Abstract
For genome-wide association studies (GWASs) and DNA sequencing studies, several powerful score-based tests, such as kernel machine regression and sum of powered score tests, have been proposed in the last few years. However, extensions of these score-based tests to more complex models, such as mixed-effects models for analysis of multiple and correlated traits, have been hindered by the unavailability of the score vector, due to either no output from statistical software or no closed-form solution at all. We propose a simple and general method to asymptotically approximate the score vector based on an asymptotically Normal and consistent estimate of a parameter vector to be tested and its (consistent) covariance matrix. The proposed method is applicable to both maximum likelihood estimation and estimating function-based approaches. We use the derived approximate score vector to extend several score-based tests to mixed effects models. We demonstrate the feasibility and possible power gains of these tests in association analysis of multiple and correlated quantitative or binary traits with both real and simulated data. The proposed method is easy to implement with a wide applicability.
Keywords: GWAS, kernel machine regression, mixed-effects models, multiple traits, SNP, sum of powered score (SPU) tests
Introduction
To detect genetic associations in genome-wide association studies (GWASs) and DNA sequencing studies, in addition to the popular univariate minimum p-value (UminP) test, many multivariate methods have been proposed to improve statistical power. Several competitive ones are score-based, such as the classic score test, a variance-component score test in kernel machine regression (KMR) [Kwee et al., 2008; Wu et al., 2010, 2011], an adaptive score test [Lin and Tang, 2011], and an adaptive sum of powered score (aSPU) test [Pan et al., 2014]. A challenge is how to extend these score-based tests to more complex models beyond the generalized linear models (GLMs) for independent data. There are several reasons to consider more complex mixed-effects models in genetic association studies. First, even for a single trait analysis, to properly account for some complex and hidden relatedness among the study subjects, or more generally for population structure or population stratification, mixed-effects models have been proposed as a general and effective approach [e.g. Zhang et al., 2010; Yu et al., 2011; Zhou and Stephens, 2014]. These mixed-effects models differ from the standard ones in that a random effect is introduced to induce correlations among all the subjects, thus requiring some special and fast algorithms for model fitting as implemented in several recent software packages. These packages do not directly output the score vector. Second, there has been increasing interest in association analysis of multiple traits, which may help gain power and shed light on pleiotropy. In addition, one may encounter correlated traits as arising from familial studies. To account for correlations among multiple traits, either marginal models (based on generalized estimating equations, GEE) [Liang and Zeger, 1986] or mixed-effects models [Breslow and Clayton, 1993] can be applied. For quantitative traits, a linear mixed-effects model (LMM) can be used, from which the score vector can be derived. Accordingly the KMR test has been extended to LMMs [Maity et al., 2012; Schifano et al., 2012]. However, it is unclear how to extend the KMR and other score-based tests to generalized linear mixed models (GLMMs) and Cox mixed-effects models, for which there is no closed-form for the score vector (since the marginal likelihood involves an integral with random effects and in general has no closed form) [Breslow and Clayton, 1993]. Although as an alternative to GLMM, marginal models/GEE can be used, from which (generalized) score-based tests can be derived [Wang et al., 2013; Zhang et al., 2014], there may be substantial differences between the two in terms of modeling assumptions, interpretation, and thus their choices [Diggle et al., 2013]. More importantly, in genetic association studies, as discussed earlier, random effects may be necessary to effectively account for population structure, prompting the use of non-linear mixed-effects models. In these situations, due to the lack of computer output or closed-form solution for the score vector, it is challenging to implement score-based tests.
In this paper we propose a simple and yet general method to approximate the score vector for any model. It is based on the asymptotics of an estimator of the parameters to be tested. It applies to both the maximum likelihood estimates (MLEs) and estimating function-based estimates. Its implementation involves only a few lines of R code. We demonstrate its use in two types of mixed-effects models, a multivariate linear mixed-effects model (mvLMM) proposed very recently for genetic association analysis of multiple quantitative traits while correcting for cryptic relatedness and population stratification [Zhou and Stephens, 2014], and a GLMM with correlated binary traits. Among others, we use both real and simulated data to illustrate possible power gains of some approximate score-based tests over the standard Wald and UminP tests.
Methods
Review: some score-based tests
Suppose U = (U1, …, Uk)T is the score vector for a set of k parameters to be tested with H0: ψ = 0. The classic score test is
which is asymptotically equivalent to the Wald test and likelihood ratio test (LRT). The UminP test that has been widely used in GWASs can be written as
where Vjj is the jth diagonal element of . Recently, a variance-component score test in KMR has been proposed for GLMs and shown to be powerful for analysis of SNP sets [Kwee et al., 2008; Wu et al., 2010, 2011]. As discussed by Pan (2011), with a linear kernel it is equivalent to the sum of squared score (SSU) test:
Pan et al (2014) proposed a family of the so-called sum of powered score (SPU) tests:
| (1) |
for a set of integers γ ≥ 1. It is easy to see that SPU(1) and SPU(2) are exactly the same as the Sum test and the SSU test respectively [Pan, 2009]; the Sum test, an example of so-called burden tests, has been shown to perform well for genetic association testing, especially for rare variants, under some situations [Li and Leal, 2008; Pan, 2009]. In addition, for an even integer γ → ∞, we have
the SPU(∞) is closely related to the UminP test (but ignoring possibly varying variances of Uj’s). Alternatively, an SPU(γ) test can be regarded as a weighted score test [Lin and Tang, 2011] with adaptive weights on each component j. In practice, since it is unknown which γ value would yield high power, we use an adaptive SPU (aSPU) test to combine the evidence across the SPU tests:
| (2) |
where PSPU(γ) is the p-value of the SPU(γ) test, and Γ contains a set of candidate values γ. Pan et al (2014) found that in many situations Γ = {1, 2, 3, …, 8, ∞} appeared to perform well, which will be used here.
In general, resampling methods can be used to obtain p-values for the SPU and aSPU tests. In this paper, we assume that the asymptotic null distribution of the score vector U ~ N(0, V) holds (under H0). Accordingly, we can generate B independent copies of the null score vector U(b), for which the B copies of the SPU test statistics can be calculated. Then the p-value of each SPU(γ) test is calculated as . Furthermore, based on the same B copies of the simulated score vector, we calculate the p-value for the aSPU test as with and .
In this paper we use the SPU and aSPU tests as examples, though other score-based tests [e.g. Wu et al., 2010; Lin and Tang, 2011; Sun et al., 2013] can be equally applied. Our main point does not depend on the choice of a specific score-based test; rather, we aim to show how to extend a score-based test to cases where there is no easy access to the score vector, as arising in below two important applications. To be concrete, we focus on detecting genetic association with single nucleotide polymorphisms (SNPs), but the proposed method is generally applicable to other problems of interest.
Two example models
Multivariate linear mixed model
A multivariate linear mixed model (mvLMM) was proposed by Zhou and Stephens (2014) to test for association with multiple phenotypes while correcting for possible population stratification. Specifically, suppose we would like to test for association between a multivariate trait and a single SNP. We first combine the n trait vectors by row such that the resulting trait matrix Y is of dimension n × k, and the jth column of Y corresponds to phenotype j while the ith row of Y corresponds to the multiple traits from the ith subject; W is an n × q design matrix for covariates (including a column of 1’s for the intercept); x = (x1, …, xn)T is an n × 1 vector of genotype scores (i.e. the counts of the minor allele) for the SNP.
The mvLMM can be written as:
| (3) |
where λ is a q × k matrix of regression coefficients for covariates; ψ is a k × 1 vector of the SNP effect sizes for the k phenotypes; G is an n × k matrix of random effects; E is an n × k matrix of random errors; K is an n × n known kinship matrix, or more generally, a genetic relatedness matrix (GRM) estimated from whole-genome genotype data, In × n is an n × n identity matrix, Vg is a k × k symmetric matrix of genetic variance components, Vε is a k × k symmetric matrix of environmental variance components, and MNn × k(0, V1, V2) denotes the n × k matrix normal distribution with mean 0, a column covariance matrix V1 of dimension n × n, and a row covariance matrix V2 of dimension k × k. The goal is to test H0: ψ = 0.
A mvLMM differs from a standard LMM in that an n × n matrix K is used to account for possible genetic relatedness among all the subjects. Since the kinship matrix K may be full and may not be block-diagonal, it means that all the subjects may be possibly correlated. However, as discussed by Zhou and Stephen (2014), the mvLMM can be rewritten more like a standard LMM in the following way. An eigen-decomposition of the relatedness matrix K can be performed as , where Uk is a n × n matrix of eigen vectors and Dk is a diagonal n × n matrix with diagonal elements corresponding to eigen values (i.e. diag(δ1, …, δn)). Then one can obtain the transformed phenotype matrix Ỹ = UkY, transformed covariate matrix W̃ = UkW, transformed SNP vector x̃ = Ukx, transformed random effect matrix G̃ = UkG and transformed residual error matrix Ẽ = UkE. After transformation, for each individual i, the transformed phenotypes given the transformed covariates and SNP follow independent (but not identical) multivariate normal distributions:
| (4) |
where for i = 1, …, n, is the ith row vector of Ỹ, is the ith row vector of W̃, x̃i is the ith element of vector x̃, is the ith row vector of G̃ and is the ith row vector of Ẽ; Var(ỹi) = δiVg + Ve ≡ Vi.
Based on model (4), one can write down the score vector:
| (5) |
where λ̂0 and V̂i,0 are obtained by fitting the null model under H0 : ỹi = λTw̃i + g̃i + ẽi.
It is quite challenging to develop a fast algorithm to fit a mvLMM. Now such an algorithm is implemented in software package GEMMA [Zhou and Stephens, 2014]. However, as for most software packages, one is not able to obtain the score vector directly from the output. A simple and practical way to obtain the score vector is, as proposed earlier, to approximate it by the MLE and its covariance estimate, both available directly from the output of GEMMA; accordingly a score-based test can be simply constructed and applied.
Generalized linear mixed model
In a familial study, we observe that in each family i, subject j has a univariate trait Yij, q covariates Wij = (Wij1, …, …, Wijq)T and p SNPs Xij = (Xij1, …, Xijp)T. We would like to test for association between the trait and the SNPs through a GLMM:
| (6) |
where g() is a link function, μij = E(Yij|Xij, Wij, bi) is the conditional mean of the trait for subject j in family i, λ = (λ1, …, λq)T is a q × 1 vector of regression coefficients for covariates Wij, ψ = (ψ1, …, ψp)T a p × 1 vector of regression coefficients for SNP set Xij, and bi is a random effect inducing correlations among the traits of the subjects from the same family.
The goal is to test H0: ψ = 0. However, in general, due to the lack of the closed-form for the marginal likelihood, there is no closed-form expression for the score vector for ψ either [Breslow and Clayton, 1993]. Hence, it is not easy to develop a score-based test for such a model. Below we propose a new method to approximate the score vector, based on which it is straightforward to construct a score-based test.
New method: approximating the score vector
Estimation via maximum likelihood
Suppose that we would like to test H0: ψ = ψ0 in the presence of nuisance parameter λ. Denote θ̂0 = (ψ0, λ̂0)T as the restricted MLE of θ = (ψ, λ)′ under H0, while θ̂ = (ψ̂, λ̂)T as the unrestricted MLE of θ = (ψ, λ)T (i.e. under H1). Partition the Fisher’s information matrix H accordingly as
Denote the whole score vector for θ as Uθ(θ) = (Uψ(θ)T, Uλ(θ)T)T. As shown by Kent (1982, the equation following (4.1)),
| (7) |
Since the consistent estimator , we have
Thus, we first fit a full model (under H1) to obtain the MLE ψ̂ and its covariance estimate , then we can approximate the score vector U and its covariance matrix accordingly. In this way, we can construct (approximate) score-based tests such as the score test, the SPU and aSPU tests. In particular, it is easy to see that the approximate score-based score test is the same as the Wald test:
Estimation via estimating functions
For estimating function-based approaches, although a generalized score test [e.g. Kent, 1982; Rotnitzky and Jewell, 1990; Boos, 1992] can be constructed, the popular statistical software may not provide direct output of such (generalized) score vectors. For easy implementation, it may be useful to approximate the (generalized) score vector by the parameter estimate and its covariance matrix. Specifically, by treating an unbiased mean 0 estimating function as a (generalized) score function and by a Taylor expansion, we still have equation (7). However, is the model-based covariance estimator, which is not consistent unless all working assumptions hold (essentially assuming that the estimating function is indeed a score function). More generally, a consistent sandwich estimator is used. Hence, we can modify the score vector approximation as
Accordingly, once we obtain the point estimate ψ̂, its model-based covariance estimated and its sandwich estimate , we can obtain an approximation to the score vector U, based on which we can construct a score-based test. Again it is easy to verify that the score test based on the approximate score is exactly the same as the Wald test.
We explored the use of such tests for marginal approaches to GLMMs for correlated binary data (i.e. GEE) [Liang and Zeger, 1986] (and to Cox regression for correlated survival data; not shown). Note that in general our proposed approximate score vector is derived based on an asymptotically normal point estimator, and thus is only asymptotically unbiased, while the generalized score vector is simply the estimating function being used and is often unbiased for finite samples; this difference leads to varying performances of an approximate score-based test and an exact generalized score test [Boos, 1992] for finite samples, though their difference diminishes as the sample size increases, as to be shown later for GEE.
Results
Example
ADNI data
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California-San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org.
We applied the methods to the ADNI-1 data consisting of 681 non-Hispanic Caucasians with both genotypic and phenotypic data. The phenotype were cortical thickness measures of some regions of interest (ROIs) in the brain; they were cross-sectionally processed using FreeSurfer by UCSF researchers [Hartig et al., 2012]. We tested on about 20 SNPs and several multivariate traits as considered in Shen et al (2010) and Zhang et al (2014). For the purpose of illustration, we only show the results for two SNPs and four multivariate traits: APOE-ε4 in gene APOE that is well known to be associated with AD, and rs7526034 on chromosome 1 (LOC199897), both were associated with multiple neuroimaging phenotypes [Shen et al., 2010]; the four multivariate traits were left and right sides of “Par” (denoted as LPar and RPar), each with 4 ROIs (inferior and superior parietal gyri, supramarginal gyrus, and precuneus), right side of “Front” with 6 ROIs (caudal midfrontal, rostral midfrontal, superior frontal, lateral orbitofrontal, and medial orbitofrontal gyri and frontal pole), right side of “LatTemp” with 3 ROIs (inferior temporal, middle temporal and superior temporal gyri). Given a large number of parameters to be estimated (> k2 with k traits) in a mvLMM, as pointed out by Zhou and Stephen (2014), only a small to moderate number of phenotypes (~ 2–10) were recommended to be used for a typical sample size for GWAS (i.e. n in thousands). Hence, with only a moderate sample size n = 681 here, we only considered a few multivariate traits containing no more than 6 univariate traits (otherwise, in addition to the questionable asymptotics, we also encountered some numerical convergence problems).
For each subject i, five covariates (Wi) were included: baseline age, gender, baseline education (in years), handedness (left or right) and baseline intracranial volume (ICV), plus an intercept term; Xi was the genotypic score of one of the above two SNPs; Yi = (Yi1, …, Yik)T was a vector of k quantitative traits (i.e. cortical thickness measures of k ROIs). For each pair of the SNP-multivariate trait, a mvLMM (3) was fitted using software GEMMA; we applied the proposed method to approximate the score vector based on the MLE and its covariance estimate, from which the approximate score-based SPU and aSPU tests were conducted. The K matrix in the mvLMM was estimated based on nearly a half million SNPs in the data (prior to fitting the model) in GEMMA. We used a step-up procedure to gradually increase the value of the simulation number B to calculate the p-values: starting from B = 104, if a p-value was no more than 5/B, we increased B to ten times of its previous value and then repeated the test until either its p-value was more than 5/B or we reached B = 107. For comparison, we also showed the results of the asymptotic Wald and Score tests, directly available from the output of GEMMA.
Analysis results
As shown in Table 1, for most SNP-multivariate trait pairs, the aSPU test gave similar results as those of the classic Wald and Score tests. However, there were a few differences. Notably, for rs7526034-RLatTemp, the aSPU test gave a more significant p-value than those of the Wald and Score tests; on the other hand, for APOE-ε4-RPar, it was the reverse. Among the SPU tests, SPU(1) usually gave most significant results, presumably because of the smaller number of univariate traits (k) and the same direction of the SNP-univariate trait associations. In summary, our results demonstrate the feasibility of using our proposed method to approximate the score vector for a complex mvLMM, and accordingly construct the score-based aSPU test, which might be more powerful in some situations (to be shown in simulations) and thus can be complementary to the standard Wald and Score tests.
Table 1.
P-values of the various mvLMM-based tests in analysis of the ADNI data.
| SNP | Test | LPar(4) | RFront(6) | RLatTemp(3) | RPar(4) |
|---|---|---|---|---|---|
| APOE-ε4 | SPU(1) | 1.2e-05 | 2.0e-07 | 2.0e-05 | 1.3e-06 |
| SPU(2) | 8.3e-02 | 2.4e-02 | 1.3e-01 | 3.8e-04 | |
| SPU(3) | 1.9e-01 | 2.4e-03 | 1.9e-02 | 1.3e-02 | |
| SPU(4) | 1.6e-01 | 7.7e-03 | 6.8e-02 | 1.1e-03 | |
| SPU(5) | 3.6e-01 | 4.3e-03 | 3.5e-02 | 2.1e-02 | |
| SPU(∞) | 2.9e-01 | 5.2e-03 | 4.8e-02 | 7.8e-03 | |
| aSPU | 3.5e-05 | 5.0e-07 | 6.0e-05 | 3.8e-06 | |
| Wald | 1.2e-05 | 1.4e-05 | 4.6e-05 | 6.0e-08 | |
| Score | 1.8e-05 | 2.3e-05 | 5.9e-05 | 1.4e-07 | |
| rs7526034 | SPU(1) | 5.7e-02 | 9.0e-04 | <1.0e-07 | 6.2e-02 |
| SPU(2) | 8.2e-01 | 5.7e-02 | 2.0e-01 | 1.4e-02 | |
| SPU(3) | 6.5e-01 | 5.8e-02 | 6.8e-02 | 4.8e-02 | |
| SPU(4) | 8.4e-01 | 8.3e-02 | 2.1e-01 | 2.6e-02 | |
| SPU(5) | 7.7e-01 | 7.5e-02 | 1.5e-01 | 4.8e-02 | |
| SPU(∞) | 8.4e-01 | 8.4e-02 | 2.7e-01 | 4.0e-02 | |
| aSPU | 1.3e-01 | 2.3e-03 | <1.0e-07 | 3.5e-02 | |
| Wald | 2.6e-01 | 1.4e-05 | 2.0e-06 | 3.5e-04 | |
| Score | 2.6e-01 | 1.6e-05 | 3.1e-06 | 4.3e-04 | |
Simulations
Simulation I: mvLMM
To mimic real data, we used the ADNI data to generate multivariate phenotypes according to the fitted the mvLMM models (3) while using the covariates and SNPs in the ADNI data too. Specifically, two SNP-phenotype pairs, rs7526034-RLatTemp and APOE-ε4-RPar were chosen; from their corresponding fitted models (3) we obtained the parameter estimates such as λ̂, ψ̂, V̂g and V̂ε. Those parameter estimates except ψ̂, which was either 0 for the null model or was scaled by a factor 1/2 to reduce the effect size of the SNP for the non-null model (since we were using a nominal significance level at 0.05), were then used to simulate the phenotypes by model (3). For each simulated dataset, as before, the MLE of ψ and its covariance estimate were obtained from GEMMA to approximate its score vector so that the SPU and aSPU tests could be applied. We used B = 1000 to calculate their p-values. As a comparison, we also used the Wald test, Score test and LRT directly output by GEMMA. Based on five thousand replicates for each set-up, we obtained the empirical Type I error and power estimates. However, we note that there were some convergence problems when running GEMMA for about 1% and 2% of simulated datasets for the two SNP-phenotype pairs respectively; our results were based on the remaining ones without any convergence problems.
As shown in Table 2, the Type I error rates for both pairs were slightly inflated for all the tests except the LRT, which had largely inflated Type I error rates. The inflation could be due to a large number of parameters to be estimated in an mvLMM with a moderate sample size.
Table 2.
Simulation I: empirical Type I error and power for two SNP-phenotype pairs, rs7526034-RLatTemp (pair 1) and APOE-ε4-RPar (pair 2), based on fitting mvLMM.
| Pair | Approximate score vector | Score | LRT | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SPU(γ) | aSPU | Wald | |||||||||
| γ = 1 | 2 | 3 | 4 | 5 | ∞ | ||||||
| 1 | Type I | 0.062 | 0.067 | 0.067 | 0.066 | 0.069 | 0.068 | 0.066 | 0.067 | 0.067 | 0.178 |
| Power | 0.722 | 0.103 | 0.195 | 0.112 | 0.139 | 0.120 | 0.621 | 0.599 | 0.591 | 0.604 | |
| 2 | Type I | 0.058 | 0.065 | 0.063 | 0.066 | 0.065 | 0.067 | 0.068 | 0.064 | 0.058 | 0.188 |
| Power | 0.644 | 0.389 | 0.335 | 0.381 | 0.336 | 0.361 | 0.643 | 0.689 | 0.674 | 0.641 | |
Since the Type I error rates based on fitting the mvLMM were slightly inflated, while it was known that there was barely any population stratification in the ADNI data [Xu et al., 2014], we fitted the corresponding model after treating K = I; however, we experienced some numerical convergence problems in fitting the mvLMM, likely due to that the two unstructured covariance matrices Vg and Vε were not identifiable (after forcing K = I). Thus we simply used function gls() in R package nlme to fit a corresponding marginal linear model with or without top 10 principal components (PCs); the PCs were extracted using Plink [Purcell et al., 2007] based on almost a half million SNPs of the 757 subjects in the ADNI data. As shown in Table 3, the Type I error rates were better controlled. Note that since the simulated data were generated with a K ≠ I, some slight inflation of a Type I error rate was expected under the incorrect assumption K = I.
Table 3.
Simulation I: empirical Type I error and power for two SNP-phenotype pairs, rs7526034-RLatTemp (pair 1) and APOE-ε4-RPar (pair 2) based on fitting marginal linear models (LMs) with or without the top 10 PCs.
| Model | Pair | Approximate score vector | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SPU(γ) | aSPU | Wald | ||||||||
| γ = 1 | 2 | 3 | 4 | 5 | ∞ | |||||
| LM, 10 PCs | 1 | Type I | 0.049 | 0.057 | 0.063 | 0.057 | 0.061 | 0.058 | 0.061 | 0.059 |
| Power | 0.742 | 0.092 | 0.212 | 0.117 | 0.158 | 0.134 | 0.648 | 0.611 | ||
| 2 | Type I | 0.050 | 0.052 | 0.050 | 0.051 | 0.048 | 0.049 | 0.052 | 0.052 | |
| Power | 0.616 | 0.347 | 0.306 | 0.347 | 0.316 | 0.339 | 0.610 | 0.664 | ||
| LM, no PCs | 1 | Type I | 0.049 | 0.059 | 0.062 | 0.057 | 0.061 | 0.057 | 0.060 | 0.057 |
| Power | 0.749 | 0.095 | 0.214 | 0.116 | 0.160 | 0.136 | 0.645 | 0.609 | ||
| 2 | Type I | 0.052 | 0.056 | 0.054 | 0.057 | 0.052 | 0.056 | 0.054 | 0.053 | |
| Power | 0.633 | 0.360 | 0.317 | 0.361 | 0.327 | 0.352 | 0.634 | 0.683 | ||
The two SNP-phenotype pairs were chosen partly because in the ADNI data analysis the aSPU test gave a more significant p-value than those of the Wald and Score tests for pair 1, while it was the opposite for pair 2 (Table 1). It was confirmed that in the simulations the aSPU test was indeed slightly more (or less) powerful than the Wald and Score tests for pair 1 (or pair 2).
Simulation II: LMM
We considered a case with unrelated individuals and multiple quantitative traits similar to those in Zhang et al (2014), for which the exact score vector could be derived. We would compare the performance of an exact score-based test with that of its approximate score-based one. For each subject i = 1, …, n, we generated his/her genotype data as in Pan (2009). Specifically, for each subject i, we first generated a latent vector Gi = (Gj1, …, Gj,p+1)′ from a multivariate Normal distribution with a first-order auto-regressive (AR-1) covariance structure with parameter ρ = 0.5: Cov(Gis,Git) = ρ|s−t|. Second, each latent element Gis was dichotomized to 0 or 1 with probability Prob(Gis = 1) as its minor allele frequency (MAF), randomly drawn from a uniform distribution. Third, we independently generated another haplotype for subject i, then combined the two haplotypes to form the genotypes for subject j. In this way, we obtained the genotypes of all the subjects.
The first SNP was chosen as the causal one with MAF randomly drawn from a uniform distribution U(0.3, 0.4), while the MAFs of the other SNPs were independently drawn from U(0.1, 0.5). For each subject i, we simulated k traits Yi = (Yi1, …, Yik) from a linear model:
| (8) |
where λ = (λ1, …, λk)T, ψ = (ψ1, …, ψk)T; with σ = 1 and R as a compound symmetry (CS) correlation matrix with the correlation parameter r = 0.3; xi is the genotype dosage of the causal SNP. Under H0 we had ψ = 0; under H1 we had ψm ≠ 0 for 1 ≤ m ≤ 5 and ψm = 0 for 5 < m ≤ k. The non-zero ψj’s were simulated from a uniform distribution U(0.2, 0.3). In this way, under H1, only the first 5 traits were associated with the causal SNP; that is, as k = 5, 10, 20, 30, 40, we gradually increased the number of the non-associated traits from 0 to 5, 15, 25 and 35.
The simulated data were fitted by a LMM:
| (9) |
where bi was a normal random effect to model the correlations among multiple traits, εij was the random error, xi was a scalar corresponding to the genetic score of the SNP nearest to the causal SNP.
We implemented both the approximate score-based and exact score-based tests. For an approximate score-based test, we fitted model (9) and used the MLE of ψ to approximate its score vector and its variance-covariance matrix. For an exact score-based test, we fitted the reduced LMM moded under H0, Yij = λj + bi + εij, to obtain the MLEs λ̂ = (λ̂1, …, λ̂k)T. Denote I as the k × k identity matrix. The exact score vector and its variance-covariance matrix can be written as:
| (10) |
where V̂i was the MLE of Vi with its diagonal elements and off-diagonal elements . Partition the score vector and its covariance into two parts corresponding to the intercept and ψ parameters respectively:
then we have the exact score vector for ψ as U2 and its covariance matrix .
To estimate the Type I error and power, 1000 datasets were independently simulated and analyzed. Each of the 1000 datasets consisted of 1000 subjects. We used B = 1000 to obtain p-values for any permutation based methods. As a comparison, we also showed the results from the UminP.
As shown in Table 4, first, regardless of the test being examined, its version based on the approximate score vector and that based on the exact score vector gave almost the same results, suggesting the high accuracy of the asymptotic approximation in this case. Second, we note that the Type I error rates were satisfactorily controlled, even for 40 traits. Third, in agreement with Zhang et al (2014), the aSPU test was more powerful than the score test for five traits, but not for other numbers of traits; both were much more powerful than single trait-based UminP test, presumably due to the former two’s combining information across the five associated traits.
Table 4.
Simulation II: empirical type I error rates and power of the approximate (“approx”) score- and exact score-based tests when multiple traits were correlated with a CS structure with correlation r = 0.3. The first five traits were associated with a causal SNP with effect size βj ~ U(0.2, 0.3); the SNP nearest to the causal SNP was tested.
| SPU(γ) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Score vector | #traits | UminP | γ = 1 | 2 | 3 | 4 | 5 | ∞ | aSPU | Score | |
| Type I | approx | 5 | 0.041 | 0.042 | 0.040 | 0.048 | 0.038 | 0.044 | 0.043 | 0.042 | 0.034 |
| 10 | 0.050 | 0.051 | 0.056 | 0.056 | 0.057 | 0.059 | 0.053 | 0.052 | 0.059 | ||
| 20 | 0.048 | 0.057 | 0.046 | 0.048 | 0.049 | 0.047 | 0.050 | 0.045 | 0.047 | ||
| 30 | 0.048 | 0.048 | 0.051 | 0.050 | 0.049 | 0.051 | 0.050 | 0.053 | 0.053 | ||
| 40 | 0.047 | 0.052 | 0.042 | 0.046 | 0.048 | 0.045 | 0.051 | 0.042 | 0.046 | ||
| exact | 5 | 0.043 | 0.045 | 0.034 | 0.042 | 0.038 | 0.043 | 0.043 | 0.039 | 0.034 | |
| 10 | 0.052 | 0.051 | 0.056 | 0.057 | 0.054 | 0.052 | 0.050 | 0.052 | 0.059 | ||
| 20 | 0.046 | 0.053 | 0.051 | 0.050 | 0.049 | 0.048 | 0.049 | 0.046 | 0.047 | ||
| 30 | 0.047 | 0.047 | 0.053 | 0.056 | 0.045 | 0.053 | 0.050 | 0.053 | 0.053 | ||
| 40 | 0.051 | 0.055 | 0.039 | 0.047 | 0.045 | 0.042 | 0.049 | 0.046 | 0.046 | ||
| Power | approx | 5 | 0.140 | 0.686 | 0.139 | 0.288 | 0.146 | 0.188 | 0.144 | 0.549 | 0.435 |
| 10 | 0.324 | 0.274 | 0.528 | 0.341 | 0.454 | 0.331 | 0.324 | 0.486 | 0.590 | ||
| 20 | 0.394 | 0.097 | 0.597 | 0.433 | 0.544 | 0.403 | 0.392 | 0.512 | 0.606 | ||
| 30 | 0.378 | 0.088 | 0.581 | 0.395 | 0.540 | 0.399 | 0.375 | 0.493 | 0.570 | ||
| 40 | 0.364 | 0.071 | 0.538 | 0.416 | 0.532 | 0.404 | 0.368 | 0.474 | 0.543 | ||
| exact | 5 | 0.138 | 0.685 | 0.133 | 0.285 | 0.141 | 0.182 | 0.141 | 0.543 | 0.434 | |
| 10 | 0.323 | 0.274 | 0.536 | 0.334 | 0.458 | 0.315 | 0.322 | 0.479 | 0.588 | ||
| 20 | 0.384 | 0.099 | 0.594 | 0.426 | 0.544 | 0.401 | 0.397 | 0.519 | 0.606 | ||
| 30 | 0.384 | 0.080 | 0.571 | 0.399 | 0.543 | 0.398 | 0.378 | 0.500 | 0.569 | ||
| 40 | 0.369 | 0.068 | 0.534 | 0.419 | 0.531 | 0.410 | 0.368 | 0.470 | 0.542 | ||
Simulation III: GLMM
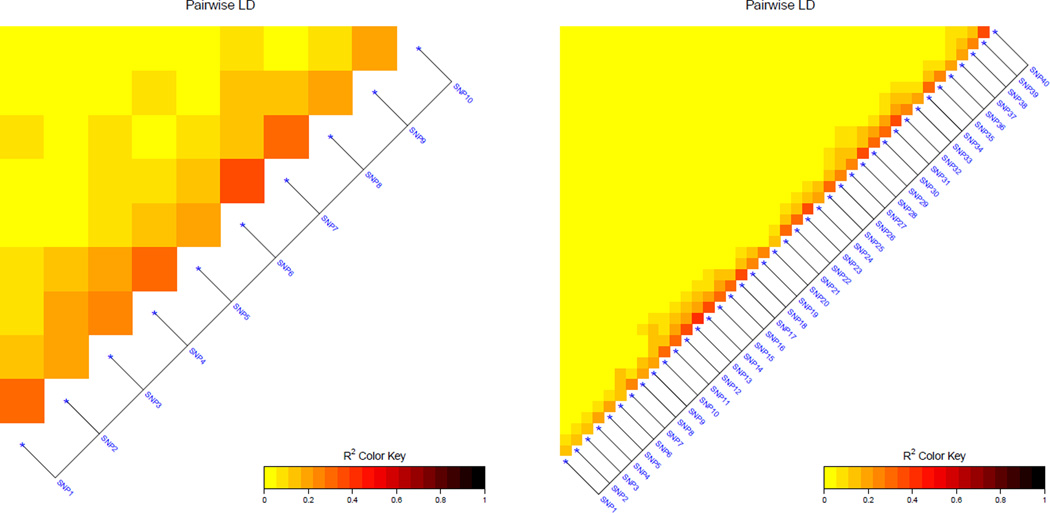
We considered a familial/trio study design with a single binary trait; since there were multiple subjects in each family, their traits might be correlated. For each of the two parents in each family i = 1, …, n, we generated their haplotypes and thus genotypes as described in the previous section (with ρ = 0.8); then their offspring’s haplotype and thus genotype data were obtained according to the Mendelian transmission. In this way, we obtained the genotype data with p + 1 SNPs for each subjects. The SNP at the center (i.e. at position p/2 + 1) was chosen as the causal one with MAF 0.3, while the MAFs of the other SNPs were independently drawn from U(0.1, 0.4). Figure 1 shows linkage disequilibrium (LD) plots for the generated SNPs (after the causal SNP was removed) based on one parent from each family.
Figure 1.
LD plots for simulated genotypes with (n = 200, p = 10) (left panel) and (n = 400, p = 20) (right panel).
For subject j in family i, denote xij as the genotypic score for the causal SNP, and Xij = (Xij1, …, Xijp)T as a vector of the genotypic scores for the p non-causal SNPs. The disease indicator Yij = 0 or 1 was generated from the below GLMM: Assuming λ0 is the background log odds ratio, ψ0 is the effect sizes, the resulting GLMM model can be written as:
| (11) |
where λ0 = −log(4) was chosen to have a 20% background disease prevalence, while ψ0 at varying effect sizes was used to investigate the empirical Type I error (ψ0 = log(1)) and power (ψ0 > log(1)). We fixed σb = 1, and considered two cases with (p = 10, n = 200) and (p = 20, n = 400). For each simulation set-up, we generated 1000 simulated datasets to estimate the empirical Type I error or power.
We fitted both a GLMM (6) and a corresponding marginal model to test H0: ψ = 0. A GLMM was fitted using function glmer() from R package lme4. Either the Laplace approximation (LA) or adaptive Gaussian quadrature (AGQ) were used to approximate the (marginal) log-likelihood. For AGQ, we specified the number of points per axis nAGQ = 25; in the manual of lme4, it was mentioned that “A model with a single, scalar random-effects term could reasonably use up to 25 quadrature points per scalar integral” [Bates et al, 2014]. For a marginal model, both a working independence and a working compound symmetry (CS) correlation structures were used in GEE. We then applied our method to approximate the score vector for the fitted GLMM and GEE models respectively, based on which we applied the SPU and aSPU tests; we used B = 1000 to calculate their p-values. As a comparison, we also showed the results from the UminP and Wald tests; recall that the Wald test is equivalent to the approximate score test.
As shown in Table 5, it seems that the Type I error rates were appropriately controlled by all the tests except the Wald test, which gave slightly inflated Type I error rates in GEE as well known in the literature [e.g. Zhang et al., 2014]. It is clear that, for the same fitted model, the aSPU test was much more powerful than the popular UminP and Wald tests. Among the SPU tests, the SPU(1) was nearly as powerful as SPU(2) and SPU(3) for a smaller number of parameters to be tested with p = 10, but it was less powerful than the latter two for p = 20. This is in agreement with the analysis and motivation of the SPU tests: for p = 10, because all the 10 SNPs were correlated with the causal SNP, the SPU(1) test was expected to be powerful, as more generally known for the burden tests; on the other hand, for p = 20, since some SNPs were barely correlated with the causal SNP, to minimize the effects on power of the non-associated SNPs, a larger γ (here γ = 3) would yield higher power for the SPU(γ) test. Among the three fitted models, the aSPU test based on GEE(CS) was more powerful than that based on the other two models, the latter of which gave similar results; the lower power of GEE(Ind) might be due to the use of the working independence correlation structure, while the GLMM was fitted by an approximate maximum likelihood (using either the LA by default, or ACQ in the function glme), leading to loss of efficiency. It is noted that the results from the LA and ACQ approximations were very close; they seemed to be a little conservative with Type I rates much lower than the nominal 0.05.
Table 5.
Simulation III: empirical Type I error (for OR=1) and power (for OR > 1) of the approximate score-based tests with correlated binary traits. The two cases were for (n = 200, p = 10) and (n = 400, p = 20). A GLMM was fitted using either the Laplace (LA) or adaptive Gaussian quadrature (AGQ) approximation; the working correlation structure in GEE was assumed to be either independent(Ind) or compound symmetry (CS).
| SPU(γ) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MoDel | Case | OR | UminP | γ = 1 | 2 | 3 | 4 | 5 | ∞ | aSPU | Wald |
| GLMM (LA) | 1 | 1 | 0.018 | 0.044 | 0.015 | 0.016 | 0.011 | 0.012 | 0.014 | 0.020 | 0.021 |
| 1.4 | 0.078 | 0.125 | 0.100 | 0.105 | 0.088 | 0.090 | 0.074 | 0.100 | 0.057 | ||
| 1.8 | 0.216 | 0.323 | 0.296 | 0.294 | 0.266 | 0.257 | 0.195 | 0.300 | 0.141 | ||
| 2.2 | 0.406 | 0.514 | 0.494 | 0.511 | 0.461 | 0.452 | 0.368 | 0.470 | 0.280 | ||
| 2.6 | 0.580 | 0.702 | 0.681 | 0.680 | 0.645 | 0.641 | 0.568 | 0.667 | 0.427 | ||
| 3 | 0.736 | 0.825 | 0.822 | 0.826 | 0.799 | 0.794 | 0.713 | 0.807 | 0.587 | ||
| 2 | 1 | 0.028 | 0.030 | 0.018 | 0.024 | 0.017 | 0.019 | 0.024 | 0.023 | 0.032 | |
| 1.4 | 0.096 | 0.157 | 0.116 | 0.138 | 0.106 | 0.108 | 0.090 | 0.131 | 0.067 | ||
| 1.8 | 0.344 | 0.424 | 0.422 | 0.467 | 0.425 | 0.431 | 0.353 | 0.414 | 0.187 | ||
| 2.2 | 0.677 | 0.668 | 0.750 | 0.776 | 0.750 | 0.755 | 0.667 | 0.748 | 0.419 | ||
| 2.6 | 0.862 | 0.834 | 0.897 | 0.914 | 0.902 | 0.904 | 0.848 | 0.893 | 0.666 | ||
| 3 | 0.950 | 0.918 | 0.964 | 0.969 | 0.968 | 0.972 | 0.938 | 0.963 | 0.825 | ||
| GLMM (AGQ) | 1 | 1 | 0.015 | 0.047 | 0.014 | 0.011 | 0.008 | 0.010 | 0.011 | 0.022 | 0.018 |
| 1.4 | 0.073 | 0.118 | 0.084 | 0.096 | 0.085 | 0.083 | 0.070 | 0.092 | 0.039 | ||
| 1.8 | 0.198 | 0.310 | 0.279 | 0.281 | 0.248 | 0.242 | 0.187 | 0.281 | 0.122 | ||
| 2.2 | 0.390 | 0.505 | 0.472 | 0.489 | 0.439 | 0.431 | 0.353 | 0.463 | 0.255 | ||
| 2.6 | 0.565 | 0.688 | 0.669 | 0.664 | 0.624 | 0.616 | 0.536 | 0.654 | 0.397 | ||
| 3 | 0.724 | 0.816 | 0.817 | 0.817 | 0.792 | 0.783 | 0.695 | 0.794 | 0.557 | ||
| 2 | 1 | 0.024 | 0.029 | 0.016 | 0.023 | 0.016 | 0.015 | 0.022 | 0.024 | 0.025 | |
| 1.4 | 0.085 | 0.149 | 0.110 | 0.133 | 0.096 | 0.103 | 0.083 | 0.122 | 0.056 | ||
| 1.8 | 0.329 | 0.407 | 0.408 | 0.450 | 0.396 | 0.415 | 0.332 | 0.412 | 0.166 | ||
| 2.2 | 0.665 | 0.667 | 0.727 | 0.764 | 0.730 | 0.737 | 0.657 | 0.744 | 0.374 | ||
| 2.6 | 0.851 | 0.833 | 0.892 | 0.910 | 0.895 | 0.901 | 0.834 | 0.888 | 0.637 | ||
| 3 | 0.947 | 0.916 | 0.962 | 0.970 | 0.968 | 0.970 | 0.928 | 0.958 | 0.810 | ||
| GEE (Ind) | 1 | 1 | 0.030 | 0.040 | 0.024 | 0.023 | 0.026 | 0.024 | 0.026 | 0.032 | 0.057 |
| 1.4 | 0.093 | 0.134 | 0.108 | 0.111 | 0.100 | 0.099 | 0.088 | 0.116 | 0.106 | ||
| 1.8 | 0.215 | 0.291 | 0.284 | 0.288 | 0.253 | 0.248 | 0.202 | 0.272 | 0.170 | ||
| 2.2 | 0.375 | 0.478 | 0.459 | 0.467 | 0.428 | 0.422 | 0.366 | 0.445 | 0.308 | ||
| 2.6 | 0.519 | 0.625 | 0.625 | 0.628 | 0.596 | 0.588 | 0.519 | 0.603 | 0.438 | ||
| 3 | 0.672 | 0.754 | 0.755 | 0.757 | 0.736 | 0.729 | 0.656 | 0.740 | 0.558 | ||
| 2 | 1 | 0.036 | 0.037 | 0.035 | 0.033 | 0.032 | 0.034 | 0.042 | 0.033 | 0.081 | |
| 1.4 | 0.113 | 0.146 | 0.128 | 0.149 | 0.134 | 0.137 | 0.110 | 0.146 | 0.125 | ||
| 1.8 | 0.323 | 0.368 | 0.368 | 0.410 | 0.377 | 0.394 | 0.320 | 0.385 | 0.229 | ||
| 2.2 | 0.589 | 0.593 | 0.662 | 0.709 | 0.677 | 0.680 | 0.579 | 0.672 | 0.427 | ||
| 2.6 | 0.775 | 0.761 | 0.829 | 0.847 | 0.826 | 0.830 | 0.770 | 0.825 | 0.619 | ||
| 3 | 0.902 | 0.865 | 0.926 | 0.939 | 0.932 | 0.928 | 0.881 | 0.925 | 0.755 | ||
| GEE (CS) | 1 | 1 | 0.044 | 0.061 | 0.041 | 0.037 | 0.035 | 0.035 | 0.034 | 0.040 | 0.065 |
| 1.4 | 0.116 | 0.141 | 0.133 | 0.136 | 0.115 | 0.117 | 0.111 | 0.133 | 0.104 | ||
| 1.8 | 0.271 | 0.366 | 0.349 | 0.358 | 0.323 | 0.319 | 0.261 | 0.337 | 0.222 | ||
| 2.2 | 0.459 | 0.576 | 0.573 | 0.577 | 0.541 | 0.538 | 0.459 | 0.547 | 0.385 | ||
| 2.6 | 0.653 | 0.730 | 0.734 | 0.740 | 0.704 | 0.698 | 0.632 | 0.738 | 0.526 | ||
| 3 | 0.800 | 0.845 | 0.862 | 0.862 | 0.847 | 0.840 | 0.780 | 0.851 | 0.670 | ||
| 2 | 1 | 0.039 | 0.041 | 0.032 | 0.035 | 0.035 | 0.038 | 0.041 | 0.038 | 0.070 | |
| 1.4 | 0.124 | 0.183 | 0.159 | 0.180 | 0.151 | 0.163 | 0.124 | 0.167 | 0.137 | ||
| 1.8 | 0.433 | 0.476 | 0.500 | 0.543 | 0.509 | 0.508 | 0.438 | 0.506 | 0.309 | ||
| 2.2 | 0.750 | 0.712 | 0.805 | 0.832 | 0.806 | 0.809 | 0.755 | 0.791 | 0.544 | ||
| 2.6 | 0.895 | 0.881 | 0.925 | 0.943 | 0.933 | 0.931 | 0.883 | 0.925 | 0.749 | ||
| 3 | 0.969 | 0.934 | 0.971 | 0.982 | 0.978 | 0.978 | 0.962 | 0.976 | 0.881 | ||
Since the proposed method of approximating the score vector is asymptotic, to further evaluate its finite-sample performance, we also applied the score-based tests based on the exact, not approximate, GEE (generalized) score formulas, as implemented in Zhang et al (2014). Comparing the test results in Tables (5) and (6), we can see that the approximate score-based tests had slight losses of power for the smaller sample size n = 200, but performed equally well as the exact score-based tests for the larger sample size n = 400.
Table 6.
Simulation III: empirical Type I error (for OR=1) and power (for OR > 1) of the GEE score-based tests with correlated binary traits. The two cases were for (n = 200, p = 10) and (n = 400, p = 20).
| SPU(γ) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Case | OR | UminP | γ = 1 | 2 | 3 | 4 | 5 | ∞ | aSPU | Score |
| GEE (Ind) | 1 | 1 | 0.036 | 0.047 | 0.037 | 0.038 | 0.031 | 0.027 | 0.034 | 0.038 | 0.042 |
| 1.4 | 0.090 | 0.129 | 0.120 | 0.117 | 0.108 | 0.106 | 0.090 | 0.117 | 0.074 | ||
| 1.8 | 0.228 | 0.295 | 0.305 | 0.303 | 0.277 | 0.264 | 0.220 | 0.284 | 0.148 | ||
| 2.2 | 0.404 | 0.474 | 0.480 | 0.489 | 0.459 | 0.450 | 0.399 | 0.462 | 0.280 | ||
| 2.6 | 0.567 | 0.630 | 0.654 | 0.658 | 0.623 | 0.614 | 0.555 | 0.625 | 0.406 | ||
| 3 | 0.698 | 0.766 | 0.786 | 0.779 | 0.759 | 0.752 | 0.694 | 0.765 | 0.544 | ||
| 2 | 1 | 0.049 | 0.046 | 0.044 | 0.038 | 0.039 | 0.042 | 0.048 | 0.051 | 0.054 | |
| 1.4 | 0.124 | 0.141 | 0.152 | 0.158 | 0.148 | 0.151 | 0.115 | 0.153 | 0.092 | ||
| 1.8 | 0.355 | 0.369 | 0.403 | 0.429 | 0.413 | 0.415 | 0.344 | 0.413 | 0.198 | ||
| 2.2 | 0.627 | 0.595 | 0.690 | 0.725 | 0.696 | 0.699 | 0.613 | 0.693 | 0.399 | ||
| 2.6 | 0.803 | 0.770 | 0.852 | 0.865 | 0.843 | 0.841 | 0.787 | 0.841 | 0.597 | ||
| 3 | 0.915 | 0.875 | 0.941 | 0.949 | 0.941 | 0.942 | 0.895 | 0.934 | 0.745 | ||
| GEE (CS) | 1 | 1 | 0.040 | 0.066 | 0.047 | 0.048 | 0.044 | 0.043 | 0.043 | 0.050 | 0.041 |
| 1.4 | 0.119 | 0.140 | 0.135 | 0.134 | 0.127 | 0.122 | 0.114 | 0.130 | 0.082 | ||
| 1.8 | 0.282 | 0.361 | 0.370 | 0.369 | 0.342 | 0.329 | 0.276 | 0.349 | 0.192 | ||
| 2.2 | 0.487 | 0.572 | 0.582 | 0.585 | 0.554 | 0.546 | 0.474 | 0.554 | 0.357 | ||
| 2.6 | 0.651 | 0.738 | 0.763 | 0.747 | 0.727 | 0.719 | 0.647 | 0.737 | 0.506 | ||
| 3 | 0.823 | 0.847 | 0.865 | 0.870 | 0.850 | 0.845 | 0.797 | 0.865 | 0.644 | ||
| 2 | 1 | 0.047 | 0.044 | 0.041 | 0.040 | 0.045 | 0.048 | 0.044 | 0.049 | 0.054 | |
| 1.4 | 0.150 | 0.172 | 0.162 | 0.183 | 0.159 | 0.168 | 0.140 | 0.168 | 0.098 | ||
| 1.8 | 0.446 | 0.465 | 0.519 | 0.543 | 0.527 | 0.525 | 0.465 | 0.512 | 0.258 | ||
| 2.2 | 0.766 | 0.712 | 0.825 | 0.832 | 0.823 | 0.823 | 0.770 | 0.816 | 0.507 | ||
| 2.6 | 0.908 | 0.879 | 0.928 | 0.947 | 0.937 | 0.942 | 0.896 | 0.931 | 0.739 | ||
| 3 | 0.973 | 0.936 | 0.975 | 0.982 | 0.981 | 0.979 | 0.967 | 0.978 | 0.866 | ||
Discussion
We have described an asymptotic approach to approximating the score vector in some complex models, such as a mvLMM or GLMM. The approximate score vector can then be used to construct any score-based tests, including kernel machine regression and adaptive sum of powered score tests for multiple traits or familial data. Using both real and simulated data, we have demonstrated such approximate score-based tests can improve power over (and control the Type I error rate better than) the standard Wald test and the UminP test that has been widely used in GWAS. The proposed approximate score vector offers a simple and general way to extend many score-based tests to other complex models, in which the score vector is unavailable from the statistical package being used. Although we have focused on the mvLMM and GLMM, we also considered the LMM, the Cox frailty model [Therneau et al., 2003] and the Cox mixed-effects model [Therneau, 2012], and reached similar conclusions (results not shown to save space); the difference between the two Cox models is that the latter includes a random-effect to account for genetic relatedness across all subjects, similar to that adopted in the mvLMM. We envision the use of the proposed approximate score vector in other models.
Our proposed general approach is a two-step procedure. In the first step, a full model including a set of parameters to be tested is fitted, then in the second step the score vector for the parameter and its covariance matrix are approximated based on the parameter estimates and their covariance matrix. In this way, a score-based test can be applied without directly calculating the score vector (and its covariance matrix), which may not be easy to derive based on existing software packages, such as for mvLMM and GLMM. Due to the nature of the proposed two-step approach, the validity of the approach depends on both that of the first step and the asymptotics. For example, if we have a familial dataset with trait-ascertained samples, then it is necessary to appropriately account for the sample ascertainment in step one, e.g. based on some family-based association testing procedures [Moerkerke et al., 2010; Zhang et al., 2012]. Furthermore, since the proposed approximation to the score vector is based on the asymptotics of the parameters to be tested, it has some limitations. First, if the sample size is too small or more generally, if the conditions for the asymptotics do not hold, e.g. in analysis of rare variants [Chen et al., 2013; Jiang et al., 2014], then it may not perform well with inflated false positives and false negatives. Second, in order to obtain a point estimate of the parameters to be tested, a full model has to be fitted, which may not be even computationally feasible if the number of the parameters to be estimated is too large relative to the sample size. Nevertheless, the proposed method offers a simple and practical way to extend many score-based tests to more complex models, for which the score vector is either unavailable from software or has no closed-form solution. In addition to testing for main effects as considered here, it will also be interesting to explore the use of the proposed method to detect gene-gene and gene-environment interactions [Tzeng et al., 2011].
Acknowledgments
This research was supported by NIH grants R01GM113250, R01HL116720 and R01DA033958, and by the Minnesota Supercomputing Institute (MSI). The authors thank Dr. Xiang Zhou for help with the use of mvLMM, and the reviewers for constructive comments.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
References
- Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1–6. 2014 http://CRAN.R-project.org/package=lme4. [Google Scholar]
- Boos DD. On generalized score test. The American Statistician. 1992;46:327–333. [Google Scholar]
- Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. Journal of the American Statistical Association. 1993;88:9–25. [Google Scholar]
- Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genetic Epidemiology. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P, Heagerty P, Liang K-Y, Zeger S. Analysis of Longitudinal Data. Oxford: 2013. [Google Scholar]
- Hartig M, Truran-Sacrey D, Raptentsetsang S, et al. ADNI: Alzheimer’s Disease Neuroimaging Initiative. San Francisco: 2012. UCSF FreeSurfer Overview and QC Ratings. [Google Scholar]
- Jiang Y, Conneely KN, Epstein MP. Flexible and Robust Methods for Rare Variant Testing of Quantitative Traits in Trios and Nuclear Families. Genetic Epidemiology. 2014;38:542–551. doi: 10.1002/gepi.21839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent JT. Robust properties of likelihood ratio tests. Biometrika. 1982;69:19–27. [Google Scholar]
- Klei L, Luca D, Devlin B, Roeder K. Pleiotropy and principal components of heritability combine to increase power for association analysis. Genetic Epidemiology. 2008;32:9–19. doi: 10.1002/gepi.20257. [DOI] [PubMed] [Google Scholar]
- Korte A, Vilhjlmsson BJ, Segura V, Platt A, Long Q, Nordborg M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nature Genetics. 2012;44:1066–1071. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. American Journal of Human Genetics. 2008;82:386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American Journal of Human Genetics. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Lin DY, Tang ZZ. A general framework for detecting disease associations with rare variants in sequencing studies. American Journal of Human Genetics. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Zhu H, Knickmeyer R, Styner M, Gilmore J, Ibrahim JG. Projection regression models for multivariate imaging phenotype. Genetic Epidemiology. 2012;36:631–641. doi: 10.1002/gepi.21658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity A, Sullivan PF, Tzeng J. Multivariate Phenotype Association Analysis by Marker-Set Kernel Machine Regression. Genetic Epidemiology. 2012;36:686–695. doi: 10.1002/gepi.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerkerke B, Vansteelandt S, Lange C. A doubly robust test for gene-environment interaction in family-based studies of affected offspring. Biostatistics. 2010;11:213–225. doi: 10.1093/biostatistics/kxp061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genetic Epidemiology. 2009;33:497–507. doi: 10.1002/gepi.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Relationship between genomic distance-based regression and kernel machine regression for multi-marker association testing. Genetic Epidemiology. 2011;35:211–216. doi: 10.1002/gepi.20567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Kim J, Zhang Y, Shen X, Wei P. A powerful and adaptive association test for rare variants. Genetics. 2014;197:1081–1095. doi: 10.1534/genetics.114.165035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotnitzky A, Jewell NP. Hypothesis testing of regression parameters in semiparametric generalized linear models for cluster correlated data. Biometrika. 1990;77:485–497. [Google Scholar]
- Schifano ED, Epstein MP, Bielak LF, Jhun MA, Kardia SL, Peyser PA, Lin X. SNP set association analysis for familial data. Genetic Epidemiology. 2012;36:797–810. doi: 10.1002/gepi.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Jr, Weiner MW, Saykin AJ, Alzheimer’s Disease Neuroimaging Initiative Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng Y, Hsu L. A unified mixed-effects model for rare-variant association in sequencing studies. Genet Epidemiol. 2013;37:334–344. doi: 10.1002/gepi.21717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, Grambsch PM, Pankratz VS. Penalized survival models and frailty. Journal of Computational and Graphical Statistics. 2003;12:156–175. [Google Scholar]
- Therneau TM. Mixed Effects Cox Models. R-package description. 2012 URL: http://cran.r-project.org/web/packages/coxme/vignettes/coxme.pdf.
- Tzeng JY, Zhang D, Pongpanich M, Smith C, McCarthy MI, Sale MM, Worrall BB, Hsu FC, Thomas DC, Sullivan PF. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. American Journal of Human Genetics. 2011;89:277–288. doi: 10.1016/j.ajhg.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee S, Zhu X, Redline S, Lin X. GEE-Based SNP Set Association Test for Continuous and Discrete Traits in Family-Based Association Studies. Genetic Epidemiology. 2013;37:778–786. doi: 10.1002/gepi.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-Set Analysis for Case-Control Genome-wide Association Studies. American Journal of Human Genetics. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Shen X, Pan W, Alzheimer’s Disease Neuroimaging Initiative Longitudinal Analysis Is More Powerful than Cross-Sectional Analysis in Detecting Genetic Association with Neuroimaging Phenotypes. PLoS ONE. 2014;9:e102312. doi: 10.1371/journal.pone.0102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wu H, Guo CY, Fox CS. Analyze multivariate phenotypes in genetic association studies by combining univariate association tests. Genetic Epidemiology. 2010;34:444–454. doi: 10.1002/gepi.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wang Y. Methods for analyzing multivariate phenotypes in genetic association studies. Journal of Probability and Statistics. 2012;2012:652569. doi: 10.1155/2012/652569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics. 2005;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nature Genetics. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nature Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu Z, Shen X, et al. Testing for association with multiple traits in generalized estimation equations, with application to neuroimaging data. NeuroImage. 2014;96:309–325. doi: 10.1016/j.neuroimage.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Langefeld CD, Grunwald GK, Fingerlin TE. Testing gene-environment interactions in family-based association studies using trait-based ascertained samples. Stat Med. 2014;33:304–318. doi: 10.1002/sim.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES. Mixed linear model approach adapted for genome-wide association studies. Nature Genetics. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]