Abstract
Endoscopy is an important tool in screening and monitoring inflammatory bowel disease (IBD); however, it is invasive, costly, and associated with risks to the patients. Recently, circulating microRNAs (miRNAs) have emerged as promising noninvasive biomarkers. We proposed that the expression of serum microRNA223 (miR-223) could be a biomarker for IBD.
Studies were conducted using serum samples from 100 patients with IBD (50 with Crohn's disease [CD] and 50 with ulcerative colitis [UC]) and 50 healthy controls. The expression of serum miR-223 was measured by quantitative reverse transcription-polymerase chain reaction. The clinical disease activity was assessed by measurement of the Crohn's disease activity index for CD and the Mayo score for UC. Endoscopies were performed and graded according to the simple endoscopic score for CD and the ulcerative colitis endoscopic index of severity scores for UC. Blood samples for the measurement of high-sensitivity C-reactive protein (hs-CRP) and erythrocyte sedimentation rate (ESR) were taken within 1 week before or after endoscopy.
Serum miR-223 expression increased 2.2 times in patients with CD and 2.8 times in patients with UC compared with the control group. Most importantly, the level of serum miR-223 was correlated with several indicators of disease activity both in CD and UC. Serum miR-223 demonstrated a higher Spearman r value than ESR and hs-CRP in detecting the disease activity of patients with IBD.
Serum miR-223 might be a promising biomarker for monitoring disease activity in IBD patients.
INTRODUCTION
Inflammatory bowel disease (IBD), which includes Crohn's disease (CD) and ulcerative colitis (UC), is believed to develop via a complex interaction between genetics, immune response, and environmental factors, and this interaction is reflected in broad gene expression changes.1–3 Owing to the natural history of repeated recurrence of IBD, the disease activity of patients would be evaluated and monitored repeatedly. However, most of these monitoring tools are invasive, time-consuming, and expensive. Thus, it is not suitable and optimal for routine clinical applications. To overcome this dilemma, lots of biomarkers in peripheral blood may be an ideal alternative choice in the screening and disease activity assessment of IBD.
MiRNAs are short single stranded non-coding RNAs, which can regulate gene expression at the posttranscriptional level and may interfere with the process of transcription.4 More than 60% of protein coding messenger RNAs were reported to be regulated by them, influencing important cellular biological functions such as proliferation, migration and invasion, signal transduction, autophagy, and apoptosis.5,6 Till now more and more data showed that patients with IBD have altered miRNA profiles both in tissues and peripheral blood compared with healthy controls.7,8
Recently, circulating miRNAs were explored to be promising potential biomarkers in the diagnosis and screening of multiple cancers.9 They were present in serum in a cell-free state, and resistant to harsh conditions and in a stable form, which made their potential use as noninvasive biomarkers.6,10–15 Furthermore, miRNAs have the benefit of being obtained with relatively minimally invasive procedures and can be rapidly quantified by quantitative-polymerase chain reaction (PCR) or microarrays. Therefore, circulating miRNAs are attractive noninvasive biomarkers with promising clinical values.
Recently, detection of miRNAs in the peripheral blood has been extended to immune disorders such as systemic lupus erythematosus and rheumatoid arthritis.16 However, few studies have been performed on the relationship between miRNAs and IBD. In the present study, our goal was to determine if serum microRNA223 (miR-223) differed between IBD patients and healthy controls and to determine if there was any correlation between miR-223 levels and other indicators of disease activity both in CD and UC.
MATERIALS AND METHODS
Serum Samples and Disease Activity
Serum was obtained from 100 patients with IBD (50 with CD and 50 with UC) and 50 healthy controls at the First Affiliated Hospital, Sun Yat-sen University. All the participants gave written informed consent, and the study protocol was approved by the Human Ethics Committee of the First Affiliated Hospital, Sun Yat-Sen University. The diagnosis of CD and UC was confirmed by standard parameters as previously described.17–19 The site of disease was defined according to the Montreal classification.20 The clinical disease activity was assessed by the measurement of the Crohn's disease activity index (CDAI) for CD and the Mayo score for UC.21,22 Endoscopies were performed and graded according to the simple endoscopic score (SES) for CD (SES-CD) scoring systems and ulcerative colitis endoscopic index (UCEIS) of severity scores for UC.22–24 Patients with infectious colitis and colorectal cancer were excluded. Individuals who had normal height and body mass index and no history of chronic diseases were recruited for the control group. Blood samples for the measurement of high-sensitivity C-reactive protein (hs-CRP) and erythrocyte sedimentation rate (ESR) were taken within 1 week before or after endoscopy. To avoid bias, all gastroenterologists performing the endoscopies were unaware of the results of the disease activity index. Serum was stored at −80°C until RNA isolation.
RNA Isolation and Reverse-Transcriptase PCR
Total RNA was isolated from 1 mL of serum using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to manufacturer's instructions. For the reverse transcription (RT) of miR-223, we prepared a reaction with the RT master mix using the Transcriptor First Stand cDNA Synthesis Kit (Roche, Germany, Penzberg).
Quantitative-PCR
The Fast Start Universal SYBR Green Master (Roche) was used to confirm the miRNA expression changes of miR-223 both in the serum and the colonic tissues (primer sequences for miR-223 forward primer: GCGGCGGTGTCAGTTTGTC; reverse primer: GTGCAGGGTCCGAGGT). The expression of miRNA was calculated relative to U6 (primers sequences for U6, forward primer: CTCGCTTCGGCAGCACA; reverse primer: AACGCTTCACGAATTTGCGT). A comparative threshold cycle method was used to compare each condition with the control.
Statistical Analysis
Experimental results are expressed as mean ± SD. Statistical analyses for quantitative RT-PCR were performed with the unpaired, 2-tailed Student t tests and one-way analysis of variance for comparing all pairs of groups (SPSS 16.0). The association between 2 variables was assessed by Spearman rank correlation coefficient (r) for nonparametric correlations. P < 0.05 was considered statistically significant.
RESULTS
Identification of Serum MiR-223 as a Signature for IBD Patients
Quantitative RT-PCR revealed that the expression of serum miR-223 was significantly increased in CD and UC patients compared with the control group. The clinical characteristics of the patients and the controls were shown in Table 1. Serum miR-223 expression showed an increase of 2.2-fold in patients with CD and 2.8-fold in patients with UC compared with the control group (Figure 1; P < 0.01).
TABLE 1.
Demographic and Clinical Characteristics of the Study Population
FIGURE 1.
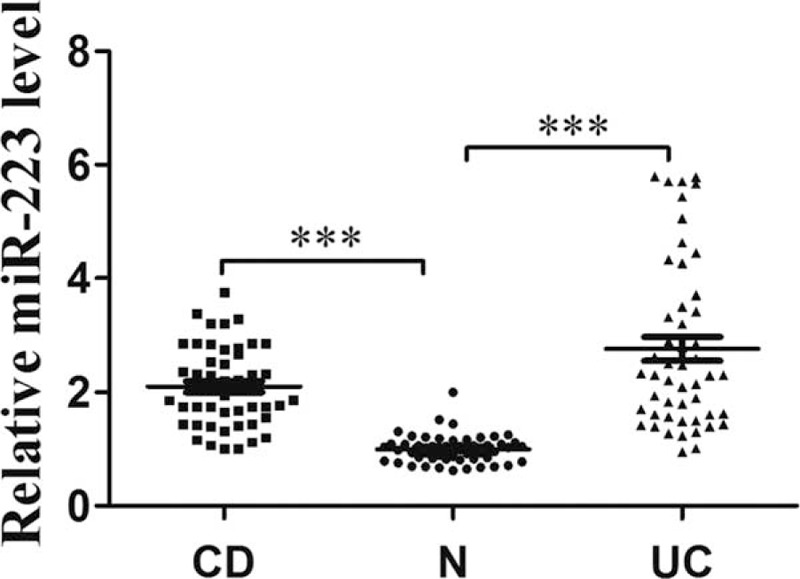
The expression of circulating miR-223 in IBD patients and healthy controls.∗∗p < 0.01. IBD = inflammatory bowel disease, miR-223 = microRNA223.
Expression Levels of Serum MiR-223 Were Correlated With Disease Activity of IBD
To identify the level of serum miR-223 expression with potential diagnostic value, we analyzed the correlation of serum miR-223 with CDAI, ESR, hs-CRP, and SES-CD in patients with CD (Figure 2), and at the same time we analyzed the correlation of serum miR-223 with the Mayo score, ESR, hs-CRP, and UCEIS in patients with UC (Figure 3). We found that the level of serum miR-223 had a positive correlation with these indicators of disease activity in both CD and UC, suggesting that the miR-223 level was associated with IBD disease activity.
FIGURE 2.
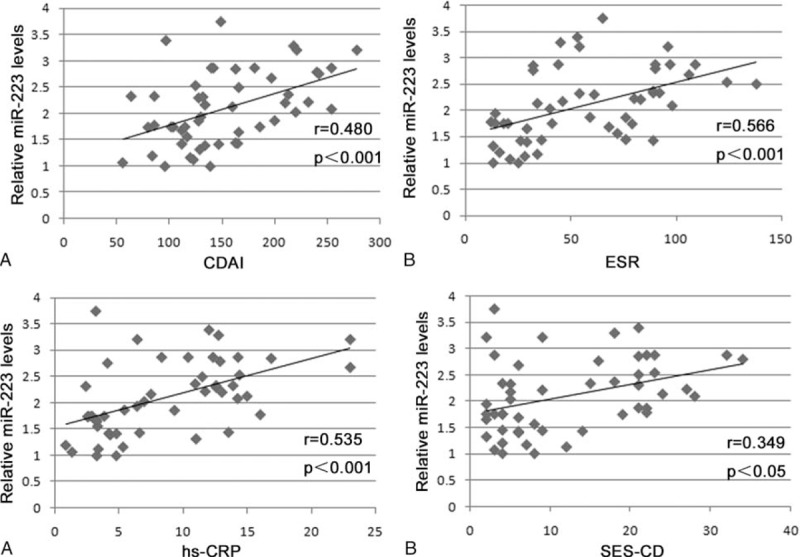
Correlation of circulating miR-223 with CDAI, ESR, hs-CRP, and SES-CD in patients with CD: (A) Spearman correlation analysis of miR-223 with CDAI in patients with CD; (B) Spearman correlation analysis of miR-223 with ESR in patients with CD; (C) Spearman correlation analysis of miR-223 with hs-CRP in patients with CD; (D) Spearman correlation analysis of miR-223 with SES-CD in patients with CD. CD = Crohn's disease, CDAI = Crohn's disease activity index, ESR = erythrocyte sedimentation rate, hs-CRP = high-sensitivity C-reactive protein, miR-223 = microRNA223, SES-CD = simple endoscopic score for CD.
FIGURE 3.
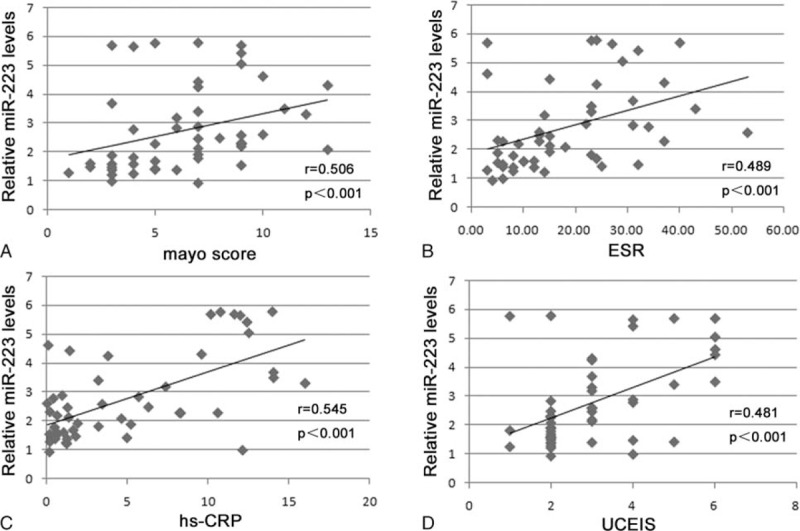
Correlation of circulating miR-223 with Mayo score, ESR, hs-CRP, and UCEIS in patients with UC: (A) Spearman correlation analysis of miR-223 with Mayo score in patients with UC; (B) Spearman correlation analysis of miR-223 with ESR in patients with UC; (C) Spearman correlation analysis of miR-223 with hs-CRP in patients with UC; (D) Spearman correlation analysis of miR-223 with UCEIS in patients with UC. ESR = erythrocyte sedimentation rate, hs-CRP = high-sensitivity C-reactive protein, miR-223 = microRNA223, UC = ulcerative colitis, UCEIS = ulcerative colitis endoscopic index of severity scores.
Predictive Values of Serum MiR-223 in IBD Patients
We investigated the correlation of miR-223, ESR, and hs-CRP with CDAI and found that compared with ESR and hs-CRP, miR-223 demonstrated a higher Spearman r value in detecting the disease activity of patients with CD (Figure 4A, B). In patients with UC, we also detected the correlation of ESR and hs-CRP with the Mayo score and UCEIS. Interestingly, we found that the correlations of hs-CRP with the Mayo score and UCEIS did not exhibit statistically significant differences (Figure 5A, B). The correlation between ESR and Mayo score also did not exhibit statistically significant differences (Figure 5C). When correlating with UCEIS, miR-223 demonstrated a higher Spearman r value (r = 0.481) than ESR (Figure 5D; r = 0.334).
FIGURE 4.
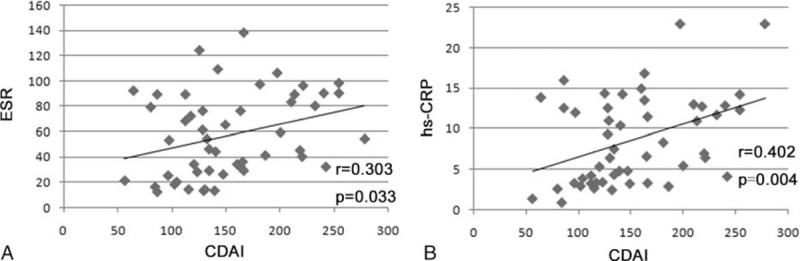
Correlation of ESR and hs-CRP with CDAI in patients with CD: (A) Spearman correlation analysis of ESR with CDAI in patients with CD; (B) Spearman correlation analysis of hs-CRP with CDAI in patients with CD. CD = Crohn's disease, CDAI = Crohn's disease activity index, ESR = erythrocyte sedimentation rate, hs-CRP = high-sensitivity C-reactive protein.
FIGURE 5.
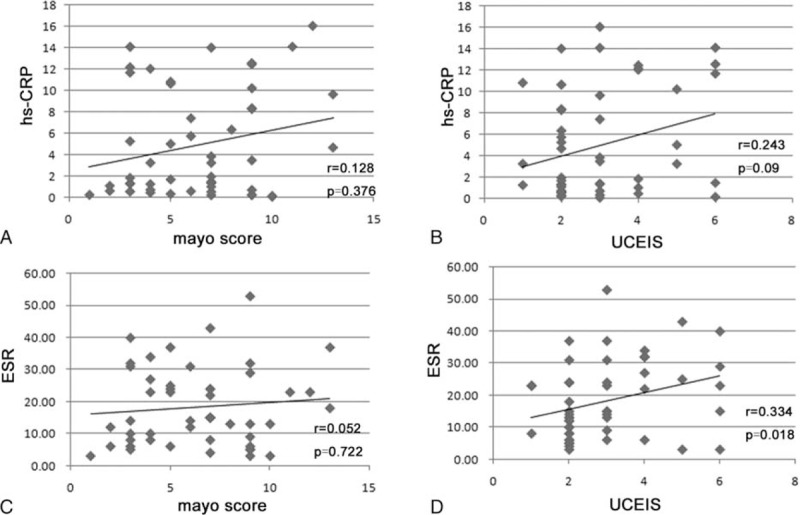
Correlation of ESR and hs-CRP with Mayo score and UCEIS in patients with UC: (A) Spearman correlation analysis of hs-CRP with Mayo score in patients with UC; (B) Spearman correlation analysis of hs-CRP with UCEIS in patients with UC; (C) Spearman correlation analysis of ESR with Mayo score in patients with UC; (D) Spearman correlation analysis of ESR with UCEIS in patients with UC. ESR = erythrocyte sedimentation rate, hs-CRP = high-sensitivity C-reactive protein, UC = ulcerative colitis, UCEIS = ulcerative colitis endoscopic index of severity scores.
DISCUSSION
The most common biomarkers for monitoring disease activity for IBD now are ESR and hs-CRP; however, they always show a relatively low specificity and high expression heterogeneity in patients with IBD.25,26 Thus, even today, the screening and monitoring of IBD remain challenging even to the most experienced physicians. Considering this, the identification of novel noninvasive IBD biomarkers would be beneficial for clinical practice. Recently, miRNA was found to be a new regulator in the etiology of many diseases, including cancer and inflammatory diseases. Whether miRNA could be used as a new biomarker in disease screening and monitoring is a hot topic in this field.
Except miRNA expression in human tissue,27 more and more studies examined miRNA expression in the peripheral blood of IBD patients.28,29 Endogenous miRNAs are stable in peripheral blood,10 and they can avoid rapid enzymatic degradation and renal clearance.30 A previous study reported that the existence of specific miRNA expression patterns was associated with IBD and their different stages, which supported the utility of miRNA as possible biomarkers.31
On the basis of our miRNA microarray data in colonic tissues (not shown here), we found that the expression of miR-223 in tissues was highly upregulated in IBD patients compared with controls, highlighting that miR-223 might play an important role in the development of IBD. However, it is not convenient to detect the miRNA expression in the tissues of IBD patients, due to the indispensable application of colonoscopy and prior bowel cleansing. It would be much easier and more convenient to detect it in serum. Therefore, we tried to explore that whether miR-223 in serum had diagnostic and/or prognostic value for patients with IBD, and at the same time to compare its value with other well-known biomarkers such as hs-CRP and ESR. In this study, we found that levels of serum miR-223 significantly increased in IBD patients compared with controls. Furthermore, we found that miR-223 had a positive correlation with these indicators of disease activity in both CD and UC, and it might be correlated much better with disease activity of CD than ESR and hs-CRP. These supported its potential role as a diagnostic and therapeutic target in future clinical application for IBD. In accordance with our study, Polytarchou et al recently confirmed that miR-223 was upregulated in the peripheral blood of UC patients compared with control subject.32 By using microRNA prediction algorithms (www.microRNA.org), we found miR-223 might target 5886 genes. Among this, we found that Claudin-8 might be a potential interesting downstream target, which is a tight junction-integral protein, playing important roles in maintaining cell polarity and permeability and is regarded as the backbone of the intestinal barrier. Further studies would be needed to confirm its role in the development of IBD.
In conclusion, the present study revealed that the level of serum miR-223 significantly increased in IBD patients compared with controls. Serum miR-223 was a more reliable method for monitoring disease activity than ESR or hs-CRP and that serum-derived miR-223 might have superior prognostic value than ESR or hs-CRP. Therefore, circulating miR-223 might serve as a new promising biomarker for IBD. However, it is necessary to confirm the results of our pilot study in larger samples with subtypes of patients according to treatment, disease duration, and localization.
Footnotes
Abbreviations: CD = Crohn's disease, CDAI = Crohn's disease activity index, ESR = erythrocyte sedimentation rate, hs-CRP = high-sensitivity C-reactive protein, IBD = inflammatory bowel disease, miRNAs = microRNAs, SES-CD = simple endoscopic score for CD, UC = ulcerative colitis, UCEIS = ulcerative colitis endoscopic index of severity scores.
HW and SZ are co-first authors.
This study was supported by grants from the National Natural Science Foundation of China (#81270473, #81470821, #81301769), the Key Project of the National 12th five-year Research Program of China (#2012BAI06B03), Guangdong Science and technology (#2014A020212128), the Fundamental Research Funds for Sun Yat-sen University (#15ykpy12), and the Pearl River S&T Nova Program of Guangzhou (#201505042032472).
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet 2001; 10:445–456. [DOI] [PubMed] [Google Scholar]
- 2.Okahara S, Arimura Y, Yabana T, et al. Inflammatory gene signature in ulcerative colitis with cDNA macroarray analysis. Aliment Pharmacol Ther 2005; 21:1091–1097. [DOI] [PubMed] [Google Scholar]
- 3.Yu Q, Zhang S, Li L, et al. Enterohepatic helicobacter species as a potential causative factor in inflammatory bowel disease: a meta-analysis. Medicine 2015; 94:e1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim DH, Saetrom P, Snove O, Jr, et al. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A 2008; 105:16230–16235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalla R, Ventham NT, Kennedy NA, et al. MicroRNAs: new players in IBD. Gut 2015; 64:504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 8.Wu F, Zikusoka M, Trindade A, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008; 135:1624–1635. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834–838. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18:997–1006. [DOI] [PubMed] [Google Scholar]
- 11.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008; 141:672–675. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids—a promising, non-invasive tool for early detection of several human diseases. FEBS Lett 2007; 581:795–799. [DOI] [PubMed] [Google Scholar]
- 14.Chim SS, Shing TK, Hung EC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 2008; 54:482–490. [DOI] [PubMed] [Google Scholar]
- 15.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010; 56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai Y, Huang YS, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus 2007; 16:939–946. [DOI] [PubMed] [Google Scholar]
- 17.Thayu M, Shults J, Burnham JM, et al. Gender differences in body composition deficits at diagnosis in children and adolescents with Crohn's disease. Inflamm Bowel Dis 2007; 13:1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubner SE, Shults J, Baldassano RN, et al. Longitudinal assessment of bone density and structure in an incident cohort of children with Crohn's disease. Gastroenterology 2009; 136:123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thayu M, Denson LA, Shults J, et al. Determinants of changes in linear growth and body composition in incident pediatric Crohn's disease. Gastroenterology 2010; 139:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006; 55:749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Best WR, Becktel JM, Singleton JW, et al. Development of a Crohn's disease activity index. National Cooperative Crohn's Disease Study. Gastroenterology 1976; 70:439–444. [PubMed] [Google Scholar]
- 22.Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med 1987; 317:1625–1629. [DOI] [PubMed] [Google Scholar]
- 23.Daperno M, D’Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn's disease: the SES-CD. Gastrointest Endosc 2004; 60:505–512. [DOI] [PubMed] [Google Scholar]
- 24.Travis SP, Schnell D, Krzeski P, et al. Developing an instrument to assess the endoscopic severity of ulcerative colitis: the Ulcerative Colitis Endoscopic Index of Severity (UCEIS). Gut 2012; 61:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest 2003; 111:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fengming Y, Jianbing W. Biomarkers of inflammatory bowel disease. Dis Markers 2014; 2014:710915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu F, Zhang S, Dassopoulos T, et al. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis 2010; 16:1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paraskevi A, Theodoropoulos G, Papaconstantinou I, et al. Circulating MicroRNA in inflammatory bowel disease. J Crohns Colitis 2012; 6:900–904. [DOI] [PubMed] [Google Scholar]
- 29.Wu F, Guo NJ, Tian H, et al. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn's disease. Inflamm Bowel Dis 2011; 17:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol 2015; 8:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iborra M, Bernuzzi F, Correale C, et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol 2013; 173:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polytarchou C, Oikonomopoulos A, Mahurkar S, et al. Assessment of circulating microRNAs for the diagnosis and disease activity evaluation in patients with ulcerative colitis by using the nanostring technology. Inflamm Bowel Dis 2015; 21:2533–2539. [DOI] [PubMed] [Google Scholar]


