Abstract
There is strong epidemiologic evidence linking chronic exposure to inorganic arsenic (iAs) to a myriad of adverse health effects, including cancer of the bladder. The present study set out to identify DNA methylation patterns associated with iAs and its metabolites in exfoliated urothelial cells (EUCs) that originate primarily from the urinary bladder, one of the targets of arsenic (As)-induced carcinogenesis. Genome-wide, gene-specific promoter DNA methylation levels were assessed in EUCs from 46 residents of Chihuahua, Mexico, and the relationship was examined between promoter methylation profiles and the intracellular concentrations of total As (tAs) and As species. A set of 49 differentially methylated genes was identified with increased promoter methylation associated with EUC tAs, iAs, and/or monomethylated As (MMAs) enriched for their roles in metabolic disease and cancer. Notably, no genes had differential methylation associated with EUC dimethylated As (DMAs), suggesting that DMAs may influence DNA methylation-mediated urothelial cell responses to a lesser extent than iAs or MMAs. Further analysis showed that 22 of the 49 As-associated genes (45%) are also differentially methylated in bladder cancer tissue identified using The Cancer Genome Atlas repository. Both the As- and cancer-associated genes are enriched for the binding sites of common transcription factors known to play roles in carcinogenesis, demonstrating a novel potential mechanistic link between iAs exposure and bladder cancer.
Keywords: Arsenic, Bladder Cancer, DNA Methylation, Epigenetics, Urothelial Cells
INTRODUCTION
Exposure to inorganic arsenic (iAs) in drinking water is of great concern globally with more than 100 million people exposed to iAs levels exceeding the World Health Organization’s guideline of 10 µg As/L.1,2 Chronic exposure to iAs has been linked to a host of detrimental health effects in humans, including impaired memory and intellectual function, heart and respiratory system disease, liver hypertrophy, and diabetes.3 In addition to these non-cancer endpoints, iAs is a known carcinogen, associated with liver, lung, prostate, skin, and urinary bladder cancers.4 Bladder cancer is of particular concern as an estimated 571,500 individuals are currently impacted by this disease in the U.S. alone5
Decades of research have uncovered several mechanisms that likely underlie iAs-induced carcinogenesis. Specifically, oxidative stress,6 altered DNA repair capacity,7 and alterations in epigenetic-mediated gene regulation8 have all been suggested as potential causal events in iAs-induced carcinogenesis. In relationship to epigenetic modifications, the following changes have been observed in response to iAs exposure: differential microRNA expression,9 posttranslational histone modifications,10 and changes in both global and gene-specific DNA methylation patterns.8,11 These iAs-associated epigenetic shifts may lead to altered expression of target genes/pathways that influence disease state such as cancer.8 Of relevance to the current study, in vitro evidence suggests that iAs exposure may cause alterations in the expression levels of critical genes, such as regulators of cell proliferation and the carcinogenic process, through changes in the DNA methylation maintenance machinery in human uroepithelial cells.12,13 Similarly genes related to cell death and proliferation have also been identified as mutated in human urothelial bladder cancer.14
Responses to iAs exposure, including inter-individual differences in disease susceptibility, are known to be tightly associated with iAs metabolism.15 In vitro evidence has also clearly demonstrated that the toxicity associated with iAs differs from iAs metabolites.16 The capacity to metabolize iAs into trivalent and pentavalent monomethylated and dimethylated arsenicals (MMAs and DMAs, respectively) differs among individuals even with the same exposure levels, influencing the relative amounts of iAs, MMAs, and DMAs excreted in urine.15 Low ratios of urinary DMAs/MMAs, which is thought to be an indicator of low iAs methylation capacity, have been correlated with an increased risk of some of the adverse health effects.17 For example, higher percentages of urinary MMAs and thus lower percentages of DMAs have been associated with increased risk of lung cancer18 and urothelial bladder cancer,19 among others diseases.17 A recent study in Chihuahua, Mexico, showed that As species retained in exfoliated urothelial cells (EUCs) can also serve as indicators of adverse health effects associated with iAs exposure.20
In the present study we set out to investigate epigenetic alterations, specifically cytosine methylation, associated with iAs exposure in an exposed human population and contrast these findings with biomarkers of an iAs-associated disease (i.e. bladder cancer), with the ultimate goal of identifying key genes and biological pathways potentially involved in iAs-induced disease. Specifically iAs and its metabolites were analyzed in relationship to promoter DNA methylation profiles in EUCs of subjects from a recently established cohort based in the Chihuahua, Mexico.20 This region is an area of concern as it is estimated that more than 450,000 people are exposed to iAs levels exceeding 50 µg/L in Mexico.21 The current research aimed to examine associations between DNA methylation profiles in EUCs with trivalent and pentavalent forms of total As (tAs), iAs, MMAs, and DMAs in the same EUCs. Relating these epigenetic changes associated with iAs exposure to a potential disease outcome (i.e. bladder cancer), a common set of differentially methylated genes was identified. Mechanistic similarities were identified between the lists of iAs-associated and bladder cancer-associated genes, where common transcription factors with known involvement in carcinogenesis are known to bind to the promoter regions of the genes. This finding may have functional implications in iAs-induced human carcinogenesis as this investigation represents a direct linkage between DNA methylation patterns and levels of iAs / iAs metabolites within the same target cells.
MATERIALS AND METHODS
Study Population and Sample Collection
All procedures involving human subjects were approved by IRBs in UNC Chapel Hill, U.S. and Cinvestav-IPN, Mexico. Individuals participating in the present study (n=46) represent a subcohort of a larger study (n=374) in Chihuahua, Mexico.20 Study participants were required to be at least 18 years of age and have at least 5 years of uninterrupted residency in the study area. All participants selected for the current investigation were Hispanic females, as the EUC counts in urine from females are significantly higher than males,20 thus producing adequate DNA sample amounts for DNA methylation analysis. Pregnant women and subjects with kidney or urinary tract infections were excluded because these medical conditions could affect iAs metabolism or purity of EUCs isolated from urine. Individuals at risk for occupational exposure to As were also excluded. Household tap water samples were collected for As analysis performed at Cinvestav-IPN (Mexico City). Study subjects were evaluated through medical examination at the Universidad Autónoma de Chihuahua. Body weight and height were recorded and used to calculate the body mass index (BMI). Spot urine samples were collected for As analysis and isolation of EUCs. Urine and EUC samples were frozen and shipped at −70°C for speciation analysis at UNC Chapel Hill.
Determination of Arsenic Species in Household Water and Urine
Concentrations of arsenic in acid-digested water samples were determined in Cinvestav-IPN (Mexico City) using hydride generation atomic absorption spectrometry (HG-AAS) with cryotrapping (CT), as previously described.22,23 Two water samples were not available from the 46 individuals in the study, thus a total of 44 water samples was available for household water analysis. Arsenic species in urine, including iAs, MMAs, and DMAs, were analyzed by HG-CT-AAS.23 Limits of detections (LODs) were determined based upon urine volumes of 200 µL per sample resulting in 0.05 ng As/mL for MMAs, 0.05 ng As/mL for DMAs, and 0.1 ng As/mL for iAs. Total As (tAs) in urine was calculated as the sum of iAs, MMAs, and DMAs. Urinary creatinine concentration was determined by colorimetric assay (Cayman Chemical Company, Ann Arbor, MI), and specific gravity was measured using a digital Atago PAL refractometer (Atago USA, Bellevue, WA).
EUC Isolation
The isolation and purity of EUCs was described in detail in our previous report from the Chihuahua cohort.20 Briefly, EUCs were isolated from urine (100 ml per subject) by centrifugation at 4°C, washed with phosphate buffered saline (PBS), and again centrifuged. EUCs were resuspended in PBS and counted. Resuspended EUCs were checked for the presence of bacteria, yeast, and red/white blood cells using a light microscope. All EUCs used in the present study were free of microbial contamination and contained <5% of red and white blood cells. The cell counts of the collected EUC pellets ranged from 269,500 to 6,049,600 (mean=944,991, median=704,300).
For the purpose of this investigation, all cells isolated from bacteria- and yeast-free urine containing <5% of red and white blood cells were defined as EUCs. As described in detail in our previous report,20 these cells originate from the epithelial lining of the urinary tract, the urothelium. Because the routine microscopy used in this study to assess the collected EUCs cannot reliably distinguish between cells of various origins, it is not possible to further characterize the types and origins of EUCs. It is important to note that the majority of the EUCs originate in the bladder epithelium, as EUCs from female urine have been shown to originate mainly from the vesical trigone area of the bladder.24,25 For this reason, EUCs are also used in clinical practice to diagnose bladder cancer.26 Further substantiating the use of EUCs, a study specifically compared gene-specific promoter methylation patterns in urine sediment DNA versus human bladder cancer tissue and found that, for all patients evaluated, the DNA methylation patterns in urine matched those in bladder cancer tissue.27 Isolated EUCs were stored at −80°C and used for analyses four weeks after collection.
Determination of As Species in EUCs
Trivalent and pentavalent As species were measured in EUC lysates using HG-CT-inductively coupled plasma-mass spectrometry (ICP-MS), as previously detailed.20,28 Concentrations of arsenite (iAsIII), arsenate (iAsV), methylarsonite (MMAsIII), methylarsonate (MMAsV), dimethylarsinite (DMAsIII), and dimethylarsinate (DMAsV) were determined and expressed in ng As/10,000 cells. EUC pellets were lysed in deionized water. The trivalent As species, iAsIII, MMAsIII, and DMAsIII, were directly measured using an aliquot of cell lysate. A separate cell lysate aliquot was treated with 2% cysteine and analyzed for total iAs (iAsIII + iAsV), MMAs (MMAsIII + MMAsV), and DMAs (DMAsIII + DMAsV). Subtracting the values of cysteine-treated samples from the untreated samples presented the concentrations of iAsV, MMAsV, and DMAsV. Total trivalent arsenic (tAsIII) was defined as the sum of iAsIII, MMAsIII, and DMAsIII. Total pentavalent arsenic (tAsV) was defined as the sum of iAsV, MMAsV, and DMAsV. The trivalent and pentavalent total arsenic species were summed to generate tAs(III+V). The instrumental LODs of the HG-CT-analysis ranged from 0.04 to 2.0 pg As. Concentrations of EUC tAs, iAs, MMAs, and DMAs were correlated with each EUC arsenic species as well as urinary tAs, iAs, MMAs, and DMAs using the Spearman Rank Correlation test (TIBCO® Spotfire®, v5.0.0).
DNA Extraction and Methylation Analysis
DNA was extracted from the exfoliated EUCs of 46 subjects using the QIAamp DNA Blood Mini Kit (Qiagen, Valenica, CA) according to manufacturer’s instruction. Enrichment of CpG-methylated DNA was performed using the MethylMiner™ Methylated DNA Enrichment Kit (Invitrogen/Life Technologies, Grand Island, NY) and 250 µg of DNA. Enriched DNA was amplified and hybridized to Affymetrix Human Promoter 1.0R arrays (Affymetrix, Santa Clara, CA) as previously described.11 The Affymetrix Human Promoter 1.0R arrays represent >25,500 human promoter regions, ~13,000 of which contain CpG islands, known targets of DNA methylation. Data were normalized using robust multichip average29 and bioinformatically summarized at the CpG island level based on the Human Genome 18 (HG18) assembly. Average methylation abundance levels mapped to gene promoter regions were used in this analysis, as previously defined.11 Specifically, in order to calculate the average methylation abundance for each gene, the methylation abundance levels for all CpG sites mapped to the same promoter region were summed and then divided by the number of CpG sites for that gene. Microarray data have been submitted to National Center for Biotechnology Information (NCBI) Gene Expression Omnibus repository30 and are available under accession number GSE58499 (www.ncbi.nlm.nih.gov/geo).
Analysis of the Association between DNA Methylation Levels and Arsenic Species Concentrations in EUCs
The associations between DNA methylation levels and As metabolite concentrations in EUCs were evaluated using separate multi-variable models for each As species. In these statistical models, DNA methylation levels were the dependent variables and EUC arsenical levels (i.e. iAsIII, iAsV, iAs(III+V), MMAsIII, MMAsV, MMAs(III+V), DMAsIII, DMAsV, DMAs(III+V), tAsIII, tAsV, and tAs(III+V)) were the independent variables, as previously described.9 The models included covariates that are plausibly related to EUC DNA methylation profiles, specifically: age (continuous variable) and BMI (continuous variable). Differential DNA methylation was defined as a significant association between DNA methylation levels and EUC metabolite levels, where the following requirements were set: (i) p-value < 0.05, and (ii) q-value < 0.10. In addition, as a result of publications that suggest that likelihood of functional change at the mRNA level is linked to methylation abundance,8 a 2-fold change in methylation abundance was required. Fold changes were calculated using the following metric: (average DNA methylation levels of the highest exposed quartile (n=11)) / (average DNA methylation levels of the lowest exposed quartile (n=11)). All genes that passed the statistical filters above were identified as differentially methylated genes associated with EUC As levels. These statistical calculations were carried out using Partek® Genomics Suite™ software (St. Louis, MO).
An additional robust regression analysis of tAs vs. methylation levels was performed using Winsorized regression, as implemented using lmWinsor in the R fda package (version 3.1.2), with the trim=0.1 option. Two analyses were carried out, one using age and BMI as covariates, and the other as a direct regression of residuals for tAs vs. residuals for methylation, after correction for the covariates. P-values for robust regression can be more significant than for standard linear regression if there are skewed observations, which can inflate the standard errors for regression coefficients.
Analysis of the Association between DNA Methylation Levels and Arsenic Species Concentrations in Urine
To identify genes with differential methylation associated with As in urine, similar statistical analyses were carried out as with As in EUCs. Similar multi-variable models were used as previously described, in which the associations between DNA methylation levels and As species in urine were evaluated using age and BMI as covariates. Separate models were used to assess each of the associations between DNA methylation levels and the summed trivalent and pentavalent arsenical levels, specifically urinary iAs(III+V), MMAs(III+V), DMAs(III+V), and tAs(III+V). The same statistical filters were used in these analyses as the analyses with EUC arsenicals, using a multiple test correction q-value filter requirement of 0.002.
Network and Disease/Functional Enrichment Analysis
Network analysis was performed to identify enriched biological pathways amongst the genes with differential methylation associated with one or more EUC arsenicals.9 Networks were algorithmically constructed based on connectivity, enabled through Ingenuity Pathway Analysis (Ingenuity Systems®, Redwood City, CA). Disease signatures and biological functions within the constructed networks were identified using the right-tailed Fisher’s Exact test, as detailed previously.9,31 Over-represented diseases/functions were defined as those that contain more targets than expected by chance using a p-value cut-off of 0.01.
Comparison of EUC As-Associated Genes to the Comparative Toxicogenomics Database (CTD)
In order to evaluate whether there is evidence that the genes with differential methylation associated with EUC As have altered gene and/or protein expression associated with As, the CTD was queried. The CTD is a manually-curated database specific for environmental contaminants and their relationships to genes, including alterations at the mRNA and protein level, collected from published toxicological and epidemiological studies. At the time of the current analysis, the CTD included over 95,000 studies to derive over 15 million toxicogenomic relationships between approximately 11,000 chemicals and 27,000 genes.32
Identifying Bladder Cancer-Associated Genes
In order to identify genes with differential methylation in human bladder cancer, a separate dataset was analyzed from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/), the product of a collaborative effort between the National Cancer Institute and the National Human Genome Research Institute to provide comprehensive data sets for use in cancer research. Publically available data for bladder urothelial carcinoma from TCGA were analyzed.
DNA methylation data were obtained for 18 individuals (the maximum number available) with both tumor and matched non-tumor tissue from the bladder epithelium. Data were filtered to remove probes with missing data for any individuals as well as probes that represent known single nucleotide polymorphisms, as detailed previously,33 after which 335,570 probes remained for analysis. The final DNA methylation files included β-values for 20,256 genes, generated using the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA). The β-value is defined as the ratio of methylated probe intensity to the intensity from both methylated and unmethylated probes, where a β-value of 1 indicates that every copy of that CpG site was methylated.34 Similar to the genome-wide analysis previously detailed, the association between DNA methylation patterns and bladder cancer disease status was evaluated using a multi-variable model. DNA methylation levels were the dependent variables and were contrasted against tumor or non-tumor status as the dichotomous independent variable. Covariates included in the model were age at initial diagnosis, diagnosis subtype (papillary or non-papillary), gender, and smoking status (binary variable). A significant relationship between tumor status and DNA methylation levels was defined for each gene by the following four requirements, which varied slightly from the iAs-associated gene requirements due to differences in the DNA methylation array platforms: (i) p-value < 0.05, (ii) q-value < 0.05, (iii) average β difference (average β value in tumors – average β value in non-tumors) ≥ 0.1 or ≤ −0.1, and (iv) a concordant β difference of ≥ 0.05 or ≤ −0.05 for ≥ 50% of the patients. β difference was calculated as (β value of the tumor specimen – β value of the non-tumor specimen) for each of the patients.
For statistical testing, a permutation test was performed comparing the n=11,837 genes evaluated for differential methylation using the Affymetrix Human Promoter assay and the Illumina Methylation BeadChip platform. A comparison between the number of total genes tested on each platform versus those that were identified as associated with EUC As and bladder cancer were compared using randomly generated lists of the same size using a Chi-squared test.
Transcription Factor Binding Enrichment Analysis
Analysis for enrichment of transcription factor binding sites within the EUC As-associated genes and the bladder cancer-associated genes was carried out using Genomatix (Genomatix Software Inc., Ann Arbor, MI) database version ElDorado 12–2013. Promoter regions were designated as 500 bp upstream and 100 bp downstream of each gene’s transcriptional start site. All differentially methylated genes and their corresponding promoter sequences were analyzed with both a minimum core and a matrix similarity of 0.95, the second highest level of sensitivity possible. Analysis of transcription factor binding enrichment was restricted to transcription factor families associated with vertebrates or general core promoter elements.
Gene-Specific Promoter Methylation Validation
In order to evaluate CpG methylation using an alternate approach, EUC DNA samples were analyzed using a CpG-methylation enrichment followed by gene-specific quantitative real time PCR (qPCR). DNA samples from five subjects within the study cohort were selected with varying levels of EUC tAs. The CpG methylation levels for two genes, specifically proteasome (prosome, macropain) subunit, beta type, 2 (PSMB2) and solute carrier family 12 (potassium/chloride transporter), member 7 (SLC12A7), were evaluated. Enrichment of CpG-methylated DNA was analyzed using the MethylMiner™ Methylated DNA Enrichment Kit (Invitrogen/Life Technologies, Grand Island, NY). For each sample, 200ng of extracted DNA was subjected to methylated DNA immunoprecipitation. Input and immunoprecipitated methylated DNA were assessed with quantitative real time PCR (qPCR) with iQ SYBR Green Supermix (Bio-Rad) using primer pairs designed within the promoter regions for PSMB2 (F: CCCCGACAAACTCCTTCTG; R: ACAGAGAAACTGGGCGTCAC) and SLC12A7 (F: TCCTGGGACTTGTTGAGGAC; R: GCGGTTTGTGTTGTGTGACT) plated in technical triplicate. qPCR cycle threshold (CT) results were generated for the following fractions: (1) Bound DNA, or gene regions that were bound to the beads with methyl-binding protein, representing methylated DNA, (2) Supernatant and wash DNA, or the fraction of DNA that was unbound, representing unmethylated DNA. In order to calculate a relative measure of methylation levels, the following equation was used: Methylation level = 100 – (Bound DNA CT – (Supernatant DNA CT + Wash DNA CT)). These methylation levels were compared to EUC tAs(III+V) levels using Spearman Rank Correlation tests.
RESULTS
Characteristics of Study Population and As Concentrations in EUCs and Urine
Drinking water samples, spot urine, and EUC samples from 46 Hispanic women of the larger Chihuahua cohort20 were analyzed within this subcohort. The average age of these women was 45 and the average BMI was 30.2. Arsenic in drinking water samples ranged from <LOD-275.4 ng As/mL, with an average of 64.1 ng As/mL and a median of 50.7 ng As/mL. Of particular note, 70% (31/44) of the drinking water samples collected were above the WHO’s recommendation of 10 µg As/L (Table 1).
Table 1. Demographic information and arsenic levels in drinking water, urine, and EUCs for the study cohort.
Population characteristics and EUC arsenic levels are also detailed for the 11 subjects with the lowest EUC tAs(III+V) levels and the 11 subjects with the highest EUC tAs(III+V) levels, representing the lowest and highest exposed quartiles when considering EUC tAs levels, respectively.
| mean, median (range) | |
|---|---|
| Age (years) | 45, 45 (20–71) |
| BMI (kg/m2) | 30.2, 29.1 (19.4–49.1) |
| Arsenic in drinking water (ng As/mL)* | 64.1, 50.7 ((<LOD**)−275.4) |
| Urine Creatinine (mg/dL) | 114.3, 110.5 (8.2–397.4) |
| Urine Specific Gravity | 1.01, 1.01 (1.00–1.04) |
| Urine Arsenicals (ng/mL) | |
| Urine iAs | 6.4, 3.8 ((<LOD)−55.3) |
| Urine MMAs | 9.4, 5.2 (0.3–58.2) |
| Urine DMAs | 49.0, 41.6 (1.7, 260.3) |
| Urine tAs | 64.7, 54.2 (2.2–373.9) |
|
Creatinine-Adjusted Urine Arsenicals (ng As/mg Creatinine) |
|
| iAs | 5.6, 3.8 ((<LOD)−45.6) |
| MMAs | 8.4, 6.1 (0.4–48.0) |
| DMAs | 44.6, 36.2 (2.7–214.6) |
| tAs | 58.6, 48.6 (3.4–308.2) |
|
Specific Gravity-Adjusted Urinary Arsenicals (ng As/SG) |
|
| iAs | 8.0, 6.7 ((<LOD)−39.1) |
| MMAs | 12.2, 8.9 (0.8–50.0) |
| DMAs | 64.7, 70.1 (3.8–223.1) |
| tAs | 84.9, 93.1 (7.3–278.9) |
| Urine Percent Arsenicals (%) | |
| iAs | 8.7, 7.6 (0.1–22.5) |
| MMAs | 14.5, 14.0 (5.0–29.3) |
| DMAs | 76.9, 79.6 (52.5, 90.8) |
| Urine Arsenical Ratios | |
| MMAs/iAs | 6.2, 1.6 (0.5, 111.4) |
| DMAs/MMAs | 6.3, 5.9 (1.8–17.0) |
| DMAs/iAs | 34.1, 10.6 (2.6–690.0) |
| EUC Percent Arsenicals (%) | |
| iAs | 69.8, 74.6 (21.0–89.5) |
| MMAs | 13.6, 12.6 (6.8–28.7) |
| DMAs | 16.6, 9.8 (2.6–71.5) |
| EUC Arsenical Ratios | |
| MMAs/iAs | 0.21, 0.18 (0.08–0.44) |
| DMAs/MMAs | 1.64, 0.66 (0.17–9.57) |
| DMAs/iAs | 0.37, 0.13 (0.03–3.41) |
| Lowest Exposed Quartile | |
| EUC tAs(III+V) (pg As/10,000 cells) | 2.14, 2.21 (0.86–3.20) |
| Age (years) | 50, 50 (27–71) |
| BMI (kg/m2) | 29.3, 29.0 (19.4–48.6) |
| Highest Exposed Quartile | |
| EUC tAs(III+V) (pg As/10,000 cells) | 33.84, 34.44 (22.38–46.13) |
| Age (years) | 42, 46 (20–71) |
| BMI (kg/m2) | 30.8, 26.0 (20.9–42.3) |
Arsenic levels in drinking water were determined using n=44 samples
LOD refers to limit of detection.
Because iAs metabolism plays a large role in iAs-associated disease etiology,17 the concentrations of various As species in EUCs were determined. Specifically, tAsIII, tAsV, tAs(III+V), iAsIII, iAsV, iAs(III+V), MMAsIII, MMAsV, MMAs(III+V), DMAsIII, DMAsV, and DMAs(III+V) in EUCs were measured for each of the 46 study subjects (Table 2). Notably, the EUC concentration of tAs(III+V), an indicator of iAs exposure, ranged from 0.9 to 46.1 pg As / 10,000 cells (mean = 14.3 pg As / 10,000 cells). iAsIII, iAsV, and MMAsIII were the major As species in EUCs; MMAsV, DMAsV and DMAsIII were only minor metabolites. The average ratios of EUC arsenicals were as follows: MMAs/iAs = 0.21, DMAs/MMAs = 1.64, and DMAs/iAs = 0.37. In contrast, DMAs was the major metabolite found in urine, followed by MMAs and iAs. The urine metabolite ratios were: MMAs/iAs = 6.2, DMAs/MMAs = 6.3, DMAs/iAs = 34.1 (Table 1). Despite the large differences in arsenical ratios, EUC arsenical concentrations were significantly (p<0.05) correlated with urine arsenical concentrations for each respective species when using either unadjusted, creatinine-adjusted, or specific gravity-adjusted urine arsenical measures (Table S1, Supporting Information). Interestingly, the most significant correlations were found between EUC As concentrations and the unadjusted urine As concentrations for all As species evaluated, tAs, iAs, MMAs, and DMAs. Also of note, the creatinine-adjusted urinary arsenical concentrations showed the least significant correlation to arsenical concentrations in EUCs. Among the EUC arsenic species there were significant (p<0.05) correlations with only one exception, namely the comparison between MMAsIII and DMAsV (p=0.05).
Table 2. Arsenic species concentrations in human EUCs.
Means and medians were calculated across all subjects (n=46).
| EUC Arsenic Species | III | V | III+V |
|---|---|---|---|
| mean, median (range in pg As / 10,000 cells) | |||
| tAs | 10.9, 8.6 (0.4–58.6) |
3.7, 2.4 (0.4–15.4) |
14.3, 11.2 (0.9–46.1) |
| iAs | 8.9, 6.7 (0.2–53.6) |
2.3, 1.1 ((<LOD)−9.6) |
10.9, 8.0 (0.5–38.9) |
| MMAs | 1.7, 1.3 (0.1–8.3) |
0.4, 0.2 ((<LOD)−2.1) |
2.1, 1.4 (0.1–10.3) |
| DMAs | 0.3, 0.2 ((<LOD)−2.5) |
1.0, 0.7 (0.1–4.6) |
1.4, 1.0 (0.2–5.3) |
Gene-Specific DNA Methylation Levels are associated with Concentrations of As Species in EUCs
The relationships between EUC As species and promoter DNA methylation levels were assessed across > 14,000 genes. A total of 49 differentially methylated genes were identified, all of which were hypermethylated in relationship to one or more As species. A total of 47 genes had promoter methylation levels associated with iAs exposure, as characterized by EUC tAs levels, in which 14 genes had promoter methylation levels associated with EUC tAsV and 43 with tAs(III+V). In addition, 3 genes had promoter methylation levels associated with EUC iAsV, 8 with iAs(III+V), 4 with MMAsIII, 0 with MMAsV, and 11 with MMAs(III+V) (Tables 3 and 4, Table S2 and S3, Supporting Information). There were no genes with methylation levels associated with EUC tAsIII, iAsIII, DMAsIII, DMAsV, or DMAs(III+V).
Table 3. Number of genes with differential methylation associated with arsenic species in EUCs.
| Arsenic Species | III | V | III+V |
|---|---|---|---|
| tAs | 0 | 14 | 43 |
| iAs | 0 | 3 | 8 |
| MMAs | 4 | 0 | 11 |
| DMAs | 0 | 0 | 0 |
Table 4. Genes (n=49) with differential methylation associated with at least one EUC arsenic species and their relationship to metabolic disease and cancer.
An “x” indicates a relationship of a gene to metabolic disease and/or cancer signaling. Genes are also specified as being hyper- or hypomethylated in bladder cancer tissue.
| Gene Symbol(s) |
Gene Name | Associated EUC Arsenical(s) |
Metabolic Disease Signaling (p=0.001) |
Cancer Signaling (p=0.002) |
Gene Promoter Hyper- / Hypomethylation in Bladder Cancer |
|---|---|---|---|---|---|
|
AK095619 / DLX6-AS1 |
DLX6 antisense RNA 1 | tAsV, tAsIII+V | |||
|
AK097754 / RGPD4-AS1 |
RGPD4 antisense RNA 1 (head to head) |
tAsIII+V, iAsIII+V, MMAsIII, MMAsIII+V |
|||
| ANKFY1 | Ankyrin repeat and FYVE omain containing 1 |
tAsIII+V | x | Hypomethylation | |
| ANPEP | Alanyl (membrane) aminopeptidase |
tAsIII+V, iAsIII+V, MMAsIII, MMAsIII+V |
x | Hypermethylation | |
| ASAP1 | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
tAsV, tAsIII+V | x | Hypomethylation | |
| ASTN2 | Astrotactin 2 | tAsIII+V | x | ||
| ATP5D | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
tAsIII+V | Hypermethylation | ||
|
AX748071 / RIIAD1 |
Regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
tAsIII+V | |||
|
BC041459 / TARID |
TCF21 antisense RNA inducing promoter demethylation |
tAsIII+V | |||
|
C10orf139 / LINC00200 |
Long intergenic non-protein coding RNA 200 |
tAsIII+V | |||
|
C5orf39 / ANXA2R |
Annexin A2 receptor | tAsV, tAsIII+V | Hypomethylation | ||
| CALHM1 | Calcium homeostasis modulator 1 |
tAsIII+V | x | Hypermethylation | |
| COL12A1 | Collagen, type XII, alpha 1 | tAsIII+V | x | ||
|
CR605298 / FENDRR |
FOXF1 adjacent non-coding developmental regulatory RNA |
tAsIII+V | |||
| EOMES | Eomesodermin | tAsIII+V, MMAsIII+V |
x | Hypomethylation | |
| FBXW8 | F-box and WD repeat domain containing 8 |
tAsIII+V | Hypomethylation | ||
| GGA2 | Golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
tAsV | x | ||
| GPNMB | Glycoprotein (transmembrane) nmb |
tAsV, tAsIII+V | x | x | |
| HIST1H3G | Histone cluster 1, H3g | tAsIII+V, MMAsIII+V |
|||
| IGFBP5 | Insulin-like growth factor binding protein 5 |
tAsV, tAsIII+V | x | x | Hypermethylation |
| IGSF21 | Immunoglobin superfamily, member 21 |
tAsIII+V | Hypermethylation | ||
|
IQWD1 / DCAF6 |
DDB1 and CUL4 associated factor 6 |
tAsV | |||
| KCNT2 | Potassium channel, subfamily T, member 2 |
tAsIII+V | x | ||
| LAMA2 | Laminin, alpha 2 | tAsIII+V, iAsIII+V, MMAsIII+V |
x | Hypermethylation | |
| LRP5L | Low density lipoprotein receptor-related protein 5-like |
tAsIII+V | |||
| LRRC40 | Leucine rich repeat containing 40 |
MMAsIII+V | x | ||
| MAFF | V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
tAsIII+V | |||
| MAPK15 | Mitogen-activated protein kinase 15 |
tAsV, tAsIII+V, iAsV | Hypermethylation | ||
| MEIS2 | Meis homeobox 2 | tAsV, tAsIII+V, iAsIII+V |
x | ||
| METTL13 | Methyltransferase like 13 | tAsV, tAsIII+V, iAsV, iAsIII+V, MMAsIII, MMAsIII+V |
|||
| NCAN | Neurocan | tAsIII+V | x | Hypermethylation | |
| NRSN1 | Neurensin 1 | tAsIII+V | x | Hypermethylation | |
| OR4Q3 | Olfactory receptor, family 4, subfamily Q, member 3 |
tAsIII+V | x | ||
| OR8S1 | Olfactory receptor, family 8, subfamily S, member 1 |
tAsV | Hypermethylation | ||
| PCDHGC5 | Protocadherin gamma subfamily C, 5 |
tAsV, tAsIII+V, iAsV, iAsIII+V, MMAsIII, MMAsIII+V |
x | ||
| PRDM2 | PR domain containing 2, with ZNF domain |
tAsIII+V | x | Hypermethylation | |
|
PRIC285 / HELZ2 |
Helicase with zinc finger 2, transcriptional coactivator |
tAsIII+V | |||
| PSMB2 | Proteasome (prosome, macropain) subunit, beta type, 2 |
tAsIII+V | x | x | Hypermethylation |
| RALGPS1 | Ral GEF with PH domain and SH3 binding motif 1 |
tAsIII+V | x | Hypermethylation | |
| SLC12A7 | Solute carrier family 12 (potassium/chloride transporter), member 7 |
tAsV, tAsIII+V, MMAsIII+V |
x | x | Hypomethylation |
| TAOK2 | TAO kinase 2 | tAsIII+V | |||
| TMSB15A | Thymosin beta 15a | tAsIII+V, iAsIII+V | |||
| TRAF3IP1 | TNF receptor-associated factor 3 interacting protein 1 |
tAsIII+V | x | ||
| UBE3C | Ubiquitin protein ligase E3C | tAsIII+V | x | ||
| USP7 | Ubiquitin specific peptidase 7 | tAsIII+V, MMAsIII+V |
x | x | Hypermethylation |
| VWA3A | Von Willebrand factor A domain containing 3A |
MMAsIII+V | Hypermethylation | ||
| YJEFN3 | YjeF N-terminal domain containing 3 |
tAsIII+V | Hypomethylation | ||
| ZBTB39 | Zinc finger and BTB domain containing 39 |
tAsIII+V | x | ||
| ZNF14 | Zinc finger protein 14 | tAsV, iAsIII+V | x |
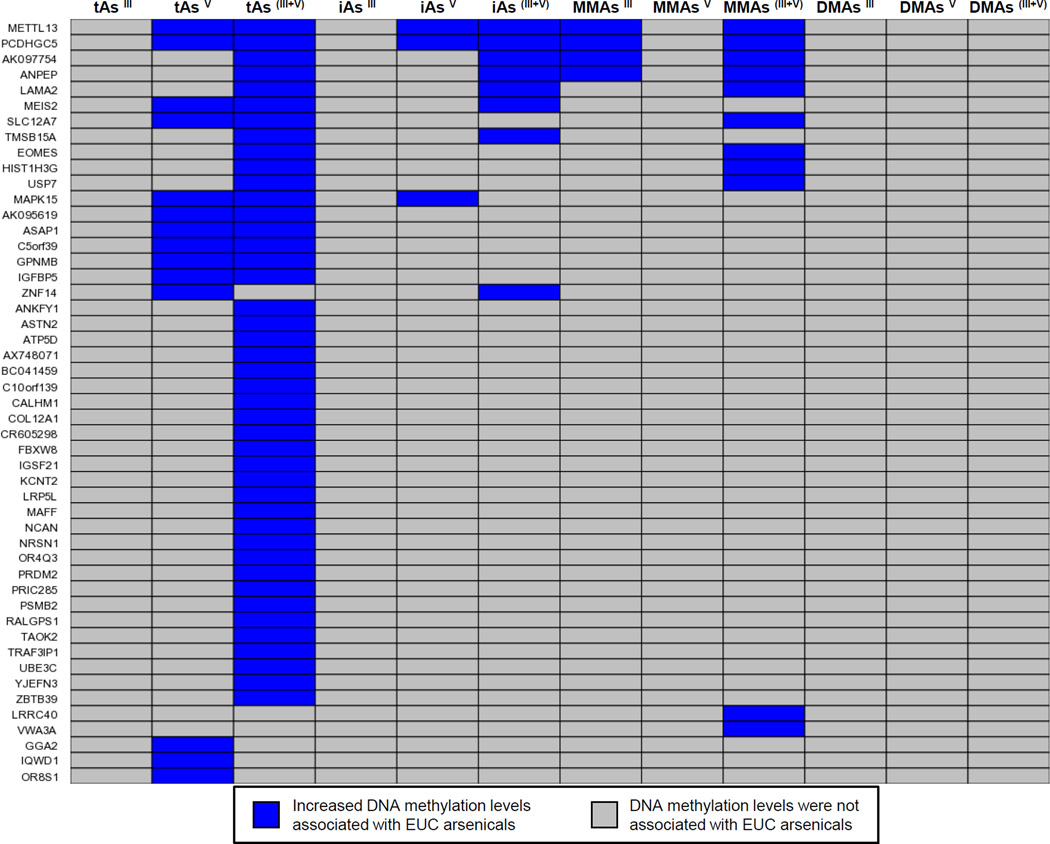
There was considerable overlap among the genes with differential methylation associated with EUC As in relationship to the As species (Figure 1, Table S2, Supporting Information). Specifically, 16% (7 out of 43) of the genes associated with tAs(III+V) were also associated with iAs(III+V) and 21% (9 out of 43) that were associated with tAs(III+V) were also associated with MMAs(III+V). Eleven genes were associated with two or more arsenical groups with summed trivalent and pentavalent species: tAs(III+V), iAs(III+V), and MMAs(III+V). These eleven genes included alanyl (membrane) aminopeptidase (ANPEP), laminin, alpha 2 (LAMA2), methyltransferase like 13 (METTL13), RGPD4 antisense RNA 1 (RGPD4-AS1, also known as AK097754), SLC12A7, and ubiquitin specific peptidase 7 (USP7), among others (Table 4). These eleven genes had fold changes in methylation greater than 2 that when calculated as percent change in methylation represent estimated changes ranging from 0.05 to 0.35 (average = 0.16) associated with tAs(III+V) (Table S3, Supporting Information). To note, while all genes associated with EUC As were hypermethylated, a less stringent statistical filter (p<0.10) would identify both hypo- and hypermethylated genes as associated with EUC As (data not shown). Because the largest number of genes had methylation levels associated with EUC tAs(III+V), descriptive statistics were reported for this As exposure as well as the covariates used in the regression model for each quartile of As exposure (Table 1).
Figure 1.
Heat map illustrating the differentially methylated genes associated with tAs, iAs, MMAs, and/or DMAs concentrations in EUCs. Genes are sorted according to those associated with the highest number of EUC arsenical groups (top) to the lowest number of arsenical groups (bottom).
To investigate potential lack of robustness in our findings of association of tAs with methylation levels, we also performed Winsorized regression using age and BMI as covariates, with 10% trimming of the observations. The results (Figure S1, Supporting Information) showed even stronger association than the original regression-based findings, indicating that the findings had not been driven by a few influential observations.
Gene-Specific DNA Methylation Levels are Associated with Concentrations of As Species in Urine
In order to compare differentially methylated genes identified using EUC As versus urinary As, the relationships between urinary As species and promoter DNA methylation levels were assessed across the 49 genes with differential methylation associated with EUC As. Of these 49 genes, 13 were associated with one or more urinary As species. Specifically, 3 genes showed promoter methylation levels associated with urinary tAs(III+V), and 13 genes showed methylation levels associated with urinary DMAs(III+V) (Table S2, Supporting Information). The three genes associated with urinary tAs(III+V), namely METTL13, SLC12A7, and RGPD4-AS1 (also known as AK095619), were also associated with urinary DMAs(III+V). Similar to the analysis using the EUC As concentrations, all of the differentially methylated genes were hypermethylated in association with urinary As.
Evaluating Gene and Protein Expression Changes using the Comparative Toxicogenomics Database
All cells from the 100-ml urine aliquots were used for DNA isolation. Because of relatively low EUC cell counts, no cells were left for RNA or protein isolation. As a result, it was not possible to directly test whether changes in gene promoter methylation were associated with function changes in transcription or protein expression. As an alternative approach, the CTD, a rich toxicological database containing thousands of published As-associated studies, was queried for known relationships between the 49 genes with differential methylation associated with EUC As and perturbations at the mRNA and protein expression level. The CTD contained information that 12 of the 49 (25%) genes have been shown to be modulated at the mRNA/protein level by iAs/iAs metabolites through in vitro and/or in vivo studies (Table S4, Supporting Information).
Metabolic Disease and Cancer Signaling Pathways are Enriched Among the As-associated Genes
An enrichment analysis identified disease signatures / biological functions to be associated (p<0.01) with the 49 genes with differential methylation related to one or more EUC As species. The two most significantly enriched disease signatures were metabolic disease (p = 0.001) and cancer (p = 0.002) (Table 4). A highly significant (p<10−9) network was constructed using the proteins encoded by the 49 genes associated with EUC As. This network contains 25 proteins encoded by hypermethylated genes related to cancer and 8 proteins encoded by hypermethylated genes associated with metabolic disease (Figure S2, Supporting Information).
Overlap between EUC Genes Differentially Methylated in Response to iAs Exposure and in Bladder Cancer
In order to generate a DNA methylation signature related to a disease outcome pertinent to iAs exposure and to EUCs, a separate database was used to identify genes differentially methylated in bladder cancer. Specifically, the TCGA repository was used to identify 7042 genes with differential methylation in human bladder cancer (Table S5, Supporting Information). Of the 7042 genes associated with bladder cancer, 5542 (79%) showed hypermethylation in the bladder tumors.
Of the 49 genes with differential methylation associated with EUC As, 22 (45%) were also associated with bladder cancer (Table 4, Table S5, Supporting Information). Permutation-based analysis demonstrates that this proportion of overlap is higher than would be expected by chance (p<0.05). Of the 22 common EUC As- and bladder cancer-associated genes, 15 were hypermethylated in bladder cancer tissue versus non-cancerous tissue, including ANPEP, LAMA2, PSMB2, and USP7.
Common Transcription Factor Binding Sites Identified in the EUC As- and Bladder Cancer-Associated Genes
Transcription factor binding site enrichment analysis of the 49 genes with differential methylation associated with EUC As revealed 22 transcription factor families with an enrichment (p<0.05). A matched gene set was queried from the genes with differential methylation associated with bladder cancer where the binding site enrichment analysis identified 27 transcription factor families (p<0.05).
Interestingly, both the EUC As- and bladder cancer-associated genes shared 21 transcription factor families in common, suggesting that common transcription factors may play a role in response to iAs exposure as well as in bladder tumors. These common transcription factor families include Sine oculis homeobox homolog 3 (SIX3), RNA polymerase II transcription factor II B (TF2B), TCF11 transcription factor (TCFF), and Two-handed zinc finger homeodomin transcription factors (ZFHX) (Table S6, Supporting Information).
PCR Promoter Methylation Results
The CpG methylation levels for two genes of interest, PSMB2 and SLC12A7, were tested with quantitative real time PCR (qPCR) using DNA from five subjects within the study cohort. The subjects spanned the range of exposure to As. These two genes were selected for further evaluation because they were both identified as genes with differential methylation associated with EUC As and bladder cancer. Similar to the genome-wide findings, the promoter methylation levels of both genes increased with increasing levels of EUC tAs(III+V) (R=0.90 for PSMB2 and R=0.60 for SLC12A7), where PSMB2 was statistically significant (p=0.04) (Figure S3).
DISCUSSION
This study is the first to characterize gene-specific DNA methylation levels associated with iAs exposure in urothelial cells primarily originating in the human urinary bladder epithelium, one of the tissues targeted in iAs-induced carcinogenesis. Here, we examined the relationship between iAs exposure and iAs metabolism and DNA methylation profiles in EUCs in individuals selected from a recently established cohort in Chihuahua, Mexico. A total of 49 differentially methylated genes were identified, all hypermethylated in relationship to tAs in EUCs (an indicator of iAs exposure) or with As metabolites, iAs and MMAs, retained in EUCs. Interestingly, differences in DNA methylation were most apparent when analyzed in the context of the sum of the trivalent and pentavalent As species (in contrast to the individual metabolites), with the greatest number of genes associated with tAs(III+V). These findings provide novel information regarding iAs exposure and metabolism and its relationship to genome-wide epigenetic modifications in urothelial cells.
Although iAs exposure has previously been associated with both hypo- and hypermethylation in leukocyte-derived DNA,33 here all 49 EUC As-associated genes showed CpG promoter region hypermethylation. Interestingly, if a less stringent statistical criteria is used, genes with both hyper- and hypomethylation are identified. Nevertheless, the observation that the most significant genes show increased methylation associated with iAs exposure is consistent with prior results in which skin lesion status in an iAs-exposed human population was associated with a general trend of gene-specific CpG hypermethylation in peripheral blood leukocytes.11 Gene-specific CpG hypermethylation is also prevalent in DNA from tumors of subjects with diseases relevant to iAs exposure, including cancer.35 Further supporting this relationship to disease, the majority (79%) of bladder cancer-associated genes also were hypermethylated. This trend for gene-specific hypermethylation associated with both iAs exposure and disease relevant to iAs exposure may represent a mechanistic link between exposure and disease as these genes may have silenced expression.
In the present study, there were no differentially methylated genes associated with EUC DMAs concentrations. This finding is consistent with a previous study where DMAs levels in urine were found to influence DNA methylation patterns in peripheral leukocytes to a lesser extent than the urinary levels of iAs and MMAs.36 Furthermore, a previous epidemiological study on an iAs-exposed population located in Taiwan found significantly higher percentages of MMAs and lower percentages of DMAs in the urine of patients with urothelial carcinoma in comparison to healthy residents.37 Together, results of this study and the previously published data suggest that accumulation of iAs and/or MMAs in urine and target tissues, which may indicate an inefficient iAs methylation, may play a significant role in both the differential DNA methylation and health risks associated with iAs exposure.
It is important to note that the data showed that the proportions and ratios of As species in EUCs differed from those present in urine, a trend that was also observed in the larger cohort.20 Specifically, iAs(III+V) were the most abundant species in EUCs, while DMAs(III+V) were the most abundant urinary metabolite in both the larger cohort20 and the subcohort analyzed in the present study. Another difference between EUC and urinary As results were the number of differentially methylated genes identified. Specifically, more genes were associated with urinary DMAs than urinary tAs, while this trend was reversed in the EUC As findings. Thus the DNA methylation associations would be influenced by the use of intra-cellular or urinary measures of As as biomarkers of exposure. These findings further suggest that the identified genes with differential methylation likely depend upon the present and proportion of As metabolites in EUCs. Regardless of these differences, it is important to note that the EUC arsenical concentrations were significantly correlated with the urinary arsenical concentrations. These findings provide important information for future investigations into DNA methylation patterns in relationship to biomarkers of iAs exposure in humans.
The 49 genes with differential methylation associated with EUC As are known to play a role in metabolic disease and cancer. This finding is intriguing, since we recently found that the accumulation of iAs and MMAs in EUCs from subjects in the Chihuahua cohort is linked to an increased prevalence of diabetes and cardiometabolic risk factors (Mendez et al., submitted). Among these 49 genes was USP7, a regulator of diabetes-related signaling.38 Of interest given known links between iAs exposure and diabetes,3,39 knockdown of USP7 in primary murine hepatocytes was shown to increase the expression of FoxO1-target gluconeogenic genes and increase glucose production.40 Furthermore, in relationship to cancer, USP7 activity plays a role in oncogenesis where elevated USP7 expression has been linked to cancers of the bladder, colon, prostate, liver, and lung.41 Notably, cancer of the bladder, prostate, liver, and lung are among the known adverse effects of iAs exposure.4 Previous studies from our lab have linked iAs exposure with alterations in gene methylation/signaling related to metabolic disease and cancer.9,11,36,42,43 The current study further supports the hypothesis that iAs and/or its metabolites may alter these key cellular signaling pathways through epigenetic modifications, specifically through the alteration of DNA methylation profiles in target tissues.
In order to determine whether the DNA methylation signature in EUCs from iAs-exposed individuals may show commonalities with the genes altered in bladder cancer, the EUC As-associated genes were compared against bladder cancer-associated genes analyzed from the TCGA repository. A total of 22 of the 49 As-associated genes (45%) were also differentially methylated in bladder cancer. Of these genes, 15 showed increased promoter methylation associated with both bladder cancer and EUC As, including USP7. In support of our findings, this gene has been previously identified to be hypermethylated in human colorectal cancer tissue.44 while many studies have linked iAs exposure with increased prevalence of bladder cancer in exposed populations,4,37,45,46 these data provide novel evidence that the epigenetic dysregulation of cancer-related genes may play an important role in the carcinogenic effects of iAs exposure.
Changes in DNA methylation levels do not always contribute to functional changes in gene expression. For example, a recent report suggests a small fraction of iAs-associated DNA methylation changes may relate to functional changes at the transcript level.33 However, DNA methylation within CpG islands located within gene promoter regions have been shown to be the most predictive of transcriptional changes,33 and as such the present analysis focused on CpG sites within gene promoter regions. As RNA and protein were not available from the limited cell number of EUCs, it was not possible to test whether the observed changes in DNA methylation were directly related to functional change in mRNA levels. Nevertheless, a total of 12 (25%) of the 49 differentially methylated genes have been previously associated with As-induced changes in mRNA/protein levels. These findings provide support for potential links between iAs-associated DNA methylation and functions changes in expression.
We have hypothesized that transcription factors, through their binding to specific regions of DNA and subsequence occupancy, may play a role in determining DNA methylation patterning that occurs in response to environment exposures, resulting in “environmental footprints”.47 The current study’s in silico evidence further supports a plausible role of transcription factor occupancy in the regulation of As exposure-associated DNA methylation patterns. A total of 21 transcription factor families were identified with enriched binding sites in the promoter regions of EUC As and bladder cancer-associated genes. This is of great interest as transcription factors are not only key molecules involved in the regulation of gene expression influenced by DNA methylation but also potential regulators of the DNA methylation profiles themselves.48 The four most significantly enriched transcription factor families, SIX3, TF2B, TCFF, and ZFHX, have known associations with human cancers.49–52 Of particular interest, dysregulation of two members of the ZFHX family, zinc finger E-box binding homeobox 1 (Zeb1) and zinc finger E-box binding homeobox 2 (Zeb2), has been associated with bladder cancer.52,53 Overall, the extensive overlap between transcription factor binding sites within genes with differential methylation associated with EUC As and bladder cancer indicates that similar transcription factors may mediate both exposure and disease-related epigenetic effects. This finding may have clinical implication as the identified transcription factors could be targeted for disease prevention associated with environmental exposure to iAs.
While this study provides an increased understanding of As exposure and its link to epigenetic effects within cells derived primarily from the bladder, it is not without limitations. The TCGA data used to identify the bladder cancer-associated genes contains data from humans with differing demographics than those used in the As analysis and was analyzed using a different methylation platform. The TCGA also includes data from individuals with invasive bladder cancer, while the EUCs were collected from apparently healthy individuals. Despite these differences, 22 genes were identified as common among the genes with differential methylation associated with EUC As and bladder cancer, representing important gene targets potentially linking iAs exposure and disease. It is likely that even more overlaps would be identified if exposure and disease status (e.g. bladder cancer) were established in the same individual and study cohort. Nevertheless, such a comparison is currently limited by the available epigenetic repositories and the scope of medical examination that is feasible in a field study, similar to the present study involving the cohort in Chihuahua.
In summary, the results from the present study demonstrate that iAs exposure and specific As metabolites retained in EUCs are associated with the altered promoter methylation of key cancer and metabolic disease associated genes. Future studies will further examine biomarkers of iAs exposure and metabolism and disease status while minimizing population variability. Taken together this research provides novel evidence of associations between iAs and its metabolites with DNA methylation profiles within EUCs that primarily originate from a human tissue directly targeted by iAs exposure, the bladder epithelium. Results from this study provide important knowledge of potential mechanistic links between environment exposure to iAs and human health outcomes.
Supplementary Material
Acknowledgments
Funding
This project was supported by the National Institutes of Health, specifically from the National Institute of Environmental Health Sciences (T32-ES007018, P30-ES010126, P42-ES005948, RO1-ES019315, R01-ES015326). Support was also provided by the UNC Nutrition Obesity Research Center (grant no. DK056350) and the Center for Environmental Health and Susceptibility (grant no. P30ES010126). This project was partially supported by the academic network “International Consortium of Research on Environmental Contaminants and their Health Effects” from PROMEP-SEP.
The authors would like to thank Nikhil Umesh and Elizabeth Martin for their assistance with the statistical analyses and Katie A. Bailey for her contributions to the sample preparation and processing.
ABBREVIATIONS
- As
arsenic
- AsIII
arsenite
- AsV
arsenate
- BMI
body mass index
- EUCs
exfoliated urothelial cells
- DMAs
dimethylated arsenic
- DMAsIII
dimethylarsinite
- DMAsV
dimethylarsinate
- EOMES
eomesodermin
- iAs
inorganic arsenic
- LAMA2
laminin alpha 2
- METTL13
methyltransferase like 13
- MMAs
monomethylated arsenic or methylarsenic
- MMAsIII
methylarsonite
- MMAsV
methylarsonate
- PSMB2
proteasome (prosome, macropain) subunit, beta type, 2
- RGPD4-AS1
RGPD4 antisense RNA 1
- SLC12A7
solute carrier family 12 (potassium/chloride transporter), member 7
- SIX3
sine oculis homeobox homolog 3
- tAs
total arsenic
- TCFF
TCF11 transcription factor
- TF2B
RNA polymerase II transcription factor II B
- USP7
ubiquitin specific peptidase 7
- ZFHX
two-handed zinc finger homeodomain transcription factors
Footnotes
The authors declare no competing financial interests.
ASSOCIATED CONTENT
Supporting Information Available
Figures showing additional genome-wide statistical results, network signaling related to EUC As, and PCR validation results. Tables detailing further arsenic measures, differentially methylated genes, CTD results, and associated transcription factors. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Uddin R, Huda NH. Arsenic poisoning in bangladesh. Oman Med. J. 2011;26:207. doi: 10.5001/omj.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. First addendum to 3rd addition. Geneva, Switzerland: WHO (World Health Organization) Press; 2006. Guidelines for drinking water quality. [Google Scholar]
- 3.Kapaj S, Peterson H, Liber K, Bhattacharya P. Human health effects from chronic arsenic poisoning--a review. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2006;41:2399–2428. doi: 10.1080/10934520600873571. [DOI] [PubMed] [Google Scholar]
- 4.NTP. Report on Carcinogens. Twelfth Edition. U.S. Department of Health and Human Services, Public Health Service, National Toxicology Program; 2011. Arsenic and Inorganic Arsenic Compounds. [Google Scholar]
- 5.N.C.I. SEER Stat Fact Sheets: Bladder Cancer. [Accessed on: 2014 Jun 12];Surveillance, Epidemiology, and End Results (SEER) Program. 2014 http://seer.cancer.gov/statfacts/html/urinb.html.
- 6.Kitchin KT, Conolly R. Arsenic-induced carcinogenesis--oxidative stress as a possible mode of action and future research needs for more biologically based risk assessment. Chem. Res. Toxicol. 2010;23:327–335. doi: 10.1021/tx900343d. [DOI] [PubMed] [Google Scholar]
- 7.Shen S, Lee J, Weinfeld M, Le XC. Attenuation of DNA damage-induced p53 expression by arsenic: a possible mechanism for arsenic co-carcinogenesis. Mol. Carcinog. 2008;47:508–518. doi: 10.1002/mc.20406. [DOI] [PubMed] [Google Scholar]
- 8.Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2:87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, Drobna Z, Currier J, Douillet C, Olshan AF, Rubio-Andrade M, Styblo M, Garcia-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: Altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 2014;55:196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, Ali E, Uddin MN, Liu X, Zoroddu MA, Gamble MV, Costa M. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol. Biomarkers Prev. 2012;21:2252–2260. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smeester L, Rager JE, Bailey KA, Guan XJ, Smith N, Garcia-Vargas G, Del Razo LM, Drobna Z, Kelkar H, Styblo M, Fry RC. Epigenetic Changes in Individuals with Arsenicosis. Chem. Res. Toxicol. 2011;24:165–167. doi: 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang YC, Hung WC, Chen WT, Yu HS, Chai CY. Sodium arsenite-induced DAPK promoter hypermethylation and autophagy via ERK1/2 phosphorylation in human uroepithelial cells. Chem. Biol. Interact. 2009;181:254–262. doi: 10.1016/j.cbi.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Huang YC, Hung WC, Chen WT, Jiang WH, Yu HS, Chai CY. Effects of MEK and DNMT inhibitors on arsenic-treated human uroepithelial cells in relation to Cyclin-D1 and p16. Toxicology Lett. 2011;200:59–66. doi: 10.1016/j.toxlet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Network C. G. A. R. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vahter M. Mechanisms of arsenic biotransformation. Toxicology. 2002;181–182:211–217. doi: 10.1016/s0300-483x(02)00285-8. [DOI] [PubMed] [Google Scholar]
- 16.Naranmandura H, Carew MW, Xu S, Lee J, Leslie EM, Weinfeld M, Le XC. Comparative toxicity of arsenic metabolites in human bladder cancer EJ-1 cells. Chem. Res. Toxicol. 2011;24:1586–1596. doi: 10.1021/tx200291p. [DOI] [PubMed] [Google Scholar]
- 17.Tseng CH. Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2007;25:1–22. doi: 10.1080/10590500701201695. [DOI] [PubMed] [Google Scholar]
- 18.Steinmaus C, Yuan Y, Kalman D, Rey OA, Skibola CF, Dauphine D, Basu A, Porter KE, Hubbard A, Bates MN, Smith MT, Smith AH. Individual differences in arsenic metabolism and lung cancer in a case-control study in Cordoba, Argentina. Toxicol. Appl. Pharmacol. 2010;247:138–145. doi: 10.1016/j.taap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YC, Su HJ, Guo YL, Hsueh YM, Smith TJ, Ryan LM, Lee MS, Christiani DC. Arsenic methylation and bladder cancer risk in Taiwan. Cancer Causes Control. 2003;14:303–310. doi: 10.1023/a:1023905900171. [DOI] [PubMed] [Google Scholar]
- 20.Currier JM, Ishida MC, González-Horta C, Sánchez-Ramírez B, Ballinas-Casarrubias L, Gutiérrez-Torres DS, Hernández Cerón R, Viniegra Morales D, Baeza Terrazas FA, Del Razo LM, García-Vargas GG, Saunders RJ, Drobná Z, Fry RC, Matoušek T, Buse JB, Mendez MA, Loomis D, Stýblo M. Associations between Arsenic Species in Exfoliated Urothelial Cells and Prevalence of Diabetes among Residents of Chihuahua, Mexico. Environ. Health Perspect. 2014;122:1088–1094. doi: 10.1289/ehp.1307756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bundschuh J, Litter MI, Parvez F, Roman-Ross G, Nicolli HB, Jean JS, Liu CW, Lopez D, Armienta MA, Guilherme LR, Cuevas AG, Cornejo L, Cumbal L, Toujaguez R. One century of arsenic exposure in Latin America: a review of history and occurrence from 14 countries. Sci. Total Environ. 2012;429:2–35. doi: 10.1016/j.scitotenv.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Del Razo LM, García-Vargas GG, Valenzuela OL, Castellanos EH, Sánchez-Peña LC, Currier JM, Drobná Z, Loomis D, Stýblo M. Exposure to arsenic in drinking water is associated with increased prevalence of diabetes: a cross-sectional study in the Zimapán and Lagunera regions in Mexico. Environ Health. 2011:10. doi: 10.1186/1476-069X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernández-Zavala A, Matoušek T, Drobná Z, Paul DS, Walton F, Adair BM, Jiří D, Thomas DJ, Stýblo M. Speciation analysis of arsenic in biological matrices by automated hydride generation-cryotrapping-atomic absorption spectrometry with multiple microflame quartz tube atomizer (multiatomizer) J Anal At Spectrom. 2008;23:342–351. doi: 10.1039/b706144g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortin F, Anghel T, Brochu P, Lemieux N. Optimizing urothelial cell preparation for the human urinary micronucleus assay. Toxicol. In Vitro. 2010;24:1821–1827. doi: 10.1016/j.tiv.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Tyler DE. Stratified Squamous Epithelium in Vesical Trigone and Urethra - Findings Correlated with Menstrual Cycle and Age. Am. J. Anat. 1962;111:319–335. doi: 10.1002/aja.1001110306. [DOI] [PubMed] [Google Scholar]
- 26.Landman J, Chang Y, Kavaler E, Droller MJ, Liu BC. Sensitivity and specificity of NMP-22, telomerase, and BTA in the detection of human bladder cancer. Urology. 1998;52:398–402. doi: 10.1016/s0090-4295(98)00219-2. [DOI] [PubMed] [Google Scholar]
- 27.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W, Westra WH, Schoenberg M, Zahurak M, Goodman SN, Sidransky D. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J. Natl. Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 28.Matoušek T, Currier JM, Trojánková N, Saunders RJ, Ishida MC, González-Horta C, Musil S, Mester Z, Stýblo M, Dědina J. Selective hydride generation-cryotrapping- ICP-MS for arsenic speciation analysis at picogram levels: analysis of river and sea water reference materials and human bladder epithelial cells. J Anal At Spectrom. 2013;28:1456–1465. doi: 10.1039/C3JA50021G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, Bhattacharya S, Kandjanapa K, Soontararuks S, Nookabkaew S, Mahidol C, Ruchirawat M, Samson LD. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007;3:e207. doi: 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Rosenstein MC, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database: update 2013. Nucleic Acids Res. 2013;41:D1104–D1114. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, Stýblo M, García-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: Identifying sites of 5-methyl cytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol. Sci. 2014;143:97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 36.Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, Garcia-Vargas G, Del Razo LM, Drobna Z, Styblo M, Fry RC. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J. Biochem. Mol. Toxicol. 2013;27:106–115. doi: 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang YK, Huang YL, Hsueh YM, Yang MH, Wu MM, Chen SY, Hsu LI, Chen CJ. Arsenic exposure, urinary arsenic speciation, and the incidence of urothelial carcinoma: a twelve-year follow-up study. Cancer Causes Control. 2008;19:829–839. doi: 10.1007/s10552-008-9146-5. [DOI] [PubMed] [Google Scholar]
- 38.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 39.James KA, Marshall JA, Hokanson JE, Meliker JR, Zerbe GO, Byers TE. A case-cohort study examining lifetime exposure to inorganic arsenic in drinking water and diabetes mellitus. Environ. Res. 2013;123:33–38. doi: 10.1016/j.envres.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Hall JA, Tabata M, Rodgers JT, Puigserver P. USP7 Attenuates Hepatic Gluconeogenesis Through Modulation of FoxO1 Gene Promoter Occupancy. Mol. Endocrinol. 2014;28:912–924. doi: 10.1210/me.2013-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem. Biophys. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 42.Bailey KA, Laine J, Rager JE, Sebastian E, Olshan A, Smeester L, Drobná Z, Styblo M, Rubio-Andrade M, Garcia-Vargas G, Fry RC. Prenatal Arsenic Exposure and Shifts in the Newborn Proteome: Interindividual Differences in Tumor Necrosis Factor (TNF)-Responsive Signaling. Toxicol. Sci. 2014;139:328–337. doi: 10.1093/toxsci/kfu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benton MA, Rager JE, Smeester L, Fry RC. Comparative genomic analyses identify common molecular pathways modulated upon exposure to low doses of arsenic and cadmium. BMC Genomics. 2011;12:173. doi: 10.1186/1471-2164-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Oh T, Chung H, Rha S, Kim C, Moon Y, Hoehn BD, Jeong D, Lee S, Kim N, Park C, Yoo M, An S. Identification of GABRA1 and LAMA2 as new DNA methylation markers in colorectal cancer. Int. J. Oncol. 2012;40:889–898. doi: 10.3892/ijo.2011.1245. [DOI] [PubMed] [Google Scholar]
- 45.Smith AH, Goycolea M, Haque R, Biggs ML. Marked increase in bladder and lung cancer mortality in a region of Northern Chile due to arsenic in drinking water. Am. J. Epidemiol. 1998;147:660–669. doi: 10.1093/oxfordjournals.aje.a009507. [DOI] [PubMed] [Google Scholar]
- 46.Karagas MR, Tosteson TD, Morris JS, Demidenko E, Mott LA, Heaney J, Schned A. Incidence of transitional cell carcinoma of the bladder and arsenic exposure in New Hampshire. Cancer Causes Control. 2004;15:465–472. doi: 10.1023/B:CACO.0000036452.55199.a3. [DOI] [PubMed] [Google Scholar]
- 47.Sanders AP, Smeester L, Rojas D, DeBussycher T, Wu MC, Wright FA, Zhou YH, Laine JE, Rager JE, Swamy GK, Ashley-Koch A, Lynn Miranda M, Fry RC. Cadmium exposure and the epigenome: Exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics. 2014;9:212–221. doi: 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 49.Mo ML, Okamoto J, Chen Z, Hirata T, Mikami I, Bosco-Clement G, Li H, Zhou HM, Jablons DM, He B. Down-regulation of SIX3 is associated with clinical outcome in lung adenocarcinoma. PLoS One. 2013;8:e71816. doi: 10.1371/journal.pone.0071816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Zhang A, Cao X, Chen J, Xia Y, Zhao H, Shen A. General transcription factor IIb overexpression and a potential link to proliferation in human hepatocellular carcinoma. Pathol. Oncol. Res. 2013;19:195–203. doi: 10.1007/s12253-012-9569-x. [DOI] [PubMed] [Google Scholar]
- 51.Steffen J, Seeger M, Koch A, Krüger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Lee H, Jun SY, Lee YS, Lee HJ, Lee WS, Park CS. Expression of miRNAs and ZEB1 and ZEB2 correlates with histopathological grade in papillary urothelial tumors of the urinary bladder. Virchows Arch. 2014;464:213–220. doi: 10.1007/s00428-013-1518-x. [DOI] [PubMed] [Google Scholar]
- 53.Majid S, Dar AA, Saini S, Deng G, Chang I, Greene K, Tanaka Y, Dahiya R, Yamamura S. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One. 2013;8:e67686. doi: 10.1371/journal.pone.0067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.