Abstract
The standard for single-cell analysis of phenotype and function in recent decades has been fluorescence flow cytometry. Mass cytometry is a newer technology that uses heavy metal ions, rather than fluorochromes, as labels for probes such as antibodies. The binding of these ion-labeled probes to cells is quantitated by mass spectrometry. This greatly increases the number of phenotypic and functional markers that can be probed simultaneously. Here, we review topics that must be considered when adapting existing flow cytometry panels to mass cytometry analysis. We present a protocol and representative panels for surface phenotyping and intracellular cytokine staining (ICS) assays.
Keywords: Mass cytometry, CyTOF, Immunophenotyping, Panel design
1 Introduction
Flow cytometry has become the method of choice in immunology for phenotypic and functional analysis of single cells, owing to its high throughput and ability to analyze multiple parameters in combination (up to 15 or so with advanced instruments). Still, the enormous complexity of immune cells makes even this degree of multiplexed readouts limiting. While it is possible to do single-cell analysis of gene expression across the entire genome [1], no technology has allowed a similar degree of comprehensive probing of proteins at the single-cell level.
By replacing fluorescent labeling of probes for flow cytometry with heavy metal ion labels, the potential for higher multiplexing is greatly enhanced. Unlike the highly overlapping emission spectra of typical fluorochromes, the readout of atomic masses by mass spectrometry is very discrete, and can span a wide mass window. Thus, the use of ion-labeled probes and mass spectrometry as a readout for flow cytometry (i.e., mass cytometry) is a conceptually attractive approach [2, 3].
The development of mass cytometry as a viable research tool for multiparameter analysis of immune cells was greatly facilitated by the availability of a commercial mass cytometer (CyTOF, DVS Sciences, Toronto; hereafter referred to as mass cytometer), along with software for conversion of the mass spectrometry signals to conventional flow cytometry standard (fcs) files. At least two laboratories have since exploited this technology to examine the heterogeneity of human immune cells in unprecedented detail [4–7].
Among the theoretical advantages of mass cytometry are (1) increased numbers of simultaneous probes that can be used, without loss of sensitivity and (2) lack of spillover between mass channels. In fluorescence flow cytometry, the number of simultaneous probes one can use is limited not only by the optical spectrum, but also by the availability of sufficiently bright fluorophores. Similarly, optical spillover between fluorochromes requires the application of compensation matrices to fluorescence flow cytometry data, increasing the complexity of analysis and the chances for errors of interpretation. Here we will address the degree to which these factors are in fact resolved by mass cytometry, and the considerations that remain.
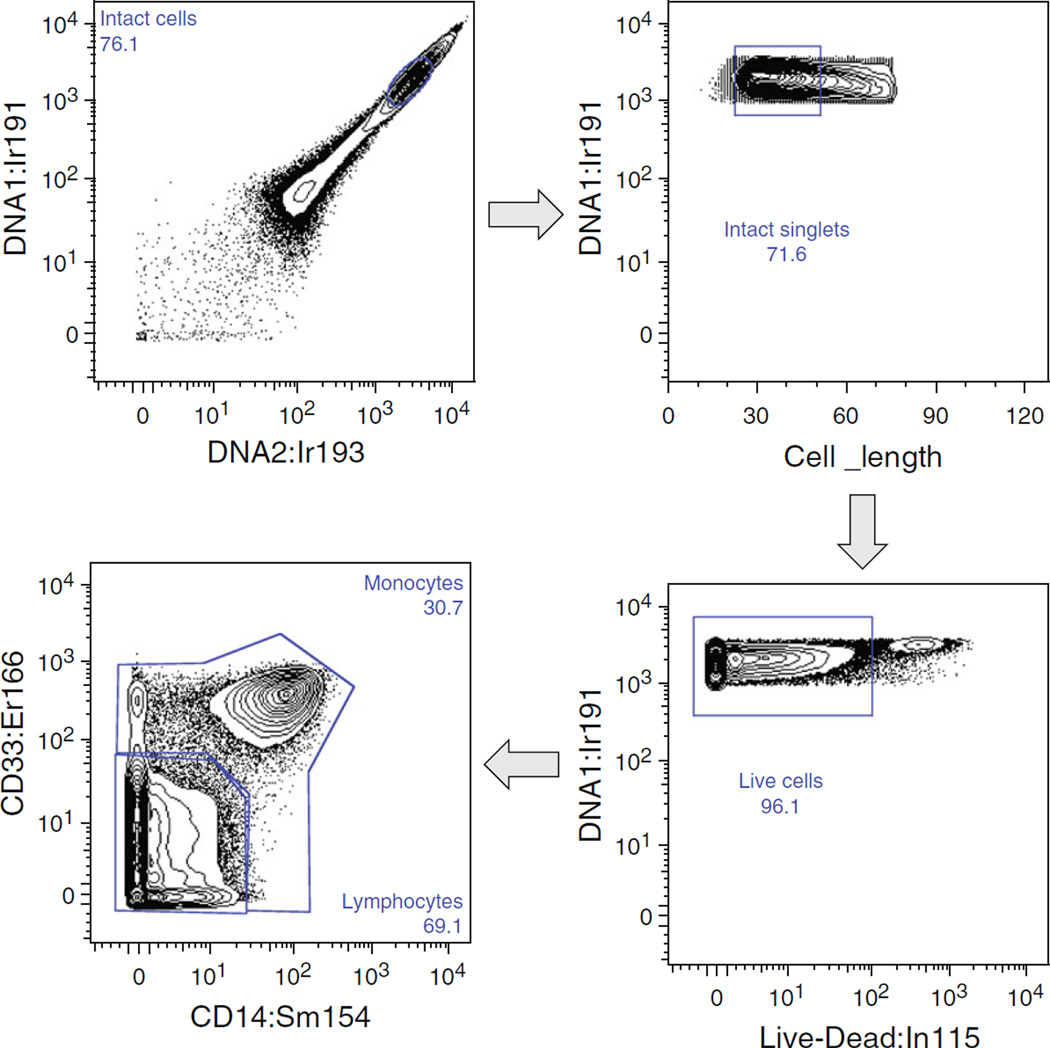
One important difference between fluorescence flow cytometers and a mass cytometer is that there is no mass cytometer analog to either forward scatter or side scatter. Therefore, there is a strict requirement for metal-labeling to discriminate cell types, or to identify cell events at all. If a cell is not labeled with at least one metal in the mass range of the mass cytometer, it will not be counted, adversely affecting percent-of-parent statistics. This is commonly addressed by labeling all cells containing DNA using iridium-containing intercalators (Atomic mass, AM 191, 193). Similarly, live-dead stains must also contain appropriate metal ions to be counted. Molecules containing both a chelator and maleimide moiety [5], or cisplatin [8] have been used as viability stains in mass cytometry. See Fig. 1 for an example of gating on iridium intercalator, viability stain, and monocytes vs. lymphocytes using CD14 and CD33.
Fig. 1.
Initial gating of a PBMC sample in CyTOF. Top, left : Gating on the dense cluster of events with strong staining for the two isotopes of Ir intercalator eliminates much of the debris, and events within the gate are referred to as “intact cells.” Top, right : Further gating on the Cell_length parameter eliminates some presumed cell aggregates, and events within this gate are referred to as “intact singlets.” Bottom, right : A thiol-reactive dye is taken up by cells with compromised membranes, so those with low staining are gated as “live cells.” Bottom, left : Initial marker-based gating is done on CD14 vs. CD33, to separate monocytes from lymphocytes. Because clean discrimination of these populations is essential to further analysis, we routinely use both markers for this purpose. All data was collected using Data Dual (Dd) calibration. The mass cytometer can give data in three different outputs: Intensity, Pulses Count, or Dual (d) mode. Dual mode is the calibration curve relating Intensity and Pulses Count, and is least dependent upon a particular machine. This calibration can be done using a commercial tuning solution containing known amounts of specified elements spanning the instrumental mass window (instrument Dual: Di), and will be saved as the calibration file until the calibration is run again. Data Dual (Dd) uses the first milliseconds of ion events from that particular sample to create the calibration, then uses that calibration to determine the cell event data. Di calibration information is written into the header of all files, regardless of whether the user specifies Di or Dd acquisition. Therefore, the data can be processed from one format to the other post-acquisition
The collection of pulse height and area (or width) information in fluorescence flow cytometers allows for reasonably efficient discrimination of cell aggregates, which differ from single cells in the ratio of these parameters. This is obviously not possible on a mass cytometer, and hence positive identification of single-cell events is more difficult. The use of the “cell length” parameter, or the amount of time over which a cell event is detected, during gating analysis only partially eliminates cell aggregates (see Fig. 1). Slowing the acquisition rate by diluting the sample reduces doublets at the cost of increased time per sample. More complex strategies can be undertaken, such as “cell barcoding” [6], followed by gating out of events that contain more than a single barcode. This strategy can be effective if the majority of aggregation occurs after the stage of barcode labeling.
The CyTOF 2 mass cytometer is currently tuned for a mass window approximately AM 89–209. The high end and the low end of the mass window have somewhat lower signal intensities (“dim channel”) compared to the middle of the mass range (“bright channel”). Maximum sensitivity is centered slightly higher than the middle of the range. The lanthanide metals are La139-Yb176, and vary in signal intensity by less than a factor of 4. Nonetheless, it is important to understand that the relative “brightness” of the channels is a function of their position within the measured mass window. Therefore, if the mass window is significantly expanded or shifted to higher or lower masses, the peak intensities will shift accordingly.
Design of Antibody Panels for Mass Cytometry
The factors to consider when pairing antibodies with metals are similar to pairing an antibody with a fluorochrome [9–11].
The expression level of the marker. Markers with higher expression can be used in “dimmer” channels. Conversely, markers with lower expression require “brighter” channels in order for the positive population to be resolved from the background. For markers induced with in vitro stimulation, those with small fold changes in expression after stimulation would require brighter metals to resolve the difference over the unstimulated sample.
The resolution needed for gating. It is often beneficial to use two markers in a bivariate plot to cleanly resolve a population of interest, rather than relying on histograms. In such cases, one bright channel and one dimmer channel are usually sufficient for resolution: for instance, B cells (CD19+ CD20+) can be cleanly gated from total CD3− cells using CD20-Dy164 (bright) vs. CD19-Nd142 (dim). In some instances, two channels of intermediate sensitivity may be needed, such as when both bivariate markers are of medium or low expression.
-
The type of expression. Some markers, such as CD27, exhibit bimodal expression: These can generally be labeled with dimmer metals, since they exhibit clearly resolved positive and negative populations, with no intermediate population.
Alternatively, other markers such as CCR7 or CD45RA have a spectrum or “smear” of expression. These often require brighter metals to allow finer distinction between important cell populations, even in bivariate plots.
If you need to redesign a panel, shifting metals up or down by one or two mass units will generally not impact signal/resolution.
Tm169 is the brightest metal for the AM 89–209 mass window for current commercially available polymer reagents/lanthanides. If labeling with Tm169 yields no signal for cells of known positivity, either: (1) the target molecule is not expressed in sufficient density for resolution by mass cytometry; or (2) the antibody is losing binding specificity upon labeling. Use of different labeling chemistry, or polymer-labeling a commercial fluorescent conjugate and checking it for binding by fluorescence analysis, can help resolve the second possibility.
Quantum dots (Qdots) and nanocrystals. Particles of hundreds to tens of thousands of metal atoms can efficiently be burned in the mass cytometry argon plasma, and the resulting ions can be quantified. Therefore, Qdots and similar nanocrystals labeled with antibodies can be one way to boost signal for a particular marker. As in fluorescence, non-specific binding needs to be carefully monitored.
Most commercial Qdots contain cadmium (usually CdSe), which lies within the mass cytometer mass window. Typically, only one Cd-Qdot can be used in a given panel: they contain natural-abundance Cd, which has eight naturally occurring isotopes (AM 106–116, 114 the most abundant). Cadmium is at the very low end of the sensitivity range and therefore would normally only be useful for markers of extremely high abundance. However, each Qdot contains thousands of Cd atoms, effectively increasing the signal of most markers to a reasonable level.
CdSe/CdTe Qdots contain tellurium (AM 120–130). However, since they also contain Cd, they cannot easily be used simultaneously with CdSe Qdots. Additionally, the xenon impurities (AM 124–136) present in the argon gas could potentially cause a noticeable background in the tellurium mass range.
Commercial InGaP Qdots contain primarily In115 (95.71% natural abundance). Cadmium lacks a 115 isotope, so these could be compatible with Cd Qdots, if Cd112 and In115 are monitored.
Potential Sources of Contaminating Signals
Due to the mass resolution of the time-of-flight separation, there is little or no spillover from one channel to the next due to the detector itself. In addition, most of the metals used in the AM 89–209 window, such as the lanthanides, Ir, Pt, In, Pd, or Cd, are seldom found in biological samples from healthy individuals. Therefore, there is no equivalent to “autofluorescence.”
However, other sources of contaminating signals must be considered, including metal and environmental impurities, and oxidation products [12].
-
Metal impurities. These can be impurities either of different elements, or of alternative isotopes of the same element; most typically, the greatest amount of impurity is seen in the next higher mass channel (“M+1”), with sometimes significant impurity in “M−1” or “M+2” as well; this is due to the nature of the isolation procedure.
The metals that are sold as part of antibody labeling kits are of very high purity (98% and higher in most cases). As a practical matter, this means that “compensation” analogous to fluorescent antibodies is not needed, as most of the signal will be of the specified mass, with little to no signal at “M+1” or another contaminating mass. However, metal salts from other commercial sources may be of lesser purity. For example, the chemistries of the lanthanides (Ln) are sufficiently similar that undesired lanthanides (often La139) can be contaminants in purchased salts since they are difficult to purify using only chemical methods. There are currently no labeling kits containing Gd157 due to purity concerns about the available salts. If using these less-pure isotopes, some caveats to consider include the following:- Consider using them for “dump” channels or exclusion markers. If only events that are negative for the label in question are subjected to further analysis, the impurities present should not cause any issues.
- Put a lower-abundance marker at a less-pure “M” so that the absolute spillover (usually up to 0.5–1% of “M” signal) is reduced. If “M” is a less pure isotope and is labeling a high-abundance marker, do not put a low-abundance marker at the M+1 position. Aim for at least medium-abundance so that positive and negative populations can still be clearly resolved if there is isotopic “spillover.”
- For channels that have significant spillover, use combinations of markers that label mutually exclusive populations. For instance, put a T-cell-specific marker at M+1 when using a B-cell-specific marker labeled with a less-pure isotope M.
- Impurities from the sample or environment. There are several sources of impurities to guard against. When in doubt, a highly diluted aliquot of the suspected stock can be injected into the mass cytometer in tuning/liquid mode and observed for contamination.
- Many laboratory dish soaps have high levels of barium (AM 130–138). Barium tends to persist even after multiple rinses. This is a problem even if these masses are not being used in the experiment, as it leads to detector aging, and can result in oxidation signals in M+16 channels (see point #3 below). It is therefore generally advised to store mass cytometry buffers in brand-new plastic or glass vessels that have never been through laboratory wash.
- Low levels of mercury, lead, or tin can sometimes be found in lab buffers, especially those made with "house" distilled water rather than reverse osmosis (e.g., MilliQ) water, or from commercial stock solutions that were not specified as metal-free. Even iodine (mass 127) is in the mass window of the mass cytometer.
- Unexpected sources of contamination include, for example, striker flints for Bunsen burners. These contain high levels of cerium (AM 136–142) and lanthanum, as well as traces of neodymium and other lanthanides.
Oxidation products. All metals exhibit some degree of oxidation in the argon plasma. This cannot be eliminated, but can be minimized with proper instrument warm-up and tuning of the current and make-up gas each day. This tuning should result in oxides <3% of maximum signal. Technically, oxide formation decreases signal at M, while increasing signal at M+16. However, it is easier to detect a small increase in oxide at M+16 than to detect a small decrease in signal at M.
Some lanthanides are more easily oxidized than others. La139 is the worst (oxide mass 155). Pr141 (oxide AM 157), Nd (oxide AM 158–166), and Gd (oxide AM 172–176) have notable levels of oxidation as well. Eu (oxide AM 167, 169) has very low oxide levels. When using a more-easily oxidized metal, it is useful to use it for markers that are relatively low-abundance compared to the marker at M+16, so that M+16 spillover (≤0.5–1% of “M” signal) is minimized.
It is important to remember that the undesired signals listed above are all a function of the signal intensity of M. Even with a less pure isotope such as Gd157, the total interference may only add up to a few percent of M signal, distributed among all spillover channels (M+1, M−1, M+16, environmental contamination, Ln, etc.). Therefore, careful pairing of marker abundance and metal signal intensity, along with metal salt purity will minimize any potential spillover. Generally, a signal of <101 Dual counts can be considered as background. Therefore, if the signal at M is <103 Dual counts, any spillover contribution would be at or below background level in the affected channels.
Qualification of Antibody Conjugates
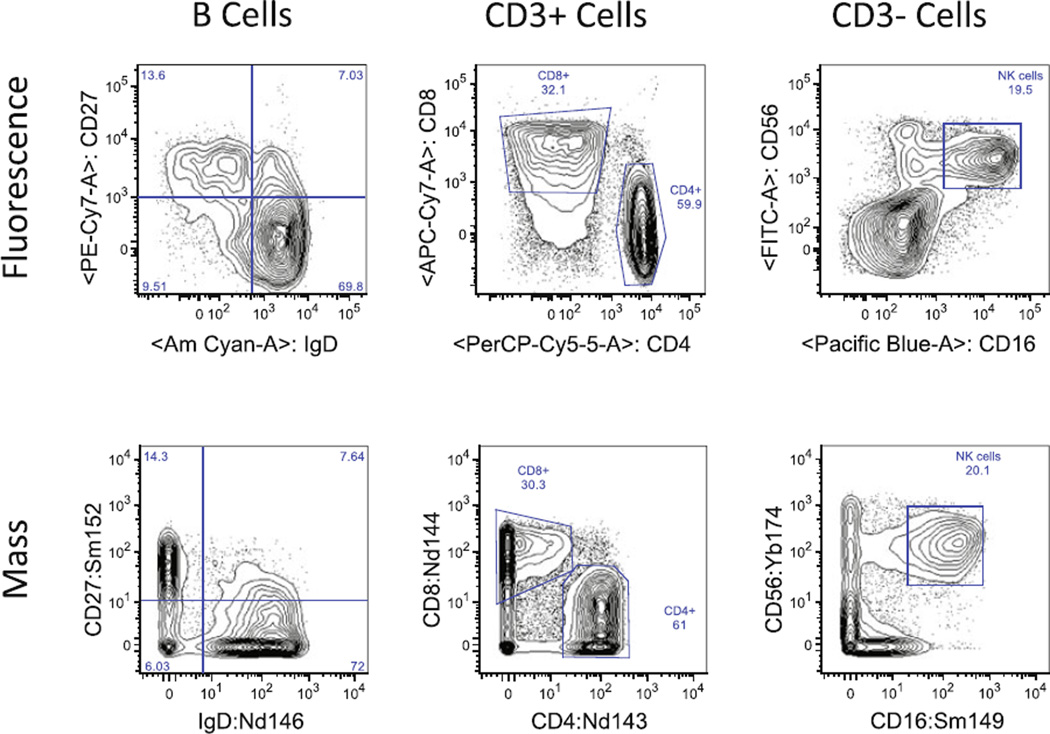
As with standard fluorescence flow cytometry, qualification of antibodies for assay purposes is critical. This is particularly relevant since comparatively few preconjugated metal–antibodies are currently commercially available. Therefore, many antibody–metal conjugates will have to be conjugated in-house by the end-user. In most cases, an antibody clone that works for fluorescence flow cytometry can be successfully conjugated for use in mass cytometry. In most of the remaining cases, another widely used clone can be substituted successfully. See Fig. 2 for some representative examples comparing various markers in fluorescence and mass cytometry. However, with the relatively young state of the field, there are occasionally markers for which no suitable antibody clone has yet been identified.
Fig. 2.
Representative comparisons of fluorescence (top row) and mass cytometry (bottom row) for the same antibody combinations on cryopreserved PBMC from the same donor. Parent populations are shown at the top of each column. Left to right : naïve and memory B cell sub-populations, CD4+ and CD8+ T cells, and NK cells. In each case, the staining patterns differ somewhat, especially with regard to background staining of negative populations; but overall frequencies of each gated population are very similar. All data was collected using Data Dual (Dd) calibration
Furthermore, we recommend that even after a successful antibody clone–metal pairing is found, each new batch of conjugated antibodies should be checked for activity against a reference before use in assays with unknown samples.
There are several points to keep in mind when testing new antibody conjugates.
The expected expression pattern of the marker. This includes: cell type (monocytes, NK cells, T cells, B cells, etc.); location (peripheral circulation, bone marrow, lymph node, gut lumen, etc.); and effects of stimulation, differentiation, or cell cycle phase. For example, one might test an antibody on stimulated cells if antigen expression is not expected on unstimulated cells.
Effects of sample processing. Some markers (e.g., CD62L, PD-1) are reduced upon cryopreservation (though these can be partially restored after resting of thawed cells). Some cell types are also lost or reduced after processing (e.g., granulocytes and dendritic cells after Ficoll gradient separation). Finally, staining after fixation and/or permeabilization can destroy epitopes. For example, many anti-CD16 antibodies lose binding after fixation. Conversely, there can be a large increase in nonspecific binding of many anti-CD56 antibodies after fixation.
-
Use of both positive and negative controls. Often, different cell types within the same sample can provide positive and negative controls for antibody staining. For example, B cells can serve as a negative control for T cell markers, etc. However, beware of limitations of this approach, as many markers are expressed by more than one type of cell, often at lower levels or in small subpopulations.
If doing two-step staining with element-labeled secondary antibodies, one should include additional controls such as: secondary antibody in the absence of the primary antibody, and secondary antibody in the presence of a known primary antibody.
Cell lines can be useful for antibody qualification (see proteinatlas.org for immunohistochemistry data for ~4300 proteins on 47 cell lines). Of course, the antigen expression on a cell line may be higher or lower than seen on primary cells. Also, data from proteinatlas.org are from samples fixed, paraffin-embedded, deparaffinized with xylene, rehydrated with ethanol, boiled in antigen-retrieval solution, stained, then read by a computer. Thus, the staining may not match that seen on fresh samples in flow cytometry.
Use of more than one donor during antibody-conjugate validation. Some donors have unusual patterns of expression or cell distribution. TCRγδ+ T cells are an example of a highly donor-dependent population. We have observed occasional donors with low or negative expression of CD33 on monocytes, or very skewed distributions of memory T-cell subsets (e.g., nearly all CD8+ T cells are CD28+ or CD28−). By use of more than one donor, false conclusions about the performance of the antibody are less likely.
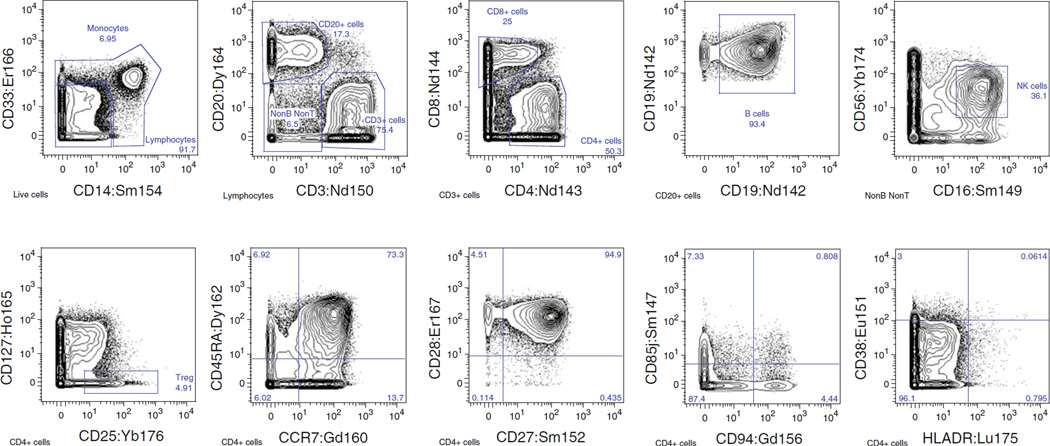
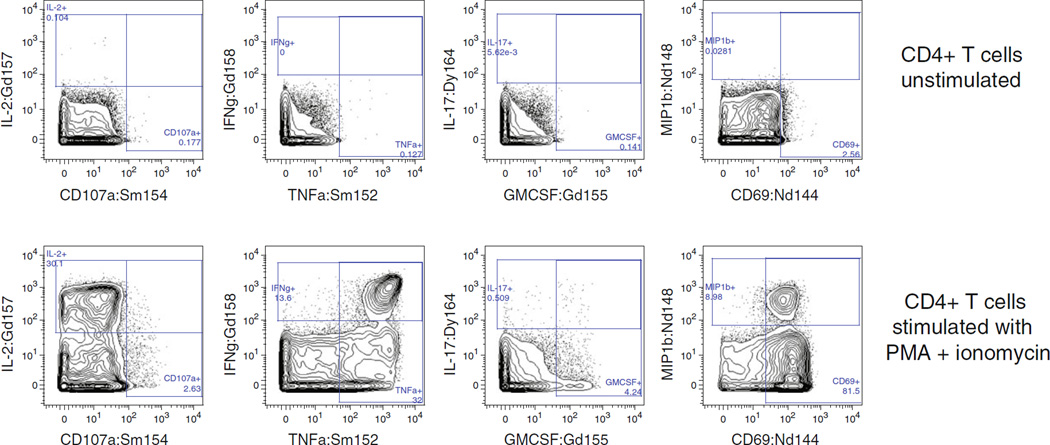
Once a panel is designed and conjugates are tested and titrated, it is advisable to test performance and reproducibility of the panel on control samples such as healthy subject PBMC. See Fig. 3 for an example of the staining pattern of PMA+ionomycin-activated normal PBMC with a selection of markers from a 38-antibody intracellular cytokine panel.
Fig. 3.
Representative staining of various cell-surface (a) and intracellular (b) markers in a 38-antibody panel that includes eight intracellular cytokines. Plots show staining from a positive control, stimulated with PMA+ionomycin, but the panel has been successfully used with antigen-specific stimulations as well. Resolution of surface and intracellular markers is generally quite good. The parent population is shown at the bottom of each plot. All data was collected using Data Dual (Dd) calibration
2 Materials
96-well round bottom plates (see Note 1).
CyFACS buffer: 0.1% BSA+ 2 mM EDTA+ 0.1% NaAzide in PBS made with MilliQ water and no heavy metal contaminants (no glass or beakers washed with soap). Filter with a 0.2 µm filter; store at room temperature.
CyPBS: PBS without heavy metal contaminants (10× PBS from Rockland). No contact with beakers or bottles washed with soap. Filter with a 0.2 µm filter; store at room temperature.
MilliQ dH2O: No contact with beakers or bottles washed with soap.
- DOTA-maleimide: B-272 from Macrocyclics.
- Use to make 5 mg/mL In115*-DOTA-maleimide, dissolved in MilliQ water. Add 100 µl 3% nitric acid per 10 mL solution to maintain low pH and store at 4 °C.
- In115* is natural-abundance indium from a high-purity metal salt.
0.1 µm spin filters: Millipore UFC30VV00.
16% Paraformaldehyde: Electron Microscopy Sciences Cat. 15710.
DVS Sciences iridium intercalator solution: 2000× or 500× stock, use at final 1×.
Saponin-based permeabilization buffer (Ebioscience Cat. 00-8333-56).
Set of MAXPAR-labeled antibodies–labeled as per MAXPAR kit from DVS Sciences (see Note 2).
3 ICS Staining Protocol
3.1 Sample Collection
Two million viable cells per well (as measured by dye exclusion method such as Vicell; rested, if desired) (see Note 3).
3.2 Cell Activation
Perform as described in Lovelace and Maecker [13].
3.3 Sample Processing
Wash 1× in CyFACS buffer (flick plate or aspirate to remove supernatant).
Make Ab cocktail in CyFACS buffer (Filter with 0.1 µm spin filter).
Resupend cells in 50 µL filtered Ab cocktail.
Incubate for 30–60 min on ice.
Wash 2× in CyFACS buffer (see Note 4).
Resuspend cells in 100 µl of 1:3000 diluted In115-DOTA maleimide in CyPBS.
Incubate for 30 min on ice.
Wash 3× in CyFACS buffer.
Resuspend in 100 µl of 2 % PFA in CyPBS (see Notes 5 and 6).
Incubate at 4°C overnight.
Wash 2× in 1× eBioscience perm buffer (1× in MilliQ water).
Make intracellular staining cocktail in 1× perm. buffer and filter with 0.1 µm spin filter.
Incubate on ice for 45 min.
Wash 3× in CyFACS buffer.
Resuspend in 2 % PFA+ 1× Ir-Interchelator in CyPBS.
Incubate for 20 min at room temperature.
Wash 1× in CyFACS buffer.
Wash 3× in MilliQ water (see Note 7).
Resuspend in MilliQ water for running on mass cytometer (see Note 8). Filter through a 25 µM cell strainer prior to acquisition (see Note 9).
To perform only surface phenotyping : Subheading 3.1 : 1 million live cells are usually sufficient. Omit stimulations in Subheading 3.2. In Subheading 3.3, omit step 13, and 2 % PFA in step 16.
3.4 Data Acquisition and Analysis
Start the machine. Warm up and tune as in Ref. [14].
Acquire data as in Ref. [14]. The length of the run will be dependent upon the volume of your sample: dilution with MilliQ water to ~1 million cells/mL is recommended for minimizing doublets as well as maximizing sample throughput (see Note 10).
Analyze data using third-party flow analysis software such as FlowJo (Treestar) or Cytobank (see Note 11 and Fig. 1). Note that some display settings may need to be altered for proper viewing.
Acknowledgments
The authors thank Sean Bendall for helpful discussions, and Sheena Gupta and Meena Malipatlolla for contributing example data. Development of this protocol was funded in part by grant 2 U19 AI057229 S4 from the National Institutes of Health.
Footnotes
Plates vs. tubes: cells can be handled in 96-well microtiter plates or in 12 × 75 mm polystyrene tubes. Deepwell plates are useful for additional volume per wash (see Note 4).
Lanthanides cannot be photobleached. Therefore, there is no need to protect antibody stocks or samples from standard lab lighting.
The current cell transmission efficiency of the mass cytometer is 20–25%, compared to 95+% for a standard fluorescence flow cytometer. Therefore, coupled with cell loss due to the suggested number of wash steps (see Note 4) greater starting numbers of cells will be required for similar cell event counts.
Due to the sensitivity of the detector, mass cytometry samples require a large number of washes to minimize nonspecific background from staining steps. While this must be balanced against the loss of cells with each wash step, reducing the number of washes below what is listed here is not recommended.
All cells that are injected into the mass cytometer have been fixed and permeabilized. This is necessary to allow the iridium intercalator to effectively enter the cell.
MilliQ water can cause improperly fixed cells to lyse (see Note 5). Therefore, ensure that your PFA is fresh. While it is not always necessary to open a new bottle/ampule, PFA should be generally protected from light and exposed to atmosphere for less than a month to be completely active.
Ultrapure (e.g., MilliQ) water is required for washes and final resuspension at the end of the staining protocol. This helps ensure that there is little or no free metal or antibody upon injection into the mass cytometer. This also ensures that there are no buffer salts carried along with the sample. While most buffer salts will not make it through the quadrupole mass filter window (AM 89–209), they will accumulate on the metal cones at the entry to the machine. Over long run-times, buffer salts can accumulate and cause the tuning to drift, particularly the Current setting.
Stained samples can be kept at 4 °C for up to a week. However, fresher samples are optimal. If samples must be stored, it is preferable that they be stored in 2 % PFA/CyPBS, or at least in CyFACS. Regardless of the storage conditions, it will be necessary to do at least one MilliQ water wash prior to resuspension in MilliQ water and injection. Do not freeze stained samples.
All samples must be filtered through 25 µm cell strainers before injecting into the mass cytometer. This will minimize the likelihood that a clog will form in the nebulizer tubing or the nebulizer itself.
While the mass cytometer is capable of acquiring cells at up to 1000 cells/s, this usually causes an unacceptable number of doublets. Event rates of approximately 300–500 cells/s strike a better balance between optimizing the number of singlet events while still allowing sample acquisition in a reasonable timeframe. The event viewing window in the software shows ~1/350 snapshot of the data actually being acquiring per second. Therefore, an average of ~1 cell event/screen refresh would be in this 300–500 cells/s range.
The number of cell events counted by the mass cytometer during acquisition is an upper limit to the number of true cell events. The software registers a cell event as metal signal in any mass channel that is a number of standard deviations above background (default = 3 S.D.). Therefore, debris or other background signal from your sample could achieve this threshold and be counted. High signal in both iridium channels (Ir191 and Ir193) represents intact cells and is a useful first gate (Fig. 1). The number of Intact cells/Singlets/Live cells in a standard sample is often 50–60% of the total initially counted by the machine.
References
- 1.Kalisky T, Quake SR. Single-cell genomics. Nat Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 2.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;36:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 4.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8(+) T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe'er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maecker HT. Multiparameter flow cytometry monitoring of T cell responses. Methods Mol Biol. 2009;485:375–391. doi: 10.1007/978-1-59745-170-3_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62:169–173. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- 11.Rundberg Nilsson A, Bryder D, Pronk CJ. Frequency determination of rare populations by flow cytometry: a hematopoietic stem cell perspective. Cytometry A. 2013;83:721–727. doi: 10.1002/cyto.a.22324. [DOI] [PubMed] [Google Scholar]
- 12.Ornatsky OI, Kinach R, Bandura DR, Lou X, Tanner SD, Baranov VI, Nitz M, Winnik MA. Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2008;23:463–469. doi: 10.1039/b710510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovelace P, Maecker HT. Multi-parameter intracellular cytokine staining. Methods Mol Biol. 2010;699:165–178. doi: 10.1007/978-1-61737-950-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leipold MD, Maecker HT. Mass cytometry: protocol for daily tuning and running cell samples on a CyTOF mass cytometer. J Vis Exp. 2012 doi: 10.3791/4398. [DOI] [PMC free article] [PubMed] [Google Scholar]