Abstract
Multidrug antiporters of the Major Facilitator Superfamily couple proton translocation to the extrusion of cytotoxic molecules. The conformational changes that underlie the transport cycle and the structural basis of coupling of these transporters have not been elucidated. Here we utilized extensive Double Electron Electron Resonance measurements to uncover the conformational equilibrium of LmrP, a multidrug transporter from L. lactis, and to investigate how protons and ligands shift this equilibrium to enable transport. We find that the transporter switches between outward-open and outward-closed conformations depending on the protonation states of specific acidic residues forming a transmembrane protonation relay. Our data can be framed in a model of transport wherein substrate binding initiates the transport cycle by opening the extracellular side. Subsequent protonation of membrane-embedded acidic residues induces substrate release to the extracellular side and triggers a cascade of conformational changes that concludes in proton release to the intracellular side.
In an increasing number of cases, antibiotic resistance of bacterial strains relies on the expression of specialized molecular pumps called multidrug transporters1. In addition to their association with a pressing clinical issue, these polyspecific exporters of cytotoxic molecules present a fascinating variation on the classic membrane transport theme. While most membrane transporters are specific to a single substrate, multidrug transporters can recognize a vast range of structurally diverse molecules and efficiently extrude them2. Furthermore, multidrug exporters share a common evolutionary origin with substrate-specific importers, indicating that a similar scaffold has evolved to either export or import substrates.
A general mechanism for membrane transport was first proposed almost fifty years ago3 and later coined the alternating access model4. In this model, the transporter alternates between two different structural states that enable binding of the substrate on one side of the membrane and subsequent release on the other. Vectorial translocation across the bilayer entails conformational changes to switch accessibility of the binding site. While a wealth of biochemical and structural data lends support to the general concept of alternating access, the underlying structural mechanics has yet to be defined for each transporter class (reviewed in ref. 5).
The Major Facilitator Superfamily (MFS) of transporters groups together ion-coupled secondary transporters of sugars, amino acids, drugs, nucleosides and a variety of organic and inorganic anions and cations6. They share a conserved common architecture consisting of two symmetry-related bundles of 6 transmembrane (TM) helices encoded by a single polypeptide. The two bundles cradle the substrate in a binding site at their interface. Crystal structures of MFS members capture conformations described as inward-open7,8, i.e. open to the intracellular medium, outward-open9 where the substrate can access the protein from the extracellular medium only, or occluded conformations10–12 shielding the substrate binding pocket from both membrane sides. Together, these structures stimulated the rocker-switch model for alternating access, wherein the N- and C-terminal halves of the protein rock back and forth against each other through rigid-body motions9,13. However, molecular dynamics simulations14,15 and biophysical studies16,17 indicate that the structural changes likely involve important local motions as well. Pioneering work from the Kaback group on lactose permease (LacY) culminated in a detailed mechanistic model developed in the framework of a high resolution structure and complemented by analysis of conformational dynamics18. In this model, LacY sequentially binds proton and substrate in an outward-facing conformation19, transitions into an occluded intermediate and then adopts an inward-facing conformation that enables substrate and proton release. Sugar binding induces proton transfer between essential glutamate residues, triggering the opening to the intracellular side and subsequent proton and sugar release20. Deprotonation which is driven by the transmembrane proton gradient, triggers the return to the outward-open conformation21,22.
In contrast, the conformational cycle of MFS multidrug resistance (MDR) exporters has not been elucidated. The only available crystal structure of an MFS-MDR exporter, that of the E. coli EmrD, profiles a transporter in a doubly occluded conformation with a hydrophobic cavity located in the bilayer11. However, hardly any functional studies exist for this transporter to enable mechanistic interpretation of the structure. Building on the LacY model, the E. coli MdfA and L. lactis LmrP multidrug antiporters have been proposed to operate in a reciprocal fashion, binding their substrates from a high-affinity, inward-open conformation and switching to a low-affinity, outward-open conformation to allow substrate release23,24. Critical to proton and substrate transport is a group of acidic residues located mainly in TM segments. Several studies demonstrated that the accessibility of these residues can be modulated by substrate binding and identified the stoichiometries of substrate to proton transport19,25,26. However, a detailed understanding of how protein conformational motion couples ion gradients to substrate translocation and the sequence of events that defines the transport mechanism are missing.
Here we address these central elements of multidrug transport through extensive Double Electron-Electron Resonance (DEER, or PELDOR: Pulsed Electron-Electron Double Resonance) analysis27–29 of LmrP, which catalyzes the export of a large variety of structurally diverse molecules, comprising antibiotics, anticancer drugs and other cytotoxic molecules2,30. These molecules are typically hydrophobic ions which enable them to partition into the bilayer while having polar moieties, often bearing one or two positive charges. Contrary to solute carriers, MDR transporters bind substrates directly from the inner leaflet of the membrane bilayer26. Several transmembrane acidic residues, namely Asp68, Asp128, Asp142, Asp235 and Glu327 are critical in the cotransport of protons and cationic substrates by LmrP31.
DEER distance distributions obtained from pairs of spin labels on the intra- and extracellular sides of LmrP revealed a highly dynamic transporter that shifts between multiple conformational states in equilibrium. This conformational equilibrium is regulated by protonation of key transmembrane residues, defining a structural switch. Together the data support a mechanistic model of drug extrusion that sheds new light on the alternating access mechanism for MFS multidrug transporters.
RESULTS
Mapping LmrP conformations with EPR probes
In order to directly follow the conformational dynamics of LmrP, we carried out DEER measurements on spin-labeled cysteine pairs28 placed on the extra- or intracellular ends of the transmembrane helices. This technique allows for direct and accurate characterization of distance distributions on purified protein, identifying the individual conformations in equilibrium under different biochemical conditions. Molecular models of LmrP reflecting potential outward-open, occluded and inward-open structural intermediates were built using the crystal structures of FucP, EmrD and LacY, respectively, as templates (Supplementary Fig. 1). The positions of cysteine pairs were chosen on the basis of these structural models and are represented on the EmrD-derived LmrP model (Supplementary Fig. 2a–c). Cysteines were introduced on a cysteine-less background (LmrP C270A) and reacted with the spin label probe 1-oxyl-2,2,5,5- tetramethylpyrroline-3-methyl-methanethiosulfonate (MTSSL). The activity of each double mutant was verified by following transport of the Hoechst 33342 substrate in inverted membrane vesicles following established procedures30 (Supplementary Fig. 3). Non-functional mutants were excluded from the DEER analysis.
Because LmrP drug transport is driven by a pH gradient, we reasoned that changes in pH should shift the conformational equilibrium of the transporter. Under physiological conditions, the extracellular medium of L. lactis is typically acidic (pH 6.5 or less), while the intracellular medium is maintained around pH 732. In order to clearly separate putative pH-dependent conformational states, we measured interhelical distances at either pH 5 or pH 8.
LmrP conformational equilibrium is modulated by pH
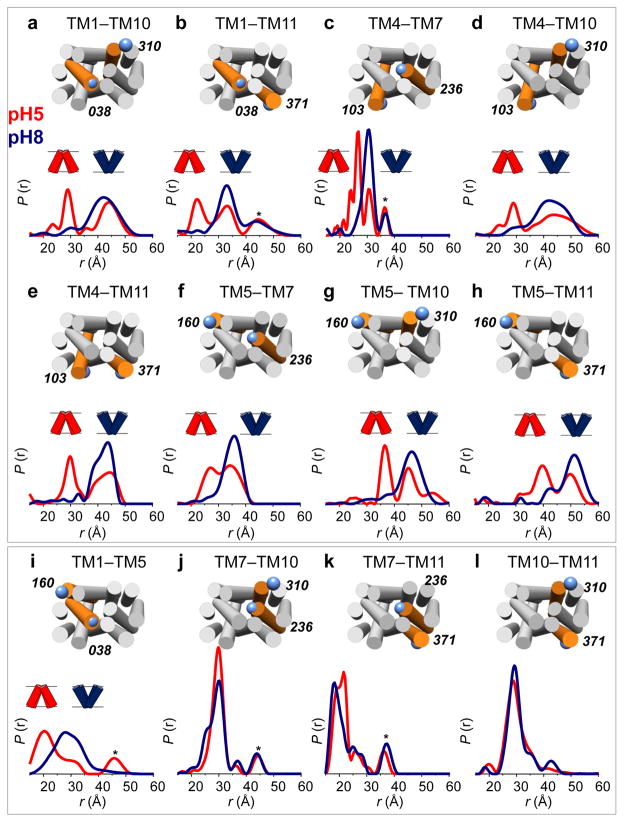
LmrP shows major structural differences on its extra- and intracellular sides at the two pH’s. Distance distributions for 12 spin label pairs on the extracellular side of the transporter reveal report large amplitude distance changes that are collectively consistent with a closing of this side of the transporter at lower pH. Distance distributions obtained at pH 8 (Figure 1, blue curves) typically show one major population, with the main peak accounting for seventy to ninety percent of the total area under the curve. At pH 5, all distances measured between the N-terminal (TMs 1–6) and C-terminal (TMs 7–12) halves of the transporter display a second population, with distances typically 10 Å shorter (Fig. 1a–h, red curves). Distances measured within the same bundle show little to no pH dependence (Fig. 1i–l), except for TM1–5 (Fig. 1i). Interestingly, the conformation observed at pH 8 is also partly populated at pH 5, indicating the coexistence of two LmrP conformers under acidic conditions. Thus, while LmrP adopts an outward-open conformation at pH 8, we observe a coordinated closing of the transporter at pH 5, producing an outward-closed conformation.
Figure 1. The extracellular side of LmrP closes at low pH.
DEER distance distributions between labeled cysteine pairs located on the extracellular ends of TM helices. At pH 8 (blue curves) a single population (i.e. >70%) is observed for all distances. At pH 5 (red curves) a significant decrease in the distances is observed for pairs between the N-terminal (TMs 1–6) and C-terminal (TMs 7–12) bundles (panels a–h), while no effect is observed for distances within the same bundle (panels i–l). Distributions were normalized: r indicates interspin distance, P(r) indicates the distance probability. Residue numbers and positions of cysteine pairs are depicted on an LmrP model based on the crystal structure of the E. coli homolog EmrD (see Methods) viewed from the extracellular side. Targeted helices are highlighted in orange with TM numbers indicated atop. Asterisks denote peaks resulting from partial aggregation observed in some samples upon concentration27.
Conformational differences are concomitantly observed at the intracellular side (Figure 2). For most distances measured between the two halves of LmrP, the observed changes indicate an overall opening on the intracellular side at pH 5 (Fig. 2a–f, red curves) compared to pH 8 (Fig. 2a–f, blue curves). Distances measured within the same bundle remain largely unaffected (Fig. 2g,h), and a concerted shortening of the intra- and extracellular distances between TM5 and TM11 is observed at pH 5 (Figs. 1h and 2f). For distances spanning both halves of LmrP, four out of six report a single conformation on the intracellular side at low pH (Fig. 2a–f, red curves), in contrast to the bimodal distributions observed for all reporters on the extracellular side (Fig. 1a–h, red curves).
Figure 2. Acidic pH opens the intracellular side of LmrP.
DEER distance measurements on the intracellular side of LmrP, performed at pH 8 (blue curves) and pH 5 (red curves). Distances were measured either between the N-terminal and C-terminal bundles (panels a–f) or within the same bundle (panels g,h). Residue numbers and positions are depicted on an LmrP model based on the crystal structure of the E. coli homolog EmrD viewed from the intracellular side. Asterisks denote aggregation peaks.
We interpret this data as revealing that LmrP is in equilibrium between at least two conformational states that alternately open/close each sides of the transporter. The fact that two or more conformations are observed on one side (e.g. extracellular distances at pH 5) while one conformation is detected on the other (e.g. intracellular distances at pH 5) indicates limited conformational coupling between the helical movements on both ends of the protein and thus suggests that the structural changes do not strictly follow rigid-body motions. Shifting the pH does not alter spin label mobility as deduced from CW-EPR line shapes (Supplementary Figs. 4 and 5) indicating that the distance changes cannot merely be attributed to spin-label rotamer rearrangements, but rather reflect TM domain movements. In addition, the position and amplitude of the distance distributions were not significantly altered by variations of the background slope that changes the RMSD between data and fits by up to 10% (see METHODS and Supplementary Fig. 6)
Protonation of acidic residues stabilizes distinct states
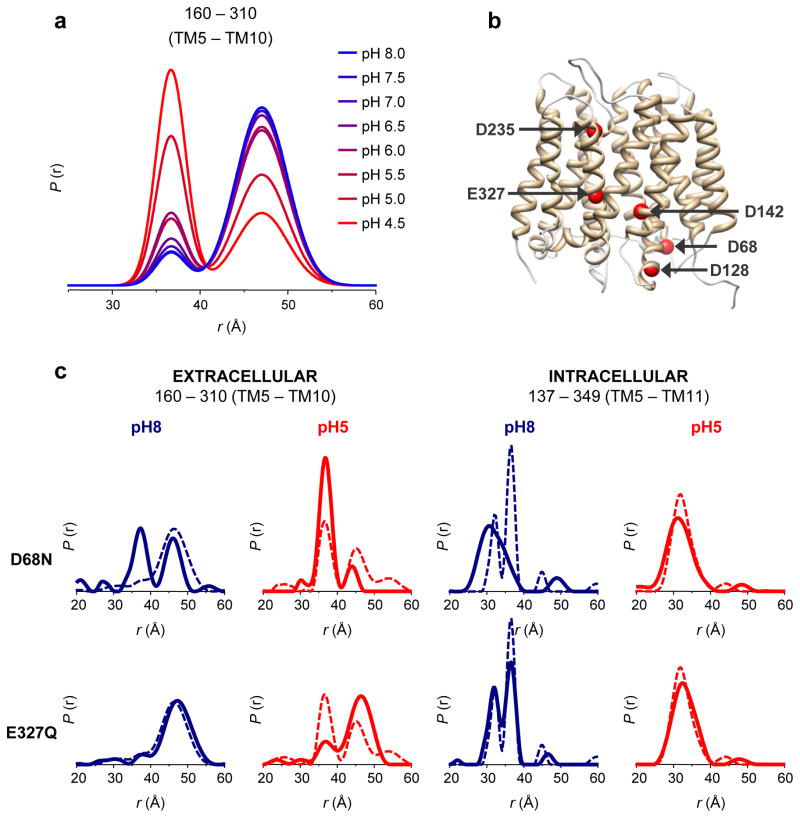
How does LmrP sense pH differences? Previous mutagenic analysis has identified a number of acidic residues that may be implicated in substrate binding and/or proton coupling31. To establish that the conformational transitions deduced from DEER distance distributions correlate with protonation of acidic side chains, we carried out a titration experiment where the extracellular TM5–TM10 distance (mutant L160C–T310C) was monitored at eight different pH values, ranging from 4.5 to 8 (Fig. 3a). A two-population fit of the distance distributions clearly shows a gradual shift in the equilibrium from an outward-open to outward-closed conformation as the pH is reduced, with the population ratio inverting at pH 4.8 (Supplementary Fig. 7).
Figure 3. The conformational equilibrium of LmrP is coupled to the protonation of acidic residues.
(a) Distance distributions of the extracellular reporter pair L160C–I310C were determined at eight different pH values, ranging from 4.5 to 8.0 (color gradient from red to blue). To quantify the variation in population ratios as a function of pH, fits were carried out assuming a two-component Gaussian distance distribution in a home design software implemented in MATLAB (see Supplementary Fig. 7 and METHODS). At low pH, the short component (centered at 36 Å) dominates the bimodal distance distributions; at high pH the long component (centered at 47 Å) is dominant. (b) Key acidic residues depicted on an EmrD-based model of LmrP: Asp68 (bottom of TM2), Asp128 (bottom of TM4), Asp142 (center of TM5), Asp235 (top of TM7), Glu327 (center of TM10). (c) Protonation mimetic of key acidic residues can block the conformational switch. Single mutations D68N and E327Q were combined with double cysteine mutants L160C–I310C and V137C–S349C, which served as extracellular and intracellular reporters. Distance measurements at pH 5 (red curves) and pH 8 (blue curves) in the absence (dashed line) and presence (full line) of each mutation reveal the structural consequence of permanent protonation of these essential acidic residues.
To identify the transmembrane acidic residues whose protonation induces conformational changes, Asp68, Asp128, Asp142, Asp235 and Glu327 (Fig. 3b) were mutated to asparagine or glutamine, thereby mimicking permanent protonation. These point mutants were then introduced into the L160C–T310C or V137C–S349C background to monitor the extracellular and intracellular conformations of LmrP, respectively (Supplementary Figs. 8 and 9). Our measurements indicate that distance distributions can be altered by the D68N or E327Q substitutions (Fig. 3c) and, to a lesser extent, the D128N substitution (Supplementary Fig. 8). We observe that the D68N mutation induces the outward-closed conformation at pH 8, a state only seen at pH 5 in the WT background. At pH 5, the D68N mutant shows almost a single outward-closed population, with the outward-open conformation being nearly absent. On the intracellular side, this mutation also locks the transporter in a pH 5-like state, with the TM5–TM11 distance showing a single distribution at pH 8 identical to that observed for WT LmrP at pH 5. We conclude that protonation of Asp68, located at the interface with the intracellular medium, stabilizes the outward-closed / inward-open conformation following a global conformational change. Thus, the reported functional importance of this aspartate33–36 reflects a critical role in driving conformational changes.
In contrast, the E327Q mutation restricts LmrP to the outward-open conformation. The short distance component, which reflects the outward-closed conformation, is almost absent in the L160C–T310C distance distribution, even at pH 5. The mutation does not alter the pH-induced shift in equilibrium at the pair monitoring the intracellular side. This indicates that the conformational equilibrium can be partly uncoupled between the extracellular and intracellular ends.
Interestingly, protonation of Asp128 (D128N) substantially increases the population of outward-closed state at pH 5, while the intracellular distance reporter is less affected (Supplementary Fig. 8). We interpret this result as suggesting that Asp128 acts cooperatively with other protonation-dependent events, since the effects of D128N are only observed at low pH. Mutation D142N has a moderate effect on the intracellular reporter at pH 8, favoring the long distance. For D235N no marked change is observed relative to the WT background.
Together our DEER data show that the conformational equilibrium between two discrete, pH-dependent structural states can be altered by protonation of single acidic residues: protonation of Glu327 stabilizes the outward-open state while protonation of Asp68 shifts the equilibrium towards the outward-closed state.
Substrate binding stabilizes the outward-open conformation
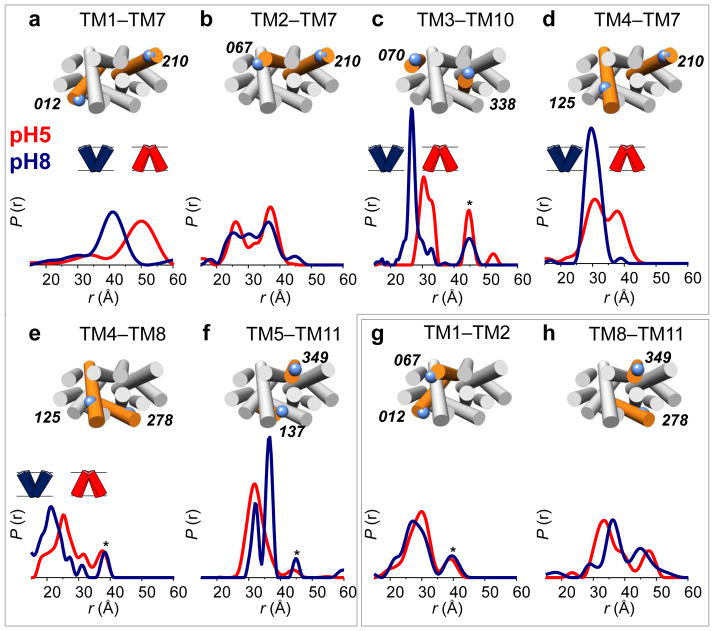
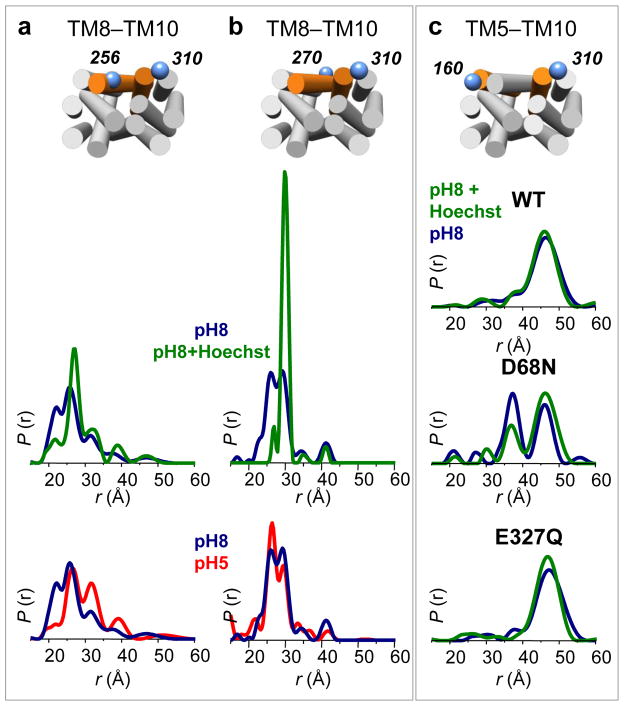
Protonation of acidic side chains must be triggered from the extracellular side and must be coupled to binding and translocation of substrate. To assess the effect of substrate binding on LmrP conformational equilibrium, we measured all interhelical distances in the presence of the well characterized LmrP substrate Hoechst 33342 at pH 8. We observed no major changes in most distance distributions (Supplementary Figs. 10 and 11), with a notable exception for spin label pairs involving TM8 (Fig. 4a,b, green curves). For these spin label pairs, substrate addition leads to narrowing of the distance distributions to a unique population, with more than 90 % of the proteins in a ligand-bound conformation. Addition of 1 mM of Kanamycin, a molecule not transported by LmrP, does not affect the distance distributions (Supplementary Fig. 12), demonstrating that these conformational changes are substrate-specific.
Figure 4. Hoechst 33342 binding restricts TM8 conformational flexibility and stabilizes the outward-open conformation.
(a–b) Distances distributions measured from the center (Cys270) and the extracellular end (I256C) of TM8 to the extracellular end of TM10 (I310C) at pH 8 (blue curves), pH 8 in the presence of 1 mM Hoechst 33342 (green curves) and pH 5 (red curves). The presence of substrate restricts the distributions to a single population, while TM8 adopts multiple conformations in apo LmrP, both at pH 5 and pH 8. (c) Effect of substrate binding (1 mM Hoechst 33342) on the extracellular conformation at pH 8 in the absence of a functional mutation; in combination with D68N; in combination with E327Q. The effect of the D68N mutation is partly reversed by the presence of the substrate.
Distance distributions involving labels on TM8 (i.e. I256C or C270) show multiple conformations, at both pH 8 and pH 5 (Fig. 4a,b, blue and red curves) suggesting a flexible helix. Consistent with this interpretation, Hoechst 33342 binding decreases spin label mobility on TM8, with the strongest effect in close proximity with the presumed ligand cavity, as proposed in the crystal structure of EmrD11 (Supplementary Fig. 13). Taken together, these results show that, under basic conditions, TM8 becomes constrained in the presence of ligand.
To identify the conformational state stabilized by substrate binding, we monitored the L160C–T310C distance (TM5–TM10) as a reporter (outward-open vs. outward-closed) in either a WT, D68N or E327Q background (Fig. 4c). At pH 8 in the presence of 1 mM Hoechst 33342, both WT and E327Q LmrP are in the outward-open conformation. Remarkably, the presence of the ligand tends to partly reverse the conformational (closing) effect of the D68N mutation increasing the population of the outward-open conformation, consistent with the conclusion that substrate binding stabilizes this state.
DISCUSSION
The main finding of this study is that the multidrug MFS transporter LmrP alternates between at least two conformations: outward-open and outward-closed, in a transition driven by the change in the protonation state of one or more transmembrane acidic residues. Therefore, this finding suggests that a proton gradient would power the structural transitions required for transport by driving the proton transfer between key acidic residues. The side chain of Asp68, which is the most conserved residue in the MFS family and has been proposed to be involved in proton transfer and lipid-protein interaction in LmrP33, would act as a master conformational switch. While this residue is located far from the putative substrate binding site, its mutation leads to transport inhibition and decreased substrate binding in a number of MFS transporters34–36. Protonation of Asp68, as the culmination of proton translocation from the extracellular side, induces an outward-closed conformation (Figure 3). The network of acidic residues important for propagation of conformational changes includes membrane-embedded Glu327, which is required for ligand binding and transport and is believed to interact directly with the substrate37. Protonation of this residue stabilizes an outward-open conformation but has little or no influence on the intracellular side. Based on these data we propose that, under gradient conditions, proton(s) will travel down from Glu327 to Asp68, leading to transition from outward-open to outward-closed and thus creating an efficient conformational switch powered by the proton-motive force. Proton passage is likely to involve other transmembrane residues of functional importance, such as Asp235, Asp142 and Asp128, as suggested by previous mutagenic studies38–40 and the moderate but notable effects observed by mimicking protonation of these residues.
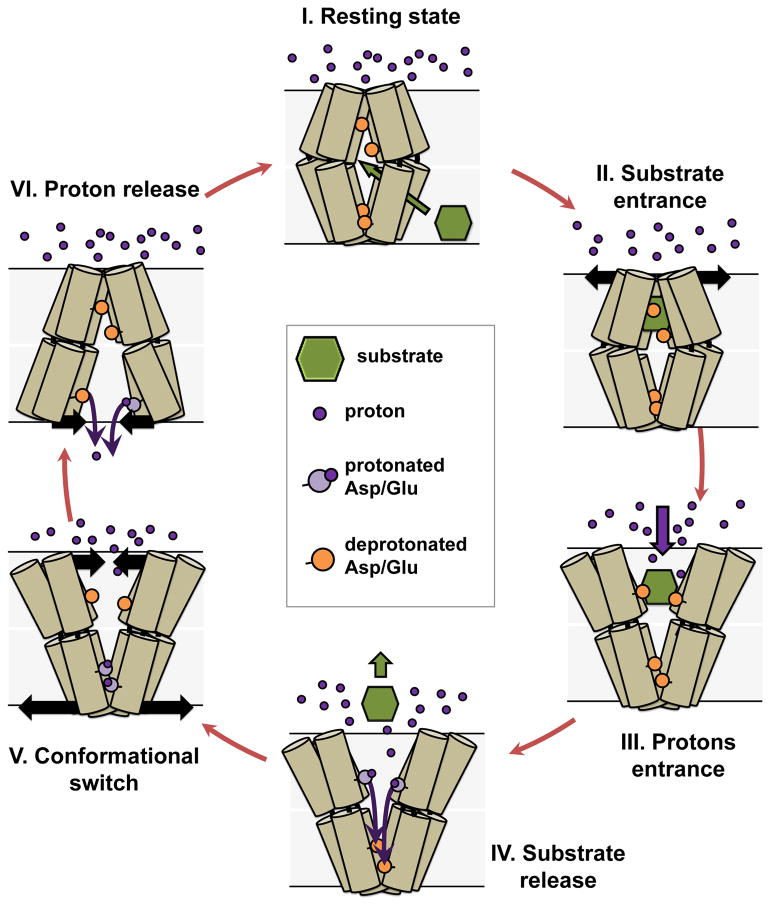
While the DEER data was obtained under constant pH conditions, it can be extended to a model of drug export by LmrP in the presence of a pH gradient (Figure 5). In this model, proton translocation along the transmembrane charge network can only occur subsequent to substrate binding in order to prevent uncoupled proton transport, which would short the transmembrane ion gradient. Therefore, the resting state (i.e. in the absence of substrate but in presence of a proton gradient) cannot be the outward-open conformation since this conformation is likely to allow proton entrance and protonation of the binding site. We propose that the resting state is a proton occluded state that nevertheless is open to substrate binding from the inner leaflet of the bilayer (state I&II in Fig. 5). Indeed, DEER distance distributions demonstrate the closing of the extracellular end of the TMs (Fig. 1) at acidic pH which is typically encountered on the extracellular side under transport conditions. The intracellular pH is neutral to slightly alkaline, which according to our data should lead to a closing of the intracellular side (Fig. 2). Because the conformational changes we identify here do not involve rigid body motion of TMs, the two sides of the transporter have structural domains that can rearrange independently. This relative decoupling of the two sides of LmrP is critical to sensing of the pH gradient and enables the transporter to adopt the occluded state, where both ends are closed to the solvent. In support of this model, MFS transporters are expected to populate a distinct occluded state5,41, an expectation borne out by multiple MFS transporter structures11,42. We attribute the absence of an occluded conformation in our DEER analysis to the uniform pH conditions of the experiments. The notion of substrate binding to the transporter directly from the bilayer originates from studies of multiple classes of multidrug transporters26,43–46. Population of an inward-facing conformation requires the protonation of Asp68. The inward-open crystal structure of LacY shows that Asp68 is solvent accessible in this conformation which, considering the neutral intracellular pH of L. lactis, would lead to rapid deprotonation. Thus, our data suggests that the inward-facing state would be transient under transport condition but is stabilized here by the homogeneous acidic pH conditions.
Figure 5. Proposed LmrP transport cycle based on DEER distance measurements.
For clarity, only six TMs of LmrP are depicted, with the extracellular side on top. The bilayer of the membrane is represented by shaded grey boxes and the transmembrane proton gradient illustrated by the large number of extracellular protons (purple spheres). (I) In the resting state, LmrP is occluded from the extracellular environment. The ligand (in green) enters from the membrane bilayer. (II) Substrate binding stabilizes the outward-open conformation. (III) The outward-open conformation exposes the binding site to extracellular protons (purple dots). (IV) Protons and/or water molecules enter the binding pocket and protonate carboxylic residues (orange circles, colored in light purple once protonated) that coordinate the substrate, thereby releasing it. (V) Protons access the acidic residues, eventually leading to protonation of D68, which induces closing of LmrP on the extracellular side and concomitant opening on the intracellular side. (VI) This opening will lead to (partial) exposure to the neutral intracellular milieu, allowing for deprotonation of Asp68 at which point the transporter resets to the resting state.
Ligand entrance to a proton-occluded conformation could occur at the interface between the N- and C-terminal halves of the protein, more specifically at the TM5–TM8 interface where we observed conformational changes in the presence of the substrate Hoechst 33342 but not of hydrophobic molecules that are not substrates. Molecular dynamics simulation of the E. coli homolog EmrD lends support to this model by suggesting that TM8 may act as a gate for ligand access47. Moreover, the interface between TM5 and TM8 has been proposed to be an important part of the substrate translocation pathway in LacY20,48. Because Hoechst 33342 binding stabilizes an outward-facing conformation (Supplementary Fig. 10), we propose that, during transport, transition to this conformation triggers proton entrance and thus protonation of the acidic residues in the binding pocket. Consistent with this model, protonation of Glu327 stabilizes the outward-open state, even under acidic condition (Fig. 3c). Therefore, opening of the extracellular end could be due, in the presence of the gradient, to the cooperative effect of substrate binding and Glu327 protonation. Protonation of the binding site will in turn reduce substrate binding affinity promoting its release into the extracellular medium (state IV). Coupling of protons and substrate could be achieved by competition for the same binding residues, as proposed for EmrE49, although a more indirect structural coupling is also feasible, as proposed recently for MdfA23 and the multidrug transporter NorM50 of the multidrug and toxic compound extrusion (MATE) family.
In our model, substrate-induced opening of the extracellular side initiates the conformational changes by enabling protons to access the network of acidic residues (Asp235-Glu327-Asp142-Asp128) across the membrane all the way to the intracellular side where protonation of Asp68 leads to closing of the extracellular side of the TM bundle (Fig. 5). At this stage of the transport cycle, both the deprotonation of Glu327 (Fig. 3) and protonation of the key residues on the intracellular end (Asp128, Asp68, Fig. 3 & Supplementary Fig. 8) favor the outward-closed/inward-open state, which is stabilized under acidic conditions (Figs. 1&2). In a cellular context, this state would expose Asp68 to the neutral pH of the cytoplasm, leading to its deprotonation, and thus intracellular proton release. Subsequently, the transporter would switch back to the resting state; one substrate has been extruded and one or several protons have been imported, in agreement with our current knowledge of extrusion stoichiometry in LmrP.
Our model differs fundamentally from existing models for MDR MFS transporters. In the current alternating access model, the substrate-bound, high affinity conformation corresponds to the outward-open state for an importer and to the inward-open conformation for an exporter, as proposed for the E. coli MFS multidrug transporter MdfA23. In the latter model, we expect the substrate to stabilize an inward-open, not an outward-open conformation which contradicts our findings. Interestingly, crystal structures of the NorM transporter bound to three different substrates all captured an outward-open state50, suggesting that such conformation is compatible with high-affinity binding, at least in the case of MATE multidrug transporters.
While the extra and intracellular parts form distinct structural domains, appropriate coupling between these domains must be realized to achieve protein function, i.e. a conformational change triggered on one side must be transmitted to the other during the transport cycle. Disruption of such coupling (as in the E327Q mutant) leads to transport impairment. As already proposed by Kaback and coworkers, reciprocity between opening on the extracellular side and the intracellular side may thus not be obligatory18.
High resolution crystal structures will be needed to adequately describe the local interactions that stabilize different LmrP conformations and in particular to elucidate how protonation of Glu327 and Asp68 trigger conformational changes. The role of Asp68 in driving conformational transitions may be divergent between MFS transporters. While our results reveal a direct role of Asp68 protonation in stabilizing the inward-facing conformation, it should be noted that for LacY, Asp68 is not described as a proton binding/release site16. Identifying the specific polar networks involved in the interactions with Asp68 may highlight conserved and divergent conformational switching mechanisms between MFS transporters. We have previously proposed that interactions between Asp68 and the lipid environment could be important in regulating substrate transport33, suggesting that the bilayer could have an important effect on LmrP conformational dynamics. Therefore, it will be critical to test the model proposed here in membrane-like systems, such as proteoliposomes or nanodiscs, to elucidate the effect of lipids on the proton-dependent conformational changes of the transporter.
A simple multidrug transport mechanism emerges from our studies. In order to be transported by LmrP, substrates must merely i) bind in the ligand pocket and ii) subsequently catalyze proton entrance (i.e. by opening of the extracellular side). The simplicity of this mechanism thus provides the basis of how structurally diverse molecules can be extruded by a single transporter. It will be particularly interesting to investigate whether this model can be generalized to other pH-gradient-driven members of the MFS family.
ONLINE METHODS
LmrP homology models
For each of the three structural templates (EmrD, PDB entry: 2GFP, LacY PDB: 2V8N and FucP PDB: 3O7Q), the sequence of LmrP was aligned using multiple alignments of LmrP and orthologs together with the sequences of the template and its orthologs. Initial alignments were generated using ClustalW53, and then manually adjusted to i) prevent insertion and deletion in the TM helices and ii) avoid introduction of charged residues facing the lipid tails. Subsequent LmrP-template sequence alignments were then used to generate molecular models with Modeller51 (see Supplementary Figs. 1&2). Figures were prepared with Chimera52.
Design & construction of the mutants
Cysteine-replacement residues were selected to be located at the extracellular or intracellular end of a chosen TM region by using the homology model, while avoiding mutation of conserved residues. The mutations were introduced in C-terminally His-tagged LmrP in a derivative of the E. coli PCR®4 Blunt-TOPO® vector (Invitrogen) by site-directed mutagenesis using the QuikChange Lightning kit (Stratagene). Unless otherwise stated, the endogenous cysteine 270 was previously replaced by an alanine using the same method. After transformation, plasmid DNA was extracted and verified by sequencing. The lmrp gene fragment containing the desired mutation was then introduced into the pHLP5-3C vector, a derivative of the L. lactis expression vector pHLP5 containing C-terminally His-tagged LmrP54.
Bacterial strains, plasmids and growth conditions
The L. lactis NZ9000 strain was used as a host for pHLP5-3C based plasmid expression, as described previously24,30,33. Briefly, cells were grown at 30 °C in M17 medium supplemented with 0.5 % glucose and 5 mg mL−1 chloramphenicol until the OD660 reached 0.8. Overexpression of LmrP mutants was then induced by addition of 1:1000 dilution of the supernatant of the nisin producing L. lactis strain NZ9700. After 2 hours of induction, cells were harvested by centrifugation at 5,000 g.
Preparation of inside-out membrane vesicles
Cells were washed in 50 mM HEPES, pH 7 and resuspended (10 mL for each L of culture) in the same buffer containing 5 mg mL−1 of lysozyme and 10 μg mL−1 of DNAse I. After 1 h incubation at 30 °C, cells were broken by 4 passes at ~15,000 psi using a high pressure homogenizer. Cell debris and undisrupted cells were subsequently removed by a 20 min centrifugation at 17,000 g. Inside-out membrane vesicles were then isolated by ultracentrifugation at 125,000 g for 1 h at 4 °C and resuspended in 50 mM HEPES pH 7, 100 mM NaCl and 10 % (v/v) glycerol (10 mL per L of cells). Vesicles were then frozen in liquid nitrogen and stored at −80 °C for further use.
Transport assay
The transport activity of the LmrP mutants was assayed as described previously30. Briefly, inside-out membrane vesicles of LmrP-expressing cells (~ 1 mg of LmrP) were incubated 5 min in transport buffer (50 mM HEPES, 2 mM MgCl2, 300 mM KCl, pH7.4) at 30 °C in the presence of 0.1 μM Hoechst 33342 (Invitrogen). Addition of 2 mM Mg2+-ATP allowed to generate a proton-motive force by activating the endogenous F0/F1 H+ - ATPase, thereby initiating LmrP transport activity. Fluorescence spectroscopy (EX 355 nm, EM 457 nm) was used to measure the rate of extrusion of the substrate Hoechst 33342 out of the membrane. The decrease of Hoechst 33342 fluorescence over time as a result of its extrusion from the membrane allows quantification of LmrP transport activity.
Protein purification and labeling
Inside-out membrane vesicles were solubilized with 0.5 % (w/v) β-dodecylmaltoside in the presence of 1 mM DTT for 1.5 h at 4 °C on a rotating wheel. The insoluble fraction was then removed by ultracentrifugation at 125,000 g for 1 h and supernatant was batch-incubated for 2 h with previously equilibrated Ni-NTA resin (25 μL resin per mL supernatant) in the presence of 10 mM imidazole. The slurry was then transferred to a column, the flow-through discarded, and the resin washed with 8 volumes of buffer A (50 mM HEPES, 100 mM NaCl, 0.05 % (w/v) β-DDM and 20 mM imidazole), after which the protein was eluted by stepwise addition of buffer B (buffer A containing 250 mM imidazole). The concentration of the protein was determined by UV absorbance measurement at 280 nm. Spin-labeling was performed by adding a 30-fold molar excess of MTSSL (Enzo Life Sciences) from a 100 μM stock solution in DMF. The reaction was kept at room temperature for 2 hours, and the process was repeated, followed by overnight incubation on ice. The protein was then run on a SDX-200 (GE Healthcare) size-exclusion chromatography column in TMA buffer (50 mM Tris-MES-Acetate, 100 mM NaCl, 10 % (v/v) glycerol and 0.05 % (w/v) β-DDM) at pH 8. For measurements performed at pH 8, the protein was directly concentrated to 100–150 μM using a 100K MWCO concentrator, after which glycerol was added to a final concentration of 23 % (v/v). For measurements performed at pH 5, an additional run on a desalting column was performed after size-exclusion chromatography and prior to concentration in order to exchange the buffer from TMA pH 8 to TMA pH 5. Where appropriate, Hoechst 33342 was added to a final concentration of 1 mM.
EPR measurements
DEER measurements were performed on a Bruker 580 pulsed ESR spectrometer operating at Q- (33.4 GHz) band using a standard four pulse protocol55. Data was collected with the samples at 83 K with 23 % (v/v) glycerol as cryoprotectant. Analysis of the DEER data to determine the distance distributions was carried in DeerAnalysis2010 or DeerAnalysis201156. The data was fit with Tikhonov regularization and L-curve determination of the optimal regularization parameter57. Optimal background correction was established by statistical analysis of the fits. Three different background slopes that change the RMSD between DEER data and the fits by up to 10% were tested. In general, the background slopes are well-constrained by the data-collection window. Thus, minimal changes in the amplitude and position of the distance distributions were observed as a consequence of variations in the background slope. (see Supplementary Fig. 6) For the few cases that tolerated uncertainties in the choice of the background slope, the corresponding changes in the distance distributions do not affect the presence of multiple components. For few samples, we observed evidence of partial protein aggregation. This was manifested in the raw DEER decays by a deviation of the baseline from a stretched exponential and the lack of an oscillation in the echo decay even at longer collection times. Aggregation results in a component at the tail end of the distance distribution. The artifactual nature of these peaks (marked with an asterisk) could thus be demonstrated by varying the measured duration of the echo intensity oscillation. For the data displayed in Fig. 3a, additional fits were carried out assuming a sum of two Gaussian distributions to describe the distance distribution. The data were fit simultaneously with the center and sigma for each Gaussian distribution the same across the data set, and the amplitude of each Gaussian distribution allowed to vary for each pH condition. In addition, the slope and depth of modulation was also allowed to vary for each pH condition. This fitting routine was implemented in MATLAB. CW EPR spectra were collected at room temperature on a Bruker EMX spectrometer (X-band) at an incident power of 10 milliwatts and 1.6-G modulation amplitude. For the double cysteine mutants, CW measurements were carried out on the same samples used in the DEER experiments, loaded in the DEER tubes, run after the DEER experiment.
Supplementary Material
Acknowledgments
We thank H. Remaut for insightful discussions and H. Koteiche and R. Steed for critically reading the manuscript.
This work was supported by a Mandat d’Impulsion Scientifique (n° F.4523.12) from the Fonds de la Recherche Scientifique (F.R.S.-F.N.R.S.), Belgium to Cedric Govaerts and by the National Institute for General Medical Sciences, the National Institutes of Health, USA GM-077659 to Hassane S. Mchaourab.
M.M. was a Research Fellow of the Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture (F.R.I.A.), Belgium and the F.R.S.-F.N.R.S.
C.M. is a Research Fellow of the F.R.I.A
C.G. is a Research Associate of the F.R.S.-F.N.R.S.
Footnotes
Author Contributions
M.M., C.M., H.S. and C.G.: experimental design. M.M and C.M: mutagenesis, expression, activity, purification and labeling experiments. M.M., R.S, S.M. and H.M.: EPR measurements. R.S and H.M.: DEER and CW data analysis. C.G. and C.M.: molecular modeling. C.G. and H.M. oversaw all aspects of the experiments and manuscript preparation. All authors participated in interpreting the data and writing the paper.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Putman M, van Veen HW, Degener JE, Konings WN. The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines. Microbiology. 2001;147:2873–80. doi: 10.1099/00221287-147-10-2873. [DOI] [PubMed] [Google Scholar]
- 3.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–70. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 4.Tanford C. Mechanism of free energy coupling in active transport. Annu Rev Biochem. 1983;52:379–409. doi: 10.1146/annurev.bi.52.070183.002115. [DOI] [PubMed] [Google Scholar]
- 5.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807:167–88. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–35. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–20. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 8.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci U S A. 2007;104:15294–8. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–8. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 10.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30:417–26. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–4. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature. 2012;490:361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 13.Lemieux MJ, Huang Y, Wang DN. The structural basis of substrate translocation by the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr Opin Struct Biol. 2004;14:405–12. doi: 10.1016/j.sbi.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Holyoake J, Sansom MSP. Conformational change in an MFS protein: MD simulations of LacY. Structure. 2007;15:873–84. doi: 10.1016/j.str.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Enkavi G, Tajkhorshid E. Simulation of spontaneous substrate binding revealing the binding pathway and mechanism and initial conformational response of GlpT. Biochemistry. 2010;49:1105–14. doi: 10.1021/bi901412a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Madej MG, Kaback HR. Helix dynamics in LacY: helices II and IV. J Mol Biol. 2010;396:617–26. doi: 10.1016/j.jmb.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Madej MG, Guan L, Nie Y, Kaback HR. An early event in the transport mechanism of LacY protein: interaction between helices V and I. J Biol Chem. 2011;286:30415–22. doi: 10.1074/jbc.M111.268433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The alternating access transport mechanism in LacY. J Membr Biol. 2011;239:85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smirnova I, Kasho V, Sugihara J, Vázquez-Ibar JL, Kaback HR. Role of protons in sugar binding to LacY. Proc Natl Acad Sci U S A. 2012;2012:6–11. doi: 10.1073/pnas.1214890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Jensen MØ, Tajkhorshid E, Schulten K. Sugar binding and protein conformational changes in lactose permease. Biophys J. 2006;91:3972–85. doi: 10.1529/biophysj.106.085993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci U S A. 2007;104:16504–9. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan L, Kaback HR. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc Natl Acad Sci U S A. 2004;101:12148–52. doi: 10.1073/pnas.0404936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fluman N, Ryan CM, Whitelegge JP, Bibi E. Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol Cell. 2012;47:777–87. doi: 10.1016/j.molcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazurkiewicz P, Driessen AJM, Konings WN. Energetics of wild-type and mutant multidrug resistance secondary transporter LmrP of Lactococcus lactis. Biochim Biophys Acta. 2004;1658:252–61. doi: 10.1016/j.bbabio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Lewinson O, et al. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc Natl Acad Sci U S A. 2003;100:1667–72. doi: 10.1073/pnas.0435544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolhuis H, et al. Energetics and mechanism of drug transport mediated by the lactococcal multidrug transporter LmrP. J Biol Chem. 1996;271:24123–8. doi: 10.1074/jbc.271.39.24123. [DOI] [PubMed] [Google Scholar]
- 27.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–46. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 28.McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–61. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou P, McHaourab HS. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys J. 2010;98:L18–20. doi: 10.1016/j.bpj.2009.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putman M, Koole LA, van Veen HW, Konings WN. The secondary multidrug transporter LmrP contains multiple drug interaction sites. Biochemistry. 1999;38:13900–5. doi: 10.1021/bi991262k. [DOI] [PubMed] [Google Scholar]
- 31.Mazurkiewicz P, Konings WN, Poelarends GJ. Acidic residues in the lactococcal multidrug efflux pump LmrP play critical roles in transport of lipophilic cationic compounds. J Biol Chem. 2002;277:26081–8. doi: 10.1074/jbc.M203141200. [DOI] [PubMed] [Google Scholar]
- 32.Andersen AZ, et al. The metabolic pH response in Lactococcus lactis: an integrative experimental and modelling approach. Comput Biol Chem. 2009;33:71–83. doi: 10.1016/j.compbiolchem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Hakizimana P, Masureel M, Gbaguidi B, Ruysschaert JM, Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for proton gradient sensing? J Biol Chem. 2008;283:9369–76. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- 34.Seol W, Shatkin AJ. Site-directed mutants of Escherichia coli alpha-ketoglutarate permease (KgtP) Biochemistry. 1992;31:3550–4. doi: 10.1021/bi00128a032. [DOI] [PubMed] [Google Scholar]
- 35.Pazdernik NJ, Matzke EA, Jessen-Marshall AE, Brooker RJ. Roles of charged residues in the conserved motif, G-X-X-X-D/E-R/K-X-G-[X]-R/K-R/K, of the lactose permease of Escherichia coli. J Membr Biol. 2000;174:31–40. doi: 10.1007/s002320001029. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi A, Someya Y, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. The role of a conserved sequence motif, GXXXXRXGRR, in a putative cytoplasmic loop between helices 2 and 3. J Biol Chem. 1992;267:19155–62. [PubMed] [Google Scholar]
- 37.Schaedler Ta, van Veen HW. A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J. 2010;24:3653–61. doi: 10.1096/fj.10-156927. [DOI] [PubMed] [Google Scholar]
- 38.Gbaguidi B, et al. Proton motive force mediates a reorientation of the cytosolic domains of the multidrug transporter LmrP. Cell Mol life Sci. 2004;61:2646–57. doi: 10.1007/s00018-004-4298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gbaguidi B, Hakizimana P, Vandenbussche G, Ruysschaert JM. Conformational changes in a bacterial multidrug transporter are phosphatidylethanolamine-dependent. Cell Mol life Sci. 2007;64:1571–82. doi: 10.1007/s00018-007-7031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazurkiewicz P, Poelarends GJ, Driessen AJM, Konings WN. Facilitated drug influx by an energy-uncoupled secondary multidrug transporter. J Biol Chem. 2004;279:103–8. doi: 10.1074/jbc.M306579200. [DOI] [PubMed] [Google Scholar]
- 41.Madej MG, Soro SN, Kaback HR. Apo-intermediate in the transport cycle of lactose permease (LacY) Proc Natl Acad Sci U S A. 2012:1–9. doi: 10.1073/pnas.1211183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirai T, Subramaniam S. Structure and transport mechanism of the bacterial oxalate transporter OxlT. Biophys J. 2004;87:3600–7. doi: 10.1529/biophysj.104.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolhuis H, et al. Multidrug resistance in Lactococcus lactis: evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J. 1996;15:4239–45. [PMC free article] [PubMed] [Google Scholar]
- 44.Shapiro AB, Ling V. Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur J Biochem. 1997;250:122–9. doi: 10.1111/j.1432-1033.1997.00122.x. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell BA, Paulsen IT, Brown MH, Skurray RA. Bioenergetics of the staphylococcal multidrug export protein QacA. Identification of distinct binding sites for monovalent and divalent cations. J Biol Chem. 1999;274:3541–8. doi: 10.1074/jbc.274.6.3541. [DOI] [PubMed] [Google Scholar]
- 46.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem. 1997;272:21964–9. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 47.Baker J, Wright SH, Tama F. Simulations of substrate transport in the multidrug transporter EmrD. Proteins. 2012;80:1620–32. doi: 10.1002/prot.24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frillingos S, Kaback HR. The role of helix VIII in the lactose permease of Escherichia coli: II. Site-directed sulfhydryl modification. Protein Sci a Publ Protein Soc. 1997;6:438–43. doi: 10.1002/pro.5560060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soskine M, Adam Y, Schuldiner S. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J Biol Chem. 2004;279:9951–5. doi: 10.1074/jbc.M312853200. [DOI] [PubMed] [Google Scholar]
- 50.Lu M, et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc Natl Acad Sci U S A. 2013;110:2099–104. doi: 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;Chapter 2(Unit 2.9) doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Putman M, van Veen HW, Poolman B, Konings WN. Restrictive use of detergents in the functional reconstitution of the secondary multidrug transporter LmrP. Biochemistry. 1999;38:1002–8. doi: 10.1021/bi981863w. [DOI] [PubMed] [Google Scholar]
- 55.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–32. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 56.Jeschke G, et al. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 57.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–95. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.