Abstract
The Auxin Binding Protein 1 (ABP1) is one of the most studied proteins in plants. Since decades ago, it has been the prime receptor candidate for the plant hormone auxin with a plethora of described functions in auxin signaling and development. The developmental importance of ABP1 has recently been questioned by identification of Arabidopsis thaliana abp1 knock-out alleles that show no obvious phenotypes under normal growth conditions. In this study, we examined the contradiction between the normal growth and development of the abp1 knock-outs and the strong morphological defects observed in three different ethanol-inducible abp1 knock-down mutants ( abp1-AS, SS12K, SS12S). By analyzing segregating populations of abp1 knock-out vs. abp1 knock-down crosses we show that the strong morphological defects that were believed to be the result of conditional down-regulation of ABP1 can be reproduced also in the absence of the functional ABP1 protein. This data suggests that the phenotypes in abp1 knock-down lines are due to the off-target effects and asks for further reflections on the biological function of ABP1 or alternative explanations for the missing phenotypic defects in the abp1 loss-of-function alleles.
Keywords: Arabidopsis, auxin, AUXIN BINDING PROTEIN 1 (ABP1), knock-down mutant, off-target
Introduction
The naturally occurring auxin, indole-3-acetic acid, plays a central role in plant growth and development alone or in orchestration with other plant hormones. Proper sensing and interpretation of fluctuating cellular auxin signals is necessary for mediating a diverse range of developmental and cell biology responses ( Enders & Strader, 2015; Grunewald & Friml, 2010; Paciorek et al., 2005; Petrasek et al., 2006). In the early screens for auxin receptors, Auxin Binding Protein 1 (ABP1) has been identified based on its ability to bind auxin with high affinity ( Hertel et al., 1972; Löbler & Klämbt, 1985) and soon became a prime candidate for an extracellular auxin receptor based mainly on electrophysiological studies utilizing antibodies against ABP1 that showed rapid ABP1-mediated modulation of plasma membrane ion transport in an early step of auxin action ( Barbier-Brygoo et al., 1989; Leblanc et al., 1999). Over the next decades, the auxin-binding activity of ABP1 has been characterized in detail by biochemical studies ( Batt et al., 1976; Napier et al., 2002; Napier & Venis, 1995; Ray et al., 1977) and its protein structure including the auxin-binding pocket has been revealed ( Woo et al., 2002). Phylogenetic studies have shown that ABP1 homologues are present in the genomes of all plant species from bryophytes to flowering plants ( Tromas et al., 2010) with more than one copy present e.g. in the genome of maize, rice, poplar or the moss Physcomitrella patens ( http://phytozome.jgi.doe.gov/pz/portal.html).
Since its discovery, however, the biological importance of the ABP1 protein as a plasma membrane auxin receptor has been a matter of debates, in part because of its predominant subcellular localization in the endoplasmic reticulum (ER) in maize where the conditions for auxin binding are unfavorable ( Habets & Offringa, 2015; Napier et al., 2002). Recently, these discussions were revived by the isolation of two new Arabidopsis abp1 knock-out alleles, abp1-c1 and abp1-TD1 ( Gao et al., 2015) that show no obvious phenotypes under standard growth conditions. The contradiction between this observation and the previously published embryo-lethal phenotypes of abp1 mutants ( Chen et al., 2001; Tzafrir et al., 2004) has recently been clarified by proving that the embryo-lethality of the originally reported alleles abp1-1 and abp1-1s was caused by disruption of the tightly-linked neighboring gene BELAYA SMERT (BSM) rather than knock-out of ABP1 ( Dai et al., 2015; Michalko et al., 2015). This correction and the demonstration of normal embryo development in the abp1 knock-outs ( Michalko et al., 2015) suggest that ABP1 plays no essential role in early Arabidopsis embryogenesis.
The ongoing discussion focuses on the relevance of ABP1 in auxin signaling and other post-embryonic auxin-related biological processes that have been demonstrated using different genetic tools, namely the conditional knock-down (KD) lines, the abp1-5 weak allele harboring a point mutation in the ABP1 auxin-binding pocket and gain-of-function alleles, all of which often provided internally consistent results ( Braun et al., 2008; Čovanová et al., 2013; David et al., 2007; Grones et al., 2015; Robert et al., 2010; Sassi et al., 2014; Tromas et al., 2013; Xu et al., 2010; Xu et al., 2014).
Conditional ABP1 KD lines SS12S6, SS12K9 and abp1-AS have been generated using two fundamentally different approaches of gene or protein down-regulation. In the SS12S6 and SS12K9 lines, ABP1 was inactivated by inducible over-expression of a recombinant immunoglobulin fragment termed single-chain fragment variable (scFv) ( Conrad & Fiedler, 1998). This construct, consisting of the heavy- and light-chain variable domains of a well-characterized anti-ABP1 monoclonal antibody mAb12 ( David & Perrot-Rechenmann, 2001; David et al., 2007; Leblanc et al., 1999) linked by a flexible peptide was additionally fused to the sequence encoding the 3‘KDEL motif to mediate scFv ER-retention in the SS12K9 line, while the SS12S-encoded scFv12 was meant to be secreted to the apoplast. In planta-produced scFv12 was able to pull down ABP1, and reciprocally immuno-precipitation of ABP1 using another antibody was shown to pull down scFv12 ( Tromas et al., 2009). An antisense approach was utilized in the abp1-AS line, where inducible over-expression of full-length ABP1 antisense cDNA led to the formation of duplexes with its sense mRNA, thus preventing ABP1 translation, and potentially also transcription by RNA interference mechanism ( Meister & Tuschl, 2004; Tufarelli et al., 2003). Both antibody- and antisense-based lines use the ethanol-inducible system, which is well established and widely used for the conditional expression of plant genes ( Deveaux et al., 2003; Roslan et al., 2001).
These three abp1 knock-down lines have been instrumental to connect ABP1 function to multiple cellular and developmental processes. For example, they showed defects in shoot and root growth ( Braun et al., 2008; Tromas et al., 2009), cell wall re-modeling ( Paque et al., 2014) or clathrin-mediated endocytosis of PIN auxin efflux carriers ( Dhonukshe et al., 2007; Robert et al., 2010). In contrast, the abp1 gain-of-function transformants promote PIN internalization both in tobacco and Arabidopsis ( Grones et al., 2015; Robert et al., 2010). Contrasting effects of ABP1 KD and gain-of-function lines were shown also in the case of auxin effect on the control of leaf epidermal pavement cells morphogenesis ( Braun et al., 2008; Nagawa et al., 2012) on ROP GTPase activation ( Xu et al., 2010) and on microtubule rearrangement ( Chen et al., 2014; Xu et al., 2014). Furthermore, analysis of ABP1 variants with mutations in the auxin-binding pocket demonstrated the importance of auxin-binding to ABP1 for its gain-of-function phenotypes ( Grones et al., 2015). Altogether, these studies provided an internally consistent picture showing involvement of ABP1 signaling in multiple physiological and cellular processes. These observations were further supported by the finding that loss-of-function mutants in TMK receptor-like protein kinases, that were recently shown to interact with ABP1 in an auxin-inducible manner, show similar phenotypes with abp1 KD mutants ( Xu et al., 2014) which was consistent with the importance of the ABP1/TMK complex-mediated auxin perception in plant development. Recent identification of wild-type looking Arabidopsis abp1 loss-of-function alleles by Gao et al. (2015) thus questions the interpretation of data obtained in the aforementioned studies.
Here, we address the missing phenotypes in the true abp1 null alleles in relation to the strong and consistent morphological defects observed in the conditional abp1 knock-down lines. We show that the morphological phenotypes in SS12S6, SS12K9 and abp1-AS can be generated in the absence of functional ABP1 protein and we discuss possible underlying causes of this.
Material and methods
Plant material and growth conditions
Arabidopsis thaliana mutants used in this study were: abp1-c1, abp1-TD1 ( Gao et al., 2015), abp1-AS, SS12K9, SS12S6 ( Braun et al., 2008; David et al., 2007). A. thaliana Col-0 wild type seeds were obtained from The Nottingham Arabidopsis Stock Centre (NASC, http://www.arabidopsis.info). For in vitro experiments, seeds were surface-sterilized with chlorine vapor, vernalized for 2 days in the dark at 4°C and grown on 1/2 MS 0.8% agar medium with or without 1% w/v sucrose (pH 5.9) on vertical Petri dishes under long day conditions (16 h light/8 h dark) or in complete darkness at 21°C. A sterilized microtube with 500 µl 5% ethanol was placed at the bottom of the plate to induce expression of abp1-AS, SS12K9 and SS12S6 constructs before germination. Plates with 5-day old etiolated or 7-day old light-grown seedlings were scanned on a flatbed scanner, phenotyped by visual examination and used for DNA/RNA extraction.
Genotyping mutants
Ethanol-inducible ABP1 down-regulating lines ( abp1-AS, SS12K9, SS12S6) were genotyped for the presence of the alcR gene encoding the transcriptional regulator of the ethanol-inducible system using primers alcR_for and alcR_rev ( Table 1). Fragments amplified from abp1-c1 with primer pairs ABP1-U409F + ABP1-586R or ABP1-5P + ABP1-586R were digested with BslI, which cuts the WT fragment once and does not cut the mutant fragment; abp1-TD1 was genotyped as described previously ( Gao et al., 2015). Genomic DNA was isolated using the CTAB extraction method. GoTaq G2 polymerase (Promega) and Bio-Rad T100 Thermal Cycler were used for PCR under following conditions: initial denaturation 5 min 98°C; 35–45 cycles (denaturation 30 s at 98°C; annealing 30 s at 55°C, elongation 1 min at 72°C); final elongation 5 min at 72°C. Restriction analysis was performed by adding the restriction enzyme directly to unpurified PCR reaction. Alternatively, Phire Plant Direct PCR Kit (Thermo Scientific by Finnzymes) and QIAquick Gel Extraction Kit (QIAGEN) were used following manufacturer’s instructions to genotype the SS12K9 x abp1-c1 line.
Table 1. Primer sequences used in this study.
| ABP1-U409F | CCTCATCACACAACAAAGTCACTC |
| ABP1-586R | GGAGCCAGCAACAGTCATGTG |
| ABP1-5P | ATGATCGTACTTTCTGTTGGTTCC |
| ABP1-2E | TTGCCAATCGTGAGGAATATTAG |
| pSKTAIL-L3 | ATACGACGGATCGTAATTTGTCG |
| AlcR F | AGAACAAAGAAAGCCAGGA |
| AlcR R | GCGTGAGAGAAAAGATGA |
| TUB2 F | TAACAACTGGGCCAAGGGACAC |
| TUB2 R | ACAAACCTGGAACCCTTGGAGAC |
Quantitative RT-PCR
Total RNA from approximately twenty 8-day old seedlings frozen in liquid nitrogen was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. 2 µg of purified total RNA were used for a reverse transcription reaction using the iScript cDNA Synthesis Kit (BioRad). qRT-PCR was performed using the LightCycler 480 SYBR Green I Master chemistry (Roche) in a LightCycler480 II thermal cycler (Ser. no. 5659, Roche) according to manufacturer’s instructions. cDNA diluted 1:10 in water was used as a template to prepare 5 µL reaction mixture (final volume). Primers used for the quantitative RT-PCR were designed using QuantPrime ( http://www.quantprime.de). The ABP1 cDNA fragment (84 bp in length) was amplified with ABP1-2E and ABP1-586R primers. Arabidopsis Tubulin beta chain 2 ( TUB2, At5g62690) amplified with TUB2-F and TUB2-R primers was used as a reference gene ( Dataset 1). Gene expression was calculated with the 2 -ΔΔCT method ( Livak & Schmittgen, 2001). Results are expressed as the average +/- standard deviation of 2 biological and three technical replicates. Sequences of primers used for genotyping and qRT-PCR analysis are listed in Table 1.
Results
Copyright: © 2016 Michalko J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
All crosses were genotyped for the presence of the alcR transcriptional regulator (first row of the gel images) which is an integral part of the ethanol-inducible cassette in abp1 knock-down lines. The presence of point mutation in abp1-c1 crosses was genotyped by restriction analysis of ABP1 PCR product as described in Gao et al. (2015) (second row of the gel images of abp1-c1 crosses). The presence of the T-DNA insertion in abp1-TD1 crosses was genotyped according to Gao et al. (2015) (second and third row of gel images of abp1-TD1 crosses). GeneRuler DNA ladder mix #0331 (Thermo Scientific) was used as a fragment size standard to determine the approximate size of DNA fragments. Fragment sizes of 1000 bp and 500 bp are indicated.
Copyright: © 2016 Michalko J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Individual samples are annotated with their position on a 384-well plate (column A), the cDNA (column B) and primer pair (column C); the Cp value of each sample is shown in column D. The experiment was performed in two biological (1 or 2 at the last position in column B) and three technical replicates. Figure 3c shows gene expression calculated with the 2 -ΔΔCT method ( Livak & Schmittgen, 2001) from values of ABP1-2E and TUB as a reference gene; using ABP1-5P and/or EF as a reference gene instead gave similar results.
Copyright: © 2016 Michalko J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Segregation of strong morphological defects in conditional abp1 knock-down alleles crossed with abp1-TD1 and abp1-c1 knock-out alleles
To investigate the contradiction between missing phenotypic defects in the loss-of-function abp1 alleles and strong morphological defects of conditional ABP1 down-regulating lines (knock-down; KD), we decided to cross both types of lines to test three possible scenarios: 1) The absence of the strong morphological defects in the abp1-c1 or abp1-TD1 alleles is caused by an adaptation of the plants to the permanent loss of the ABP1 function, which compensates for this deletion; 2) the strong morphological phenotypes induced in the KD lines do not require functional ABP1 and are caused by off-target effects; or 3) both abp1-TD-1 and abp1-c1 lines contain background mutation(s) that suppress the phenotypes caused by the absence of ABP1.
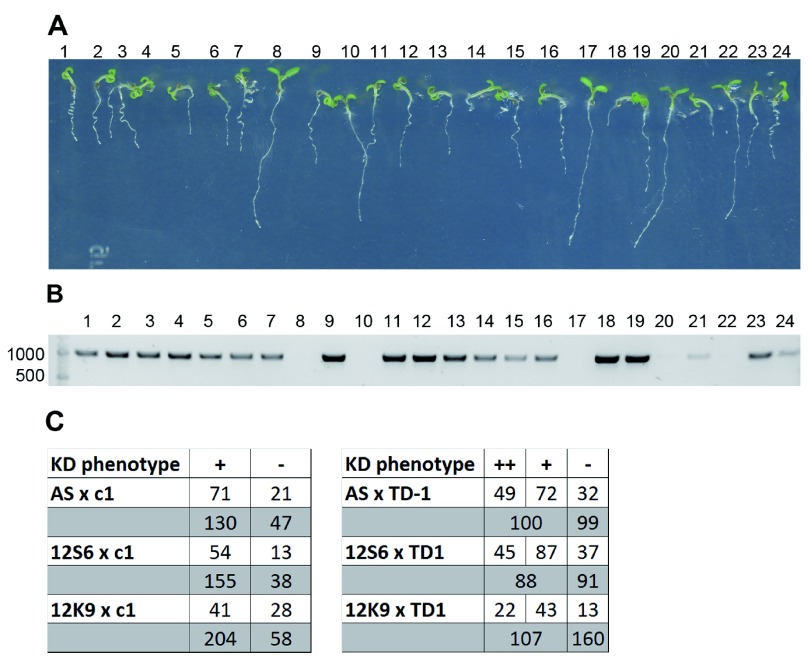
We crossed each of the conditional lines with abp1-TD1 and abp1-c1 null mutants and with an ABP1-WTc1 line as a control and analyzed seedling phenotypes of ethanol induced F2 segregating plants ( Figure 1a). We hypothesized that in case of an adaptive process, the conditional abp1 KD phenotypes (short wavy roots and epinastic cotyledons) would not be manifested in homozygous abp1 null background, resulting in a 9/16 KD and 7/16 WT phenotype segregation ratio. If the inducible phenotypes in the KD lines are independent of ABP1, these phenotypes will be manifested even in the absence of the functional ABP1 gene, thus resulting in a classic Mendelian 3/4 KD and 1/4 WT phenotype segregation ratio. In case of the presence of background suppressive mutation(s), the KD phenotype segregation ratio would lie anywhere between 3/16 (dominant suppressor mutation closely linked to the ABP1 locus) and 3/4 (recessive mutation with low penetrance and no linkage to ABP1) ( Supplementary Figure 1).
Figure 1. Strong morphological defects in conditional abp1 knock-down lines correlate with the presence of the ethanol-inducible cassette and segregate normally when crossed with abp1-c1 knock-out allele.
( A) abp1-AS × abp1-c1 F2 plants grown for 7 days in the presence of 5% ethanol segregate strong morphological defects characteristic of the abp1 conditional knock-down (KD) alleles approximately in a 3:1 ratio. ( B) alcR-specific PCR bands amplified from the genomic DNA of abp1-AS × abp1-c1 F2 plants shown in ( A) demonstrate that the KD phenotype is caused by the presence of the ethanol-inducible insertion. ( C) Phenotypes of the scFv12-based KD lines segregate similarly in F2 crosses with abp1-c1, while altered segregation ratios can be observed in F2 of all three KD alleles crossed to abp1-TD1, which is most apparent in seedlings grown for 5 days in the dark (grey background).
Segregation of the morphological phenotypes in the F2 plants from different crosses is summarized in Figure 1b. These observations show that strong phenotypes in both the abp1 antisense-based and the scFv12-based conditional knock-down lines segregate approximately 75% in the F2 crosses with abp1-c1. This observation favors the scenario that the strong morphological defects in the KD lines are not influenced by the presence or absence of the functional ABP1 gene copy. The F2 phenotypic segregation is however shifted in favor of WT-looking plants in all three KD lines crossed to abp1-TD1. This segregation shift may be ascribed to partial transcriptional silencing of the ethanol-inducible constructs due to the presence of multiple 35S promoters/enhancers in the constructs themselves as well as the tandem T-DNA insertion in abp1-TD1.
We genotyped all analyzed F2 plants for the presence of the alcR transcriptional regulator, which is an integral part of the ethanol-inducible system and verified that the observed morphological defects were indeed correlating with the presence of the ABP1 KD constructs ( Figure 1c). About 5% of seedlings from all lines showed WT phenotype despite being positive for the presence of alcR or vice versa. As this phenomenon was independent of ABP1 genetic background and could not be reproduced in F3 progeny ( Supplementary Figure 2), we put it down to biological variability and/or occasional silencing of the ethanol-inducible constructs.
Strong morphological defects in conditional abp1 knock-down alleles can be manifested in homozygous abp1 knock-out alleles
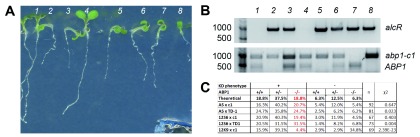
To investigate whether the abp1 KD phenotypes can be observed in the absence of a functional copy of the At4g02980 ABP1 gene we further genotyped the respective abp1 mutations in F2 seedlings of all crosses ( Figure 2). As summarized in Figure 2c, in all crosses we were able to identify multiple homozygous abp1 mutants that showed the strong KD phenotype following ethanol induction. This analysis demonstrates that strong morphological phenotypes in abp1 antisense-based ( abp1-AS) and scFv12 antibody-based ( SS12S6, SS12K9) conditional KD lines can be generated also in a null abp1 background.
Figure 2. Mendelian segregation of strong ethanol-inducible phenotypes in the F2 generation of abp1 knock-out × knock-down crosses is independent of abp1 mutant background.
( A) Representative abp1-AS × abp1-c1 F2 plants, ( B) PCR products amplified from their genomic DNA and ( C) segregation ratios from all crosses show that the ethanol-inducible phenotypes segregate independently of the presence of abp1 knock-out alleles following approximately Mendelian rules for di-hybrid crosses. Homozygous abp1 knock-out mutants with the inducible KD phenotype could be found in all crosses (plants 2,5,8 in ( A) and ( B), red numbers in ( C)), suggesting that the phenotype does not require a functional ABP1 gene. Strong deviations from the expected Mendelian segregation were detected in the SS12K9 × abp1-c1 cross, indicating genetic linkage between ABP1 locus and the inserted ethanol-inducible scFv construct.
In case of the crosses SS12K9 × abp1-c1 and SS12K9 × abp1-TD1 we observed a lower level of allelic segregation between the abp1 mutations and the KD construct in their F2 progeny ( Figure 2c). Out of 28 genotyped plants with WT phenotype, 24 (85.7%) were homozygous for abp1 mutation and did not contain the ethanol-inducible KD cassette. These results point towards genetic linkage between these two loci, most likely caused by the positional effect of the KD cassette located close to the ABP1 locus on the chromosome 4. Nevertheless, some level of genetic recombination was happening between the two loci in the crosses as demonstrated by the identification of three F2 SS12K9 × abp1-c1 plants showing KD phenotype that were homozygous for abp1-c1 mutation ( Figure 2c). This analysis confirms that also SS12K9 conditional KD construct can generate strong morphological phenotypes in the homozygous abp1 knock-out alleles despite the insertion position being linked to the ABP1 locus. Altogether these data are consistent with results obtained by the other crosses and further support that morphological phenotypes in the abp1 knock-down lines can be generated in the absence of the functional ABP1.
Analysis of F3 generation confirms SS12K9-induced strong morphological defects in absence of ABP1 function
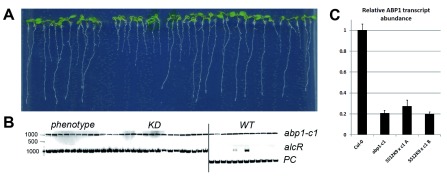
Next we tested the occurrence of the strong KD-induced morphological phenotypes in the absence of the functional ABP1 in the next generation by analyzing the F3 progeny of two SS12K9 × abp1-c1 plants showing strong KD phenotype. We confirmed that the F3 progeny was homozygous for the abp1-c1 mutation and segregated the ethanol-inducible construct approximately in a 3:1 ratio ( Figure 3b). After induction with ethanol, the analyzed F3 population of the SS12K9 × abp1-c1 plant A segregated into 27 plants (67.5%) with KD phenotype and 13 WT looking plants (32.5%) ( Figure 3). The F3 population of plant B segregated into 18 plants with KD phenotype (81.2%) and 4 WT looking plants (18.2%) (data not shown). Genotyping of all F3 plants with ethanol-inducible phenotypes revealed that they contain KD construct in the homozygous abp1-c1 background ( Figure 3b). Notably, among the 17 analyzed WT looking F3 seedlings we also identified two plants that contain the ethanol-inducible construct in homozygous abp1-c1 background ( Figure 3b) suggesting that in these plants the functionality of the construct was affected, most probably by its silencing. Nonetheless, the majority of the plants containing the ethanol-inducible construct generated the strong morphological phenotypes even in the abp1 -/- homozygous background.
Figure 3. Strong inducible knock-down phenotypes in the absence of functional ABP1 gene confirmed in the SS12K9 × abp1-c1 F3 progeny.
( A) Representative seedlings of the ethanol-induced F3 progeny of one of the SS12K9 × abp1-c1 F2 plants (plant A) that showed KD phenotype in the absence of the functional ABP1. All F3 seedlings manifesting KD phenotype were homozygous for abp1-c1 mutation. ( B) Genotyping of the plants shown in A. The image is assemled from different regions of two gels that were copy-pasted next to each other in order to save space. ( C) qRT-PCR analysis of KD-phenotype positive F3 seedlings of both lines revealed that ABP1 transcript levels are reduced by about 80% like in the original abp1-c1 mutant. Altogether these data confirm that in the abp1 down-regulating lines the KD phenotype can be manifested without the ABP1 function. In ( C) average of two biological and three technical replicates +/- SD is shown. PC- positive control.
We also analyzed the ABP1 expression in WT, abp1-c1 and SS12K9 × abp1-c1 F3 seedlings by quantitative RT-PCR just to verify that introducing KD alleles does not influence, in any way, the ABP1 expression ( Figure 3c). We observed ca. 80% decrease in ABP1 transcript levels in abp1-c1. We assume that this difference - somewhat surprising, since the CRISPR-induced small deletion does not necessarily decrease transcript levels - is probably caused by the decreased stability of the mutant mRNA. SS12K9 × abp1-c1 F3 plants positive for the KD phenotype and homozygous for abp1-c1 showed the same 80% decrease in ABP1 transcription.
In summary, the phenotypic, genotypic and expression analyses consistently showed that all three conditional abp1 knock-down alleles can generate strong morphological defects also in the absence of the functional ABP1 protein.
Discussion
Strong morphological phenotypes in abp1 conditional knock-down alleles are not caused by ABP1 down-regulation
All three available conditional abp1 knock-down alleles have been extensively characterized and used to link number of developmental and cellular processes to the ABP1-mediated signaling (for overview, see Grones & Friml, 2015). They are based on two unrelated strategies for down-regulation of the protein’s functionality: the antisense ( abp1-AS) and the scFv12 monoclonal antibody expression ( SS12S6, SS12K9), which suppress the protein functionality by entirely different mechanisms and at different levels ( Tromas et al., 2009). All three lines showed consistent and reproducible results in a number of different laboratories and a number of developmental, physiological and cellular processes.
Nonetheless, our analysis, made possible by the newly available abp1 knock-out lines ( Gao et al., 2015), strongly suggests that these observed and described effects are not caused by conditional down-regulation of the ABP1. This is supported by the fact that all three constructs show the same strong conditional phenotypes in two different homozygous abp1 null alleles. This means that even in the absence of the functional ABP1 protein, the ethanol-inducible constructs are inducing phenotypic defects that were originally ascribed to the down-regulation of ABP1. Therefore, results generated using these lines need to be critically re-interpreted.
Possible modes of action of abp1 conditional knock-down lines
All three types of abp1 KD Arabidopsis lines generate indistinguishable morphological phenotypes. How it is possible that independent lines using fundamentally different approaches for functional down-regulation of a unique target would have in fact the same off-target effects; we do not know. One possible explanation is that the morphological defects are an artifact of the ethanol-inducible expression system. However, control lines generated in parallel using the same vector and expressing the UIDA reporter gene did not exhibit any significant growth and developmental alterations ( Braun et al., 2008). Furthermore, a number of authors have used the same ethanol-inducible system to control the expression of distinct genes and to the best of our knowledge, there are no reports describing similar phenotypes by using the ethanol-inducible system for other genes in other studies ( Battaglia et al., 2006; Deveaux et al., 2003; Laufs et al., 2003; Peaucelle et al., 2008; Roslan et al., 2001). This system was also used to successfully rescue mutant defects after ethanol induction of gene expression e.g. for LEAFY ( Maizel & Weigel, 2004) or for N-myristoyltransferase ( Pierre et al., 2007) indicating that it is not responsible per se of the phenotypes observed with the ethanol inducible ABP1 AS and scFv12 constructs. In tobacco plants and BY-2 cells, tetracycline de-repressible promoter-driven expression of the SS12S and SS12K constructs resulted in similar growth defects as their ethanol-inducible expression in Arabidopsis ( Braun et al. 2008; David et al., 2007), suggesting that the observed phenotypes are tightly correlated to the scFv12 action . The expression of the scFv12 in the cytosol had however no effect on cell proliferation in BY2 cells indicating that expression of scFv12 per se is not sufficient to generate severe phenotypes whatever its cellular localisation and that scFv12 effects are correlated to its secretion and/or retention in the ER that are known location of ABP1 ( David et al., 2007).
Another possibility is that both the antisense- and antibody-based lines have off-target(s) either on the very same gene(s) or elements of a common genetic pathway. Such a hypothesis would be supported by strict similarities in the phenotypes resulting from ABP1 antisense and scFv12 expression and by the fact that opposite and auxin-related defects were observed in both constitutive and conditional gain-of-function Arabidopsis transgenic plants as well as transitionally expressing tobacco cells ( Grones et al., 2015; Robert et al., 2010). ABP1 is placed within the superfamily of cupins based on the presence of cupin-like motifs HXH(X) 11G and P(X) 4H(X) 3N (where X is any amino-acid residue) and a β-barrel jellyroll fold subunit structure ( Dunwell et al., 2004; Woo et al., 2002). The epitope recognized by the scFv12 might be present in proteins belonging to this functionally highly diverse protein superfamily. On the other hand, the sequence similarity of even the closest ABP1 homologues in Arabidopsis does not seem to be sufficiently high to be targeted by the abp1-AS constructs, thus this explanation is unlikely as well.
We also cannot completely rule out that the WT phenotype of the abp1 knock-out mutants is caused by suppressor mutation(s). However, we do not consider it very likely, as this would imply that the similar mutation(s) or mutations with similar effects are present in the genetic background of both abp1-c1 and abp1-TD1, which are independent alleles from independent mutant collections.
In summary, we do not understand how it is possible that the used abp1 knock-down alleles generate the similar strong morphological phenotypes also in absence of the functional ABP1 protein. All possible explanations we could come up with are unlikely, including common off-targets in abp1 antisense and antibody KD lines or common suppressor mutations in two different abp1 knock-out alleles. Thus, more experimentation is needed to figure out what really happens in the different abp1 KD lines and how it is possible that they independently generate phenotypes that are so consistent. Whatever the explanation at the end will be, in light of the presented data it seems obvious that these lines do not act solely by down-regulating the ABP1 function, despite the accumulation of well-fitting data from independent and complementary approaches. It is a sobering realization that even when you use independent approaches with all standard controls performed, there is no real guarantee that the observations will not lead you amiss.
Data availability
The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Michalko J et al.
Data associated with the article are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication). http://creativecommons.org/publicdomain/zero/1.0/
F1000Research: Dataset 1. Scans of ethanol-induced F2 seedlings of crosses (A) SS12S6 × abp1-c1, (B) SS12S6 × abp1-TD1, (C) abp1-AS × abp1-c1, (D) abp1-AS × abp1-TD1, (E) SS12K9 × abp1-c1 and (F) SS12K9 × abp1-TD1 that were used for phenotyping and genotyping ( Figure 1 and Figure 2)., 10.5256/f1000research.7654.d110722 ( Michalko et al., 2016a).
F1000Research: Dataset 2. Agarose gel images from the PCR genotyping of the F2 crosses (A) SS12S6 × abp1-c1, (B) SS12S6 × abp1-TD1, (C) abp1-AS × abp1-c1, (D) abp1-AS × abp1-TD1, (E) SS12K9 × abp1-c1 and (F) SS12K9 × abp1-TD1 ( Figure 3), 10.5256/f1000research.7654.d110723 ( Michalko et al., 2016b).
F1000Research: Dataset 3. Source qPCR data ( Figure 3c), 10.5256/f1000research.7654.d110724 ( Michalko et al., 2016c).
Acknowledgements
We would like to thank Mark Estelle and Yunde Zhao for providing abp1-c1, abp1-TD1 and abp1-WTc1 seeds. We thank Emeline Huault for technical assistance.
Funding Statement
This work was supported by ERC Independent Research grant (ERC-2011-StG-20101109-PSDP to JF). JM internship was supported by the grant “Action Austria – Slovakia”. MG was supported by the scholarship "Stipendien der Stipendienstiftung der Republik Österreich". Work by EH and CPR were supported by ANR blanc ANR-14-CE11-0018.
[version 1; referees: 1 approved
Supplementary material
Supplementary Figure 1. Theoretical genotype and phenotype segregation in F2 progeny of the abp1 knock-out x knock-down cross.
Expected genotype and phenotype segregation ratios for three possible scenarios are shown. S/s = ethanol-inducible cassette positive/negative, A/a = wild-type ABP1/ abp1 knock-out. Genotypes manifesting wild-type phenotype are shown on white background, KD phenotype on green background, genotypes that might exhibit both WT and KD phenotypes are on pale green background.
Supplementary Figure 2. Restoration of the ethanol-inducible phenotype in the progeny of the F2 WT-looking plant from the cross SS12K9 × abp1-c1 containing knock-down cassette and wild-type ABP1 version.
In the presence of 5% ethanol, F3 progeny show homogenous KD phenotype indicating that silencing of the construct might be responsible for wild-type phenotype of this plant in the F2 generation.
References
- Barbier-Brygoo H, Ephritikhine G, Klämbt D, et al. : Functional evidence for an auxin receptor at the plasmalemma of tobacco mesophyll protoplasts. Proc Natl Acad Sci U S A. 1989;86(3):891–895. 10.1073/pnas.86.3.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt S, Wilkins MB, Venis MA: Auxin binding to corn coleoptile membranes: Kinetics and specificity. Planta. 1976;130(1):7–13. 10.1007/BF00390838 [DOI] [PubMed] [Google Scholar]
- Battaglia R, Brambilla V, Colombo L, et al. : Functional analysis of mads-box genes controlling ovule development in Arabidopsis using the ethanol-inducible alc gene-expression system. Mech Dev. 2006;123(4):267–276. 10.1016/j.mod.2006.01.002 [DOI] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, et al. : Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell. 2008;20(10):2746–2762. 10.1105/tpc.108.059048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, et al. : ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 2001;15(7):902–911. 10.1101/gad.866201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H, et al. : Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature. 2014;516(7529):90–3. 10.1038/nature13889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad U, Fiedler U: Compartment-specific accumulation of recombinant immunoglobulins in plant cells: an essential tool for antibody production and immunomodulation of physiological functions and pathogen activity. Plant Mol Biol. 1998;38(1–2):101–109. 10.1023/A:1006029617949 [DOI] [PubMed] [Google Scholar]
- Čovanová M, Sauer M, Rychtář J, et al. : Overexpression of the auxin binding protein1 modulates PIN-dependent auxin transport in tobacco cells. PLoS One. 2013;8(7):e70050. 10.1371/journal.pone.0070050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhang Y, Zhang D, et al. : Embryonic lethality of Arabidopsis abp1-1 is caused by deletion of the adjacent bsm gene. Nat Plants. 2015;1: 15183. 10.1038/nplants.2015.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KM, Couch D, Braun N, et al. : The auxin-binding protein 1 is essential for the control of cell cycle. Plant J. 2007;50(2):197–206. 10.1111/j.1365-313X.2007.03038.x [DOI] [PubMed] [Google Scholar]
- David KM, Perrot-Rechenmann C: Characterization of a tobacco Bright Yellow 2 cell line expressing the tetracycline repressor at a high level for strict regulation of transgene expression. Plant Physiol. 2001;125(4):1548–1553. 10.1104/pp.125.4.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, et al. : The ethanol switch: a tool for tissue-specific gene induction during plant development. Plant J. 2003;36(6):918–930. 10.1046/j.1365-313X.2003.01922.x [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, et al. : Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17(6):520–527. 10.1016/j.cub.2007.01.052 [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S: Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65(1):7–17. 10.1016/j.phytochem.2003.08.016 [DOI] [PubMed] [Google Scholar]
- Enders TA, Strader LC: Auxin activity: Past, present, and future. Am J Bot. 2015;102(2):180–196. 10.3732/ajb.1400285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, et al. : Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci U S A. 2015;112(7):2275–2280. 10.1073/pnas.1500365112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grones P, Chen X, Simon S, et al. : Auxin-binding pocket of ABP1 is crucial for its gain-of-function cellular and developmental roles. J Exp Bot. 2015;66(16):5055–5065. 10.1093/jxb/erv177 [DOI] [PubMed] [Google Scholar]
- Grones P, Friml J: Auxin transporters and binding proteins at a glance. J Cell Sci. 2015;128(1):1–7. 10.1242/jcs.159418 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J: The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 2010;29(16):2700–2714. 10.1038/emboj.2010.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets ME, Offringa R: Auxin Binding Protein 1: A Red Herring After All? Mol Plant. 2015;8(8):1131–1134. 10.1016/j.molp.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Hertel R, Thomson KS, Russo VE: In-vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta. 1972;107(4):325–340. 10.1007/BF00386394 [DOI] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, et al. : Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development. 2003;130(4):785–796. 10.1242/dev.00295 [DOI] [PubMed] [Google Scholar]
- Leblanc N, David K, Grosclaude J, et al. : A novel immunological approach establishes that the auxin-binding protein, Nt-abp1, is an element involved in auxin signaling at the plasma membrane. J Biol Chem. 1999;274(40):28314–28320. 10.1074/jbc.274.40.28314 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔ CT method. Methods. 2001;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Löbler M, Klämbt D: Auxin-binding protein from coleoptile membranes of corn ( Zea mays L.). I. Purification by immunological methods and characterization. J Biol Chem. 1985;260(17):9848–9853. [PubMed] [Google Scholar]
- Maizel A, Weigel D: Temporally and spatially controlled induction of gene expression in Arabidopsis thaliana. Plant J. 2004;38(1):164–71. 10.1111/j.1365-313X.2004.02027.x [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T: Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. 10.1038/nature02873 [DOI] [PubMed] [Google Scholar]
- Michalko J, Dravecká M, Bollenbach T, et al. : Embryo-lethal phenotypes in early abp1 mutants are due to disruption of the neighboring BSM gene [version 1; referees: 3 approved]. F1000Res. 2015;4:1104. 10.12688/f1000research.7143.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Glanc M, Perrot-Rechenmann C, et al. : Dataset 1 in: Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000Research. 2016a. Data Source [DOI] [PMC free article] [PubMed]
- Michalko J, Glanc M, Perrot-Rechenmann C, et al. : Dataset 2 in: Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000Research. 2016b. Data Source [DOI] [PMC free article] [PubMed]
- Michalko J, Glanc M, Perrot-Rechenmann C, et al. : Dataset 3 in: Strong morphological defects in conditional Arabidopsis abp1 knock-down mutants generated in absence of functional ABP1 protein. F1000Research. 2016c. Data Source [DOI] [PMC free article] [PubMed]
- Nagawa S, Xu T, Lin D, et al. : ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 2012;10(4):e1001299. 10.1371/journal.pbio.1001299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier RM, David KM, Perrot-Rechenmann C: A short history of auxin-binding proteins. Plant Mol Biol. 2002;49(3–4):339–348. 10.1023/A:1015259130955 [DOI] [PubMed] [Google Scholar]
- Napier RM, Venis MA: Auxin action and auxin-binding proteins. New Phytol. 1995;129(2):167–201. 10.1111/j.1469-8137.1995.tb04291.x [DOI] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, et al. : Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435(7046):1251–1256. 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- Paque S, Mouille G, Grandont L, et al. : AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in Arabidopsis. Plant Cell. 2014;26(1):280–295. 10.1105/tpc.113.120048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, et al. : Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol. 2008;18(24):1943–1948. 10.1016/j.cub.2008.10.065 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, et al. : PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312(5775):914–918. 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Pierre M, Traverso JA, Boisson B, et al. : N-myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell. 2007;19(9):2804–2821. 10.1105/tpc.107.051870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PM, Dohrmann U, Hertel R: Characterization of naphthaleneacetic Acid binding to receptor sites on cellular membranes of maize coleoptile tissue. Plant Physiol. 1977;59(3):357–364. 10.1104/pp.59.3.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, et al. : ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143(1):111–121. 10.1016/j.cell.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, et al. : Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 2001;28(2):225–35. 10.1046/j.1365-313X.2001.01146.x [DOI] [PubMed] [Google Scholar]
- Sassi M, Ali O, Boudon F, et al. : An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr Biol. 2014;24(19):2335–2342. 10.1016/j.cub.2014.08.036 [DOI] [PubMed] [Google Scholar]
- Tromas A, Braun N, Muller P, et al. : The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS One. 2009;4(9):e6648. 10.1371/journal.pone.0006648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Paponov I, Perrot-Rechenmann C: AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci. 2010;15(8):436–446. 10.1016/j.tplants.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Tromas A, Paque S, Stierlé V, et al. : Auxin-binding protein 1 is a negative regulator of the SCF TIR1/AFB pathway. Nat Commun. 2013;4:2496. 10.1038/ncomms3496 [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, et al. : Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34(2):157–165. 10.1038/ng1157 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, et al. : Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135(3):1206–1220. 10.1104/pp.104.045179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo EJ, Marshall J, Bauly J, et al. : Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 2002;21(12):2877–2885. 10.1093/emboj/cdf291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, et al. : Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 2014;343(6174):1025–1028. 10.1126/science.1245125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, et al. : Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell. 2010;143(1):99–110. 10.1016/j.cell.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]