Abstract
The molecular mechanism of stop codon recognition by the release factor eRF1 in complex with eRF3 has been described in great detail; however, our understanding of what determines the difference in termination efficiencies among various stop codon tetranucleotides and how near-cognate (nc) tRNAs recode stop codons during programmed readthrough in Saccharomyces cerevisiae is still poor. Here, we show that UGA-C as the only tetranucleotide of all four possible combinations dramatically exacerbated the readthrough phenotype of the stop codon recognition-deficient mutants in eRF1. Since the same is true also for UAA-C and UAG-C, we propose that the exceptionally high readthrough levels that all three stop codons display when followed by cytosine are partially caused by the compromised sampling ability of eRF1, which specifically senses cytosine at the +4 position. The difference in termination efficiencies among the remaining three UGA-N tetranucleotides is then given by their varying preferences for nc-tRNAs. In particular, UGA-A allows increased incorporation of Trp-tRNA whereas UGA-G and UGA-C favor Cys-tRNA. Our findings thus expand the repertoire of general decoding rules by showing that the +4 base determines the preferred selection of nc-tRNAs and, in the case of cytosine, it also genetically interacts with eRF1. Finally, using an example of the GCN4 translational control governed by four short uORFs, we also show how the evolution of this mechanism dealt with undesirable readthrough on those uORFs that serve as the key translation reinitiation promoting features of the GCN4 regulation, as both of these otherwise counteracting activities, readthrough versus reinitiation, are mediated by eIF3.
Keywords: programmed stop codon readthrough, termination, eRF1, tetranucleotide, GCN4, uORF
INTRODUCTION
The accuracy of translation of genetic information from genes into proteins that occurs on the ribosome is highly dependent on the precise decoding of the genetic code, which is contained in individual mRNAs. The sequence of nucleotide triplets in the right open reading frame determines the sequence of amino acid residues in the corresponding polypeptide and, with a few exceptions, such as for example, ambiguous decoding of the same codon by two different sense tRNAs or various types of programmed frameshifting (Dinman 2012), this basic rule is relatively strictly observed; translation has a misincorporation rate of 10−3 to 10−4 (Zaher and Green 2009). Nearly the same applies to recognition of all three stop codons by the release factor eRF1 that enters the A-site of the 80S ribosome in complex with the GTP-binding protein eRF3 (for review, see Jackson et al. 2012; von der Haar and Valasek 2014). There are some specific cases, however, when a stop codon does not signal the proper end of translation, which can thus continue beyond to the next stop codon. This phenomenon is called stop codon readthrough or nonsense suppression and occurs when a near-cognate aminoacyl-tRNA (nc-tRNA) or a suppressor tRNA is incorporated at a given stop codon (Dabrowski et al. 2015). It can be “spontaneous” and thus relatively infrequent (the order of spontaneous stop codon leakiness is UGA > UAG > UAA); or it can be programmed to C-terminally extend the protein of interest as a response to, for example, specific environmental changes demanding an alteration of the corresponding protein's properties (for more details, see Dinman 2012). Stop codon readthrough can also occur at a premature termination codon (PTC) within the coding region of a given gene, which is desirable because it can prevent the action of a nonsense-mediated decay (NMD) pathway by ensuring the synthesis of a full-length protein (Keeling et al. 2014). In fact, there are many diseases caused by PTCs that often occur in the termination nonfavorable context, making them good substrates for drug-stimulated programmed readthrough (Linde and Kerem 2008). Mainly from this point of view this topic is of high importance also for medical reasons (Lee and Dougherty 2012).
Taking all this into account, it is important to investigate which nc-tRNAs can decode individual stop codons and under what circumstances. Namy's group has only recently made great progress in this direction by experimentally demonstrating in vivo that tyrosine or glutamine preferentially incorporates at UAA (with similar frequencies), tyrosine predominantly inserts at UAG, and tryptophan or cysteine at UGA (Blanchet et al. 2014). Lysine and arginine were also shown to incorporate at UAA/UAG and UGA stop codons, respectively, and glutamine at UAG, but at very low rates. Soon after that, Jacobson's group extended these findings by a comprehensive in vivo analysis of termination readthrough from a PTC under different readthrough-inducing conditions (Roy et al. 2015). Their work identified exactly the same amino acid residues that are being inserted during readthrough, only with the predominant frequencies of incorporation for glutamine and not for tyrosine at UAG. However, what determines which of the amino acid residues that are specific for each stop codon incorporates at a given stop codon at a given frequency is not known. It was previously demonstrated that the identity of a nucleotide immediately following any stop codon (the +4 nucleotide) fine-tunes its termination efficiency; i.e., determines its permissiveness for readthrough (for an overview, see Dabrowski et al. 2015). For example, efficiency of readthrough on UGA determined by the +4 nucleotide identity follows this order of tetranucleotides: UGA-C > UGA-A > UGA-G > UGA-U (Beznosková et al. 2015). In fact, C at the +4 position is the strongest readthrough inducer among all four bases at all three stop codons (Dabrowski et al. 2015). Hence it is possible that in addition to the stop codon, it is also the +4 base that influences which nc-tRNA gets preferentially accepted by the A-site during readthrough.
We recently showed that one of the key translation initiation factors—eIF3—interacts with pretermination complexes (pre-TCs) and controls translation termination and readthrough (Beznosková et al. 2013). In particular, it interferes with the eRF1 ability to recognize the third/wobble position of any programmed stop codon leading to the rejection of the eRF1–eRF3.GTP complex from pre-TCs. At the same time, eIF3 promotes incorporation of nc-tRNAs with a mismatch at the same position and thus represents one of the key players in programmed stop codon readthrough (Beznosková et al. 2015). For UGA there are two nc-tRNAs with the mismatch at the third/wobble position, Trp- and Cys-tRNAs. The third and last nc-tRNA with the wobble mismatch, Tyr-tRNA, is shared by the UAA and UAG stop codons. Incorporation of all of these three nc-tRNAs is promoted by eIF3 and, in fact, we provided evidence that efficient readthrough at UGA is enabled exclusively by the former two nc-tRNAs with the mismatch at the third/wobble position (Beznosková et al. 2015). This does not seem to apply to the other two stop codons, however, because other studies based on different approaches that were mentioned above showed that in addition to Tyr-tRNA, UAA and UAG can be also recoded by Gln- and Lys-tRNAs; i.e., nc-tRNAs with the mismatch at the first position (Blanchet et al. 2014; Roy et al. 2015). The UGA stop codon has two nc-tRNAs with the first position mismatch, Gly- and Arg-tRNA, the latter of which was also suggested to insert at the A-site with low frequencies by these authors; however, it did not do so in our hands (Beznosková et al. 2015).
An interesting aspect of the specific role of eIF3 in promoting readthrough is that it may actually interfere with another peculiar role of eIF3 in promoting translation reinitiation (REI) (Valášek 2012). REI is a gene-specific mechanism of translational control exerted by some short upstream ORFs (uORFs) to fine-tune the expression of the main ORF. It requires the small ribosomal subunit to remain associated with mRNA even after termination on these so called REI-permissive uORFs in order to resume scanning on the same mRNA molecule until the 40S ribosome reinitiates at the next AUG. eIF3 was shown to play a critical role in this process by making contacts with both the 40S subunit as well as sequences upstream of REI-permissive uORFs to stabilize their mutual interaction (Szamecz et al. 2008; Munzarová et al. 2011; Gunišová and Valášek 2014). Hence both processes—readthrough and REI—are dependent on the physical presence of eIF3 in termination complexes, yet from the regulatory point of view they act against each other. In other words, increased readthrough will prevent REI. Since to our knowledge virtually nothing is known about readthrough on short uORFs, it would be of interest to investigate the mutual functional relationship of readthrough and REI during termination on uORFs that are set in regulatory networks relying on high levels of REI. This is the case of, for example, the GCN4 mRNA (Gunišová and Valášek 2014), where the integrated regulatory output provided by its four short uORFs could be largely influenced by increased readthrough.
Here, we specifically focused on the leakiest stop codon of all three (UGA) that, when featuring as a PTC, responds relatively unpredictably to various types of anti-PTC treatment (Linde and Kerem 2008; Lee and Dougherty 2012). Our genetic experiments suggest that the key determinant of the highest readthrough levels displayed specifically by the UGA-C tetranucleotide (Bonetti et al. 1995; Beznosková et al. 2015) is the impaired ability of eRF1 to unambiguously recognize the stop codon when it is followed by the cytosine base. Importantly, this “cytosine-specific termination effect” has a general character because it is manifested also on the UAA and UAG stop codons. In addition, we also show that the identity of the +4 base determines the preference of nc-tRNAs for a given UGA-N tetranucleotide. And last but not least, we shed light on readthrough on short uORFs at which the eIF3-promoted REI activity meets but does not collide with the eIF3-promoted, potentially antagonistic, programmed readthrough. Hence we bring new insights into ribosomal decoding rules and translational control that can be used for better qualitative predictions of medically relevant nonsense suppressions.
RESULTS
Cytosine immediately following any stop codon specifically interferes with the eRF1 decoding in vivo
It has been shown by us and others that the nucleotide immediately following the stop codon largely determines the efficiency of readthrough, with cytosine allowing significantly the highest readthrough levels of all four bases regardless of the nature of the stop codon (Bonetti et al. 1995). However, this effect has never been explained. While this manuscript was under preparation, Ramakrishnan's group showed that guanine at the +4 position after any stop codon is pulled into the A-site and stabilized by stacking interaction with G626 of 18S rRNA (Brown et al. 2015). The authors proposed that this stacking would be more stable for purines, explaining the statistical bias at the +4 position in eukaryotes. However, from all four possibilities, only one, G at the +4 position, features in their structures with different stop codons, and, more importantly, the order of the termination leakiness (stop codon readthrough) determined by the +4 base is C > A > G > U. This means that both supposedly best terminating purines are right in between two pyrimidines, one of which terminates worse but the other even better. Hence, this novel observation still does not fully explain the differences in termination efficiency originating from the identity of the nucleotide following the stop codon.
We thus asked whether it could be caused by some specific decoding properties of the release factor eRF1 (encoded by SUP45) that might somehow sense the identity of the +4 nucleotide and perhaps interfere with or specifically accommodate its stacking interaction with 18S rRNA G626. To address this question, we first analyzed UGA as the most readthrough permissive stop codon and used two temperature-sensitive mutants of eRF1 that were shown to increase stop codon readthrough in the past. In particular, we used a sup45-M48I mutant that interferes with stop codon decoding (Bertram et al. 2000) and a sup45-Y410S mutant that directly disrupts the eRF1–eRF3 interaction (Akhmaloka et al. 2008). As shown before, the efficiency of readthrough in wild-type cells with respect to the nature of the +4 nucleotide followed this order: C > A > G > U (Beznosková et al. 2015), and both eRF1 mutants expectedly increased readthrough for all four bases at the +4 position compared to wild-type (Fig. 1A). However, whereas neither of the eRF1 mutants showed any genetic interaction with A, G and U at the +4 position (the fold increase of readthrough normalized to UGA-U was comparable to that seen in SUP45 wild-type suggesting that none of these three nucleotides interferes with the stop codon decoding by eRF1), the presence of cytosine produced a robust additive phenotype with both mutants (Fig. 1B). Importantly, essentially the same results were obtained when we subsequently examined two remaining stop codons, UAA and UAG, set in all four possible tetranucleotide combinations (Fig. 1C,D). Hence, we propose that this additive effect originates from the compromised ability of eRF1 to properly recognize any stop codon when it is followed specifically by the cytosine base, which would explain why cytosine at +4 position promotes the highest readthrough levels of all four bases regardless of the nature of the stop codon (Bonetti et al. 1995).
FIGURE 1.
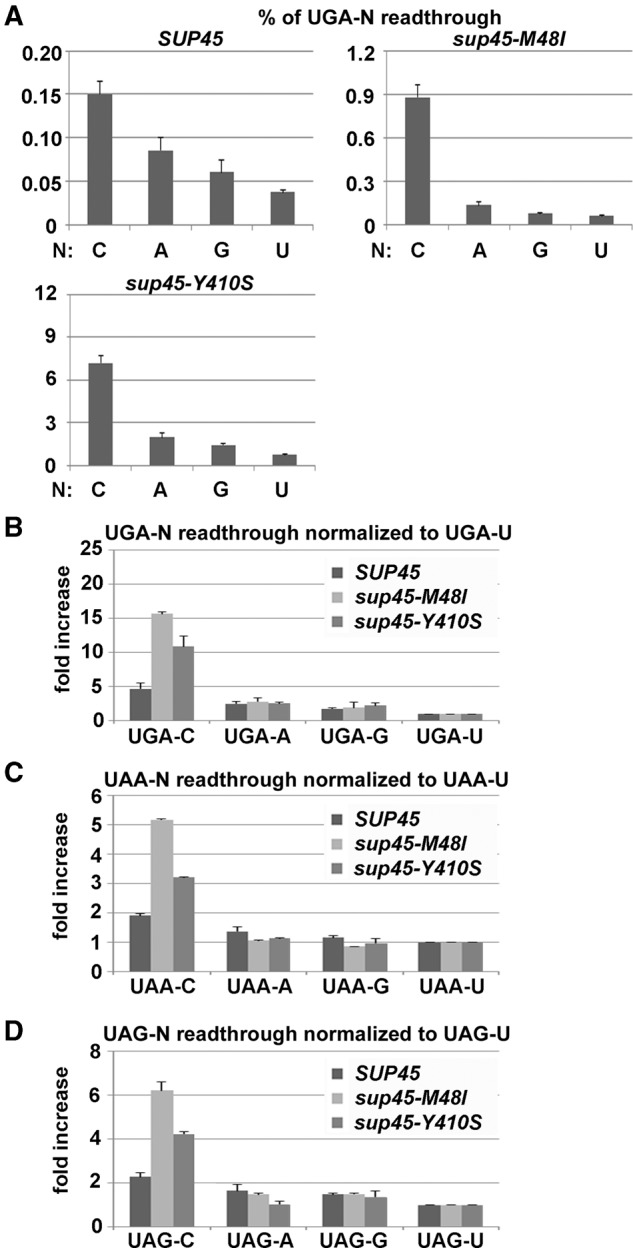
Cytosine immediately following any stop codon interferes with the eRF1 decoding in vivo. (A) Stop codon readthrough measured at all four UGA-N termination tetranucleotides in wt cells (SUP45) and two eRF1 mutants (sup45-M48I and sup45-Y410S). The 74D-694, L2327, and L2521 strains were grown in SD and processed for stop codon readthrough measurements using standard dual luciferase readthrough reporter constructs YEp-R/T-CAAC-L; YEp-R/T-UGAC-L; PBB75; PBB76; and PBB77, as described in Materials and Methods. Readthrough values are represented as mean ± SD from quintuplicates (n = 5) and each experiment was repeated at least three times. (B) Normalization of readthrough measurements from panel A; values measured for UGA-U of each of the four strains were set to one. (C) Same as in panel A, except that all four UAA-N termination tetranucleotides were examined. (D) Same as in panel A, except that all four UAG-N termination tetranucleotides were examined.
UGA-A and UGA-G tetranucleotides are preferentially read through by tryptophan and cysteine nc-tRNAs, respectively
Since cytosine was the only nucleotide at the +4 position displaying the additive effect with eRF1 mutants, we next asked what determines the differences in termination efficiency among the remaining UGA-A, -G, and -U tetranucleotides. We hypothesized that the identity of the +4 nucleotide could influence the stop codon decoding ability of nc-tRNAs that compete for the A-site occupancy with eRF1. If true, overexpression of a nc-tRNA that preferentially incorporates at a given tetranucleotide will increase the fold change of readthrough at this specific tetranucleotide more dramatically than an overexpressed nc-tRNA that is not preferred by this tetranucleotide. Since we focused this study on UGA and in our previous work we demonstrated that efficient readthrough at UGA is enabled exclusively by nc-tRNAs with a mismatch at the third, wobble position (Beznosková et al. 2015), we aimed our attention at tryptophan [tW(CCA)G1] and cysteine [tC(GCA)P1] tRNAs (Fig. 2A), overexpressed them individually in TIF35 wild-type versus tif35-KLF mutant strains (TIF35 encodes the g subunit of yeast eIF3), and measured the luciferase activities on all four UGA-N tetranucleotides. Note that the tif35-KLF mutant is defective in promoting programmed stop codon readthrough and thus the difference between TIF35 wild type and this mutant indicates the degree of programmed readthrough (Beznosková et al. 2015).
FIGURE 2.
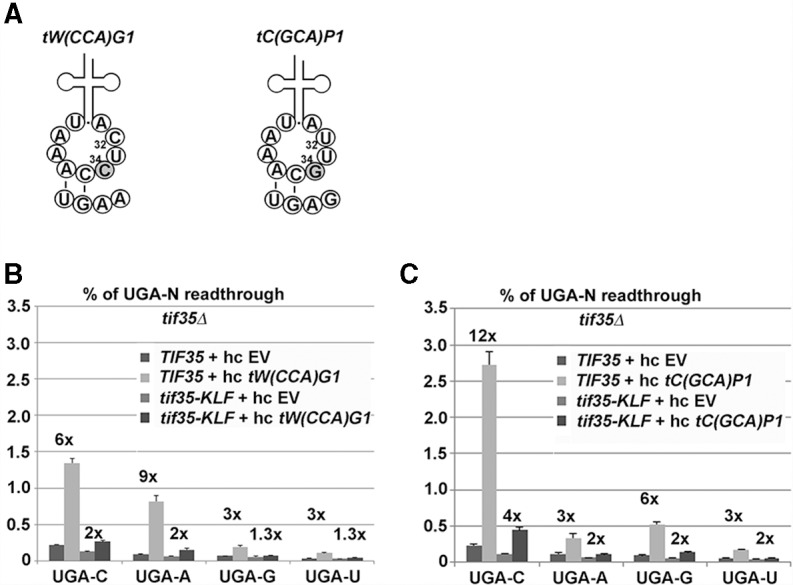
The UGA-A tetranucleotide is preferentially read through by tryptophan nc-tRNA and the UGA-C, and UGA-G tetranucleotides are preferentially read through by cysteine nc-tRNA. (A) Schematics of Trp and Cys nc-tRNAs base-paring with the indicated stop codon tetranucleotides. Only the nucleotides of the anticodon loop are shown with the third stop codon base N34 (in gray) and N32 indicated. (B) Effect of the eIF3 presence in pre-TCs on UGA-N readthrough upon increased gene dosage of tryptophan nc-tRNA. The PBH140 derivatives bearing TIF35 wt and tif35-KLF mutant alleles were transformed with either empty vector (EV) or high copy (hc) tW(CCA)G1 and the resulting transformants were grown and processed for stop codon readthrough measurements as described in Figure 1. (C) Effect of the eIF3 presence in pre-TCs on UGA-N readthrough upon increased gene dosage of cysteine nc-tRNA. Essentially the same as in B, except that hc tC(GCA)P1 was used in place of hc tW(CCA)G1.
Overexpression of Trp-tRNA increased the overall readthrough efficiency with all four tetranucleotides and this effect was in all cases dependent on intact eIF3 (Fig. 2B, compare fold differences between TIF35 wt versus mutant cells). The lowest increase (by approximately threefold) was seen with UGA-U and with UGA-G, which suggests that these two tetranucleotides incorporate this particular nc-tRNA with the same low preference. The UGA-C tetranucleotide displayed an intermediate increase (by approximately sixfold); i.e., twice above the UGA-U and -G levels. We ascribe this effect to the specific interaction between UGA-C and eRF1 mentioned above, which allows the highest fold change of readthrough (by approximately twofold) among all four tetranucleotides under physiological conditions (Fig. 1A). The highest, strongly eIF3-dependent increase of readthrough (by approximately ninefold) was upon Trp-tRNA overexpression observed with the UGA-A tetranucleotide, which is with respect to the basal readthrough efficiency otherwise comparable to UGA-G (Fig. 1A). These results thus suggest that tW(CCA)G1 has a lot higher preference for recognition of the A-containing tetranucleotide over the other three, and that the difference in the basal readthrough efficiency on UGA-G or -U is not caused by better incorporation of Trp-tRNA.
Overexpression of Cys-tRNA showed essentially the same “low preference” increase of readthrough (by threefold) as Trp-tRNA with UGA-U and, this time, with UGA-A instead of the UGA-G tetranucleotide (Fig. 2C). UGA-G conversely displayed an increase in readthrough levels by approximately sixfold and UGA-C by ∼12-fold. Note that the 12-fold increase with UGA-C cannot solely account for the readthrough increasing effect of the C4–eRF1 specific interaction and hence we propose that Cys-tRNA preferentially recognizes the C- and G-containing tetranucleotides. Unfortunately, our approach does not allow the dissection of the degree of specific contribution of the C4–eRF1 interaction versus the Cys-tRNA-driven preference for the UGA-C tetranucleotide in the overall effect. Our observation that Cys-tRNA increased the fold exchange of readthrough at UGA-G only sixfold compared to ninefold for Trp-tRNA at UGA-A is probably due to the fact that Cys-tRNA is a weaker competitor with eRF1 for the UGA stop codon between the two nc-tRNAs, and/or that it is naturally less abundant tRNA in our genetic background than Trp-tRNA. Importantly, all effects were dependent on intact eIF3, as expected.
In contrast, control experiments with arginine [tR(UCU)E] and glycine [tG(UCC)O] tRNAs, which are also near-cognate for UGA but with the mismatch at the first position and, at least in our experimental setup, do not incorporate at UGA even when overexpressed (Beznosková et al. 2015), showed no preference for any tetranucleotide (Supplemental Fig. S1). Notably, Northern blotting revealed that all four nc-tRNAs that were overexpressed in this study show increased levels by ∼1.5- through sevenfold, depending on the number of isogenes encoding a corresponding nc-tRNA, with the control Arg and Gly tRNAs displaying ∼1.9- and sevenfold increases, respectively (Supplemental Fig. S2). To rule out the “genetic background” effects, we examined two additional genetically unrelated strains and observed consistent tetranucleotide-specific preference of UGA-A for tryptophan and UGA-G for cysteine nc-tRNAs (Supplemental Fig. S3); please note varying fold increases among the tested strains most probably reflecting different endogenous levels of at least these two nc-tRNAs in these backgrounds. Hence we conclude that in contrast to the UGA-U tetranucleotide, UGA-A and UGA-G tetranucleotides are preferentially read through by tryptophan and cysteine nc-tRNAs, respectively, which is the fact that may markedly contribute to the differences in termination efficiency among these three tetranucleotides. Our findings also indicate that the frequency of preferential incorporation of nc-tRNAs at corresponding stop codons or PTCs will most probably vary with varying endogenous levels of individual nc-tRNAs in individual cell types.
Neither the eRF1 decoding ability nor the geometry of the decoding pocket determines the UGA-N tetranucleotide preference for specific nc-tRNAs
To rule out that the observed UGA-N tetranucleotide preference for nc-tRNAs is caused by structural changes that different tetranucleotides may impose on the geometry of the decoding pocket, we measured the effect of overexpression of nc-tRNAs in the presence of 200 µg/mL paromomycin. The miscoding agent paromomycin disables ribosomal discrimination against nc-tRNAs by specific altering of the geometry of the A-site codon decoding pocket, so that eRF1 can no longer actively sense the correct Watson–Crick base-pairing geometry (Bidou et al. 2012). In TIF35 wild-type cells bearing an empty vector (EV), paromomycin increased readthrough with all four tetranucleotides by a similar fold, as expected (Supplemental Fig. S4). In paromomycin-treated cells overexpressing the Trp-tRNA, however, the highest increase in readthrough compared to cells bearing EV was seen with the UGA-A and the lowest with UGA-G tetranucleotides (Fig. 3A). Conversely, cells overexpressing the Cys-tRNA displayed the highest increase in readthrough—compared to EV—with UGA-G and -C, and the lowest with UGA-A tetranucleotides (Fig. 3B). The fact that the use of paromomycin had practically no effect on the tetranucleotide preference of both nc-tRNAs suggests that it is the specific nature of these tRNAs and not the geometry of the decoding pocket that enables them to selectively sense the nature of the base occurring at the +4 position. To support this suggestion even further, we overexpressed these nc-tRNAs in sup45-M48I, which is known to directly impair the stop codon decoding and observed virtually the same effects as in the previous two set-ups (Fig. 4), with the exception of UGA-U that, for some reason, showed increased readthrough in this particular mutant (see also Fig. 1A). In detail, the UGA-A tetranucleotide allowed the highest levels of readthrough with tW(CCA)G1 overexpressed (∼4.5-fold), whereas UGA-G (and to a smaller degree also UGA-C) had the same effect with tC(GCA)P1 overexpressed (approximately six- and fourfold). Hence we conclude that the observed UGA-N tetranucleotide preference of nc-tRNAs with a mismatch at the wobble position is highly specific, at least for the termination leakiest UGA stop codon, and most probably reflects some intrinsic tetranucleotide decoding properties of these tRNAs that have not been observed before.
FIGURE 3.
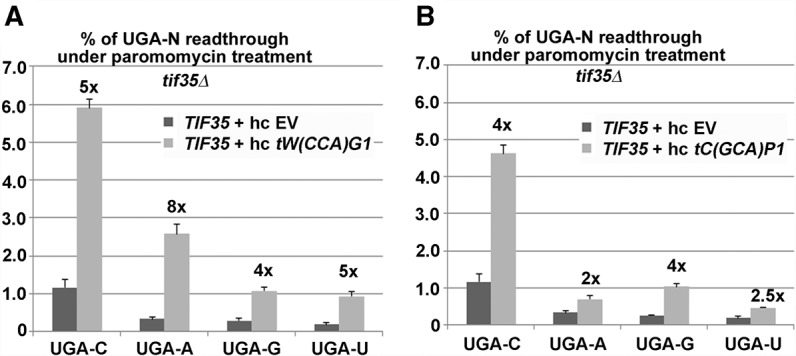
Effect of the miscoding agent paromomycin on hc tryptophan or cysteine nc-tRNA incorporation at UGA-N termination tetranucleotides. (A) The PBH140 derivative bearing TIF35 wt was transformed with either empty vector (EV) or hc tW(CCA)G1, and the resulting transformants were grown in SD with 200 µg/mL paromomycin for 6 h and processed for stop codon readthrough measurements as described in Figure 1. (B) Essentially the same as in A, except that hc tC(GCA)P1 was used in place of hc tW(CCA)G1.
FIGURE 4.
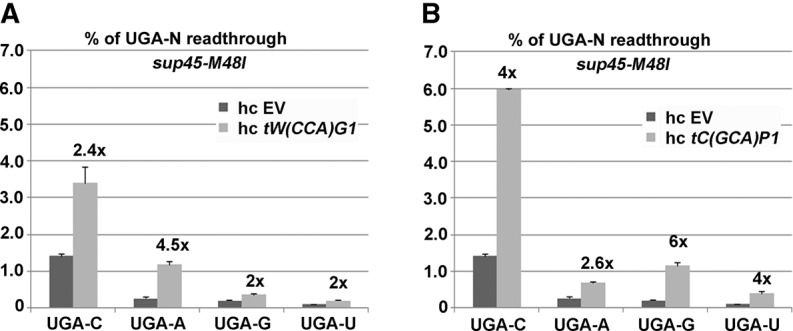
Effect of the eRF1 (sup45-M48I) mutation with impaired decoding ability on hc tryptophan or cysteine nc-tRNA incorporation at UGA-N termination tetranucleotides. (A) The L2327 was transformed with either empty vector (EV) or hc tW(CCA)G1 and the resulting transformants were grown and processed for stop codon readthrough measurements as described in Figure 1. (B) Essentially the same as in A, except that hc tC(GCA)P1 was used in place of hc tW(CCA)G1.
To understand what these properties might be, we compared primary sequences of the anti-codon loop of both tW(CCA)G1 and tC(GCA)P1 and observed the only difference at base 32, indeed with the exception of the anticodon itself (base 34) (Fig. 2A). The tryptophan tW(CCA)G1 tRNA has methylated C (Cm) at position 32, whereas the cysteine tC(GCA)P1 tRNA carries U, which is during maturation modified to become pseudouridine (Pineyro et al. 2014). We hypothesized that by swapping these nucleotides we may possibly change the tetranucleotide preference of these two nc-tRNAs, provided that position 32 plays a critical role in it. However, the C32U replacement rendered the tW(CCA)G1 tRNA inactive in our assay when in high copy (Supplemental Fig. S5). In addition, it produced a modest dominant negative slow growth phenotype (data not shown), suggesting that C32 is critically required for a proper function of this nc-tRNA in general. The U32C replacement of tC(GCA)P1 had virtually no effect on its tetranucleotide preference (Supplemental Fig. S5). Hence the question of what determines the intrinsic ability of nc-tRNAs to sense the nucleotide content of the fourth position remains open.
eIF3-promoted readthrough does not interfere with the eIF3-promoted reinitiation on REI-permissive uORF1 from the GCN4 mRNA leader
We previously proposed that following elongation, eIF3 associates with any pretermination complex regardless of the character of the stop codon, and its context, however, acts to promote readthrough only when the stop codon is programmed (Beznosková et al. 2015). At the same time, eIF3 was—thanks to its favorable location on the solvent-exposed side of the 40S ribosome (Valášek et al. 2003; Erzberger et al. 2014)—demonstrated to be one of the few initiation factors that is retained on 80S ribosomes translating short upstream ORFs even after subunit joining (Pöyry et al. 2004; Szamecz et al. 2008; Munzarová et al. 2011). More specifically, if the short uORF is not longer than 5–10 codons, eIF3 does not dissociate from the 40S subunit but remains 80S-bound even during elongation until the 80S ribosome reaches the stop codon to become a pre-TC. Some uORFs make use of this specific eIF3 ability and by engaging their 5′ sequences in a specific interaction with the N-terminal domain of the a/TIF32 subunit of eIF3 formed during termination, they prevent the small ribosomal subunit from being recycled. This enables the 40S subunit to subsequently resume scanning downstream until the next AUG has been encountered for translation REI on the same mRNA molecule. The intriguing fact that the eIF3 presence in pre-TCs formed at stop codons of short uORFs can theoretically stimulate readthrough and REI at the same time, but both activities are by their nature antagonistic, prompted us to investigate their prospective mutual interference with the help of the yeast transcriptional activator GCN4.
The GCN4 mRNA leader contains four short uORFs that together constitute a very sophisticated mechanism of translational control in response to numerous stresses, such as, for example, amino acid starvation (Hinnebusch 2005; Valášek 2012). In particular, uORFs 1 and 2 allow highly efficient resumption of scanning and subsequent REI downstream, whereas REI only modestly permissive uORF3 and REI-nonpermissive uORF4 complete the termination process by recycling both ribosomal subunits to prevent GCN4 derepression under normal growth conditions (Gunišová and Valášek 2014). Under starvation conditions, dramatically decreasing the levels of the Met-tRNA.eIF2.GTP ternary complex (TC), the majority of 40S ribosomes that have resumed scanning past uORFs 1 and 2 will skip uORFs 3 and 4 due to the low TC levels, and eventually rebind the TC on their way from uORF4 to the GCN4 gene to derepress its expression. Hence, the roles of uORF1 and uORF4 especially are the key for the whole regulatory mechanism, with uORF2 serving only as the uORF1 back-up in the case of increased leaky scanning (Gunišová and Valášek 2014).
A possibility that programmed readthrough further increases the complexity of this regulation has never been considered before. As hinted above, increased readthrough on uORFs 1 and 2 evoked by the presence of eIF3 in pre-TCs could counteract their intrinsically high permissiveness for REI. In other words, if both uORF1 and uORF2 displayed higher rates of readthrough, eIF3 would dissociate from 80S ribosomes elongating past their natural stop codons, and the pre-TCs formed at the next stop codon would thus be fully recycled. This would in turn prevent full derepression of GCN4 expression under starvation conditions. Increased readthrough on the ultimate uORF4 could, on the other hand, make the whole regulatory system even tighter because the readthrough interference with REI on this specific uORF would further diminish the already low REI activity that uORF4 under nonstarvation conditions allows.
Inspection of the 6-nt-long sequences following stop codons of these four uORFs indicated that the stop codon of one of them (namely uORF4) could be in fact highly programmed (the 6-nt-long “stop codon context” is a critical determinant of programmed readthrough [Namy et al. 2001; von der Haar and Tuite 2007]). Hence we first monitored readthrough efficiency of the UGA stop followed by the 6-nt context corresponding to all four uORFs and to the GCN4 main ORF. We took the GCN4 stop codon context as a negative control because it has a readthrough nonpermissive character with basal activity comparable to our negative controls (Beznosková et al. 2015), and because it is a genuine part of this regulatory system. Our standard pTH477 construct with the UGA-C stop codon context, which is known to allow relatively high levels of readthrough (Bonetti et al. 1995), was chosen as a positive control for programmed readthrough. All our measurements were carried out under nonstarvation conditions because the major effect of starvation is the reduction of the TC levels with little to no effect on resumption of scanning efficiencies of all four uORFs per se. Hence we assumed that their readthrough efficiencies would not be largely affected too. Our luciferase assay revealed that uORF4 is indeed subject to programmed readthrough as it displayed the eIF3-dependent ∼12-fold higher activity than the GCN4 stop codon context and approximately twofold higher than our UGA-C control (Fig. 5A). This increase is most likely too high to be explained merely by the presence of cytosine at the +4 position. The contexts of the remaining three uORFs also allowed an increase (by ∼2.5-fold) in readthrough compared to GCN4, most probably owing to A at the +4 position compared to U in the case of GCN4, with mild but still significant dependency on intact eIF3. This modestly programmed character could actually suggest that the presence of eIF3 in pre-TCs specifically in the case of uORF1 might indeed interfere with its eIF3-mediated ability to allow resumption of scanning for REI downstream. It is therefore possible that some mechanism evolved to prevent this undesirable interference between eIF3-mediated readthrough versus resumption of scanning to keep the control of GCN4 expression as tight as possible. For example, other sequences besides the 3′ adjacent 6-nt-long stop codon context could nullify this effect. Interestingly, the uORF1 coding region and the 3′ sequence immediately following its stop codon were demonstrated in the past to be absolutely essential for its function in REI (Grant and Hinnebusch 1994). To explore the possibility that the uORF1 coding sequence including its stop codon modifies the readthrough effect of its 3′ sequence context, we reexamined the efficiency of readthrough of all four uORFs comprising their genuine stop codons plus six preceding and six following nucleotides. Interestingly, we found that readthrough of the key REI-permissive ORF1 dropped down significantly approaching the minimal readthrough levels obtained with the corresponding GCN4 sequences surrounding its stop codon (Fig. 5B). In addition, uORF1 readthrough also lost its programmed character defined by the KLF mutant (Supplemental Fig. S6). This is remarkable because generally the influence of the 5′-adjacent sequence on the efficiency of readthrough is considered to be more subtle compared to the effect of the 3′-sequence context (Lee and Dougherty 2012). The other REI-permissive uORF2 that is, however, serving only as the uORF1 back-up showed no significant changes (Fig. 5B). Since both uORF1 and uORF2 terminate at the same UAA stop codon, it is highly likely that the readthrough potential of the uORF1 3′ adjacent sequence was specifically eliminated by its coding sequence and not by the replacement of the programmed UGA stop codon. uORF4 remained programmed as expected (Supplemental Fig. S6), even though its overall readthrough efficiency was reduced, most probably due to the UGA for UAA stop codon replacement (Fig. 5). Here, in contrast to uORF1, the action of the readthrough-promoting eIF3 complex is desirable to keep the uORF4 potential to resume scanning post its translation to a minimum, as proposed above. uORF3 displayed no dramatic change only it lost its programmed character like uORFs 1 and 2 (Fig. 5; Supplemental Fig. S6). Together these results indicate that the key uORF1 naturally evolved to ensure as efficient termination as possible to be able to promote potent REI despite the constant presence of the readthrough-stimulating eIF3 complex. Our data thus point at an interesting phenomenon where two counter-acting regulatory processes exist, both are mediated by the same protein factor, yet their mutual interference is prevented in favor of only one of them.
FIGURE 5.
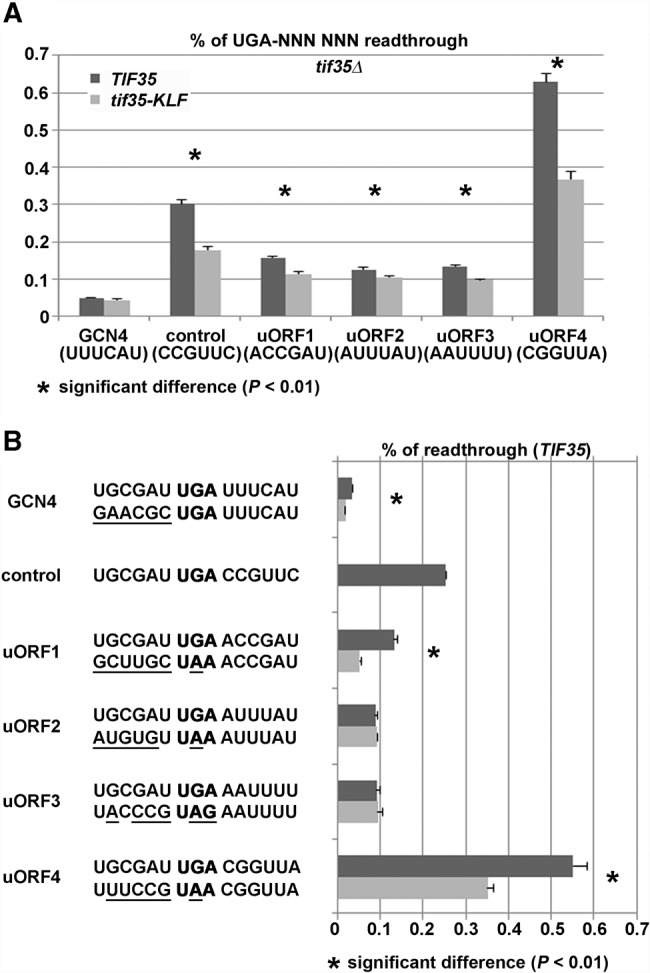
eIF3-promoted readthrough does not interfere with the eIF3-promoted REI on REI-permissive uORF1 from the GCN4 mRNA leader. (A) The 6-nt-long context immediately following stop codons of all four uORFs from the GCN4 mRNA leader allows eIF3-dependent readthrough with varying efficiency. The PBH140 derivatives bearing TIF35 wt and tif35-KLF mutant alleles were grown in SD and processed for stop codon readthrough measurements using standard dual luciferase readthrough reporter constructs pTH460; pTH477; PBB135; PBB136; PBB137; PBB138; and PBB139 as described in Materials and Methods. Changes in the measured readthrough values between TIF35 and tif35-KLF cells were analyzed by the Student's t-test (mean ± SD; n = 6) and shown to be statistically significant only for those cases marked with the asterisk (P < 0.01). (B) The 6-nt-long sequence preceding the stop codon of uORF1 nullifies the eIF3-mediated stimulation of stop codon readthrough on this REI-permissive uORF. The PBH140 derivative bearing the TIF35 wt allele was grown in SD and processed for stop codon readthrough measurements using standard dual luciferase readthrough reporter constructs pTH460; pTH477; PBB135; PBB136; PBB137; PBB138; PBB139; PBB140; PBB141; PBB142; PBB143; and PBB144 as described in Materials and Methods. Changes in the measured readthrough values between selected constructs were analyzed by the Student's t-test (mean ± SD; n = 6) and shown to be statistically significant only for those cases marked with the asterisk (P < 0.01).
DISCUSSION
The major goals of our study were to investigate (i) what is the molecular mechanism by which the +4 base influences the readthrough efficiency, (ii) if there is a preference of nc-tRNAs for incorporation into individual stop codons during readthrough, and finally if these two phenomena are interdependent. Despite the fact that several studies suggested that the sequences surrounding the stop codon had no impact on the identity or proportion of amino acids incorporated during the readthrough process (see for example the most recent one by Blanchet et al. 2014), we thought that there were still several hints suggesting the opposite. For example, numerous reports have described that the surrounding nucleotide context had a major effect on termination efficiency (Bonetti et al. 1995; Mottagui-Tabar et al. 1998; Namy et al. 2001). Especially the identity of a nucleotide following the termination codon (position +4) markedly impacts the readthrough permissiveness of all three stop codons by an unknown mechanism (Dabrowski et al. 2015) that could be in principle connected with nc-tRNA preference for a particular stop codon in a specific tetranucleotide context.
Since UGA is the least stringent of all stop codons with respect to termination accuracy and we and others recently showed that there are only two nc-tRNAs (Trp and Cys) that can efficiently incorporate at UGA (Blanchet et al. 2014; Beznosková et al. 2015), we focused our attention on UGA and these two nc-tRNAs with the mismatch at the third position. A very recent study revealed that besides Trp and Cys, also Arg can relatively efficiently incorporate at UGA (Roy et al. 2015); however, Arg-tRNA with the mismatch at the first position does not work as the specific nc-tRNA in our experimental set-up, because it does not incorporate at the UGA stop codon even when overexpressed (Beznosková et al. 2015). Thus we used it as a negative control together with Gly-tRNA, mispairing with UGA also only at the first position. Our detailed analysis of the preference of Trp-tRNA and Cys-tRNA for the UGA stop codon set in all four possible tetranucleotides (UGA-C, -A, -G, and –U) revealed that it is in fact the identity of the +4 base, which is the main determinant of this preference. UGA-U showed the lowest readthrough, as expected, with no preference for any nc-tRNA when overexpressed. UGA-C displayed the highest readthrough, also as expected, with a preference for Cys-tRNA. A strong preference for the Cys-tRNA was also observed with UGA-G, whereas the UGA-A tetranucleotide showed a strong preference for Trp-tRNA (Fig. 2). Considering that other experiments showed a lack of correlation between absolute tRNA abundance and translation elongation rates (Pechmann and Frydman 2013), we should be able to rule out that our nc-tRNA overexpression readthrough data are nonspecifically influenced by changes in elongation rates. In addition, control nc-tRNAs with a mismatch at the first position did not incorporate at UGA at the frequency above the background and expectedly showed no preference for any tetranucleotide whatsoever (Supplemental Figs. S1, S2).
What determines the observed preference of nc-tRNAs for different tetranucleotides on the molecular level remains to be investigated. Our experiments with paromomycin and the eRF1 mutant defective in decoding (Figs. 3, 4) showing little to no impact on the context-specific preference for nc-tRNAs at least rule out the contribution of the altered geometry of the decoding pocket or some noncanonical influence of eRF1. Hence it is very likely that the nc-tRNAs carry by themselves some specific features that markedly contribute to the decoding mechanism by sensing the nature of the base immediately following the stop codon. In theory these features could be represented by unique bases or some tRNA-specific modifications that the individual tRNAs undergo during maturation. In an effort to address this important question, we tested if the major “nc-tRNA preference discriminator” lies in the anticodon loop, in particular in the N32 base, but obtained inconclusive results (Supplemental Fig. S5). We also selected all nonessential genes shown to be responsible for differential modification of tryptophan and cysteine tRNAs in the past (Pineyro et al. 2014), and measured readthrough efficiency in the corresponding EUROSCARF deletion mutants. None of these mutants, however, produced any significant effect on the observed context-dependent preference for the Trp and Cys nc-tRNAs (data not shown), though we cannot rule out that we missed some modification(s) or their combination(s) that might be the key in this discriminator effect. Actually, the most obvious feature in which these two tRNAs differ is the anticodon itself, in particular N34—the third position base. It undoubtedly occurs directly in the decoding site and thus could be involved in some kind of a contact with the neighboring fourth stop codon base of the stop codon, especially in the light of the most recent observations that the stop codon tetranucleotide adopts a stable U-turn-like geometry that pulls the fourth base into the A-site and thus shortens the mRNA (Matheisl et al. 2015). For example, tryptophan's Cm34 could have some specific influence during the UGA-A decoding, whereas cysteine's G34 during UGA-G decoding (Fig. 2A). Interestingly, recent studies suggested that the position 3 codon–anticodon mispairings could be possible with multiple nonstandard Watson–Crick pairs (A-C > G-G > A-G) (Blanchet et al. 2014; Roy et al. 2015). Hence we speculate that when a nc-tRNA occurs in the A-site with the stop codon tetranucleotide adopting the U-turn, these nonstandard Watson–Crick pairs might form between the fourth stop codon base and the nc-tRNA's N34 to drive the observed “nc-tRNA preference.” New structural studies with various combinations of stop codons in different contexts bound by corresponding nc-tRNAs are very much needed to resolve this important problem.
The presence of cytosine at the +4 base position shows two effects: (i) the highest readthrough levels of all four bases at this position regardless of the stop codon identity, as shown before (Dabrowski et al. 2015); and (ii) the specific preference for Cys-tRNA in the case of the UGA stop codon. Using two eRF1 mutants with different termination defects, we showed that the first effect could arise from a poorer UGA stop codon recognition by wild-type eRF1, which holds true also for the remaining two stops (Fig. 1). Why it is so we do not know; however, the eRF1 role in this C4-specific effect is consistent with an earlier suggestion that the most robust readthrough stimulating effect of C4 could be linked to interactions of mRNA with the translational machinery rather than to interactions of the stop codon with nc-tRNAs (Phillips-Jones et al. 1995). In support, crosslinking experiments demonstrated that the +4 nucleotide in mRNA interacts with eRF1 (Bulygin et al. 2002) and, consistently, the essential K residue from the critical TASNIKS motif in the NTD of eRF1 was in a recent structural study proposed to lie in the proximity of the +4 nucleotide (des Georges et al. 2014). Our results could thus imply that the contact between the K residue and cytosine at the +4 position is qualitatively different from contacts with the other three bases in the same position, which makes it the least stringent base for efficient termination. Alternatively, it is possible that the newly observed stacking interaction between the +4 base and G626 (Brown et al. 2015) could play a specific role in the C4 effect due to the weakest (i.e., the least stable) stacking of C4 with G626 of 18S rRNA. On top of this general effect, it appears that Cys-tRNA can specifically sense C4 when occurring in the context of the UGA stop codon to incorporate more efficiently at this tetranucleotide. This effect is most probably separable from the eRF1 effect; i.e., driven specifically by some intrinsic feature(s) of the Cys-tRNA by itself that remains to be determined experimentally. Sorting out the individual contributions of all possible players in the C4 effect is especially interesting because this most programmed UGA-C tetranucleotide among all is vastly underrepresented in genomes across the species (McCaughan et al. 1995), which could mean that it is predominantly utilized as a trigger for programmed stop codons.
We also analyzed the readthrough efficiency on short uORFs that are critical for maintaining the tight translational control of a main gene by allowing high rates of translational REI, such as in the case of uORF1 from the mRNA leader of the GCN4 transcriptional activator (Hinnebusch 2005; Valášek 2012). We revealed that even though the 3′ context of the uORF1 stop codon showed increased rates of readthrough dependent on eIF3, its 5′ context evolved in a way to practically completely eliminate any readthrough whatsoever (Fig. 5). Importantly, this effect was found to be highly specific only for the key uORF1 feature of the whole GCN4 regulatory network. The last uORF from this network, uORF4, which serves as a barrier in front of the GCN4 gene allowing negligible REI to prevent GCN4 expression under normal growth conditions, gained conversely relatively high readthrough efficiency further fortifying its blocking role. Since both readthrough and REI rely on the eIF3 presence in the pre-TC, our results provide an interesting insight into how natural selection prevented one eIF3-promoted mechanism going by its nature against the other eIF3-promoted mechanism to interfere with the overall regulatory process. These results appear to be the exception to the rule that the 5′ adjacent sequences of the stop codon have a much smaller impact on readthrough efficiency compared to the 3′ context sequences (Lee and Dougherty 2012).
Taken together, our findings (i) demonstrate the importance of the +4 base for the preferred incorporation of nc-tRNAs at corresponding stop codons, and (ii) at least partially explain the highest termination leakiness of all three stop codons with cytosine at their +4 position compared to the other three bases. The readthrough phenomenon concerns not only regular stop codons but also nonsense mutations within coding regions; i.e., PTCs. In fact, it was reported that nonsense mutations account for ∼11% of all described gene lesions causing human-inherited disease and ∼20% of disease-associated single-base pair substitutions affecting gene coding regions (Mort et al. 2008). Hence we believe that our findings reported here, as well as the most recent insights by others (Blanchet et al. 2014; Brown et al. 2015; Roy et al. 2015), may potentially help with the design of new drugs or other effectors to specifically promote readthrough on various PTCs to prevent pathological effects of only partially synthetized proteins. The problem with most of the clinical trials is the efficiency of PTC-readthrough therapies, which is unfortunately not always satisfactory mainly due to unexpected discrepancies in response to the currently used drugs, which are in a majority of cases aminoglycosides (Linde and Kerem 2008; Lee and Dougherty 2012). Our experiments with nc-tRNA's overexpression indicate that changing an intracellular level of a selected endogenous nc-tRNA may in turn change the readthrough specificity of individual tetranucleotides, such as, for example, the case of Cys-tRNA changing the C > A > G > U order of readthrough efficiency to C > G > A > U (Fig. 2B). Taking a look from a different angle, Roy and colleagues recently proposed, but did not experimentally prove, that changing the sequence context and other parameters known to affect readthrough (e.g., the identity of the most 3′ nucleotide to the PTC) will most likely determine the frequency with which an amino acid gets inserted at the PTC (Roy et al. 2015). Hence, it could be proposed that the use of, for example, paromomycin may lead to the observed unexpected variability due to (i) the differing identity of the +4 base following the same type of a PTC, and (ii) differing intracellular levels of endogenous nc-tRNAs among different cell types or organisms. For instance, a paromomycin treatment of the UGA-A PTC in a background with high levels of tryptophan nc-tRNA will have a more potent impact on this PTC suppression than in a background with low tryptophan nc-tRNA levels or when set in the UGA-G context (Fig. 3A). Hence an informed choice of a particular drug that would be best suited to a given PTC mutation of medical interest should include not only the knowledge of the identity of the premature stop codon and its context, but also the knowledge of the intracellular levels of tRNAs that work as near-cognates for the PTC of interest.
MATERIALS AND METHODS
Yeast strains and plasmids
The lists and descriptions of plasmids and yeast strains used throughout this study (summarized in Supplemental Tables S1–S3) can be found in the Supplemental Material.
Stop codon readthrough assays
The majority of stop codon readthrough assays in this study were performed using a standard bicistronic reporter construct bearing a Renilla luciferase gene followed by an in-frame firefly luciferase gene. Separating the two genes is either a tetranucleotide termination signal (UGA-C) or, for control purposes, the CAA sense codon followed by cytosine. In indicated cases the termination signal and/or the following nucleotide context was modified. It is noteworthy that this system avoids possible artifacts connected to the changes in the efficiency of translation initiation associated with the NMD pathway (Muhlrad and Parker 1999), because both Renilla and firefly enzymes initiate translation from the same AUG codon. For further details, see Keeling et al. (2004). All experiments and data analysis were carried out according to the Microtiter plate-based dual luciferase protocol developed by Merritt et al. (2010) and commercially distributed by Promega. Readthrough values are represented as mean ± SD from quintuplicates (n = 5) and each experiment was repeated at least three times.
Northern blot analysis
The Quick RNA miniprep from yeast using glass beads for cell lysis was performed as previously described in Cross and Tinkelenberg (1991). The RNAs were kept in RNase-free water, run on a Criterion Precast Gel 15% TBE-Urea, 1.0 mm (Bio-Rad) and transferred to the 0.45 nylon transfer membrane (Nytran SPC, Whatman). Custom-made 5′ 32P—labeled oligonucleotides were used as probes. Signals were captured in Fuji MS phosphor storage screens, scanned with a Molecular Imager FX (Bio-Rad), and quantified with NIH ImageJ.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to David Bedwell and Sebastian Leidel for material and advice, the Libor Krásný laboratory members, and in particular Michaela Šiková for her help with Northern blotting, Tobias von der Haar for critical reading of the manuscript, and all our laboratory members for fruitful discussions. This research was supported by the Centrum of Excellence of the Czech Science Foundation P305/12/G034, Wellcome Trust grant 090812/B/09/Z (both to L.S.V.), and Charles University in Prague, project GA UK no. 323415 (to P.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054452.115.
Freely available online through the RNA Open Access option.
REFERENCES
- Akhmaloka, Susilowati PE, Subandi, Madayanti F. 2008. Mutation at tyrosine in AMLRY (GILRY like) motif of yeast eRF1 on nonsense codons suppression and binding affinity to eRF3. Int J Biol Sci 4: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznosková P, Cuchalová L, Wagner S, Shoemaker CJ, Gunišová S, Von der Haar T, Valášek LS. 2013. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet 9: e1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznosková P, Wagner S, Jansen ME, von der Haar T, Valášek LS. 2015. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res 43: 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidou L, Allamand V, Rousset JP, Namy O. 2012. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol Med 18: 679–688. [DOI] [PubMed] [Google Scholar]
- Blanchet S, Cornu D, Argentini M, Namy O. 2014. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res 42: 10061–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B, Fu L, Moon J, Bedwell DM. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 251: 334–345. [DOI] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. 2015. Structural basis for stop codon recognition in eukaryotes. Nature 524: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulygin KN, Repkova MN, Ven'yaminova AG, Graifer DM, Karpova GG, Frolova LY, Kisselev LL. 2002. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett 514: 96–101. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. 1991. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65: 875–883. [DOI] [PubMed] [Google Scholar]
- Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. 2015. Translational readthrough potential of natural termination codons in eucaryotes—the impact of RNA sequence. RNA Biol 12: 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Hashem Y, Unbehaun A, Grassucci RA, Taylor D, Hellen CU, Pestova TV, Frank J. 2014. Structure of the mammalian ribosomal pre-termination complex associated with eRF1•eRF3•GDPNP. Nucleic Acids Res 42: 3409–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD. 2012. Control of gene expression by translational recoding. Adv Protein Chem Struct Biol 86: 129–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CH, Cimermancic P, Boehringer D, Sali A, Aebersold R, et al. 2014. Molecular architecture of the 40S•eIF1•eIF3 translation initiation complex. Cell 158: 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Hinnebusch AG. 1994. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol 14: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunišová S, Valášek LS. 2014. Fail-safe mechanism of GCN4 translational control—uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res 42: 5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2012. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86: 45–93. [DOI] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Xue X, Gunn G, Bedwell DM. 2014. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet 15: 371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Dougherty JP. 2012. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol Ther 136: 227–266. [DOI] [PubMed] [Google Scholar]
- Linde L, Kerem B. 2008. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet 24: 552–563. [DOI] [PubMed] [Google Scholar]
- Matheisl S, Berninghausen O, Becker T, Beckmann R. 2015. Structure of a human translation termination complex. Nucleic Acids Res 43: 8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. 1995. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci 92: 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt GH, Naemi WR, Mugnier P, Webb HM, Tuite MF, von der Haar T. 2010. Decoding accuracy in eRF1 mutants and its correlation with pleiotropic quantitative traits in yeast. Nucleic Acids Res 38: 5479–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA. 2008. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29: 1037–1047. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Tuite MF, Isaksson LA. 1998. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur J Biochem 257: 249–254. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. 1999. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol Biol Cell 10: 3971–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzarová V, Pánek J, Gunišová S, Dányi I, Szamecz B, Valášek LS. 2011. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Hatin I, Rousset JP. 2001. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. 2013. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Jones MK, Hill LS, Atkinson J, Martin R. 1995. Context effects on misreading and suppression at UAG codons in human cells. Mol Cell Biol 15: 6593–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineyro D, Torres AG, de Pouplana LR. 2014. Biogenesis and evolution of functional tRNAs. In Fungal RNA biology (ed. Sesma A, von der Haar T), pp. 233–267. Springer International Publishing, Switzerland. [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Leszyk JD, Mangus DA, Jacobson A. 2015. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc Natl Acad Sci 112: 3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, Nielsen KH, Burela L, Hinnebusch AG, Valášek L. 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22: 2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek LS. 2012. ‘Ribozoomin’—translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 13: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Mathew A, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Tuite MF. 2007. Regulated translational bypass of stop codons in yeast. Trends Microbiol 15: 78–86. [DOI] [PubMed] [Google Scholar]
- von der Haar T, Valasek LS. 2014. mRNA translation: fungal variations on a eukaryotic theme. Springer International Publishing, Switzerland. [Google Scholar]
- Zaher HS, Green R. 2009. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 746–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


