2 complex via direct interaction with the GYF2 subunit. Mutational studies reveal that the interaction occurs through conserved tetraproline motifs of TTP. Mutation of TTP tetraproline motifs and knockout of 4EHP in mouse embryonic fibroblasts resulted in deficiencies in TTP-mediated mRNA repression at both the translational and mRNA turnover levels. These findings are consistent with a role of 4EHP in TTP-mediated translation repression as was very recently reported (Tao and Gao 2015). Moreover, our study identifies GYF2 as the factor that bridges 4EHP to TTP via interaction with the characteristic TTP tetraproline motifs, and suggests that the 4EHP-GYF2 complex links translation repression by TTP with mRNA decay.
RESULTS
The 4EHP-GYF2 translation repression complex associates with TTP
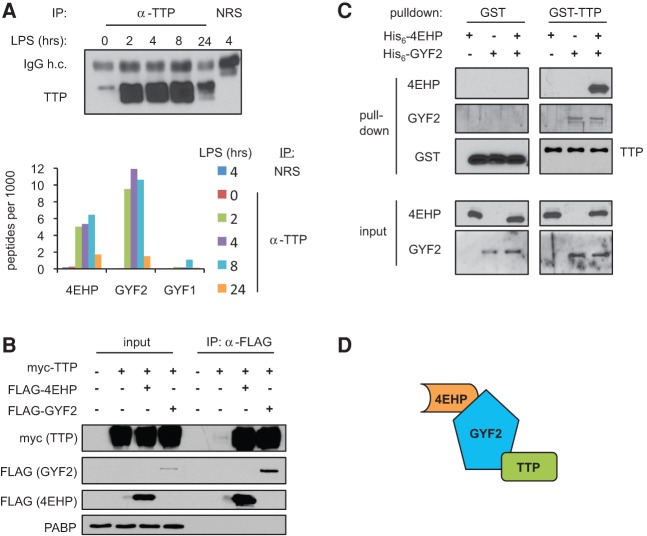
To identify candidate TTP cofactors, we performed immunoprecipitation (IP) against endogenous TTP at different time points during a lipopolysaccharide (LPS)-stimulated innate immune response in mouse RAW264.7 macrophages and subjected samples to liquid chromatography followed by tandem mass spectrometry (LC-MS/MS). IPs were performed in the presence of RNase A to prevent copurification of proteins that associate with TTP in an RNA-dependent manner. Anti-TTP IP prior to LPS induction where TTP levels are low (t = 0), and IP using normal rabbit serum (NRS) served as negative controls (Fig. 1A). As expected, we observed all components of the CCR4-NOT deadenylase complex in association with TTP, with the exception of NOT4, which is often absent from CCR4-NOT complexes (Supplemental Fig. S1; Lau et al. 2009). In addition, multiple 14-3-3 proteins and hnRNP F were also detected in association with TTP consistent with previous observations (Johnson et al. 2002; Chrestensen et al. 2004; Reznik et al. 2014) as were subunits of phosphatase 2A (PP2A), consistent with previous reports of TTP dephosphorylation by PP2A (Sun et al. 2007). Intriguingly, in addition to previously identified TTP-associated factors, our LC-MS/MS analysis identified the eIF4E homolog 4EHP and the 4EHP-associated GYF-domain protein GYF2 in association with TTP (Fig. 1A). The GYF2-paralog GYF1 was also found associated with TTP but only at modest levels. These associations were all specific to TTP as they were minimal in both of the negative control IPs (Fig. 1A; Supplemental Fig. S1). Not all factors known to complex with TTP were captured by this approach; for example, decapping and exosome components, previously observed in complex with TTP by co-IP (Chen et al. 2001; Lehner and Sanderson 2004; Lykke-Andersen and Wagner 2005), were not detected above the background in our IP LC-MS/MS data.
FIGURE 1.
TTP interacts with the 4EHP-GYF2 complex. (A, top) Western blot for TTP in samples immunoprecipitated with anti-TTP or normal rabbit serum (NRS) from mouse macrophage RAW264.7 cells treated with lipopolysaccharide (LPS) for various lengths of time as indicated. (IgG h.c.) IgG heavy chain. (Bottom) Graph showing the number of peptides for 4EHP, GYF2, and GYF1 detected per 1000 of total detected peptides in LC-MS/MS analyses of the IPs. (B) Western blots of input and anti-Flag IPs from HEK293T cells cotransfected with myc-TTP and Flag-4EHP or Flag-GYF2. Lysates were treated with RNase A prior to IP. (C) Western blots monitoring His6-4EHP and His6-GYF2 interaction with GST or GST-TTP in in vitro pull-down assays using glutathione Sepharose beads. GST and GST-TTP were detected using anti-GST; 4EHP and GYF2 were detected using anti-4EHP and anti-GYF2, respectively. (D) Proposed model of interaction between TTP and the 4EHP-GYF2 complex. TTP directly binds GYF2, which is known to interact with 4EHP.
TTP interacts with the 4EHP-GYF2 complex via a direct interaction with GYF2
To test whether the 4EHP-GYF2 complex serves as a cofactor for TTP in mRNA repression, we first sought to further characterize the association between TTP and the 4EHP-GYF2 complex. Of the two GYF proteins, we focused entirely on GYF2 given the low level of peptide recovery for GYF1 in our IP-LC-MS/MS assays. Consistent with our IP-LC-MS/MS data, myc-tagged mouse TTP could be observed to co-purify in an RNA-independent manner with Flag-tagged mouse 4EHP and GYF2 when transiently coexpressed in human embryonic kidney (HEK) 293T cells (Fig. 1B).
To test whether the interaction between TTP and 4EHP-GYF2 is direct, we produced glutathione-S-transferase (GST)-tagged mouse TTP and His6-tagged mouse 4EHP and GYF2 recombinantly in Escherichia coli for use in in vitro pull-down assays. This uncovered an interaction between TTP and GYF2 that was independent of 4EHP (Fig. 1C). A weak interaction could also be observed between TTP and 4EHP in some experiments, but this interaction was strongly enhanced by GYF2 (Fig. 1C; Supplemental Fig. S2). Therefore, complex formation between TTP and 4EHP-GYF2 is mediated primarily by a direct interaction between TTP and GYF2 with a possible minor contribution from an additional TTP-4EHP interaction (Fig. 1D).
GYF2 interacts with TTP via TTP tetraproline motifs
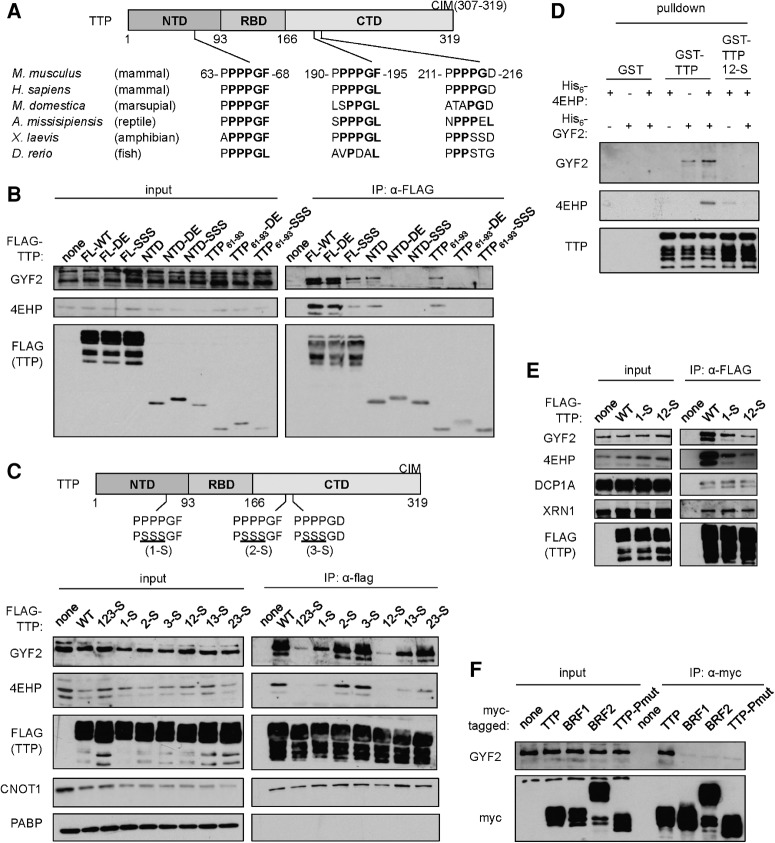
Next, to map the domain(s) of TTP important for interaction with 4EHP-GYF2, we transiently expressed domain truncation mutants of mouse TTP in HEK293T cells and monitored for effects on association with 4EHP and GYF2 by co-IP. The N-terminal domain (NTD) of TTP was necessary and sufficient for 4EHP and GYF2 association, as deletion of the TTP NTD (ΔNTD) resulted in loss of 4EHP and GYF2 association, whereas the TTP NTD alone retained 4EHP-GYF2 association (Fig. 2). The CIM sequence of TTP, responsible for interaction with the CCR4-NOT complex (Fabian et al. 2013), did not contribute to 4EHP-GYF2 binding. Further deletion analyses revealed that the last 33 amino acids of the TTP NTD (TTP61–93) are sufficient for association with 4EHP and GYF2 (Supplemental Fig. S3).
FIGURE 2.
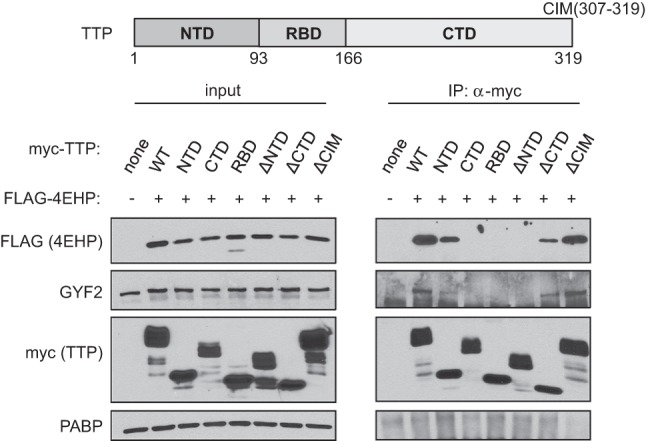
The TTP N-terminal domain is necessary and sufficient for association with 4EHP and GYF2. Western blots for indicated factors in input and anti-myc IP fractions from RNase A-treated extracts of HEK293T cells transiently expressing indicated myc-tagged TTP variants and Flag-tagged 4EHP. A schematic of TTP is shown at the top indicating the N-terminal domain (NTD), the RNA-binding domain (RBD), the C-terminal domain (CTD), and the CNOT-interacting motif (CIM).
GYF domains are known as protein interaction domains with specificity for the amino acid sequence PPPGφ, with φ representing a hydrophobic residue (Freund et al. 1999; Kofler et al. 2005). Mammalian TTP contains three tetraproline motifs, two of which are followed by GF or GL, matching the PPPGφ consensus (Fig. 3A). Sequence alignment of mammalian TTP with TTP from other vertebrates revealed strong conservation of a PPPGφ motif overlapping with the first tetraproline motif, which is located near the C-terminal end of the TTP NTD (aa 63–68), within the minimal region of TTP associating with 4EHP-GYF2 (Fig. 3A). Tetraproline motif 2 residing in the TTP CTD is less conserved but also matches a PPPGφ consensus in many vertebrates including human and mouse, whereas tetraproline motif 3 is poorly conserved and fails to match a PPPGφ consensus (Fig. 3A). Mutation of the conserved prolines of tetraproline motif 1 to serines dramatically decreased 4EHP and GYF2 association with full-length mouse TTP as well as with the TTP NTD and TTP61–93 (Fig. 3B). Mutation of the GF sequence that follows the tetraprolines to DE reduced association of the TTP NTD and TTP61–93 with 4EHP-GYF2, but had little effect in the context of full-length TTP, suggesting that this mutation is less disruptive to the interaction.
FIGURE 3.
TTP interacts with GYF2 through tetraproline motifs 1 and 2. (A) Sequence alignment of the tetraproline motifs of mouse TTP with corresponding regions of TTP from other vertebrates. Amino acids matching the PPPGφ GYF-binding consensus are highlighted in bold. (B) Western blots for proteins indicated on the left in input and anti-Flag IP samples from RNase A-treated extracts of HEK293T cells transiently expressing indicated Flag-tagged TTP variants. The first PPPPGF motif of TTP was mutated to PSSSGF (SSS) or PPPPDE (DE) in the contexts of full-length TTP (FL), the TTP NTD, or the 33 amino acids surrounding the motif (TTP61–93). (C) Same as panel B, with each of the three tetraproline motifs of Flag-tagged TTP mutated as indicated in the schematic above (1-S, 2-S, 3-S), and all combinations of those mutations. (D) Western blot for an in vitro pull-down assay similar to Figure 1C, with the addition of GST-tagged 12-S mutant of TTP. (E) Western blots of input and anti-Flag IP samples similar to panel C. (F) Western blots of input and anti-myc IP samples from RNase A-treated extracts of HEK293T cells transiently expressing myc-tagged mouse TTP, BRF1, or BRF2. A mutant version of mouse TTP with all prolines of the first tetraproline motif and the immediate downstream four prolines replaced with alanines (Pmut) was also included.
Combining P to S mutations in tetraproline motifs 1, 2, and 3 revealed that motif 2 contributes to 4EHP-GYF2 association, as mouse TTP containing mutations in both of motifs 1 and 2 showed decreased association with 4EHP-GYF2 as compared with TTP mutated in motif 1 alone (Fig. 3C). In contrast, we observed no contribution from motif 3, consistent with the absence of a hydrophobic residue following the PPPG sequence in this motif (Fig. 3A). The importance of TTP tetraproline motifs 1 and 2 for the interaction with GYF2 was confirmed in vitro, as P to S mutations in these motifs resulted in loss of GYF2 interaction (Fig. 3D). In contrast, tetraproline mutations did not impair association of TTP with previously identified mRNA decay TTP cofactors CNOT1 (Fig. 3C), DCP1A, and XRN1 (Fig. 3E). Consistent with the tetraproline motifs of TTP serving as the primary site of interaction with the 4EHP-GYF2 complex, we observed little association of GYF2 with TTP paralogs BRF1 and BRF2, which lack PPPGφ motifs (Fig. 3F).
Tetraproline motifs 1 and 2 and 4EHP contribute to TTP-mediated mRNA repression
To test the functional importance of TTP-GYF2-4EHP complex formation, we turned to mouse embryonic fibroblasts (MEFs), which express TTP endogenously upon serum stimulation. First, we took advantage of TTP knockout (TTP−/−) MEFs (Lai et al. 2006) to compare the ability of wild-type and mutant TTP proteins to repress an ARE-containing luciferase reporter mRNA. Wild-type or mutant TTP was transiently coexpressed in the TTP−/− MEFs with a firefly luciferase reporter containing the ARE of human GM-CSF mRNA. Renilla luciferase lacking an ARE served as a normalization control. As expected, wild-type TTP greatly repressed firefly luciferase expression (Fig. 4A; Supplemental Fig. S4). Measurement of mRNA levels revealed that repression of the luciferase reporter, for reasons that are unknown, occurs entirely at the translational level (Supplemental Fig. S4). Mutations in tetraproline motifs 1 and 2 reduced the ability of TTP to repress the luciferase ARE-reporter, despite this protein being expressed at similar levels as WT TTP (Fig. 4B). As expected, deletion of the CIM domain also reduced TTP activity. The absence of a complete loss of TTP function associated with mutations in the tetraproline and CIM motifs is consistent with the ability of TTP to recruit multiple repression complexes via different domains (Fig. 3; Lykke-Andersen and Wagner 2005; Fabian et al. 2013). Combining the tetraproline and CIM mutations modestly decreased TTP activity over the two single mutants (Fig. 4A), although this effect was not found statistically significant (P = 0.18).
FIGURE 4.

TTP mutated in tetraprolines 1 and 2 is impaired in its ability to repress a luciferase reporter ARE-mRNA. (A) Luciferase luminescence assays from TTP−/− MEFs transiently coexpressing wild-type or mutant TTP with two reporters, one encoding firefly luciferase (F-Luc) containing in its 3′ UTR the AU-rich element (ARE) from human GM-CSF mRNA, and one encoding Renilla luciferase (R-Luc) without an ARE as an internal control. For each sample, F-Luc activity was normalized to R-Luc and values were normalized to the wild-type TTP-transfected condition, which was set as 1. Error bars represent standard error of the mean (SEM) (n = 4). P-values were calculated using Student's t-test (paired, two-tailed). (B) Representative Western blot of samples used in panel A. PABP was used as the loading control. (C) Luciferase assays in 4EHP+/+ and 4EHP−/− MEFs similar to the experiment in panel A except in the absence of cotransfected TTP and the absence or presence of transiently expressed 4EHP and GYF2 as indicated. Error bars represent SEM (n = 3).
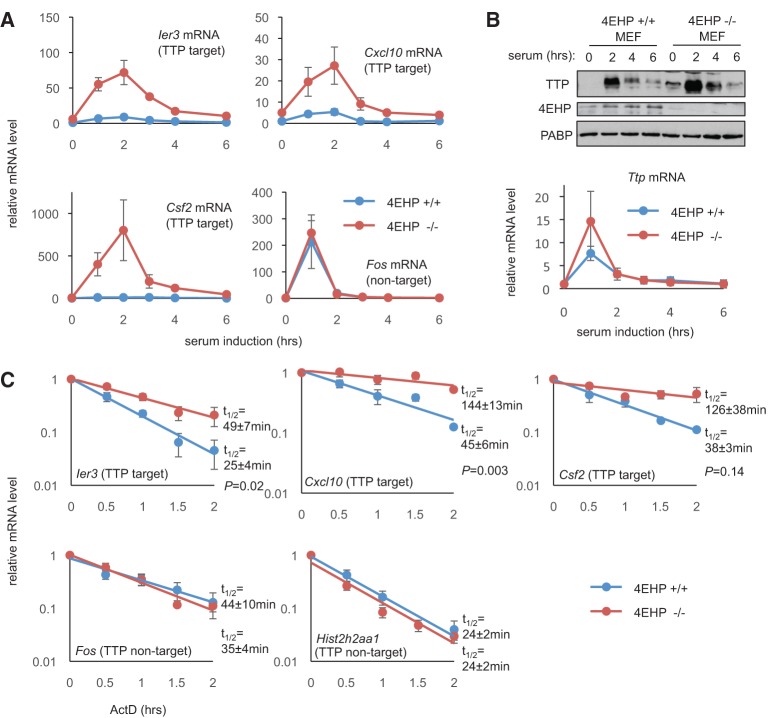
To more directly test the importance of 4EHP in ARE-mRNA repression, we turned to 4EHP−/− MEFs (Morita et al. 2012). Consistent with 4EHP-GYF2 playing a role in ARE-mRNA repression, increased luciferase activity is observed from the firefly luciferase ARE-reporter in 4EHP−/− MEFs as compared to MEFs from 4EHP+/+ littermates (Fig. 4C). Importantly, reintroduction of exogenous 4EHP and GYF2 into the knockout cell line partially rescued this increase in expression; GYF2 was coexpressed with 4EHP in this rescue experiment because GYF2 levels were previously reported to be low in the 4EHP−/− MEFs (Morita et al. 2012). These findings are consistent with a recent report of 4EHP playing a role in TTP-mediated translation repression (Tao and Gao 2015).
Endogenous TTP-target mRNAs are stabilized in 4EHP knockout MEFs
We next tested the importance of 4EHP for repression of endogenous TTP-target mRNAs using the 4EHP−/− MEFs (Morita et al. 2012). Serum addition is a well-described stimulation that induces TTP and TTP-target mRNAs in MEFs (Lai et al. 1990, 2006). We therefore monitored the induction of known TTP-target mRNAs during a time course of 6 h of serum stimulation following serum starvation in 4EHP−/− MEFs as compared to the MEFs from 4EHP+/+ littermates (Lai et al. 2006). This revealed strongly enhanced induction in 4EHP−/− MEFs of Ier3, Csf2, and Cxcl10 mRNAs, all ARE-containing mRNAs known to be regulated by TTP (Fig. 5A). In contrast, Fos mRNA, which is regulated in a TTP-independent manner (Stoecklin et al. 2001; Lai et al. 2006), was unaffected by 4EHP knockout. The increased induction of TTP-target mRNAs in 4EHP−/− MEFs occurred despite TTP mRNA and protein induction being similar, and possibly slightly higher, in the 4EHP−/− as compared to the 4EHP+/+ MEFs (Fig. 5B).
FIGURE 5.
4EHP knockout MEFs show increased induction and stability of TTP-target mRNAs. (A) Quantification of Ier3, Cxcl10, Csf2, and Fos mRNA levels using qRT-PCR during serum induction of MEFs from 4EHP+/+ (blue) and 4EHP−/− (red) littermates. Gapdh mRNA was used as an internal control for normalization and values were normalized to the values for 4EHP+/+ MEFs at t = 0. Error bars represent SEM (n = 3). (B, top) Western blots for TTP, 4EHP, and PABP during the serum induction time course described in panel A. (Bottom) qRT-PCR quantification of Ttp mRNA levels during the serum induction time course with error bars as in panel A. (C) Decay assays using actinomycin D transcriptional shutoff. mRNA levels were quantified by qRT-PCR and normalized to Gapdh mRNA, with values at t = 0 set to 1. Error bars represent SEM (n = 3). Calculated half-lives ± SEM of each mRNA species (assuming an infinite Gapdh mRNA half-life) are listed in minutes. P-values were calculated using Student's t-test (paired, two-tailed).
The observation of increased accumulation of TTP-target mRNAs in 4EHP−/− MEFs suggested a possible effect of 4EHP on mRNA decay. Indeed, actinomycin D-mediated transcription shutoff mRNA decay assays performed 2 h after serum induction revealed that Ier3, Csf2, and Cxcl10 mRNAs are all stabilized in 4EHP−/− as compared to the 4EHP+/+ littermate MEFs (Fig. 5C). In contrast, Fos and histone H2A (Hist2h2aa1) mRNAs, which undergo rapid decay independently of TTP, did not show reduced decay rates in 4EHP knockout MEFs. Collectively, our observations suggest that the 4EHP-GYF2 complex is an important cofactor in the repression of ARE-mRNAs by TTP and that 4EHP-GYF2 contributes to both translation repression and mRNA decay.
DISCUSSION
In this study, we have identified the 4EHP-GYF2 translation repression complex as a cofactor of TTP (Fig. 6). TTP associates with the 4EHP-GYF2 complex via direct interaction with GYF2 as evidenced by in vitro pull-down assays using E. coli produced proteins (Figs. 1C, 3D). The conserved first and second tetraproline motifs of TTP are necessary for maintaining direct interaction with GYF2, both in transfected mammalian cells and in in vitro pull-down assays (Fig. 3). The 4EHP-GYF2 complex contributes to TTP-mediated mRNA repression as evidenced by the derepression of an ARE-containing luciferase reporter mRNA upon mutation of TTP tetraproline motifs and knockout of 4EHP and by the accumulation and stabilization of TTP-target mRNAs induced during serum activation of 4EHP−/− MEFs (Figs. 4, 5). Given the evolutionary conservation of TTP tetraproline motifs (Fig. 3A), the role of the 4EHP-GYF2 complex in TTP function is likely conserved in vertebrates.
FIGURE 6.
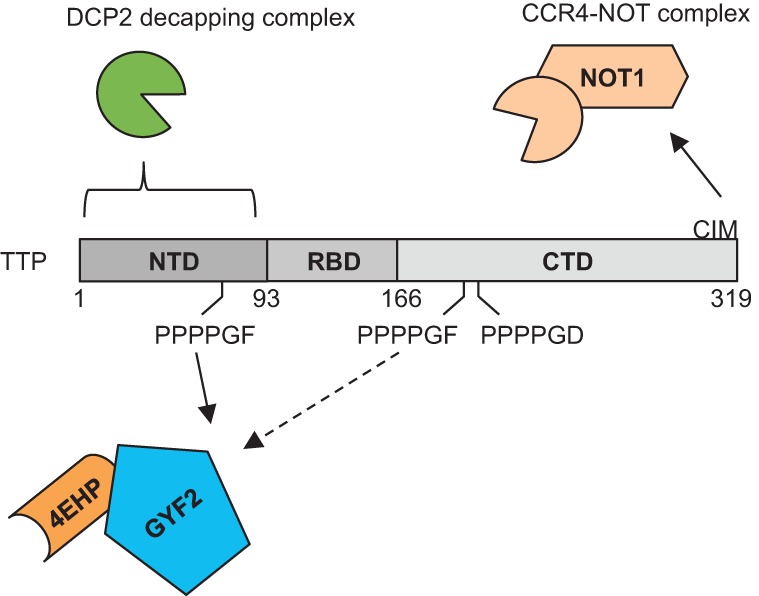
TTP recruits multiple corepressors to repress target mRNAs. Schematic showing TTP motifs and domains contributing to mRNA repression. The CIM recruits the CCR4-NOT deadenylation complex via direct interaction with CNOT1. The NTD of TTP associates with the DCP2 decapping complex. Our work demonstrates the conserved tetraproline motifs of TTP recruiting the 4EHP-GYF2 complex.
How does the 4EHP-GYF2 complex contribute to TTP activity? In addition to activating mRNA degradation, TTP has been demonstrated to promote translation repression, which might be the predominant mechanism of TTP-mediated repression in some conditions (Qi et al. 2012; Schott et al. 2014). 4EHP was recently reported to stimulate translation repression mediated by TTP (Tao and Gao 2015), which is consistent with the reported function of Drosophila 4EHP and the mammalian 4EHP-GYF2 complex in translation repression (Cho et al. 2005; Morita et al. 2012). Consistent with this, we found that mutation in tetraproline motifs 1 and 2 reduced the ability of TTP to associate with 4EHP-GYF2 (Fig. 3) and to repress a luciferase ARE-reporter (Fig. 4). 4EHP knockout also resulted in derepression of the luciferase ARE-reporter (Fig. 4C), but in addition, triggered accumulation and stabilization of TTP-target mRNA in serum-activated MEFs (Fig. 5). Therefore, in addition to stimulating translation repression, 4EHP-GYF2 also contributes to TTP-mediated mRNA degradation. 4EHP-GYF2 might link repression of translation initiation with mRNA degradation directly, for example, by interfering with the eIF4F complex to promote a remodeling step that helps expose the mRNA cap to decapping or by recruitment of unknown mRNA decay factors. Alternatively, the increased accumulation and stabilization of TTP-target mRNAs in serum-stimulated 4EHP knockout MEFs could be a secondary effect of a failure in TTP-mediated translation repression, for example, if increased translation of TTP-target mRNAs results in delayed attenuation of the serum activation response. In contrast to TTP, we observed no dramatic up-regulation of 4EHP and GYF2 during LPS stimulation of RAW macrophages or serum stimulation of MEFs (Fig. 5B; Supplemental Fig. S5A). Moreover, unlike the association with the CCR4-NOT complex (Marchese et al. 2010; Clement et al. 2011), we found no effect of the phosphatase 2A (PP2A) inhibitor okadaic acid on the association of TTP with 4EHP-GYF2 (Supplemental Fig. S5B). Thus, accumulation of TTP-4EHP-GYF2 complexes during LPS- or serum-stimulation is likely regulated by changes in TTP levels rather than by TTP phosphorylation or regulation of 4EHP or GYF2.
Our findings add a new layer of complexity to TTP-mediated mRNA regulation. In previous studies, TTP has been found to associate with and activate the exosome (Chen et al. 2001) and the DCP2 decapping (Lykke-Andersen and Wagner 2005) and CCR4-NOT deadenylase complexes (Lykke-Andersen and Wagner 2005; Fabian et al. 2013). Consistent with the ability to activate multiple mRNA repression and degradation factors, multiple domains and motifs of TTP have been found to contribute to TTP function (Johnson and Blackwell 2002; Lykke-Andersen and Wagner 2005) including the tetraproline motifs as identified in this study (Fig. 6). This ability to engage multiple mRNA repression and degradation machineries appears to be an emerging general principle in mRNA regulation by RBPs. For example, UPF1, the central component of the nonsense-mediated decay pathway, is known to associate with multiple degradation factors (Ohnishi et al. 2003; Fukuhara et al. 2005; Eberle et al. 2009). The microRNA-guided RISC complex can also both activate degradation and repress translation (Huntzinger and Izaurralde 2011). Similarly, PUF proteins are known to repress and degrade mRNAs by recruitment of multiple cofactors (Goldstrohm et al. 2006; Weidmann and Goldstrohm 2012). Thus, the ability to recruit multiple repression factors might be an important general feature of RBPs that promote mRNA repression. This could serve as a mechanism to efficiently activate sequential steps in mRNA repression and degradation. Alternatively, cofactors could be individually sufficient for mRNA repression, allowing the RBPs to carry out mRNA repression and/or degradation in different tissues using different available cofactors.
MATERIALS AND METHODS
Plasmid constructs
Coding sequences (CDS) of mouse TTP, BRF1, BRF2, 4EHP, and GYF2 were amplified from cDNA reverse-transcribed using Superscript II (Invitrogen) from total RNA of NIH/3T3 cells, and inserted into pcDNA3-based mammalian expression vectors pcDNA3-myc and pcDNA3-Flag (Lykke-Andersen 2002) and bacterial expression vectors pET-his (Lykke-Andersen et al. 2000) and pGEX-4T1 (Amersham). Sequences encoding domain truncations of TTP used in Figures 2, 3 and Supplemental Fig. S3 were inserted into pcFlag-NMS2 (Lykke-Andersen et al. 2000), which fused the domains with MS2 coat protein to add additional size. Firefly luciferase and Renilla luciferase CDS were subcloned from pGL2 and pRL (Promega), respectively, into pcDNA3-myc. A 62-bp AU-rich element of human GM-CSF (Voeltz and Steitz 1998) was inserted into the 3′ UTR of pcDNA3-myc-Luc. Plasmid sequences are available upon request.
Cell culture
RAW264.7 cells and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) with 10% fetal bovine serum (FBS). Mouse embryonic fibroblasts were cultured in DMEM containing 1 mM sodium pyruvate (Gibco) with 10% FBS and 2 mM l-glutamine (Gibco). In the experiments shown in Figure 5, cells were washed two times with PBS and grown in DMEM containing 0.5% FBS for a 24-h serum starvation period. Cells were then serum-induced with DMEM containing 10% FBS for indicated amounts of time. In mRNA decay assays (Fig. 5), 2 h after serum induction, 10 μg/mL actinomycin D was added to stop transcription. The decay time course was started after 35 min, and samples were harvested into TRIzol at indicated time points.
Antibodies and Western blotting
Western blots were performed with the following antibodies at the indicated concentrations: rabbit polyclonal anti-Myc (Sigma-Aldrich, C3956; 1:1000), rabbit polyclonal anti-Flag (Sigma-Aldrich, F7425; 1:1000), rabbit polyclonal anti-TTP (Sigma-Aldrich, T5327; 1:500), rabbit polyclonal anti-PABP (Abcam, ab21060; 1:1000), rabbit polyclonal anti-CNOT1 (Proteintech, 14276-1-AP; 1:200), rabbit polyclonal anti-GIGYF2 (Santa Cruz, sc-134708; 1:50), rabbit polyclonal anti-EIF4E2 (GeneTex, GTX103977; 1:200), and mouse monoclonal anti-GST (Abgent, AM1011a; 1:600).
Coimmunoprecipitation assays
For coimmunoprecipitation assays, cells in 10-cm plates were transfected with 5 μg of the indicated 4EHP, GYF2, TTP, BRF1, or BRF2 constructs, using TransIT 293 reagent according to manufacturer's protocol (Mirus). After 48 h, cells were washed with PBS, pelleted by centrifugation at 1000g for 5 min, and lysed in 1 mL of ice-cold hypotonic gentle lysis buffer (10 mM Tris–HCl pH 7.5, 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 1 mM PMSF, 1 μM aprotinin, and 1 μM leupeptin). Lysates were incubated on ice with 50 μg/mL RNase A for 10 min, and then the NaCl concentration was increased to 150 mM and incubated for another 10 min on ice. The supernatant was separated from debris by centrifugation at 4°C and 21,130g for 15 min, and added to 50 µL of anti-Flag M2-agarose (Sigma) or anti-cMyc agarose (Sigma). Beads were washed eight times with 1 mL NET2 (50 mM Tris–HCl pH 7.5, 150 mM NaCl, and 0.05% Triton X-100) and resuspended in 50 µL 2× SDS loading buffer (100 mM Tris–HCl pH 6.8, 4% SDS, 20% glycerol, 0.1% bromophenol blue, and 200 mM DTT) for separation by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting using standard procedures. 0.5% of total lysates and 22.5% of IP elutions were loaded for analyses.
TTP-IP and liquid chromatography/tandem mass spectrometry (LC-MS/MS)
Twenty-four micrograms of rabbit anti-TTP (Sigma-Aldrich, T5327) or normal rabbit serum were incubated with 50 mg of protein A Sepharose CL-4B beads (Fisher) overnight in 1.2 mL NET2. RAW264.7 cells were treated with 100 ng/mL of lipopolysaccharides (LPS) for indicated lengths of time (0, 2, 4, 8, or 24 h). For each time point, three 15-cm plates of cells were lysed with 4 mL hypotonic gentle lysis buffer, treated with RNase A and adjusted to 150 mM NaCl as described above for coimmunoprecipitation assays, added to protein A Sepharose-coupled antibodies, and washed. Beads were then eluted three times with 500 μL 0.1 M glycine pH 2.6. Eluants were pooled and mixed with 20% trichloroacetic acid (TCA) at −20°C overnight. After thawing on ice, protein samples were centrifuged at 21,130g for 30 min at 4°C. Pellets were then washed with 10% TCA and three times −20°C prechilled acetone, spinning at 4°C (21,130g) for 15 min between washes. Pellets were air-dried and stored at −80°C. LC-MS/MS was performed as described previously (Erickson et al. 2015), with one biological and triplicate technical repeats.
Bacterial protein purification and in vitro pull-down assays
For bacterial expression of mouse TTP, GYF2, and 4EHP, corresponding pET-his or pGEX-4T1 plasmids were transformed into E. coli BL21(DE3) cells containing the pRI952 plasmid encoding rare arginine and isoleucine tRNAs (Nielsen et al. 1999). Bacteria were cultured in 200 mL LB broth medium at 37°C to OD600 of 0.5, and then transferred to lower temperature and induced with isopropyl β-d-1-thiogalactopyranoside (IPTG). IPTG concentration, temperature, and time of incubation differed for individual proteins as follows: GST: 0.1 mM IPTG, 15°C, 16 h; GST-TTP: 0.3 mM IPTG, 25°C, 3 h; GST-TTP-12S: 0.1 mM IPTG, 15°C, 16 h; His6-GYF2: 0.4 mM IPTG, 15°C, 8 h; His6-4EHP: 0.1 mM IPTG, 15°C, 16 h. Cells were pelleted at 2300g for 10 min, resuspended in 5 mL TKET buffer (10 mM Tris–HCl pH 7.5, 100 mM KCl, 0.1 mM EDTA, 0.05% Triton X-100) with 1 mM PMSF, sonicated at 4°C for 8 × 30 sec, with 30-sec intervals, added Triton X-100 to 0.5%, nutated for 15 min, and centrifuged at 11,000g for 15 min to remove debris. Supernatant was then allowed to flow through columns of 700 μL glutothinone Sepharose 4B (GE) or 700 μL Ni-NTA agarose beads (Qiagen) twice. Columns were then washed three times with 4 mL of TKET, and eluted with 500 μL of Tris–HCl pH 7.5 with 100 mM KCl and 20 mM glutathione for GST proteins or 500 μL of TKET containing 200 mM imidazole for His6-4EHP. Elutants were dialyzed overnight at 4°C against 400 mL of PBS, and protein concentration was measured using a Bradford-based Bio-Rad Protein Assay system and freshly made bovine serum albumin (BSA) standards. His6-GYF2 was purified under denaturing conditions, following the same steps, except lysis buffer is changed to TKET containing 1 mM PMSF, 20 mM imidazole, and 6 M guanidine-HCl; cells were incubated at room temperature for 30 min for lysis instead of sonication; wash buffer is changed to TKET containing 8 M urea and 20 mM imidazole; elution buffer is changed to TKET containing 8 M urea and 200 mM imidazole
For in vitro pull-down assays, 5 μg His6-GST or GST-TTP were nutated with 25 µL of glutathione Sepharose for 2 h at 4°C, washed three times with TKET, and then nutated with 5 μg of His6-4EHP and/or His6-GYF2 in 500 μL TKET at 4°C. After 2 h, beads were washed four times with 700 μL TKET, and resuspended in 2× SDS loading buffer, to be analyzed by SDS-PAGE and Western blotting.
Luciferase assays
MEFs were plated at ≈20% confluency in 22-mm-diameter tissue culture wells in 1 mL DMEM/10% FBS. Twenty-four hours later, 0.2 μg pcDNA3-myc-F-Luc-ARE, 0.05 μg pcDNA3-myc-R-Luc, 0.3 μg pSuper.puro (OligoEngine), 0.4 μg pcDNA3-myc, and 0.05 μg of Flag-tagged TTP wild-type or mutant constructs were transfected by TransIT-X2 following manufacturer's protocols (Mirus). Twenty-four hours later, 5 μg/mL of puromycin was added for another 24 h to select for transfected cells. Cells were then lysed in 250 μL of 1× Passive Lysis Buffer (Promega) for 20 min at room temperature and 10 μL of each lysate was assayed for firefly and Renilla luciferase activities using Dual-Luciferase Reporter Assay (Promega) reagents in a NOVOstar microplate reader. In the experiments shown in Figure 4C, 0.3 µg of pcDNA3-Flag or 0.15 μg pcDNA3-Flag-4EHP and 0.15 μg pcDNA3-Flag-GYF2 were cotransfected instead of TTP constructs. P-values were calculated with two-tailed paired Student's t-test.
Quantitative qRT-PCR
Three micrograms of total RNA prepared from cells using TRIzol (Invitrogen) was treated with 0.1 U/μL DNase I (Invitrogen AM2222) in 20 μL RNase-free H2O (Ambion) with 2 U/μL RNaseOUT (Invitrogen) at 37°C for 30 min. RNA was subsequently extracted with phenol:chloroform:isoamyl alcohol (50:49:1), ethanol precipitated, washed with 70% ethanol, and dissolved in 15 µL of RNase-free H2O. One microgram of the DNase-treated RNA was reverse-transcribed using random hexamers with Superscript III according to manufacturer's protocols (Invitrogen). The corresponding cDNA was used for qPCR quantification using Fast SYBR Green Master Mix (Applied Biosystems) on a StepOnePlus System (Applied Biosystems). For each qRT-PCR reaction, fivefold serial dilutions of cDNA were used to calculate PCR efficiency (E), and standard curve of log10 of dilution factor versus Ct (threshold cycle) was plotted to determine the linear range of Ct values. For E ranged between 1.8 and 2.1, and Ct within the linear range, relative mRNA levels were calculated with the formula . Non-RT controls were monitored to confirm DNase treatment was complete. The following DNA oligos were used (at 285 nM). F-Luc_F: CTTCGCCAAAAGCACTCTG; F-Luc_R: GAGCCCATATCCTTGTCGTATC; R-Luc_F: TGGAGCCATTCAAGGAGAAG; R-Luc_R: TGTAGTTGCGGACAATCTGG; TTP-F: CGGAGGACTTTGGAACATAAAC; TTP-R: GGAGTTGCAGTAGGCGAAGTAG; GAPDH_F: CATGGCCTTCCGTGTTCCTA; GAPDH_R: CCTGCTTCACCACCTTCTTGA T; IER3_F: GCGCGTTTGAACACTTCTC; IER3_R: CAGAAGATGATGGCGAACAG; CXCL10_F: CTAGCTCAGGCTCGTCAGTTC; CXCL10_R: TGGGAAGATGGTGGTTAAGTTC; CSF2_F: TGAACATGACAGCCAGCTACTAC; CSF2_R: ACTTGTGTTTCACAGTCCGTTTC; FOS_F: GAATGGTGAAGACCGTGTCAG; FOS_R: GTCTCCGCTTGGAGTGTATC; HIST2H2AA_F: AAGCTGCTGGGCAAAGTG; HIST2H2AA_R: ACTTGCCCTTCGCCTTATG.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Perry Blackshear and Nahum Sonenberg for TTP−/− and 4EHP−/− MEFs, respectively. We thank members of the J.L.-A. laboratory for critical input on this work. This work was supported by grant R01 GM077243 from the Foundation for the National Institutes of Health to J.L.-A. and a New Scholar award from the Ellison Medical Foundation to E.J.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054833.115.
REFERENCES
- Brooks SA, Blackshear PJ. 2013. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta 1829: 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95: 1891–1899. [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. 2005. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121: 411–423. [DOI] [PubMed] [Google Scholar]
- Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J Biol Chem 279: 10176–10184. [DOI] [PubMed] [Google Scholar]
- Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. 2004. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene 23: 8673–8680. [DOI] [PubMed] [Google Scholar]
- Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. 2011. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol 31: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J 19: 3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55. [DOI] [PubMed] [Google Scholar]
- Erickson SL, Corpuz EO, Maloy JP, Fillman C, Webb K, Bennett EJ, Lykke-Andersen J. 2015. Competition between decapping complex formation and ubiquitin-mediated proteasomal degradation controls human Dcp2 decapping activity. Mol Cell Biol 35: 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. 2013. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 20: 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915. [DOI] [PubMed] [Google Scholar]
- Freund C, Dotsch V, Nishizawa K, Reinherz EL, Wagner G. 1999. The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat Struct Biol 6: 656–660. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539. [DOI] [PubMed] [Google Scholar]
- Goss DJ, Kleiman FE. 2013. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA 4: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol 26: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, et al. 2010. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol 11: 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blackwell TK. 2002. Multiple tristetraprolin sequence domains required to induce apoptosis and modulate responses to TNF α through distinct pathways. Oncogene 21: 4237–4246. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J Biol Chem 277: 18029–18036. [DOI] [PubMed] [Google Scholar]
- Joshi B, Cameron A, Jagus R. 2004. Characterization of mammalian eIF4E-family members. Eur J Biochem 271: 2189–2203. [DOI] [PubMed] [Google Scholar]
- Kofler M, Motzny K, Freund C. 2005. GYF domain proteomics reveals interaction sites in known and novel target proteins. Mol Cell Proteomics 4: 1797–1811. [DOI] [PubMed] [Google Scholar]
- Lai WS, Stumpo DJ, Blackshear PJ. 1990. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem 265: 16556–16563. [PubMed] [Google Scholar]
- Lai WS, Kennington EA, Blackshear PJ. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 23: 3798–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. 2006. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26: 9196–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT. 2009. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453. [DOI] [PubMed] [Google Scholar]
- Lehner B, Sanderson CM. 2004. A protein interaction framework for human mRNA degradation. Genome Res 14: 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AR, Dean JL. 2010. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem 285: 27590–27600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of elF4G. Mol Cell 3: 707–716. [DOI] [PubMed] [Google Scholar]
- Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, Alain T, Fonseca BD, Karashchuk G, Bennett CF, et al. 2012. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol 32: 3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200. [DOI] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Sonenberg N. 1994. Insulin-dependent stimulation of protein-synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371: 762–767. [DOI] [PubMed] [Google Scholar]
- Pfeiffer JR, Brooks SA. 2012. Cullin 4B is recruited to tristetraprolin-containing messenger ribonucleoproteins and regulates TNF-α mRNA polysome loading. J Immunol 188: 1828–1839. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W. 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi MY, Wang ZZ, Zhang Z, Shao Q, Zeng A, Li XQ, Li WQ, Wang C, Tian FJ, Li Q, et al. 2012. AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol Cell Biol 32: 913–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P, She MP, Parker R. 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell 45: 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik B, Clement SL, Lykke-Andersen J. 2014. hnRNP F complexes with tristetraprolin and stimulates ARE-mRNA decay. PLoS One 9: e100992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom E, Kim HC, Gingras AC, Marcotrigiano J, Favre D, Olsen H, Burley SK, Sonenberg N. 1998. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem 273: 13104–13109. [DOI] [PubMed] [Google Scholar]
- Sandler H, Kreth J, Timmers HT, Stoecklin G. 2011. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res 39: 4373–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J, Reitter S, Philipp J, Haneke K, Schafer H, Stoecklin G. 2014. Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet 10: e1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stoeckle P, Lu M, Muehlemann O, Moroni C. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA 7: 1578–1588. [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. 2008. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem 283: 11689–11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Stoecklin G, Van Way S, Hinkovska-Galcheva V, Guo RF, Anderson P, Shanley TP. 2007. Tristetraprolin (TTP)- 14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-α mRNA. J Biol Chem 282: 3766–3777. [DOI] [PubMed] [Google Scholar]
- Tao X, Gao G. 2015. Tristetraprolin recruits eIF4E2 to repress translation of ARE-containing mRNAs. Mol Cell Biol 35: 3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. 1996. A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4: 445–454. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Steitz JA. 1998. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol 18: 7537–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Gross JD, Wagner G, McCarthy JE. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 11: 503–511. [DOI] [PubMed] [Google Scholar]
- Weidmann CA, Goldstrohm AC. 2012. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol Cell Biol 32: 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell 1: 863–872. [DOI] [PubMed] [Google Scholar]
- Zhang LB, Prak L, Rayon-Estrada V, Thiru P, Flygare J, Lim B, Lodish HF. 2013. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature 499: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.